Abstract
Background and Purpose
Malignant glioma is a highly infiltrative malignancy that causes variable disruptions to the structure and function of the cerebrovasculature. While many of these structural disruptions have known correlative histopathologic alterations, the mechanisms underlying vascular dysfunction identified by resting state BOLD (Blood Oxygen Level Dependent) imaging are not yet known. The purpose of this study was to characterize the alterations that correlate with a BOLD biomarker of vascular dysregulation.
Materials and Methods
Thirty-two stereotactically localized biopsies were obtained from CE (n=16) and NE (n=16) regions during open surgical resection of malignant glioma in 17 patients. Pre-operative resting-state BOLD fMRI was used to evaluate relationships between radiographic and histopathologic characteristics. Signal intensity for a BOLD biomarker was compared with scores of tumor infiltration and microvascular proliferation as well as total cell and neuronal density.
Results
Biopsies corresponded to a range of BOLD signals, ranging from relatively normal (Z = -4.79) to markedly abnormal (Z = 8.84). Total cell density was directly related to BOLD signal abnormality (p = 0.013, R2 = 0.19) while neuronal labeling index was inversely related to BOLD signal abnormality (p = 0.016, R2 = 0.21). The BOLD signal abnormality was also related to tumor infiltration (p = 0.014) and microvascular proliferation (p = 0.045).
Conclusion
The relationship between local, neoplastic characteristics and a BOLD biomarker of vascular function suggests that local effects of glioma cell infiltration contribute to vascular dysregulation.
INTRODUCTION
Malignant glioma is a highly infiltrative malignancy that causes variable disruptions to the structure and function of the cerebrovasculature1. Most prominent among these is a breakdown of the BBB, associated with the main tumor mass1-3 and clinically evident through the extravasation of gadolinium-conjugated contrast media4. However, other vascular deficits are identifiable in the tumor’s infiltrative margin. Vasogenic edema, represented by T2-FLAIR hyperintensity, is the most frequently utilized abnormality for directing surgical and radiation treatments in non-enhancing (NE) areas. Recent application of functional imaging has identified additional physiologic consequences of malignant glioma infiltration, with decreased vascular reactivity noted on hypercapnia challenges5-7 and reduced cortical activation intensity and volume using task-based fMRI8-11. Tumor vessel angiogenesis is a frequently cited and plausible contributor to such deficits within the contrast-enhancing (CE) tumor mass5, 7, 9, 10. However, areas of decreased vascular reactivity and reduced cortical activation have also been identified in lower grade gliomas, indicating that even sparsely infiltrated regions of brain parenchyma could suffer similar functional impairment5. Regardless, the variable presence, distribution, and degree of these abnormalities leave their relationship to tumor infiltration uncertain.
A recent report by Chow et al12 identified a biomarker of glioma-related vascular dysregulation using resting state BOLD fMRI. The mean BOLD time series extracted from normal, tumor-free brain of the contralesional hemisphere represents normal cardiac pulsations and respiration-related changes in arterial carbon dioxide (CO2)13-15 and can be used as a temporal signature of normal vascular reactivity16. Similarly, the mean BOLD time series extracted from CE tumor represents abnormal tumor-related vascular function. By combining these temporal signatures, the spatial extent of glioma-related vascular deficits can be illustrated by identifying voxels of NE brain parenchyma that behave more like the CE tumor and less like the normal contralesional brain tissue. This technique demonstrated that vascular dysregulation can extend beyond the CE tumor core into the NE infiltrative margin. Furthermore, the presence of non-overlapping regions between the BOLD biomarker and T2-FLAIR hyperintensity, or BOLD-FLAIR mismatch, suggests discordance between deficits in neurovascular function and a breakdown of the BBB. However, the absence of a histologic correlate in Chow et al leaves substantial uncertainty as to whether this vascular dysregulation is related directly to tumor infiltration or to indirect effects of the tumor. For example, such dysfunction could be directly related to local tumor effects, such as a disruption in the structural, physical, or ultrastructural properties of the brain by invading glioma cells11. Alternatively, distal, indirect causes, such as mass effect from the tumor or vasogenic edema, disruption of shared venous drainage, or damage to axons could also explain these BOLD abnormalities8, 12, 17.
The purpose of this study was to characterize the histopathological alterations underlying the BOLD biomarker of vascular dysfunction in malignant gliomas. We compared BOLD dynamics from resting state fMRI to neoplastic characteristics of a set of stereotactically localized biopsies. We hypothesize that the degree of disruption of vascular function measured by the BOLD biomarker is related to the severity of histopathologic abnormalities associated with tumor infiltration.
METHODS
Patient Selection
We performed a retrospective analysis of our institutional database of patients consented for an IRB-approved study for the procurement of MRI-localized biopsies during open surgical resection. Seventeen patients with pathology-proven, treatment-naïve glioblastoma (n = 14) or Grade III, IDH wild-type, astrocytoma (n = 3) and pre-operative BOLD fMRI were identified. Average age was 63.8 years (range 48-82), and there were 5 males and 12 females. Thirty-two tissue biopsies were obtained from the CE (n = 16) and the NE (n = 16) regions of tumor during open surgical resection. Automated cell counting was successfully performed on 31 H&E stains and 28 NeuN stains.
Image Acquisition
Prior to tumor resection, patients underwent standard-of-care imaging (including T1-weighted, gadolinium enhanced (T1+Gd), T2-FLAIR, and DWI) as well as a resting state BOLD fMRI scan on one of four hospital scanners (Philips Intera 1.5T, Philips Achieva 3T, GE Twinspeed HDx 1.5T, or GE Excite HDX 3T). The range of scanning parameters for T1 sequences was: TR = 10-2000ms, flip angle = 13-90, TE = 2-20. For FLAIR sequences the ranges were: TR = 8000-11000ms, flip angle = 90, TE = 104-337ms, TI = 2200-2800ms. BOLD parameters were: TR = 1000-2400ms, flip angle = 72-90, TE = 20-34ms, # volumes = 150-200. The scanning parameters for each patient are listed in Supplementary Table 1. Contrast-enhanced images were acquired with IV gadobenate dimeglumine (MultiHance, Bracco Diagnostics, Monroe Township, NJ) with a weight-based dose of 0.2 mL/kg. The time between IV injection and contrast-enhanced imaging was 5 minutes. BOLD images were obtained before contrast-enhanced imaging.
