Abstract
Impaired cardiac muscle growth and aberrant myocyte arrangement underlie congenital heart disease and cardiomyopathy. We show that cardiac-specific inactivation of the homeobox transcription factor Prox1 results in disruption of the expression and localisation of sarcomeric proteins, gross myofibril disarray and growth retarded hearts. Furthermore, we demonstrate that Prox1 is required for direct transcriptional regulation of structural proteins α-actinin, N-RAP and Zyxin which collectively function to maintain an actin-α-actinin interaction as the fundamental association of the sarcomere. Aspects of abnormal heart development and manifestation of a subset of muscular-based disease have previously been attributed to mutations in key structural proteins. Our study demonstrates an essential requirement for direct transcriptional regulation of sarcomere integrity, in the context of enabling fetal cardiomyocyte hypertrophy, maintenance of contractile function and progression towards inherited or acquired myopathic disease.
Keywords: Prox1, heart development, myocardium, sarcomere, hypertrophy, myopathy
INTRODUCTION
Cardiac muscle integrity and function is dependent upon the maintenance of a rhythmically contracting meshwork of myofibrils, the basic structural and functional unit of which is known as the sarcomere. Sarcomeres are multi-protein complexes comprising interlacing myosin (thick) and actin (thin) filaments bordered by Z-discs. Several proteins important for the stability of sarcomeric structure are found in the Z-disc which not only has a structural role for cross-linking thin filaments and transmitting contractile force but also provides a vital interface for signal transduction and biomechanical sensing (Frank et al., 2006; Pyle and Solaro, 2004).
There is an absolute requirement for cardiac function during embryogenesis in mammals and as such the sarcomeric components are expressed very early in development and are correctly localised in the myofibrils by the time the linear heart tube begins to contract (Ehler et al., 1999). As development progresses, the heart increases in mass not only by cardiomyocyte hyperplasia but also through a recently recognized fetal phase of physiological hypertrophy (Hirschy et al., 2006); a process dependent upon myofibril disassembly, reassembly (Ahuja et al., 2004) and elongation (Hirschy et al., 2006). Despite insight into the morphogenetic events which accompany myofibrillogenesis and hypertrophic growth very little is known about the precise regulation of these processes during development. Furthermore, the importance of appropriate assembly and maintenance of the myofibrillar apparatus is underscored by the fact that defects in terminal differentiation and arrangement of contractile protein filaments are associated with a number of cardiac myopathies (Engel, 1999; Gregorio and Antin, 2000; Seidman and Seidman, 2001). Although many of these disorders are caused by mutations in components of the sarcomere (Ariza et al., 1995), or by perturbations in the associated calcium-dependent signalling pathways (Frey et al., 2004; Molkentin et al., 1998), insight into the precise role of dysfunctional regulatory proteins in the generation of these pathological conditions is limited and in the context of regulation of cardiac muscle structural proteins is largely unknown.
The homeobox transcription factor Prox1 has previously been shown to have essential roles during murine lymphatic, hepatocyte, retinal and pancreatic development (Dyer et al., 2003; Harvey et al., 2005; Sosa-Pineda et al., 2000; Wigle and Oliver, 1999). Multiple lines of evidence suggest a putative role for Prox1 during cardiac morphogenesis. Prox1 is expressed in the developing heart (Oliver et al., 1993; Rodriguez-Niedenfuhr et al., 2001; Tomarev et al., 1996; Wigle and Oliver, 1999) and embryos deficient in Prox1 die at around E14.5; a critical time point in development when lethality can be correlated with, and often results from, grossly reduced cardiac performance (Dyson et al., 1995). Moreover, in a specific genetic background, a proportion of Prox1-heterozygote mice fail to survive and become cyanotic soon after birth (Harvey et al., 2005); a phenotype consistent with impaired blood circulation and heart defects. Clues as to a potential role in cardiac muscle ultrastructure came from a study in zebrafish whereby Prox1 was shown to regulate terminal differentiation and myofibrillar organisation in the skeletal muscle lineage (Baxendale et al., 2004; Roy et al., 2001).
Here we reveal how Prox1 functions transcriptionally upstream of sarcomere assembly, myofibril organization and fetal cardiomyocyte growth. Accordingly, hearts which lack Prox1 display a range of cardiac anomalies, including severe sarcomeric and myofibril disruption and impaired myocyte hypertrophy. We provide evidence that Prox1 activity is required for normal expression and localisation of multiple sarcomeric components and that it directly regulates α-actinin, N-RAP (nebulin-related anchoring protein) and Zyxin, essential structural proteins, required for stabilising actin within the thin filaments and ultimately in establishing cardiomyocyte elongation and coordinated muscle contraction. These results, therefore, provide important insight into the molecular mechanisms that govern the ultrastructure and growth of cardiac muscle during development and highlight how transcriptional mis-regulation of myofibril assembly may underlie cardiac hypotrophy and myopathic disease.
MATERIALS AND METHODS
Mouse Strains and Histology
All mouse strains used have been described previously: Prox1-null (Wigle et al., 1999), Prox1loxP (Harvey et al., 2005), Nkx2.5CreKI (Moses et al., 2001) and MLC2vCreKI (Chen et al., 1998). Prox1loxP/loxP mice were crossed with Nkx2.5CreKI;Prox1loxP or MLC2vCreKI;Prox1loxP in order to generate Nkx2.5CreKI;Prox1loxP/loxP (Prox1Nkx) or MLC2vCreKI;Prox1loxP/loxP (Prox1MLC) embryos, respectively. Embryos/hearts were dissected in PBS, fixed in 4% paraformaldehyde (PFA), dehydrated, embedded in paraffin and sectioned at 15 μm. Sections were haematoxylin and eosin stained.
Immunohistochemistry and TUNEL Assays
Embryos were treated as above and sectioned at 10 μm. Immunohistochemistry was performed using anti-Prox1 (Reliatech) and anti-phosphohistone H3 (Upstate) antibodies, developed using a standard streptavidin-HRP method and counterstained with haematoxylin. DNA fragmentation was detected by TUNEL assay according to the manufacturer’s protocol (Promega).
Immunofluorescence and Confocal Microscopy
E10.5–E14.5 whole mount hearts were dissected in PBS and fixed overnight in 4% PFA. E18.5 hearts were dissected, embedded in wax and sectioned as described above. Immunofluorescence and confocal microscopy was performed as described previously (Ehler et al., 1999). The samples were analysed on a Zeiss LSM 510 confocal microscope equipped with argon and helium neon lasers using a 63x/1.4 lens. Image processing was done using the Zeiss software and Photoshop. Cell outlines were traced on sections, blinded to genotype, using Image J (http://rsb.info.nih.gov/ij/) to assist in assessing cell shape and calculating cell area as an indication of fetal hypertrophy.
Antibodies
Sarcomeric α-actinin (clone EA-53), β-catenin, vinculin and smooth muscle α-actin (clone 1A4; Sigma), cardiac α-actin (Progen), titin (T12; Prof D. Fürst), desmin (clone D33; DAKO), MHC (A4.1025; DSHB), cardiac MyBPC (Prof T. Obinata), CD31 (PECAM; BD Pharmingen), Prox1 (Reliatech) and GAPDH (Chemicon). For western blots a rabbit anti-sarcomeric α-actinin antibody was used (Prof D. Fürst).
Transmission Electron Microscopy
E13.5/E18.5 hearts were dissected and fixed in 3% glutaraldehyde (EM Grade, Agar Scientific) in 0.1 M Sodium cacodylate and 5 mM CaCl2 pH7.4. The hearts were processed in a Lynx automated tissue processor (Australian Biomedical) and embedded in resin. All sectioning was carried out on a Reichert Ultracut S ultramicrotome. Sections were imaged using a Philips CM 10 transmission electron microscope and images collected using Kodak Megaview II and SIS Keenview digital imaging systems and SIS software. Four Prox1Nkx and four control hearts were examined for each stage.
