Abstract
Purpose
Triple-negative breast cancers (TNBCs) are associated with a worse prognosis and patients with TNBC have fewer therapeutic options than patients with non-TNBC. Recently, the IRE1α-XBP1 branch of the unfolded protein response (UPR) was implicated in TNBC prognosis on the basis of a relatively small patient population, suggesting the diagnostic and therapeutic value of this pathway in TNBCs. In addition, the IRE1α-XBP1 and hypoxia-induced factor 1 α (HIF1α) pathways have been identified as interacting partners in TNBC, suggesting a novel mechanism of regulation. To comprehensively evaluate and validate these findings, we investigated the relative activities and relevance to patient survival of the UPR and HIF1α pathways in different breast cancer subtypes in large populations of patients.
Materials and Methods
We performed a comprehensive analysis of gene expression and survival data from large cohorts of patients with breast cancer. The patients were stratified based on the average expression of the UPR or HIF1α gene signatures.
Results
We identified a strong positive association between the XBP1 gene signature and estrogen receptor–positive status or the HIF1α gene signature, as well as the predictive value of the XBP1 gene signature for survival of patients who are estrogen receptor negative, or have TNBC or HER2+. In contrast, another important UPR branch, the ATF4/CHOP pathway, lacks prognostic value in breast cancer in general. Activity of the HIF1α pathway is correlated with patient survival in all the subtypes evaluated.
Conclusion
These findings clarify the relevance of the UPR pathways in different breast cancer subtypes and underscore the potential therapeutic importance of the IRE1α-XBP1 branch in breast cancer treatment.
INTRODUCTION
Triple-negative breast cancers (TNBCs) represent a breast cancer subpopulation lacking estrogen receptor (ER), progesterone receptor (PR), and HER2 expression.1 Patients with TNBC have a worse prognosis than those with non-TNBC, suggesting more aggressive biologic behavior. The unfolded protein response (UPR) is an adaptive process to alleviate endoplasmic reticulum stress during tumor growth and proliferation.2 One major UPR signaling pathway, IRE1α leading to activation of XBP1, is implicated in multiple types of cancers, including breast cancer.3
A recent study reported that TNBC cell lines and primary tumor samples from patients with TNBC express higher levels of the activated form of XBP1 than non-TNBC.4 Furthermore, this study also revealed that in TNBC cells, XBP1 coregulates HIF1α target genes, and that both XBP1 and HIF1α gene signatures were associated with poor prognosis in TNBC, but not in patients with ER+ breast cancer. In contrast, studies have revealed that XBP1 expression correlated with activity of the ER pathway. For example, the expression of ESR1, GATA3, and XBP1 were highly correlated in breast cancer.5,6 In addition, XBP1 enhances the transactivation activity of ERa7 and was identified as an estrogen-responsive gene.8 Recently, our laboratory identified that doxorubicin, a widely used chemotherapeutic agent in breast cancer treatment, is a potent inhibitor of XBP1 activation.9 Therefore, clarifying the role of XBP1 in breast cancer prognosis would not only improve the application of existing treatment strategy but also would support the preclinical development of therapy on the basis of modulating XBP1 activity.10,11
In this study, we comprehensively investigated the activity of XBP1, its relationship with the ER status as well as the HIF1α pathway, and its prognostic value in a large group of patients with breast cancer. We also performed similar analyses on another critical UPR axis, the ATF4/CHOP pathway, to query the role of the different UPR signaling branches in breast cancer prognosis.