Biopsy Acquisition
All tissue sampling was done within the course of the normal surgical plan, provided it posed no additional risk to the patient. Samples were taken from the CE tumor core in all cases and the NE region when within the planned surgical trajectory. Biopsies were obtained prior to surgical debulking to minimize the effects of brain shift and deformation. Frameless stereotactic guidance was provided by a volumetric T1+Gd scan uploaded to a neuronavigation interface (Brainlab Curve, Brainlab AG, Feldkirchen, Germany). Biopsy location was recorded by screen captures of the neuronavigation interface, allowing for downstream determination of each biopsy’s Cartesian coordinates. We have recently shown that this method results in spatial accuracy to within ~2mm18. Biopsies that were taken from radiologically-defined areas of necrosis or those that were predominantly histologically necrotic (>75% of the tissue sample), were excluded from the analysis.
Histopathological Analysis
The samples were fixed in 10% (vol/vol) formalin and embedded with paraffin for histopathologic analysis. Five micron sections were stained with H&E. Seven micron sections were immunostained with NeuN antibody (MilliporeSigma MAB377, Darmstadt, Germany). Qualitative histologic characteristics of neoplasm were evaluated by two blinded neuropathologists (GZ, PC). H&E stained sections were reviewed for the degree of tumor infiltration and scored as no tumor present (score = 0), infiltrating tumor (score = 1), or highly cellular tumor (score = 2). The tumor score was a composite of several features, including cytologic atypia and cell density. H&E slides were also reviewed for the degree of microvascular proliferation, scored as delicate microvasculature (normal vascular architecture; score = 0), simple hyperplastic structures (less than three circumferential layers with patent lumen; score = 1), or complex microvascular hyperplasia (three or more endothelial layers or glomeruloid-type vessels; score = 2).
Total cell density was calculated using an automated cell-counting algorithm18. Briefly, the H&E stained sections were digitalized using a Leica SCN400 system (Leica Biosystems Inc, Buffalo Grove, IL), with hematoxylin-stained nuclei counted in iteratively processed high-powered fields (HPFs). Total cell density was computed as the median number of hematoxylin-stained cells across all HPFs which contained at least one nucleus. To calculate the labeling index for NeuN, each immunoperoxidase stained slide was digitized at low-power 10x magnification and a semi-automated algorithm was used to determine overall NeuN index. For each slide, user-defined RGB (three-channel) and size thresholds were determined by visual inspection of 9 randomly generated low-power 10x fields (LPFs). Based on these statistics, the algorithm subsequently identified all immunoperoxidase stained cells in each LPF and computed the neuronal fraction by dividing the surface area of stained cells by the total surface area of all tissue in the LPF. The NeuN index was defined as the 90% quantile of fractions across all LPFs that contain at least one stained cell.
Screenshot Registration and Coordinate Determination
Screenshots of the neuronavigation interface showing the biopsy location were superimposed onto a co-registered T1+Gd image. Co-registration of the screenshots to the T1+Gd image in 3D space was done using custom software written in Matlab. The registration pipeline first correlated the pixel intensities of the screenshot to the voxel intensities of each volumetric T1 slice and the slice with peak correlation was identified as the biopsy. A correlation between the screenshot and the biopsy slice was then taken at different x and y offsets and the peak correlation was used to identify the correct x and y offset. Finally, each biopsy location on the screenshot was labeled with a cross-hair. To identify this location, a digital cross-hair was correlated to the screenshot at different x and y offsets and the peak correlation was used to identify the x and y coordinates of the biopsy site. All co-registered screenshots were confirmed by visual inspection.
Tumor masks
All structural images were co-registered to the T1w+Gd image with highest spatial resolution using rigid body registration (FLIRT19; 6 degrees of freedom) and all masks were created in the high-resolution reference image space. Three dimensional masks of the CE tumor were hand drawn on high resolution T1w+Gd images by a board-certified neuroradiologist (CIH). The boundaries of masks were drawn to be as specific for CE intra-axial tumor as possible. Ambiguous voxels at the border of CE tumor and NE brain tissue and at the border of CE tumor and necrotic core were excluded from the mask. Areas of linear enhancement determined to represent vessels were excluded from the mask. Extra-axial spread of tumor along the dura or ventricular surface was also excluded from the tumor mask. The control masks were generated by co-registering the T1+Gd image to MNI152 space and transforming a contralesional hemisphere mask, created in MNI152 space, to the patient’s reference space (i.e. the high-resolution T1+Gd image) using nearest neighbor interpolation. The contralesional control masks were then visually inspected by a board-certified neuroradiologist (CH) to ensure no CE tumor or abnormal FLAIR hyperintense tissue was included in the mask. To generate the functional masks, BOLD scans were registered to the high-resolution reference image (FLIRT; 6 degrees of freedom) and the inverse transform was applied to the high-resolution masks to put them into functional space. Because the BOLD data was collected at a lower spatial resolution than the structural data, the transformation of the masks resulted in partial voluming. To minimize any influence of partial voluming from non-enhancing portions of the surrounding tissue, the CE masks were thresholded such that the fraction of the CE tumor was at least 0.95. Similarly, the control masks were thresholded such that the fraction of NE tissue was at least 0.95. All original and transformed masks were visually inspected by a board-certified radiologist (CIH) to confirm accuracy.