Western Blotting
E13.5 heart lysates were prepared in RIPA buffer. Western blotting was performed using standard methods. Scanning densitometry was performed and signal quantified using Scion Image (Scion Corporation) and Image J (http://rsb.info.nih.gov/ij/).
Quantitative Real-Time PCR Analysis
mRNA was isolated from E12.5 hearts using the Micro FastTrack 2.0 kit (Invitrogen), according to the manufacturer’s instructions. Reverse transcription was performed using Superscript III RT (Invitrogen), according to the manufacturer’s instructions. qRT-PCR analysis was performed on an ABI 7000 Sequence Detector (Applied Biosystems) using SYBR Green (Quantitect™ SYBR® Green PCR Kit, Qiagen). Data was normalised to Hprt expression and analysed using DART-PCR (Peirson et al., 2003). P-values were obtained using Student’s T-Test (n=9). Complementary DNA PCR primers were obtained from Primer Bank (http://pga.mgh.harvard.edu/primerbank/) or designed using Primer Express software (version 2.0, Applied Biosystems); details of primer sequences can be found in Supplementary Table 1A.
RNA in situ hybridisation on embryonic sections
E13.5 embryos were fixed in 4% paraformaldehyde, embedded in paraffin wax and sectioned at 15 μm. RNA in situ hybridisation on sections was performed as previously described (Moorman et al., 2001), using a Digoxigenin-labelled antisense riboprobe specific for Nppa (Kuo et al., 1997).
ChIP-on-chip
E12.5 hearts were dissected in PBS/0.3% Triton X-100 and cross-linked for 3 hours at RT in 1.8% formaldehyde, homogenised in lysis buffer and sonicated. 60 μg chromatin lysate was used per IP with 10 μg anti-Prox1 antibody (Reliatech) in ChIP dilution buffer at 4ºC overnight. A no antibody ‘IP’ was performed as a control. Immune complexes were pulled down with Protein A/G beads washed, resuspended in TE and incubated at 65ºC overnight to reverse cross-links, after which DNA was purified. 10 ng DNA was blunt-ended and unidirectional adapters were ligated overnight at 16ºC. Adapter-ligated DNA was amplified by PCR. Experimental conditions, buffer composition, adapter sequences and PCR conditions can be found in the Supplemental Material. 7.5 μg amplified chromatin was fragmented and end labelled using GeneChip® WT Double-Stranded DNA Terminal Labeling Kit (Ayymetrix), hybridised to GeneChip® Mouse Promoter 1.0R Arrays (Affymetrix) then stained using GeneChip® Hybridization, Wash, and Stain Kit (Affymetrix). ChIP data was analysed using CisGenome (v1.0_beta) (http://www.biostat.jhsph.edu/~hji/cisgenome/index.htm). Three independent ChIP and no antibody control reactions were performed. Standard PCR to confirm ChIP of the enhancer elements was carried out using primers as outlined in Supplementary Table 1B.
EMSAs
Electrophoretic Mobility Shift Assays (EMSAs) were carried out as described previously (Angelo et al., 2000). Briefly in vitro translated (IVT) Prox1 or nuclear extracts from P19Cl6 cells (Habara-Ohkubo, 1996) transfected with Flag-Prox1 were incubated with 32P-labelled oligonucleotide in binding buffer at room temperature for 30 min. Overlapping 60 bp oligonucleotides, which spanned the entire binding elements for Actn2, Nrap and Zyxin as identified by ChIP-on-chip, were used in preliminary binding reactions to narrow down each element to a single 60 bp oligonucleotide, sequences highlighted in red in Fig. S9. Cold oligonucleotide (10x excess) was used for competition binding assays. Anti-Flag (M2; Sigma; 4.6 μg) was used to supershift bound Prox1 in the transfected P19Cl6 cell extracts. Each binding reaction was run through an 8% polyacrylamide gel, which was then dried and subjected to autoradiography.
Reporter transactivation assays
P19CL6 cells were maintained in standard P19 culture conditions (McBurney et al., 1982). Transfections were carried out using Effectene™ reagent as described previously (Hill and Riley, 2004). Briefly, duplicate wells of P19CL6 cells were transfected with a reporter in which luciferase was downstream of putative Prox1 binding elements from either Actn2, Nrap and Zyxin and a chick α-cardiac actin minimal promoter (Hill and Riley, 2004) with or without pcDNA3-Prox1 (250 ng) and a β-actin-β-galactodsidase expressing plasmid to normalise luciferase activity for transfection efficiency. Luciferase and β-gal activity were assayed 48 hours post transfection as previously described (Hill and Riley, 2004).
RESULTS
Prox1 is expressed in the embryonic heart
In the first instance we characterised Prox1 expression during murine embryonic heart development, comparing multiple stages (E10.5–14.5) and determining cell specific expression, compared to the restricted analyses reported previously (Oliver et al., 1993; Wigle and Oliver, 1999). Immunostaining revealed that Prox1 is expressed in the entire myocardium of the atria, ventricles and outflow tract from around E10.5 (Fig. 1). At E12.5 and E14.5 Prox1 expression is notably in the interventricular septum and myocardium (Figure 1B–E), but is excluded from the smooth muscle of the outflow tract (not shown). Furthermore, Prox1 is also expressed in the outer endocardial layer of the atrioventricular endocardial cushion leaflets (Fig. 1E, F), and the mitral valve leaflets (Fig. 1F, H), but is absent from the cushion mesenchyme (Fig. 1F–G). Confocal analysis confirmed the predicted nuclear localization of Prox1 in atrial and ventricular myocytes (Fig. S1A–D).
Figure 1. Prox1 is expressed in the myocardium of the developing heart.
Immunofluorescence on E10.5 (A) and E12.5 (B, C) frontal sections using an antibody for Prox1. Prox1 is expressed throughout the myocardium of the presumptive left and right ventricles and outflow tract at E10.5 (A) and is localised to both atrial and ventricular myocardium at E12.5 (B, C). Immunohistochemistry on E14.5 frontal sections, illustrates continued expression throughout the entire myocardium (D), in the interventricular septum (E), endocardium of the atrioventricular canal endocardial cushions (F, G) and mitral valve leaflets (F, H). Prox1 is absent from cushion mesenchyme (F, G); boxed panels (e–h) show no primary antibody controls for corresponding panels (E–H). Abbreviations: lv, left ventricle; rv, right ventricle; ivs, interventricular septum; ec, endocardial cushion; en, endocardium; mes, mesenchymeL; oft; outflow tract. Scale bars: A, B, 50 μm; C, 25 μm; D, 50 μm; E, 25 μm; F–H, 10 μm
Heart development is abnormal in Prox1-null mice
Mice that are homozygous null for Prox1 die between E14.5 and E15 (Wigle and Oliver, 1999). The cause of lethality in Prox1-null embryos has not been determined. Previously ink injections were used to assess the developing vasculature and extrapolated to conclude “normal” cardiac output (Wigle and Oliver, 1999), however, this assay cannot determine appropriate function of the developing heart or yield insight into any phenotypic (organ wide or cell-specific) abnormalities. Therefore, loss of the cardiac expression of Prox1 is a potential contributing factor to embryonic lethality. To address this we initially examined surviving postnatal day 5 (P5) Prox1-heterozygous mice and observed, at a gross anatomical level, that the hearts were reduced in size (by an average of 30%) compared to wildtype littermate controls (Fig. S2A). Histological analysis revealed a range of cardiac anomalies including hypoplastic ventricular walls, loss of muscle striation, a disorganised interventricular septum and abnormally persistent muscle surrounding the aorta (Fig. S2B, D). Analysis of Prox1-null embryos at E13.5 and E14.5 embryos revealed that mutant hearts were significantly smaller (by up to 50%) compared to wildtype littermates (Fig. S2E, F) and, consistent with the phenotype in the P5 heterozygotes, we observed low resolution myocardial disarray, including a disrupted interventricular septum (Fig. S2G, H).