MATERIALS AND METHODS
Data Sets
Microarray expression profiling data of 51 breast cancer cell lines with defined ER/PR/HER2 status13,20 were downloaded from the University of California Santa Cruz Cancer browser. The RNAseq (n = 1,182; Illumina HiSeq; Illumina, San Diego, CA) and microarray (n = 597, a subset of the 1,182 patients; Agilent G4502A_07_3 array; Agilent Technologies, Santa Clara, CA) data with the clinical information of the patients with breast cancer were downloaded from The Cancer Genome Atlas (TCGA) database through the University of California Santa Cruz Cancer browser. The clinical properties of the 1,182 patients are summarized in the Data Supplement. Microarray expression and clinical data of the 1,809 patients14 were downloaded from Kaplan-Meier Plotter website (www.kmplot.com). The ER status of the tumors in this data set was previously defined based on gene expression analysis, as previously described.14 To de fine the TNBC and non-TNBC status of the patient tumor samples in this data set, we used the same analytical approach as previously described.21 Affymetrix HG-U133A probe sets (Thermo Fisher Scientific, Waltham, MA) were used for the three genes and the corresponding threshold values were 205225_at (ESR1, 926.9917), 208305_at (PGR, 31.7232), and 216836_s_at (ERBB2, 4652.536).
Statistical Analysis
Members of the gene signatures used in this study are listed in the Data Supplement. All signature and expression data were normalized as Z-scores with a mean of 0 and standard deviation of 1. Differences in average expression (microarray or RNAseq) level of the gene signatures by ER and TNBC status were assessed using the nonparametric Wilcoxon rank-sum test. mRNA exression levels of FOXA1, ESR1, and GATA3 were first individually normalized (mean, 0; standard deviation, 1) and then averaged together before comparing with either the XBP1 gene signature expression or XBP1 gene expression using regression. Correlations and tests for statistical significance of a linear trend were evaluated using Pearson’s correlation matrix. Cox proportional hazards regression models were fit to breast cancer survival data, and the nonparametric log-rank test was used to test for significant differences in relapse-free survival (RFS) between subtypes of patients with high or low levels of average XBP1, ATF4/CHOP, or HIF1α gene signature expression. Patient survival data up to 150 months were used for the analysis.
Cell Culture and Viability Assay
MDA-MB-231, Hs578t, MDA-MB-468, MCF7, T47D, and BT474 cells were purchased from American Type Culture Collection (Manassas, VA) and maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin, and cells were cultured at 37°C with 5% CO2. Doxorubicin was purchased from Sigma-Aldrich (St. Louis, MO) and 3,6-dimethyl acetylenedicarboxylate (DMAD) was synthesized as described previously.11 For cell viability assay, 3,000 cells per well were plated into 96-well plates and treatment started 12 hours after plating. After 72 hours of treatment, XTT reagent (American Type Culture Collection) was added to the wells, then cells were incubated for 2 hours, and cell viability was calculated with the following formula: absorbance = A475nm(test) – A475nm(blank) – A660nm(test) using a BioTek Synergy H1 plate reader (BioTek, Winooski, VT).
RESULTS
First, we tested the initial conclusion of the previous study4 that basal-like breast cancer cell lines, which consist primarily of TNBC cells, express higher levels of the spliced/activated form of XBP1 compared with those of the luminal type, which consist primarily of ER+ cells.4 We found the average expression of the same XBP1 gene signature used by the previous investigators to measure XBP1 activity4 was actually higher in the same set of luminal (n = 4) than basal-like (n = 6) cell lines, albeit without statistical significance (Fig 1A). Interestingly, the average expression of an ATF4/CHOP signature representing another branch of the UPR pathway12 was significantly higher in the basal-like subtype (Fig 1B). To evaluate the activity of these stress response pathways comprehensively, we compared the average expression of the same XBP1 and ATF4/CHOP gene signatures in a large panel of 51 breast cancer lines.13 We confirmed that the average expression of the XBP1 gene signature was significantly higher in ER+ compared with ER− cell lines, consistent with previous reports (Fig 1C).5–8 However, there was no statistically significant difference in the XBP1 signature between TNBC and non-TNBC cell lines (Fig 1C). In contrast, expression of the ATF4/CHOP signature was higher in ER− or TNBC than in ER+ or non-TNBC cell lines, respectively (Fig 1C).
Fig 1.