Image Pre-Processing of BOLD Data
All functional data were processed with FMRIB Software Library (FSL version 5.0.6)19 and Matlab (2012b, MathWorks, Natick, MA) software. Each functional image was motion corrected, slice timing corrected, spatially smoothed (Gaussian filter, full width at half maximum (FWHM)=5mm), and high-pass filtered (100 sec), and skull stripped. Independent components analysis (MELODIC20) was performed – each scan resulted in ~40-90 components. The components were visually inspected and any components related to scanner noise or head movement artifacts were removed. To further minimize the effect of head motion on BOLD signal intensity, the motion parameters were regressed out of the data before further regression was performed with the tumor and control ROI time series. The 18 motion regressors used in the regression model consisted of three translations, three rotations, the first derivative of each, and the corresponding quadratic terms. The residual from this model was then used for all subsequent regression analyses.
BOLD dynamics
To create the BOLD masks, we followed the procedure described in Chow et al12. We first made the simplifying assumption that there are two functional tissue classes within the brain, tumor and healthy tissue, which are represented by the CE portion of the tumor and the contralesional hemisphere, respectively. Mean time series were extracted from the masks defined by the CE tumor and from the contralesional control hemisphere, by averaging the BOLD time series of all voxels within each mask. Multiple linear regression was performed for all brain voxels using the two regressor model, i.e., the CE tumor mean time series and the contralesional control mean time series, and their first derivatives to allow for variation in timing. The output of this analysis, and the metrics used to make our conclusions, consisted of parameter estimates (beta weights) and z-statistic values (beta weights normalized by the residual variance) for each voxel. The difference between the two z-statistic images (i.e. ZCE − Zcontrol) is used as the BOLD biomarker representing vascular function, as described in Chow et al12. Specifically, strongly negative values (blue) represent areas of normal vascular function, i.e. the voxel time series is more similar to the control ROI; strongly positive values (red) represent areas of vascular dysregulation, i.e. the voxel time series is more similar to the CE tumor.
Analysis of Histopathologic and Radiologic Correlation
All statistical tests were performed using the Statistics Toolbox in Matlab (2012b, MathWorks). Linear regression was used to evaluate the relationships between BOLD signal intensity and quantitative histopathologic features (total cell density and NeuN labeling index). The parameter estimate for the CE regressor decreases as a function of distance from the CE mask due to (1) tumor-related vascular dysregulation and (2) non-specific effects of distance i.e. voxels in close proximity have more similar intensities than voxels that are far apart. To control for non-specific effects of distance, we included a distance regressor, the measured distance from the biopsy location to the nearest CE voxel, in the regression model of cellularity vs BOLD and NeuN vs BOLD. To assess the relationship between qualitative histologic features (degree of tumor infiltration and microvascular proliferation) and the BOLD signal, a one-way analysis of variance (ANOVA) was performed.
RESULTS
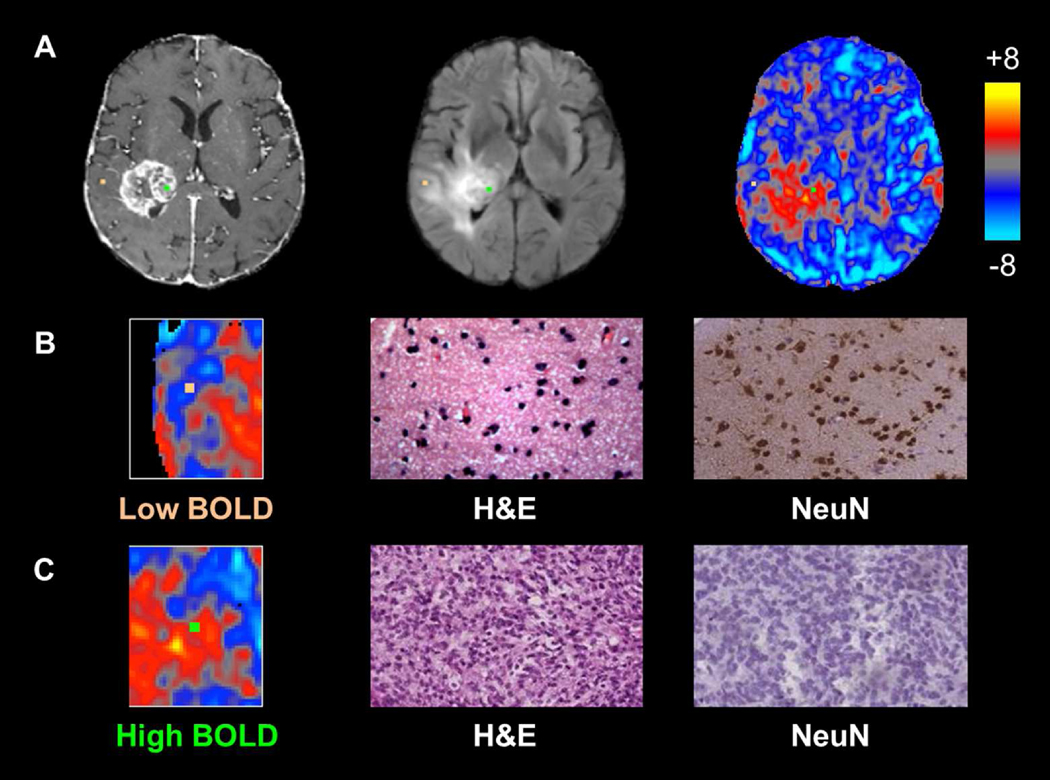
Biopsies demonstrated a range of BOLD signal intensities, based on their projection onto z-statistic maps, ranging from relatively normal (Z = -4.79) to markedly abnormal (Z = 8.84). Differences in the degree of signal abnormality appeared to be related to certain histopathologic characteristics within individual tissue samples. Representative biopsies from a single patient are shown in Figure 1, which illustrates tissue samples taken from areas of differing BOLD signal (Fig 1A). Tissue corresponding to a negative BOLD value (Z = -1.90; Fig 1B) demonstrated normal cell density, high NeuN staining, non-infiltrated gray matter, and normal, delicate microvasculature. In contrast, a sample corresponding to a highly positive BOLD signal (Z = 4.44; Fig 1C) contained hypercellular tissue, little neuronal staining, highly cellular tumor, and complex microvascular hyperplasia.