Prox1 is an essential regulator of heart development
Conventional Prox1-null embryos have a lymphatic phenotype arising from defects in endothelial cell budding as early as E11.5 (Wigle and Oliver, 1999) which may adversely affect cardiovascular development. In order to confirm a primary role for Prox1 in the developing heart, we generated a cardiac-specific knockout of Prox1 by crossing a conditional homozygous floxed Prox1 strain (Prox1loxP/loxP; (Harvey et al., 2005) with two cardiac-expressing Cre strains: Nkx2.5CreKI (designated Prox1Nkx), which directs expression of Cre recombinase throughout the majority of cardiomyocytes from E7.5 (Moses et al., 2001) and MLC2vCreKI (designated Prox1MLC) in which Cre is expressed specifically in ventricular cardiomyocytes from E8.75 (Chen et al., 1998). The two Cre strains were employed, therefore, to target Prox1 globally throughout the entire myocardium and in a subpopulation of ventricular myocardium, thus acting as respective internal controls for a cardiomyocyte-specific loss-of-Prox1 function. Prox1 protein levels in Prox1Nkx and Prox1MLC E13.5 hearts were analysed by western blot on whole heart lysates, followed by densitometry, which confirmed an approximate three-fold reduction in Prox1 protein levels in the cardiac-specific knockout hearts compared to littermate control hearts (Prox1loxP or Cre allele only; Fig. 2A). Whole mount and histological sections of Prox1Nkx and Prox1MLC embryos were examined between E10.5 and E18.5. No apparent phenotype was evident between E10.5 and E12.5 (not shown), despite expression of Prox1 in the heart at these stages of development. From E12.5 onwards Prox1Nkx and Prox1MLC embryos developed an array of cardiac defects. At E13.5 and E14.5 the Prox1Nkx and Prox1MLC conditional mutants recapitulated the cardiac phenotype in the conventional knockout embryos described above. Prox1Nkx embryos were slightly growth restricted (on average 5% smaller) compared to control littermates, with cranial haemorrhaging. Both Prox1Nkx and Prox1MLC embryos had oedema, of varying severity (Fig. 2B), indicative of inadequate cardiac function. The hearts of both Prox1-conditional mutants were up to 30% smaller compared to controls (Fig. 2C–E). Furthermore, in Prox1Nkx hearts the right atrium was reproducibly expanded (by up to 2-fold) and blood-filled, never appearing to empty appropriately, irrespective of fixation stage during the cardiac cycle, suggesting there may be impaired blood flow through the heart (Fig. 2D). Moreover, in the Prox1MLC hearts there was reduced expansion of the left ventricle (25% reduced chamber size) consistent with haemodynamic obstruction (Fig. 2E). Haematoxylin and eosin stained frontal sections through E14.5 and E13.5 hearts (Prox1Nkx, Fig. 2F–K; Prox1MLC, not shown) revealed small thin-walled ventricles, disrupted myocardium with a highly disorganised interventricular septum (Fig. 2H, I). In addition, membraneous ventricular septal defects (VSD) were observed in Prox1Nkx embryos (Fig. 2J, K) which were unlikely to be due to the disrupted myocardium, but possibly related to additional endocardial cushion defects (not shown). By E18.5 the overall area of ventricular myocardium was significantly smaller in Prox1Nkx hearts, in particular that of the right ventricle. Moreover the myocardium was disorganised and less compacted (reduced by up to 50%) and the surface of the ventricles irregular, compared to littermate controls, although both compact and trabecular layers did appear to form (Fig. 2L–N). Consistent with the predominantly right-sided defects in Prox1Nkx embryos, the pulmonary trunk was reduced in diameter, often half the size of the aorta, suggesting impaired outflow tract remodelling. At E18.5 the membraneous VSD was still evident in Prox1Nkx embryos, and we also observed muscular VSDs (Fig. 2N) that likely result from the reduced level of myocardial compaction in the ventricles.
Figure 2. Cardiac-specific loss of Prox1 perturbs embryonic and heart development.
Western blots of E13.5 control, Prox1Nkx and Prox1MLC individual isolated heart lysates using antibodies for Prox1 and GAPDH and quantification of protein levels, normalised to GAPDH, using scanning densitometry (A). Bright-field whole mount left lateral views of E14.5 Prox1Nkx, Prox1MLC and control (co) littermate embryos (B) and frontal views of hearts in control (C), Prox1Nkx (D) and Prox1MLC (E) embryos. Frontal sections through E14.5 control (F, H) and Prox1Nkx (G, I) embryos, E13.5 isolated control (J) and Prox1Nkx (K) hearts, and E18.5 isolated control (L) and Prox1Nkx (M, N) hearts. Prox1 protein levels are reduced to around a third of control levels in Prox1-conditional hearts (A). Prox1Nkx mutants are small with oedema and cranial haemorrhaging and Prox1MLC mutants reveal extensive oedema (B; white arrowheads). Prox1Nkx hearts are hypoplastic with dilation of the right atrium (white dotted lines in C and D; G) and Prox1MLC hearts are hypoplastic with reduced left ventricular expansion (E). Sections through Prox1Nkx hearts reveal myocardial disarray, particularly in the interventricular septum (H, I). By E18.5 Prox1Nkx hearts are rounded in shape and smaller than control hearts, notably the right ventricle, the ventricular wall surface is irregular (black arrowheads in M, N), there is reduced compaction (black lines in L, M) and muscular septal defects (asterisk in N). Also note the membranous ventricular septal defect observed in Prox1Nkx hearts (black arrow; K, M). Abbreviations: lv, left ventricle; rv, right ventricle; ra, right atrium; ivs, interventricular septum; ec, endocardial cushion. Scale bars: B, 5 mm; C–E, F–G, J–K, 50 μm; H–I, 10 μm; L–N, 1 mm
We observed variable and incomplete levels of Prox1 deletion in the developing heart (Fig. 2A), attributed to the mosaic nature of Cre expression, previously described for both the Nkx2.5CreKI and MLC2vCreKI strains (Smart et al., 2007). Analysis of both Prox1Nkx and Prox1MLC mutant hearts confirmed the specificity of the phenotype at the level of the myocardium and this was further supported by immunostaining for markers of vascular endothelial and smooth muscle cells (PECAM and SMαA, respectively) which revealed normal coronary vessel development (extensive differentiation of both vascular cell types) in the mutant hearts as compared to controls (Fig. S3A–D). Due to the myocardial phenotypic similarities observed in both models, the term Prox1-conditional will be used to describe both the Prox1Nkx and Prox1MLC hearts unless specifically stated otherwise.
Sarcomeric integrity is disrupted in Prox1-conditional hearts
At a gross level, determined by low resolution histological analyses, loss of Prox1 function in the heart appeared to lead to myocardial disarray manifested via disorganisation of the myofibrils. To investigate the apparent disruption of the myocardial architecture and the lack of striation observed in the ventricular wall in more detail, the expression and localisation of sarcomeric markers was analysed by high resolution confocal microscopy following immunostaining of whole mount embryonic hearts. The relative distribution of the major components of the sarcomere is illustrated in Fig. S4A. In the first instance, co-localisation of Prox1 and selected sarcomeric markers (actin and sarcomeric α-actinin) was confirmed in E13.5 control hearts (Fig. S1A–D). Prox1-conditional E13.5 and E14.5 whole mount hearts were stained with phalloidin, to visualise F-actin and the arrangement of the thin filaments, and immunostained with antibodies to sarcomeric myosin heavy chain (MHC) and cardiac myosin binding protein C (MyBP-C) to visualise the thick filament architecture. Additionally, hearts were immunostained with antibodies to sarcomeric α-actinin, a critical Z-disc protein that cross-links sarcomeric actin and is involved in many signalling transduction pathways; myomesin, which localises to the M-band where it interacts with myosin and titin providing elasticity and stability to the sarcomere, and β-catenin, which localises to the adherens junctions and demarcates cell-cell contacts (Fig. 3; see also Figures S4A and S5A–F). In Prox1-conditional hearts at E13.5 we consistently observed severe global disruption of thin filament, thick filament, Z-disc and M-band organisation in the ventricular myocardium compared to littermate controls (Fig. 3A-I, L); there was little or no striation and the punctate expression patterns of actin, sarcomeric MHC, MyBP-C, myomesin and sarcomeric α-actinin were severely disrupted. Variation in severity of the disruption of actin filaments and α-actinin, was evident (Fig. S6) consistent with mosaicism in the level of Prox1 knockdown (Fig. 2A; western analysis for Prox1 expression with scanning densitometry normalised to GAPDH). We classified overall phenotype severity based on the following three criteria revealed by the confocal ultrastructure analysis: i) cardiomyocyte cell shape (Image J; see Materials and Methods), ii) sarcomeric striation and iii) myofibrilar organisation/cell alignment; defects in one of these categories were classed as mild of which we recorded 14.3% of Prox1Nkx (n=56) and 18.2% of Prox1MLC mutants (n=41); defects in all three criteria were classed as severe: 85.7% of Prox1Nkx (n=56) and 81.8% of Prox1MLC mutants (n=41) at E13.5. Despite variation in severity of phenotype, the nature of the specific defects common to both the Prox1Nkx and Prox1MLC mutants, presented as muscle ultrastructure anomalies, confirmed a cardiomyocyte-autonomous role for Prox1. The severe phenotype was incompatible with embryonic survival, whereas incomplete deletion of Prox1 resulting in the mild phenotype contributed to a low incidence of survival of Prox1-conditional mutants to post-natal stages (5/111 Prox1Nkx and 0/64 Prox1MLC). Additionally, we occasionally observed thin filament and Z-disc disruption and loss of striation in Prox1Nkx atrial myocardium (not shown), which is consistent with the expression patterns of Prox1 and Nkx2.5. No disruption was observed in β-catenin localisation (Fig. S5A–F), suggesting that the intracellular portion of the adherens junctions is not affected.
Figure 3. All structural components of the sarcomere are severely disrupted in Prox1-conditional myocardium.
Confocal sections of immunostained E13.5 whole mount hearts from control (co; A–D, I–K) and Prox1Nkx (E–H, L–N) embryos. Actin thin filaments and myosin thick filaments lack organisation and are not striated in Prox1-conditional hearts as visualised by phalloidin staining (green; A, E) and immunostaining for sarcomeric MHC (red; B, F), respectively. Immunostaining for the thick filament component cardiac MyBP-C further demonstrates thick filament disorganisation (green; C, G). M-band disruption is demonstrated by immunostaining for myomesin (red; D, H). Z-disc disruption in Prox1Nkx hearts is revealed by immunostaining for sarcomeric α-actinin (red; I, L), titin N-terminus (green; J, M) and desmin (red; K, N). Quantitative real-time PCR (qRT-PCR) for sarcomere component genes on E12.5 Prox1Nkx hearts (O). Western blots of E13.5 control and Prox1Nkx individual (half) heart lysates using antibodies for Prox1 (non-specific band highlighted by black arrowhead), sarcomeric α-actinin, sarcomeric MHC and GAPDH, and quantification of protein levels normalised to GAPDH using scanning densitometry (P). Abbreviations: ns, non-specific.
To specifically evaluate Z-disc integrity in Prox1-conditional hearts, we used antibodies raised against the N-terminus of titin (Z-disc localised) and to desmin. Titin is a giant protein that spans half the width of a sarcomere, with its C-terminus localised at the M- band and N-terminus at the Z-disc, where it interacts with sarcomeric α-actinin. Desmin is an intermediate filament protein that also interacts with sarcomeric α-actinin at Z-discs, providing a lateral connection between Z-discs of adjacent myofibrils (Fig. S4A). Prox1-conditional E13.5 hearts were triple stained for titin (Fig. 3J, M), desmin (Fig. 3K, N) and phalloidin (not shown), which demonstrated that a loss of Prox1 results in a severe disruption of Z-disc organisation. The full extent of sarcomeric protein misregulation in Prox1-conditional hearts is summarised in Fig. S4B.
Given that Prox1 is a transcription factor, we sought to determine potential alterations to the expression levels of myofibril and Z-disc components which may underlie the ultrastructure defects. E12.5 and E13.5 Prox1-conditional and control isolated hearts were examined by quantitative real time (qRT)-PCR and western blotting, respectively. qRT-PCR was carried out on genes encoding the sarcomere components that were shown to be mislocalised in Prox1-conditional myocardium. α-cardiac actin (Actc1), sarcomeric α-actinin (Actn2) and β-MHC (Myh7) were significantly downregulated and MyBP-C (Mybpc3) was significantly upregulated (Fig. 3O). Titin (Ttn) and desmin (Des) were consistently downregulated but to largely varying levels, which led to the change not being statistically significant (Fig. 3O). Nonetheless, these results confirm the extensive sarcomere dysgenesis in the Prox1-conditional mutant hearts both in terms of appropriate organisation of the myofibrils and at the level of the expression of genes which encode key sarcomere proteins in the developing myocardium. Additionally, we analysed expression of sarcomere components at the protein level in E13.5 individual hearts. In accordance with the changes in gene expression, sarcomeric α-actinin and sarcomeric MHC were down-regulated in Prox1-conditional hearts (Fig. 3P). Levels of desmin, β-catenin and vinculin were unchanged (not shown), which alongside the confocal immunofluoresence for β-catenin (Fig. S5A–F), confirmed that cell-cell contacts through adherens type junctions are unaltered between neighbouring Prox1-conditional cardiomyocytes. Subsequently we examined the relationship between the level of Prox1 reduction and α-actinin expression further by taking the same heart sample for analysis using both western blotting and immunostaining and observed a direct correlation between the degree of Prox1 knock-down and reduced sarcomeric α-actinin levels (Fig. 3O, P). This suggests that Prox1 activity may regulate the expression of sarcomeric α-actinin and, moreover, that a modest yet significant reduction (0.7 fold) in α-actinin is sufficient to contribute to the myofibrillar disarray in the conditional mutant hearts (Fig. 3I, L). Furthermore, this analysis demonstrated a definitive association between the level of Prox1 expression and the severity of the myocardial phenotype (Fig. 3O, P; Fig. S6).
Transmission electron microscopy was carried out to examine the sarcomere ultrastructure disruption at higher resolution in both E13.5 and E18.5 Prox1Nkx myocardium. At both developmental stages examined, we observed variation in severity of sarcomere ultrastructure defects in Prox1Nkx ventricular myocardium (Fig. 4A–F). Z-disc material was either completely absent (Fig. 4B) or relatively dense but inappropriately organised (Fig. 4D). There was also associated misalignment of the M-bands with thick and thin filament disarray, suggesting reduced Prox1 function impacts globally on sarcomeric organisation (Fig. 4D; see discussion). Further examination of the cell-cell contacts as a possible source of disruption to the muscle ultrastructure, revealed both intact adherens type junctions (Fig. S5G, H), consistent with the β-catenin immunofluoresence data (Fig. S5A–F), and intact desmosomes (Fig. S5I, J) in the Prox1-conditional mutant hearts. Despite the variation in the phenotype, in the most severely affected cases we consistently observed a total lack of sarcomeric organisation at both E13.5 and E18.5 (Fig. 4E, F). Therefore, in combination, the confocal and electron microscopy studies implicate Prox1 in organising components of the sarcomere and, furthermore, suggest that Prox1 is involved in maintaining myofibril ultrastructure throughout cardiogenesis.
Figure 4. Electron micrographs of muscle ultrastructure defects in Prox1-conditional myocardium.
Transmission electron microscopy (TEM) on E13.5 (A, B, E) and E18.5 (C, D, F) control (A, C) and Prox1Nkx (B, D, E, F) hearts confirms the sarcomeric disruption in Prox1Nkx ventricular myocardium; note (C) and (D) are the same orientation and plane of section. There can be total loss of Z-disc (Z) material and intact M-band (M; A), an accompanying disruption of the M-B and (B), or disruption to the thick and thin filament alignment (dashed lines; C, D), associated with Z-disc disorganisation (T), whilst in the most severely affected hearts TEM reveals complete myofibril disarray (E, F). Scale bars: A–F, 500 nm.