Figure 1. Individual biopsies from areas of intact and disrupted vascular function exhibit distinct histopathologic characteristics.
Panel A shows biopsy locations overlaid onto T1 post-contrast, FLAIR, and BOLD images with correlative histopathology in panels B and C. The sample from relatively normal BOLD signal, representing normal vascular function (orange, Panel B), shows normal cellularity and high NeuN staining, suggestive of non-infiltrated gray matter, whereas the sample from a region of BOLD signal abnormality (green, Panel C) shows tumor tissue and no neuronal staining.
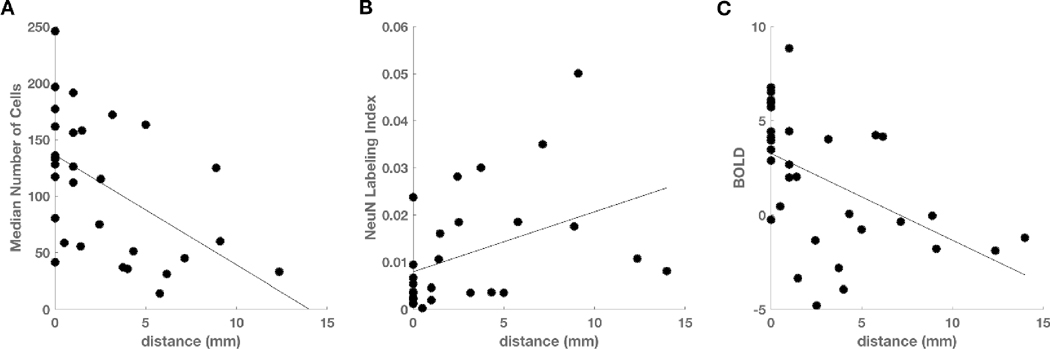
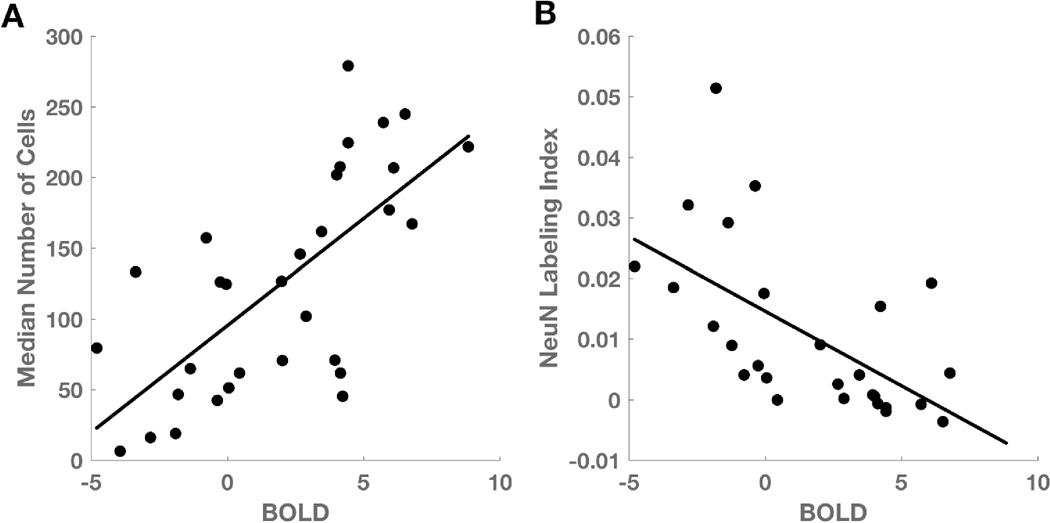
Among all biopsies, linear regression analysis demonstrated a significant inverse relationship between total cell density and distance from the nearest CE voxel (p = 0.003, R2 = 0.27; Fig 2A) and a positive relationship between neuronal density and distance (p = 0.032, R2 = 0.17; Fig 2B). The BOLD signal also shows an inverse relationship with distance (p = 0.004, R2 = 0.25; Fig 2C). Linear regression between BOLD signal intensity and total cell density, with distance to nearest CE voxel as a confounding variable, showed a positive relationship (p = 2 × 10-5, R2 = 0.48; Fig 3A). A similar analysis showed that the NeuN labeling index was negatively related to BOLD signal intensity (p = 0.001, R2 = 0.35; Fig 3B). If BOLD values in white matter were much larger than in gray matter, then it is possible that the negative relationship between BOLD and neuronal density could be trivially explained by the fact that gliomas are more likely to be found in white matter; that is, white matter which frequently contains gliomas could have, on average, high BOLD but low NeuN, whereas gray matter, which infrequently contains gliomas could have, on average, low BOLD and high NeuN. To test this hypothesis, we compared the BOLD values in gray (mean = -2.9, SD = 0.76) and white (mean = -2.4, SD = 0.83) matter on the contralesional control hemisphere. Gray voxels were significantly more negative than white voxels (t-test, p = 0.001, t = 4.00), however, the difference in BOLD due to tissue type (gray – white = - 0.54, SD = 0.56), cannot explain the very large range of BOLD values across the biopsy samples (max – min = [8.85] – [-4.79] = 13.64).
Figure 2. Effect of distance from contrast enhancement on cellularity, neuronal density, and BOLD.
(A) Cell density decreases with distance to the nearest contrast enhancing voxel (n = 31, p = 0.003). (B) Neuronal density increases with distance to contrast enhancement (n = 27, p = 0.032). (C) Intensity of the BOLD biomarker decreases with distance to contrast enhancement (n = 32, p = 0.004).
Figure 3. BOLD signal abnormality is related to total cell and neuronal density.
(A) After controlling for the effect of distance on the BOLD signal, median cell density is positively related to BOLD z-statistic (n = 31, p = 2 × 10-5). (B) NeuN labeling index is inversely related to BOLD z-statistic (n = 27, p = 0.001).