Prox1 is essential for physiological fetal hypertrophy
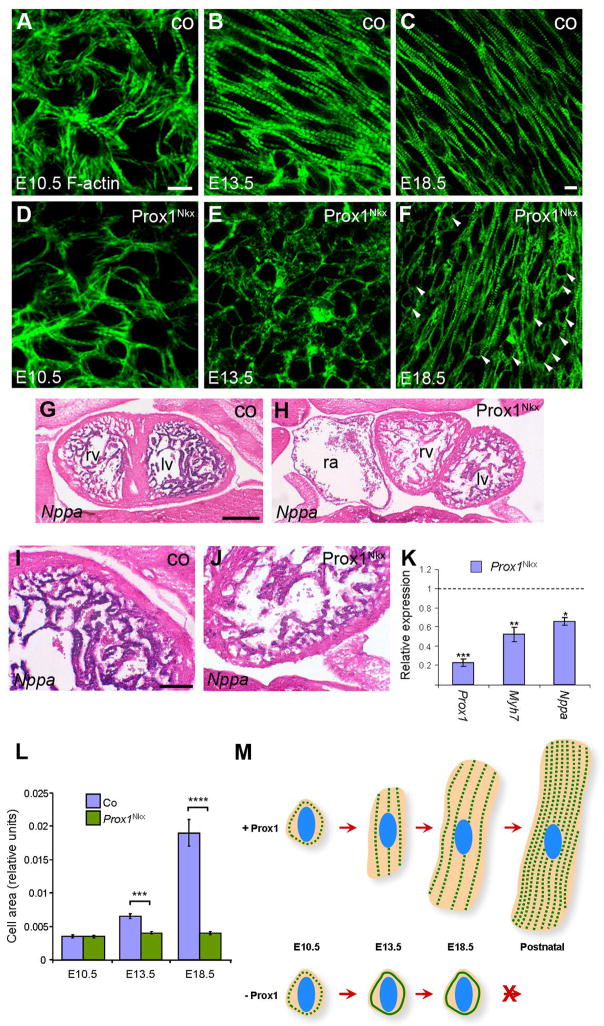
We next investigated whether Prox1 plays a role in either the initial stages of myofibrillogenesis or in the maintenance of appropriate myofibril structure throughout later stages of cardiac development. To this end we immunostained whole mount Prox1-conditional hearts at E10.5, E11.5 and E12.5 for sarcomeric α-actinin and phalloidin to pinpoint exactly when the myocardial defects were first arising. In the Prox1Nkx mutants Prox1 levels were reduced from early cardiac crescent stages (E7.5) onwards (Moses et al., 2001) and yet at E10.5 and E11.5 there was no sarcomeric disruption and the myofibrils appeared to have assembled correctly (Fig. 5A, D), whereas, by E12.5 the first signs of myofibril disorganisation were observed (Fig. S7A–D). The fact that sarcomere defects do not manifest until E13.5 and then persist throughout the rest of development suggests that Prox1 is not required for initiation of myofibrillogenesis but for subsequent maintenance of appropriate sarcomeric structure and stability. In addition, we observed, in E13.5 Prox1-conditional hearts, that cardiomyocyte morphology and distribution were noticeably altered; the cardiomyocytes were more rounded in shape, disorganised and distributed in an apparently random pattern compared to the control hearts, in which the cardiomyocytes had begun to elongate and align in parallel (Fig. 3A–N). Consistent with this, we observed a significant increase in α-actinin expression in wild type hearts between E9.5 and E12.5, indicative of a requirement for sarcomeric-dependent cell growth, post recruitment of cells and at the onset of physiological hypertrophy (Fig. S7E). We, therefore, proceeded to examine sections of E18.5 Prox1-conditional and control hearts by staining with phalloidin and immunostaining for sarcomeric α-actinin to determine whether there was a failure in fetal cardiomyocyte physiological hypertrophy. Accordingly, we observed that a large proportion of cardiomyocytes are still rounded at E18.5, having failed to undergo hypertrophic growth and acquire the characteristic rod shape of mature cardiomyocytes (Fig. 5C, F). Hypertrophy in adult hearts is induced by a large variety of stimuli but the consistent end point is re-expression of a fetal gene program, including Nppa (Anf), Myh7 (β-MHC) and Nppb (BNP)(Molkentin et al., 1998). During development Nppa, Myh7 and Nppb are markers of myocardial differentiation and chamber expansion, but since differentiation per se is accompanied by concomitant increase in cardiomyocyte cell growth we examined the expression levels of Nppa and Myh7 as surrogate markers of fetal hypertrophy. qRT-PCR on E12.5 hearts and in situ hybridisation on E13.5 embryo sections, revealed that both genes were found to be significantly down-regulated in Prox1Nkx hearts (Fig. 5G–K) indicating that not only was there markedly reduced fetal hypertrophic growth in the absence of Prox1, but also suggesting that Prox1-conditional hearts are not hypoplastic, but hypotrophic. Morphometric measurements of cell size revealed that cardiomyocytes in Prox1Nkx hearts failed to enlarge during development, compared to littermate controls, confirming a defect in hypertrophic growth as opposed to an alteration in cell shape (Fig. 5L). To confirm there was no defect in proliferation and/or apoptosis we performed immunostaining using an antibody against phospho-histone H3, a marker of cells in mitosis, and Terminal deoxynucleotidyl Transferase Biotin-dUTP Nick End Labeling (TUNEL), respectively. No decrease in proliferation or increase in apoptosis was observed between E10.5 and E13.5 (Fig. S8). Therefore, in the absence of Prox1, although laid down appropriately in early heart development, the sarcomere structure is subsequently not maintained resulting in myofibril disruption and loss of striation and a failure of cardiomyocyte hypertrophic growth and maturation (Fig. 5M).
Figure 5. Prox1 is required for fetal cardiomyocyte hypertrophy.