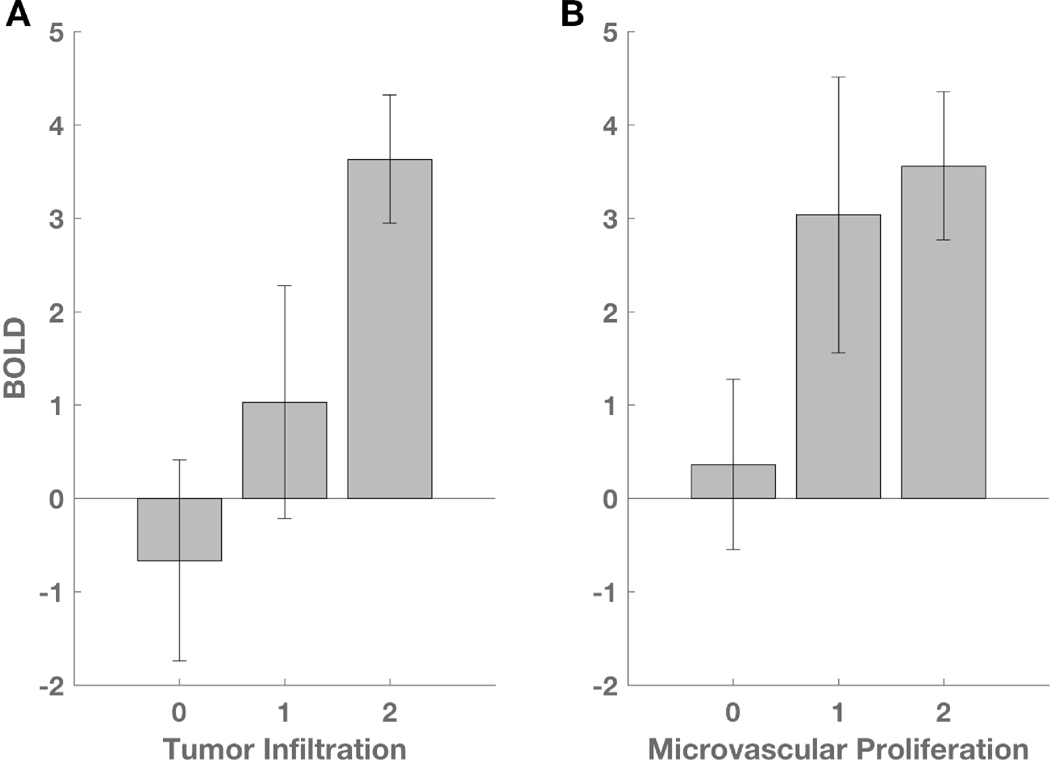
The distributions of BOLD signal abnormality related to tumor infiltration and microvascular proliferation are summarized in Figure 4. Across all biopsies, both the tumor infiltration score (ANOVA: F(2,29) = 4.95, p = 0.014) (Fig 4A) and the microvascular proliferation score (ANOVA: F(2,29) = 3.46, p = 0.045) (Fig 4B) were significantly related to the BOLD signal abnormality. A post-hoc Student’s t-test showed a significant difference in BOLD signal between the group of samples with no detectable tumor (score = 0) and those with highly cellular tumor (score = 2; p = 0.003) or the combination of infiltrating and highly cellular tumor (score 1 or 2; p = 0.029). Similarly, a t-test showed a significant difference in BOLD signal between the group of samples with no identifiable microvascular proliferation (score = 0) and those with marked vascular hyperplasia (score = 2; p = 0.038) or the combination of simple and complex vascular hyperplasia (score 1 or 2; p = 0.027).
Figure 4. BOLD signal abnormality is related to tumor infiltration and microvascular proliferation.
(A) ANOVA demonstrates a significant relationship between BOLD z-statistic and tumor infiltration (p = 0.014). Scores of 0, 1, and 2, represent no detectable tumor, infiltrating tumor, and highly cellular tumor, respectively. (B) ANOVA demonstrates a significant relationship between BOLD z-statistic and microvascular proliferation (p = 0.045). Scores of 0, 1, and 2, represent normal vascular architecture, simple hyperplastic structures, and complex microvascular hyperplasia, respectively.
DISCUSSION
The diffuse infiltration and histologic heterogeneity of malignant glioma pose a distinct challenge to the identification of these tumors’ toxic effects on the structure and function of the cerebrovasculature. The integrity of the vasculature in and around the main mass of the tumor is typically assessed with standard of care imaging that uses T1-weighted contrast enhanced and FLAIR sequences1-4. Regions of abnormal contrast enhancement associated with the main tumor mass typically correspond with elevated CBV, CBF, and microvascular permeability on perfusion-weighted imaging21-27. Interestingly, there are few vascular abnormalities in the NE, FLAIR-hyperintense peritumoral regions on perfusion imaging21-27, despite the recognized infiltration of tumor cells into these areas28-31. Additional evidence suggests that edema surrounding the tumor originates directly from the CE tissue, passively diffuses into the NE regions, and may therefore not be associated with BBB breakdown in the NE tissue32, 33.
Several intriguing studies have, however, identified decreased vascular reactivity outside CE regions of glioma5, 7, 34. In turn, it has been postulated that infiltrating glioma cells disrupt neurovascular coupling35, 36, impairing both normal hypercapnia-induced elevations in CBF and CBV and the vascular reactivity of cortical tissue, particularly within gray matter. However, without any histopathologic correlation to these radiologic findings, it has been unclear whether infiltrating glioma cells contribute directly to these abnormalities. The results of the present study, by relating histopathologic and radiologic data, demonstrate that the BOLD biomarker of vascular dysfunction is directly associated with tumor infiltration. We found markedly abnormal tissue in areas of BOLD signal abnormality and relatively unaffected brain tissue in areas of relatively normal BOLD signal. Moreover, the BOLD biomarker is directly related to total cell density and inversely related to neuronal density, and is correlated to scores of tumor infiltration and microvascular proliferation.
The spatial distribution of the BOLD biomarker is also consistent with a decreasing cell density gradient with distance from the tumor core28. The biomarker decreases as a function of distance from the CE tumor core, replicating our previous findings12. We found that the effect of distance is similar between BOLD, cellularity, and neuronal density, suggesting that the detectable infiltrating margins of the tumor span approximately 1 – 2 cm beyond the contrast enhancement, comparable to the commonly used 2 – 3 cm margins for radiation treatment37. More importantly, the relationship between the BOLD biomarker and the cell counts (i.e. cellularity and neuronal density) remains significant even after including distance from the CE core as a covariate, indicating that the BOLD data can explain additional variance of tumor heterogeneity unrelated to a simple density gradient.
The mechanisms by which glioma influences vascular function are unknown. Several potential mechanisms have been proposed as broad, or long distance, effects of glioma on vascular function. For example, (1) mass effect and consequent venous compression could alter rates of blood flow over large regions of surrounding brain tissue8, 17; (2) shared venous drainage could allow signal from blood draining from the tumor core to predominate even in the peritumoral area12; (3) the effects of aberrant metabolite and neurotransmitter production found in gliomas may extend to normal cortical vessels, inducing vascular dysregulation at indeterminate distances from invading tumor cells; and (4) the tumor may affect fibers of passage in its vicinity, inducing necrosis or structural changes with detrimental downstream effects, consistent with findings of multi-domain cognitive dysfunction in some glioblastoma patients that are not easily attributable to local injury from the tumor38 and the disruption of global resting state networks39-42.
In contrast, local tumor effects may also contribute to the vascular dysfunction seen with the BOLD biomarker. For example, invading glioma cells could perturb the structure of the neurovascular unit by compromising direct contact between vascular endothelial cells and astrocytes1, 3, 43. Alternatively, astrocytic mediation of the vascular response may be modulated directly by glioma cells’ own neurotransmitters44 or by affecting the action of normal signaling molecules.
A BOLD fMRI biomarker of vascular dysfunction has a number of potential clinical applications that could warrant further investigation. BOLD, with its sensitivity to local effects of infiltrating glioma, could be another useful adjunct in guiding surgery beyond the CE boundary, especially in light of the growing body of evidence favoring aggressive resection45, 46. For the same reason, it may be useful in targeting adjuvant radiation therapy towards affected areas unamenable to surgical resection. In the same vein, investigating the BOLD dynamics in areas of residual disease may offer insight into the mechanisms of disease recurrence and progression — it is possible that disease recurrence is more attributable to compromised vascular function than residual tumor burden alone. Thus, application of the BOLD biomarker in post-treatment surveillance could offer new insight into patterns of glioblastoma spread, assess response to new therapies, and segment areas of tumor recurrence.
There are, however, several limitations to our conclusions. There is no way to ensure perfect fidelity between the physical biopsy location and the biopsy site as represented by frameless stereotaxy during an open tumor resection. As per the study protocol, attempts were made to mitigate the effects of intraoperative brain shift within the limits of the surgical plan by minimizing intraoperative use of mannitol, CSF diversion, or hyperventilation prior to biopsy. Most importantly, all biopsy sampling was performed prior to tumor debulking. Nevertheless, it is possible that the BOLD value extracted from the presumed biopsy site could be displaced by one or more voxels from the true biopsy location. However, we recently showed that our sampling method has registration accuracy to within ~2mm18, suggesting this type of error would have minimal effects on our results. Additionally, we did not have rigid control of scanning parameters due to the retrospective nature of this study; consequently, contrast-to-noise and signal-to-noise could vary significantly across individuals. However, all structural data were reviewed by a neuroradiologist and assessed to be clearly interpretable for creating tissue masks. Similarly, variability in scanning parameters certainly affected the signal-to-noise properties of the BOLD data. Despite this variability, we showed a significant relationship between BOLD data and histological features, suggesting that future prospective studies that are able to hold scanning parameters constant could decrease the variance of this relationship. Finally, masks were constructed by a single neuroradiologist, resulting in some subjectivity in mask boundaries. However, our previous work12 has demonstrated that the vascular dysregulation maps are not sensitive to small changes in mask variability (e.g. using gray matter versus white matter as the contralesional control mask resulted in two BOLD maps that had a correlation of 0.96).
CONCLUSION
Vascular dysregulation, as represented by a resting state BOLD biomarker, is seen both within and well beyond the margins of contrast enhancement in malignant glioma. Many factors likely influence this dysfunction in NE regions. However, the statistically significant correlation between tumor infiltration and the degree of BOLD abnormality suggest that infiltrating glioma cells contribute to vascular dysfunction in both CE and NE regions.
Supplementary Material
Acknowledgments
Grant support: This project was funded in part by the 2015 Research Scientist Award from the American Society of Neuroradiology (JG) and James S. McDonnell Foundation, Irving Institute for Clinical and Translational Research: Pilot Research Award (PDC).
Support from Nvidia included 1 Titan X GPU card (12GB) to run deep learning code to perform cell counting.
Dr. Chang’s salary is supported by the NIH (NIBIB) T32 Training Grant, T32EB001631.
ABBREVIATIONS
- CE
contrast-enhancing
- NE
non-enhancing
- BOLD
blood oxygenation level dependent
- HPF
high-powered field
- LPF
low-powered field
- BBB
blood brain barrier
Footnotes
Presentation: Preliminary results from this work were presented in an oral presentation at the Eastern Atlantic Medical Student Research Forum at the University of Miami in March, 2016. Further results have been accepted for poster presentation at the Society of Neuro-Oncology Annual Meeting in November, 2016.