Phalloidin staining to visualise actin filaments on E10.5 (A, D), E13.5 (B, E) and E18.5 (C, F) control (co; A–C) and Prox1Nkx (D–F) whole mount (A, B, D, E) and sections through (C, F) isolated hearts. At E10.5 Prox1Nkx cardiomyocytes are developing normally and the appropriate ultrastructure is laid down (A, D). From E13.5 onwards Prox1Nkx cardiomyocytes fail to grow by hypertrophy but remain small, rounded cells that do not acquire the characteristic rod shape (arrows; E, F). In situ hybridisation for Nppa transcripts on frontal sections of E13.5 control (G, I) and Prox1Nkx (H, J) embryos. There is greatly reduced Nppa expression in Prox1Nkx myocardium (H, J), confirmed by qRT-PCR (K) on E12.5 isolated hearts. β-MHC (Myh7) was also found to be downregulated by qRT-PCR (K). Morphometric analysis of cell shape (Image J; http://rsbweb.nih.gov/ij/) confirmed a lack of increase in cell size through impaired elongation and hypertrophic growth in Prox1Nkx cardiomyocytes during development, excluding the possibility that the rounded cells simply reflect an alteration in cell shape (L). Representation of fetal cardiomyocyte hypertrophic growth throughout development from rounded polygonal shaped cells at E10.5 that elongate as the myofibrils grow by addition of sarcomeres to form mature postnatal cardiomyocytes with the characteristic rod shape and aligned myofibrils, whilst in the absence of Prox1 sarcomere striation is lost, myofibrils do not align and cardiomyocytes do not grow by hypertrophy (M). Abbreviations: lv, left ventricle; rv, right ventricle; ra, right atrium. In (K, L) data are represented as mean +/− SEM; * p<0.001; ** p<0.003; *** p<9E-07 (K); *** p<7E-08; ****p<3E-12 (L). Scale bars: B–G, 10 μm; H–I, 50 μm; J–K, 20 μm
Prox1 directly regulates fundamental components of the sarcomere
To gain specific insight into the molecular mechanism(s) by which Prox1 regulates myofibril organisation we sought to identify direct downstream targets of Prox1 in the developing heart. Until now no in vivo target of Prox1 has been identified, nor has a definitive consensus Prox1 DNA binding site been determined. Therefore, we combined chromatin immunoprecipitation (ChIP), against endogenous Prox1, followed by microarray analysis (ChIP-on-chip). ChIP was performed on control Prox1loxP/loxP whole embryo lysates at E12.5. A parallel assay was performed with no antibody as a control. The whole embryo ChIP and the no antibody samples were checked by qRT-PCR for enrichment of a positive control and a previously identified in vitro target of Prox1, Fgfr- 3 (Shin et al., 2006) and against a negative control, Cyp7a1 (Qin et al., 2004). The ChIP chromatin was then hybridised to GeneChip® Mouse Promoter 1.0R Arrays (Affymetrix). ChIP-on-chip revealed putative Prox1-bound enhancer regions of genes encoding the Z-disc protein α-actinin (Actn2) and the myofibrillar and adherens junction proteins N-RAP (Nrap) and Zyxin (Zyx) both of which directly interact with α-actinin in the Z-disc (Fig. 6A). ChIP of the three enhancer elements was confirmed by PCR (Fig. S9A). Genes such as Actc1, Nppa and Myh7 which we revealed to be significantly down-regulated by qRT-PCR (Figs 3O, 4K) were represented on the Promoter 1.0R Arrays but were not detected following ChIP. We analysed the Actn2, Nrap and Zyx target sequences for predicted transcription factor binding sites using MatInspector Professional (http://genomatix.gsf.de); 40–50 transcription factor binding sites were predicted per sequence, including Pax6, a transcription factor that together with Prox1 co-regulates the Sox2 promoter in vitro (Lengler et al., 2005). No putative Prox1 or Prospero (Drosophila homolog of Prox1) binding sites were identified in any of the isolated regions, although the Prox1 sites described to date are highly degenerate and predicted based on a Prospero consensus (Cook et al., 2003; Lengler et al., 2005; Shin et al., 2006). The lack of core consensus motif within the three Prox1 bound enhancer elements identified for Actn2, Nrap and Zyxin, despite the conservation of each element across species, is consistent with results obtained from unbiased screens of genome-wide conserved regulatory sequence variants (Pennacchio et al., 2006).
Figure 6. Prox1 directly regulates structural proteins.
α-actinin, N-RAP and Zyxin. Sarcomere and sarcomere-related protein genes Actn2 (sarcomeric α-actinin), Nrap and Zyx (Zyxin) were identified as potential downstream targets of Prox1 by ChIP-on-chip. For each locus the genomic region pulled-down by IP with an anti-Prox1 antibody is indicated by the yellow box, the closest gene is labelled and degree of conservation is shown. The conservation patterns are based on phastCons scores downloaded from The University of California Santa Cruz (UCSC; http://genome.ucsc.edu; A). EMSAs with in vitro translated (lane demarcated IVT) Prox1 and 32P-labelled oligonucleotides (60 bp; B) identified from each of the Actn2, Nrap, Zyxin putative Prox1-binding elements (Fig. S9) isolated via the ChIP-on-chip shown in (A). A 10-fold excess of cold oligonucleotide was used in competitive assays in each case as evidence of specific binding (lane demarcated C). EMSAs with nuclear extracts from P19Cl6 cell lysates either untransfected (lanes 1–3) or transfected with Flag-Prox1 (lanes 4–6) and 32P-labelled elements as in (B). Lanes 1 and 4 are lysate alone, lanes 2 and 5 are lysate plus an α-Flag antibody and lanes 3 and 6 are α-Flag alone controls. Note the evidence of a supershift in lane 5 compared to lane 4 for each of the Actn2, Nrap and Zyxin elements, as highlighted by the white arrowheads and indicative of specific binding by Flag-Prox1. The presence of a comparatively weak band in lanes 1 and 2 in each case represent binding by endogenous Prox1 which is expressed in P19Cl6 cells (data not shown). In vitro transcription assays to demonstrate Prox1 transactivation of a luciferase reporter downstream of the Actn2, Nrap, Zyxin putative Prox1-binding elements and minimal reporter; note the significant activation by Prox1 of the Actn2, Nrap and Zyxin reporters (D). Real time qRT-PCR for Nrap and Zyxin confirmed reduced expression of these factors in a Prox1-deficient background (E), as was previously determined for Actn2 (Fig. 3O). In (D, E) data are presented as mean +/− SEM; * p<0.05; ** p<0.001; *** p<0.003; **** p<9E-07.
The ChIP data was subsequently validated by EMSAs with overlapping probes from each of the three enhancer regions and in vitro translated Prox1 or lysates from P19Cl6 cells overexpressing Prox1 (Fig. S9B–D). Cold probe competition gel shifts (Fig. 6B) and antibody supershifts (Fig. 6C) of the refined oligonucleotide sequences (60 bp) confirmed specific Prox1 binding to the enhancers within Actn2, Nrap and Zyx. Prox1-induced transcription via the elements of all three factors was demonstrated by co-transfection and reporter gene activation assays (Actn2 15-fold; Nrap 32-fold; Zyxin 9-fold activation; Fig. 6D). Real time qRT-PCR for Nrap and Zyx (Fig. 6E) confirmed reduced expression of these factors in a Prox1-deficient background, as was previously determined for Actn2 (Fig. 3O).
DISCUSSION
Failure of Prox1-conditional cardiomyocytes to grow and to maintain sarcomere organisation throughout development results in hypotrophic hearts that are insufficient to sustain life beyond birth. We observed disruption of the sarcomere at the level of both gene expression and protein localisation and have identified Prox1-bound enhancer elements for key sarcomere-associated genes, indicating that Prox1 regulates myofibrillar organisation both directly in terms of protein expression, and indirectly via intermediate factors which control sarcomere protein localisation and integration.
Prox1 deficiency impacts directly on sarcomeric components that facilitate Z-disc and thin filament interaction. Reduced expression of both Nrap and Zyx in Prox1-conditional hearts, and the demonstration that these two genes are direct transcriptional targets of Prox1, is highly significant in terms of maintaining Z-disc stability. N-RAP has been proposed to act as a catalytic scaffold for the association of thin filament actin and Z-disc α-actinin during myofibrillogenesis (Dhume et al., 2006), and Zyxin also interacts with α-actinin to facilitate actin assembly and organization (Crawford et al., 1992; Frank et al., 2006). During myofibrillogenesis, N-RAP and Zyxin are associated with cell-cell contacts that make up the developing intercalated discs, which ultimately mature during post-natal stages (Perriard et al., 2003). Our confocal and transmission electron microscopy (TEM) observations that cell junctions (β-catenin-positive adherens-type junctions and desmosomes) are appropriately established and remain intact, between neighbouring cardiomyocytes in Prox1-conditional mutants, suggests that the role of Prox1 is primarily to regulate Nrap and Zyx to facilitate cross-linking between actin and α-actinin in the Z-disc as one of the fundamental associations of the sarcomere (Fig. S4A). Actn2 is also implicated in this study as a direct target of Prox1 and the modest, yet significant, reduction in the expression levels of Actn2 in a Prox1 mutant background, correlating with the Z-disc disruption, underlines the critical role of α-actinin in maintaining sarcomere integrity. Moreover, since Zyxin-null mice are viable (Hoffman et al., 2003), although the hearts have not been examined in detail for histological defects, the phenotype we describe represents a cumulative effect on the actin-α-actinin interaction: directly via α-actinin expression and localization and through interactions with co-factors, N-RAP and Zyxin.