References
- 1.Watkins S, Robel S, Kimbrough IF, et al. Disruption of astrocyte-vascular coupling and the bloodbrain barrier by invading glioma cells. Nat Commun. 2014;5:4196. doi: 10.1038/ncomms5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Persidsky Y, Ramirez SH, Haorah J, et al. Blood-brain barrier: structural components and function under physiologic and pathologic conditions. J Neuroimmune Pharmacol. 2006;1:223–236. doi: 10.1007/s11481-006-9025-3. [DOI] [PubMed] [Google Scholar]
- 3.Cuddapah VA, Robel S, Watkins S, et al. A neurocentric perspective on glioma invasion. Nat Rev Neurosci. 2014;15:455–465. doi: 10.1038/nrn3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28:1963–1972. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 5.Pillai JJ, Zaca D. Comparison of BOLD cerebrovascular reactivity mapping and DSC MR perfusion imaging for prediction of neurovascular uncoupling potential in brain tumors. Technol Cancer Res Treat. 2012;11:361–374. doi: 10.7785/tcrt.2012.500284. [DOI] [PubMed] [Google Scholar]
- 6.Hsu YY, Chang CN, Jung SM, et al. Blood oxygenation level-dependent MRI of cerebral gliomas during breath holding. J Magn Reson Imaging. 2004;19:160–167. doi: 10.1002/jmri.10447. [DOI] [PubMed] [Google Scholar]
- 7.Zaca D, Jovicich J, Nadar SR, et al. Cerebrovascular reactivity mapping in patients with low grade gliomas undergoing presurgical sensorimotor mapping with BOLD fMRI. J Magn Reson Imaging. 2014;40:383–390. doi: 10.1002/jmri.24406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holodny AI, Schulder M, Liu WC, et al. The effect of brain tumors on BOLD functional MR imaging activation in the adjacent motor cortex: implications for image-guided neurosurgery. AJNR Am J Neuroradiol. 2000;21:1415–1422. [PMC free article] [PubMed] [Google Scholar]
- 9.Hou BL, Bradbury M, Peck KK, et al. Effect of brain tumor neovasculature defined by rCBV on BOLD fMRI activation volume in the primary motor cortex. Neuroimage. 2006;32:489–497. doi: 10.1016/j.neuroimage.2006.04.188. [DOI] [PubMed] [Google Scholar]
- 10.Jiang Z, Krainik A, David O, et al. Impaired fMRI activation in patients with primary brain tumors. Neuroimage. 2010;52:538–548. doi: 10.1016/j.neuroimage.2010.04.194. [DOI] [PubMed] [Google Scholar]
- 11.Schreiber A, Hubbe U, Ziyeh S, et al. The influence of gliomas and nonglial space-occupying lesions on blood-oxygen-level-dependent contrast enhancement. AJNR Am J Neuroradiol. 2000;21:1055–1063. [PMC free article] [PubMed] [Google Scholar]
- 12.Chow DS, Horenstein CI, Canoll P, et al. Glioblastoma Induces Vascular Dysregulation in Nonenhancing Peritumoral Regions in Humans. AJR Am J Roentgenol. 2016;206:1073–1081. doi: 10.2214/AJR.15.14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shmueli K, van Gelderen P, de Zwart JA, et al. Low-frequency fluctuations in the cardiac rate as a source of variance in the resting-state fMRI BOLD signal. Neuroimage. 2007;38:306–320. doi: 10.1016/j.neuroimage.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy K, Birn RM, Handwerker DA, et al. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage. 2009;44:893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birn RM, Murphy K, Bandettini PA. The effect of respiration variations on independent component analysis results of resting state functional connectivity. Hum Brain Mapp. 2008;29:740–750. doi: 10.1002/hbm.20577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu P, Li Y, Pinho M, et al. Cerebrovascular reactivity mapping without gas challenges. Neuroimage. 2017;146:320–326. doi: 10.1016/j.neuroimage.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holodny AI, Schulder M, Liu WC, et al. Decreased BOLD functional MR activation of the motor and sensory cortices adjacent to a glioblastoma multiforme: implications for image-guided neurosurgery. AJNR Am J Neuroradiol. 1999;20:609–612. [PMC free article] [PubMed] [Google Scholar]
- 18.Chang P, M HR, Bowden SG, Chow DS, Gill BJA, Ung TH, Samanamud J, Englander ZK, Sonabend AM, Sheth SA, McKhann GM, II, Sisti MB, Schwartz LH, Lignelli A, Grinband J, Bruce JN, Canoll P. A Multiparametric Model for Mapping Cellularity in Glioblastoma Using Radiographically Localized Biopsies. AJNR Am J Neuroradiol. 2017 doi: 10.3174/ajnr.A5112. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkinson M, Beckmann CF, Behrens TE, et al. Fsl. Neuroimage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging. 2004;23:137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- 21.Calli C, Kitis O, Yunten N, et al. Perfusion and diffusion MR imaging in enhancing malignant cerebral tumors. Eur J Radiol. 2006;58:394–403. doi: 10.1016/j.ejrad.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 22.Hakyemez B, Erdogan C, Bolca N, et al. Evaluation of different cerebral mass lesions by perfusion-weighted MR imaging. J Magn Reson Imaging. 2006;24:817–824. doi: 10.1002/jmri.20707. [DOI] [PubMed] [Google Scholar]
- 23.Tyler JL, Diksic M, Villemure JG, et al. Metabolic and hemodynamic evaluation of gliomas using positron emission tomography. J Nucl Med. 1987;28:1123–1133. [PubMed] [Google Scholar]