Perturbation of the actin-α-actinin association, directly explains the Z-disc anomalies in Prox1-null hearts. Whilst we cannot entirely exclude additional effects of loss of Prox1 function, normal levels of cardiomyocyte proliferation and apoptosis and the specificity of phenotype at the level of sarcomeric maintenance suggests that the latter is the primary defect in Prox1-conditional mutants. Moreover, thin filament-Z-disc disruption will feedback directly onto the thin and thick filament arrangement of the sarcomere as observed at the level of TEM, resulting in a more global disorganization of myofibrils (Fig. S4B). The latter may also explain the observed M-band defects, but equally these may relate more directly to mis-regulation of Nrap in the Prox1-mutant background, since N-RAP associates with the M-bands of maturing myofibrils where it acts as a catalytic scaffold (Lu et al., 2005).
Prox1 is not required for the initial stages of myofibrillogenesis since the phenotypic defects do not begin to manifest until E12.5; despite Prox1 expression in the myocardium of the heart at earlier stages (E10.5) of development and potential Cre-mediated knockdown of Prox1 occuring from E7.5 onwards. Sarcomere proteins are expressed immediately prior to the onset of beating and, once integrated into mature myofibrils, tend to have a relatively long half-life that varies between 3 and 10 days (Martin, 1981). Moreover, between E8.25 and E10.5 the developing heart increases in mass primarily through addition of cells from the second cardiac lineage (Zaffran et al., 2004). Therefore, there may be little requirement for newly synthesised structural proteins during these early stages of heart development. The initial activation of sarcomeric proteins is clearly carried out by alternate and as yet unidentified, transcriptional pathways with the role of Prox1 confined to regulating sarcomere maintenance and stability from E10.5 onwards, when all populations of cardiac cells have been acquired and the developing heart grows by a combination of both cardiomyocyte hyperplasia and hypertrophy. The fact that we observed significantly impaired hypertrophic growth following loss of Prox1 is secondary to the primary defect of disrupted assembly of sarcomere proteins. Developing cardiomyocytes elongate in a unidirectional manner by addition of sarcomeres to the existing myofibrils, the timing of which corresponds precisely with the onset of myocardial disruption and failure of the cells to elongate in Prox1-conditional myocardium (refer to model in Fig. 5M).
In conclusion, Prox1 is essential for maintenance and maturation of the sarcomere in developing cardiomyocytes, which in turn is critical for hypertrophic growth and maturation of the embryonic myocardium. The identification of structural proteins, α-actinin, N-RAP and Zyxin, as direct targets of Prox1 suggests that mis-regulation of critical sarcomere components and their interacting protein partners is the primary cause of myofibril disruption in Prox1-conditional myocardium. A number of other studies have described roles for transcription factors in initiating or maintaining cardiac muscle ultrastructure, during development and disease, most notably SRF (Balza, Jr. and Misra, 2006; Nelson et al., 2005) GATA4, Nkx2.5 and MEF2 (Akazawa and Komuro, 2003) and Calcineurin/N-FAT (Bourajjaj et al., 2008; Heineke and Molkentin, 2006), however, to the best of our knowledge no study to-date has demonstrated direct transcriptional regulation of structural proteins in vivo.
Aberrant terminal differentiation and improper assembly of contractile protein filaments are associated with a number of cardiac myopathies (Engel, 1999; Gregorio and Antin, 2000; Seidman and Seidman, 2001). Many of these disorders are caused by mutations in components of the myofibrillar apparatus itself including β-MHC, troponins T and I, titin and α-tropomyosin (Alcalai et al., 2008; Chang and Potter, 2005) or perturbations in the associated calcium-dependent signalling pathways (Frey et al., 2004; Molkentin et al., 1998). However, a large proportion of cardiomyopathy remains unexplained with no mutations found in sarcomere or sarcomere-related proteins. Our study not only provides novel insight into transcriptional regulation of cardiomyocyte ultrastructure and hypertrophy during development, but implicates Prox1 as a critical regulatory factor which may underlie the pathology of both inherited and acquired myopathic disease.
Supplementary Material
Acknowledgments
We would like to thank P. Scambler and members of the Molecular Medicine Unit for insightful comments. We would like to thank K. Venner for technical assistance with electron microscopy, A. Cook for advice on E18.5 heart analysis, C. Shang for advice on the ChIP protocol and D. Kelberman for advice on the EMSA protocol. We would also like to thank T. Fürst for the titin antibody & D. Obinata for the MyBP-C and α-actinin antibodies. This work is funded by the British Heart Foundation and the Medical\ Research Council.
References
- Ahuja P, Perriard E, Perriard JC, Ehler E. Sequential myofibrillar breakdown accompanies mitotic division of mammalian cardiomyocytes. J Cell Sci. 2004;117:3295–3306. doi: 10.1242/jcs.01159. [DOI] [PubMed] [Google Scholar]
- Akazawa H, Komuro I. Roles of cardiac transcription factors in cardiac hypertrophy. Circ Res. 2003;92:1079–1088. doi: 10.1161/01.RES.0000072977.86706.23. [DOI] [PubMed] [Google Scholar]
- Alcalai R, Seidman JG, Seidman CE. Genetic basis of hypertrophic cardiomyopathy: from bench to the clinics. J Cardiovasc Electrophysiol. 2008;19:104–110. doi: 10.1111/j.1540-8167.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- Angelo S, Lohr J, Lee KH, Ticho BS, Breitbart RE, Hill S, Yost HJ, Srivastava D. Conservation of sequence and expression of Xenopus and zebrafish dHAND during cardiac, branchial arch and lateral mesoderm development. Mech Dev. 2000;95:231–237. doi: 10.1016/s0925-4773(00)00334-8. [DOI] [PubMed] [Google Scholar]
- Ariza A, Coll J, Fernandez-Figueras MT, Lopez MD, Mate JL, Garcia O, Fernandez-Vasalo A, Navas-Palacios JJ. Desmin myopathy: a multisystem disorder involving skeletal, cardiac, and smooth muscle. Hum Pathol. 1995;26:1032–1037. doi: 10.1016/0046-8177(95)90095-0. [DOI] [PubMed] [Google Scholar]
- Balza RO, Jr, Misra RP. Role of the serum response factor in regulating contractile apparatus gene expression and sarcomeric integrity in cardiomyocytes. J Biol Chem. 2006;281:6498–6510. doi: 10.1074/jbc.M509487200. [DOI] [PubMed] [Google Scholar]
- Baxendale S, Davison C, Muxworthy C, Wolff C, Ingham PW, Roy S. The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat Genet. 2004;36:88–93. doi: 10.1038/ng1280. [DOI] [PubMed] [Google Scholar]
- Bourajjaj M, Armand AS, da Costa Martins PA, Weijts B, van der NR, Heeneman S, Wehrens XH, De Windt LJ. NFATc2 is a necessary mediator of calcineurin-dependent cardiac hypertrophy and heart failure. J Biol Chem. 2008 doi: 10.1074/jbc.M801296200. [DOI] [PubMed] [Google Scholar]
- Chang AN, Potter JD. Sarcomeric protein mutations in dilated cardiomyopathy. Heart Fail Rev. 2005;10:225–235. doi: 10.1007/s10741-005-5252-6. [DOI] [PubMed] [Google Scholar]
- Chen J, Kubalak SW, Chien KR. Ventricular muscle-restricted targeting of the RXRalpha gene reveals a non-cell-autonomous requirement in cardiac chamber morphogenesis. Development. 1998;125:1943–1949. doi: 10.1242/dev.125.10.1943. [DOI] [PubMed] [Google Scholar]