- 24.Schramm P, Xyda A, Klotz E, et al. Dynamic CT perfusion imaging of intra-axial brain tumours: differentiation of high-grade gliomas from primary CNS lymphomas. Eur Radiol. 2010;20:2482–2490. doi: 10.1007/s00330-010-1817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noguchi T, Yoshiura T, Hiwatashi A, et al. Perfusion imaging of brain tumors using arterial spinlabeling: correlation with histopathologic vascular density. AJNR Am J Neuroradiol. 2008;29:688–693. doi: 10.3174/ajnr.A0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tourdias T, Rodrigo S, Oppenheim C, et al. Pulsed arterial spin labeling applications in brain tumors: practical review. J Neuroradiol. 2008;35:79–89. doi: 10.1016/j.neurad.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Cha S, Yang L, Johnson G, et al. Comparison of microvascular permeability measurements, K(trans), determined with conventional steady-state T1-weighted and first-pass T2*-weighted MR imaging methods in gliomas and meningiomas. AJNR Am J Neuroradiol. 2006;27:409–417. [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly PJ. Computed tomography and histologic limits in glial neoplasms: tumor types and selection for volumetric resection. Surg Neurol. 1993;39:458–465. doi: 10.1016/0090-3019(93)90031-u. [DOI] [PubMed] [Google Scholar]
- 29.Earnest Ft, Kelly PJ, Scheithauer BW, et al. Cerebral astrocytomas: histopathologic correlation of MR and CT contrast enhancement with stereotactic biopsy. Radiology. 1988;166:823–827. doi: 10.1148/radiology.166.3.2829270. [DOI] [PubMed] [Google Scholar]
- 30.Greene GM, Hitchon PW, Schelper RL, et al. Diagnostic yield in CT-guided stereotactic biopsy of gliomas. J Neurosurg. 1989;71:494–497. doi: 10.3171/jns.1989.71.4.0494. [DOI] [PubMed] [Google Scholar]
- 31.Nagashima G, Suzuki R, Hokaku H, et al. Graphic analysis of microscopic tumor cell infiltration, proliferative potential, and vascular endothelial growth factor expression in an autopsy brain with glioblastoma. Surg Neurol. 1999;51:292–299. doi: 10.1016/s0090-3019(98)00056-1. [DOI] [PubMed] [Google Scholar]
- 32.Pronin IN, Holodny AI, Petraikin AV. MRI of high-grade glial tumors: correlation between the degree of contrast enhancement and the volume of surrounding edema. Neuroradiology. 1997;39:348–350. doi: 10.1007/s002340050421. [DOI] [PubMed] [Google Scholar]
- 33.Pronin IN, McManus KA, Holodny AI, et al. Quantification of dispersion of Gd-DTPA from the initial area of enhancement into the peritumoral zone of edema in brain tumors. J Neurooncol. 2009;94:399–408. doi: 10.1007/s11060-009-9872-x. [DOI] [PubMed] [Google Scholar]
- 34.Iranmahboob A, Peck KK, Brennan NP, et al. Vascular Reactivity Maps in Patients with Gliomas Using Breath-Holding BOLD fMRI. J Neuroimaging. 2016;26:232–239. doi: 10.1111/jon.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal S, Sair HI, Pillai JJ. The Resting-State Functional Magnetic Resonance Imaging Regional Homogeneity Metrics-Kendall’s Coefficient of Concordance-Regional Homogeneity and Coherence-Regional Homogeneity-Are Valid Indicators of Tumor-Related Neurovascular Uncoupling. Brain Connect. 2017;7:228–235. doi: 10.1089/brain.2016.0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agarwal S, Lu H, Pillai JJ. Value of Frequency Domain Resting-State Functional Magnetic Resonance Imaging Metrics Amplitude of Low-Frequency Fluctuation and Fractional Amplitude of Low-Frequency Fluctuation in the Assessment of Brain Tumor-Induced Neurovascular Uncoupling. Brain Connect. 2017;7:382–389. doi: 10.1089/brain.2016.0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi SH, Kim JW, Chang JS, et al. Impact of Including Peritumoral Edema in Radiotherapy Target Volume on Patterns of Failure in Glioblastoma following Temozolomide-based Chemoradiotherapy. Sci Rep. 2017;7:42148. doi: 10.1038/srep42148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartolomei F, Bosma I, Klein M, et al. How do brain tumors alter functional connectivity? A magnetoencephalography study. Ann Neurol. 2006;59:128–138. doi: 10.1002/ana.20710. [DOI] [PubMed] [Google Scholar]
- 39.Maesawa S, Bagarinao E, Fujii M, et al. Evaluation of resting state networks in patients with gliomas: connectivity changes in the unaffected side and its relation to cognitive function. PLoS One. 2015;10:e0118072. doi: 10.1371/journal.pone.0118072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esposito R, Mattei PA, Briganti C, et al. Modifications of default-mode network connectivity in patients with cerebral glioma. PLoS One. 2012;7:e40231. doi: 10.1371/journal.pone.0040231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris RJ, Bookheimer SY, Cloughesy TF, et al. Altered functional connectivity of the default mode network in diffuse gliomas measured with pseudo-resting state fMRI. J Neurooncol. 2014;116:373–379. doi: 10.1007/s11060-013-1304-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park JE, Kim HS, Kim SJ, et al. Alteration of long-distance functional connectivity and network topology in patients with supratentorial gliomas. Neuroradiology. 2015 doi: 10.1007/s00234-015-1621-6. [DOI] [PubMed] [Google Scholar]
- 43.Hillman EM. Coupling mechanism and significance of the BOLD signal: a status report. Annu Rev Neurosci. 2014;37:161–181. doi: 10.1146/annurev-neuro-071013-014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Araque A, Parpura V, Sanzgiri RP, et al. Tripartite synapses: glia, the unacknowledged partner. Trends Neurosci. 1999;22:208–215. doi: 10.1016/s0166-2236(98)01349-6. [DOI] [PubMed] [Google Scholar]
- 45.Hervey-Jumper SL, Berger MS. Maximizing safe resection of low- and high-grade glioma. J Neurooncol. 2016;130:269–282. doi: 10.1007/s11060-016-2110-4. [DOI] [PubMed] [Google Scholar]
- 46.Duffau H. Long-term outcomes after supratotal resection of diffuse low-grade gliomas: a consecutive series with 11-year follow-up. Acta Neurochir (Wien) 2016;158:51–58. doi: 10.1007/s00701-015-2621-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.