- Cook T, Pichaud F, Sonneville R, Papatsenko D, Desplan C. Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila. Dev Cell. 2003;4:853–864. doi: 10.1016/s1534-5807(03)00156-4. [DOI] [PubMed] [Google Scholar]
- Crawford AW, Michelsen JW, Beckerle MC. An interaction between zyxin and alpha-actinin. J Cell Biol. 1992;116:1381–1393. doi: 10.1083/jcb.116.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhume A, Lu S, Horowits R. Targeted disruption of N-RAP gene function by RNA interference: a role for N-RAP in myofibril organization. Cell Motil Cytoskeleton. 2006;63:493–511. doi: 10.1002/cm.20141. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 2003;34:53–58. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- Dyson E, Sucov HM, Kubalak SW, Schmid-Schonbein GW, DeLano FA, Evans RM, Ross J, Jr, Chien KR. Atrial-like phenotype is associated with embryonic ventricular failure in retinoid X receptor alpha −/− mice. Proc Natl Acad Sci U S A. 1995;92:7386–7390. doi: 10.1073/pnas.92.16.7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler E, Rothen BM, Hammerle SP, Komiyama M, Perriard JC. Myofibrillogenesis in the developing chicken heart: assembly of Z-disk, M-line and the thick filaments. J Cell Sci. 1999;112:1529–1539. doi: 10.1242/jcs.112.10.1529. [DOI] [PubMed] [Google Scholar]
- Engel AG. Myofibrillar myopathy. Ann Neurol. 1999;46:681–683. doi: 10.1002/1531-8249(199911)46:5<681::aid-ana1>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Frank D, Kuhn C, Katus HA, Frey N. The sarcomeric Z-disc: a nodal point in signalling and disease. J Mol Med. 2006;84:446–468. doi: 10.1007/s00109-005-0033-1. [DOI] [PubMed] [Google Scholar]
- Frey N, Barrientos T, Shelton JM, Frank D, Rutten H, Gehring D, Kuhn C, Lutz M, Rothermel B, Bassel-Duby R, et al. Mice lacking calsarcin-1 are sensitized to calcineurin signaling and show accelerated cardiomyopathy in response to pathological biomechanical stress. Nat Med. 2004;10:1336–1343. doi: 10.1038/nm1132. [DOI] [PubMed] [Google Scholar]
- Gregorio CC, Antin PB. To the heart of myofibril assembly. Trends Cell Biol. 2000;10:355–362. doi: 10.1016/s0962-8924(00)01793-1. [DOI] [PubMed] [Google Scholar]
- Habara-Ohkubo A. Differentiation of beating cardiac muscle cells from a derivative of P19 embryonal carcinoma cells. Cell Struct Funct. 1996;21:101–110. doi: 10.1247/csf.21.101. [DOI] [PubMed] [Google Scholar]
- Harvey NL, Srinivasan RS, Dillard ME, Johnson NC, Witte MH, Boyd K, Sleeman MW, Oliver G. Lymphatic vascular defects promoted by Prox1 haploinsufficiency cause adult-onset obesity. Nat Genet. 2005;37:1072–1081. doi: 10.1038/ng1642. [DOI] [PubMed] [Google Scholar]
- Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- Hill AA, Riley PR. Differential regulation of Hand1 homodimer and Hand1-E12 heterodimer activity by the cofactor FHL2. Mol Cell Biol. 2004;24:9835–9847. doi: 10.1128/MCB.24.22.9835-9847.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschy A, Schatzmann F, Ehler E, Perriard JC. Establishment of cardiac cytoarchitecture in the developing mouse heart. Dev Biol. 2006;289:430–441. doi: 10.1016/j.ydbio.2005.10.046. [DOI] [PubMed] [Google Scholar]
- Hoffman LM, Nix DA, Benson B, Boot-Hanford R, Gustafsson E, Jamora C, Menzies AS, Goh KL, Jensen CC, Gertler FB, et al. Targeted disruption of the murine zyxin gene. Mol Cell Biol. 2003;23:70–79. doi: 10.1128/MCB.23.1.70-79.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Lengler J, Bittner T, Munster D, Gawad A, Graw J. Agonistic and antagonistic action of AP2, Msx2, Pax6, Prox1 AND Six3 in the regulation of Sox2 expression. Ophthalmic Res. 2005;37:301–309. doi: 10.1159/000087774. [DOI] [PubMed] [Google Scholar]
- Martin AF. Turnover of cardiac troponin subunits. Kinetic evidence for a precursor pool of troponin-I. J Biol Chem. 1981;256:964–968. [PubMed] [Google Scholar]
- McBurney MW, Jones-Villeneuve EM, Edwards MK, Anderson PJ. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature. 1982;299:165–167. doi: 10.1038/299165a0. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- Nelson TJ, Balza R, Jr, Xiao Q, Misra RP. SRF-dependent gene expression in isolated cardiomyocytes: regulation of genes involved in cardiac hypertrophy. J Mol Cell Cardiol. 2005;39:479–489. doi: 10.1016/j.yjmcc.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Oliver G, Sosa-Pineda B, Geisendorf S, Spana EP, Doe CQ, Gruss P. Prox 1, a prospero-related homeobox gene expressed during mouse development. Mech Dev. 1993;44:3–16. doi: 10.1016/0925-4773(93)90012-m. [DOI] [PubMed] [Google Scholar]
- Peirson SN, Butler JN, Foster RG. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:e73. doi: 10.1093/nar/gng073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Perriard JC, Hirschy A, Ehler E. Dilated cardiomyopathy: a disease of the intercalated disc? Trends Cardiovasc Med. 2003;13:30–38. doi: 10.1016/s1050-1738(02)00209-8. [DOI] [PubMed] [Google Scholar]
- Pyle WG, Solaro RJ. At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function. Circ Res. 2004;94:296–305. doi: 10.1161/01.RES.0000116143.74830.A9. [DOI] [PubMed] [Google Scholar]
- Qin J, Gao DM, Jiang QF, Zhou Q, Kong YY, Wang Y, Xie YH. Prospero-related homeobox (Prox1) is a corepressor of human liver receptor homolog-1 and suppresses the transcription of the cholesterol 7-alpha-hydroxylase gene. Mol Endocrinol. 2004;18:2424–2439. doi: 10.1210/me.2004-0009. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Niedenfuhr M, Papoutsi M, Christ B, Nicolaides KH, von Kaisenberg CS, Tomarev SI, Wilting J. Prox1 is a marker of ectodermal placodes, endodermal compartments, lymphatic endothelium and lymphangioblasts. Anat Embryol (Berl) 2001;204:399–406. doi: 10.1007/s00429-001-0214-9. [DOI] [PubMed] [Google Scholar]
- Roy S, Wolff C, Ingham PW. The u-boot mutation identifies a Hedgehog-regulated myogenic switch for fiber-type diversification in the zebrafish embryo. Genes Dev. 2001;15:1563–1576. doi: 10.1101/gad.195801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104:557–567. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- Shin JW, Min M, Larrieu-Lahargue F, Canron X, Kunstfeld R, Nguyen L, Henderson JE, Bikfalvi A, Detmar M, Hong YK. Prox1 promotes lineage-specific expression of fibroblast growth factor (FGF) receptor-3 in lymphatic endothelium: a role for FGF signaling in lymphangiogenesis. Mol Biol Cell. 2006;17:576–584. doi: 10.1091/mbc.E05-04-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N, Risebro CA, Melville AA, Moses K, Schwartz RJ, Chien KR, Riley PR. Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature. 2007;445:177–182. doi: 10.1038/nature05383. [DOI] [PubMed] [Google Scholar]
- Sosa-Pineda B, Wigle JT, Oliver G. Hepatocyte migration during liver development requires Prox1. Nat Genet. 2000;25:254–255. doi: 10.1038/76996. [DOI] [PubMed] [Google Scholar]
- Tomarev SI, Sundin O, Banerjee-Basu S, Duncan MK, Yang JM, Piatigorsky J. Chicken homeobox gene Prox 1 related to Drosophila prospero is expressed in the developing lens and retina. Dev Dyn. 1996;206:354–367. doi: 10.1002/(SICI)1097-0177(199608)206:4<354::AID-AJA2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- Wigle JT, Oliver G. Prox1 Function Is Required for the Development of the Murine Lymphatic System. Cell. 1999;98:769–778. doi: 10.1016/s0092-8674(00)81511-1. [DOI] [PubMed] [Google Scholar]
- Zaffran S, Kelly RG, Meilhac SM, Buckingham ME, Brown NA. Right ventricular myocardium derives from the anterior heart field. Circ Res. 2004;95:261–268. doi: 10.1161/01.RES.0000136815.73623.BE. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.