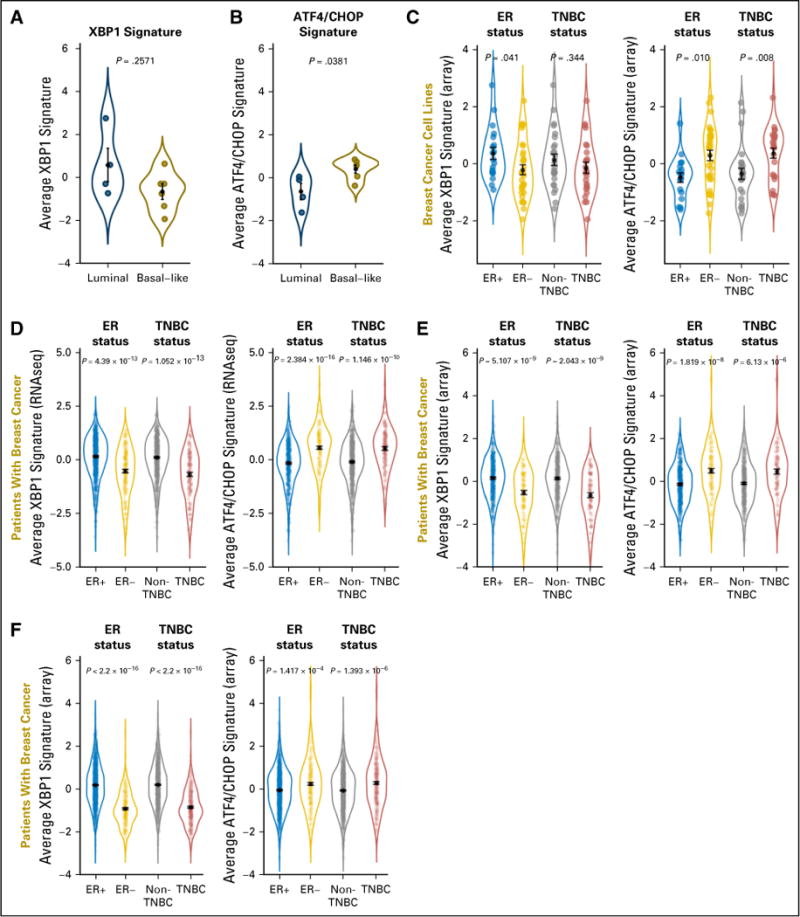
Expression levels of the XBP1 and ATF4/CHOP gene signatures in breast cancer cell lines and patients. Using the same four luminal and six basal-like cell lines reported by Chen et al,4 (A) average expression of the XBP1 gene signature was higher in the luminal type, although not statistically significant, whereas (B) average expression of the ATF4/CHOP signature was significantly higher in the basal-like type. (C) Average expression of the two gene signatures in 51 commonly used breast cancer cell lines on the basis of microarray analysis. The cell line collection consists of 19 estrogen receptor (ER)+ and 32 ER−, or 25 triple-negative breast cancer (TNBC) and 26 non-TNBC cell lines defined on the basis of mRNA/protein levels of ER/progesterone receptor/HER2. Data are combined in violin plots. (D) The same analysis on RNAseq data of patients with breast cancer from The Cancer Genome Atlas database (n = 600 ER+ and n = 179 ER−, or n = 125 TNBC and n = 647 non-TNBC). (E) The same analysis on microarray data of patients with breast cancer from The Cancer Genome Atlas data base (n= 404 ER+ and n =118 ER−, or n= 91 TNBC and n=431 non-TNBC). (D, E) Only patients with known status for ER/progesterone receptor/HER2 defined experimentally were used for analysis. (F) The same analysis on microarray data from a curated collection of 1,809 patients with breast cancer (n = 1,505 ER+ and n = 304 ER−, or n = 341 TNBC and n = 1,468 non-TNBC). The ER status was defined previously and the TNBC status was defined by computational analysis, as described in Materials and Methods. For all panels, P values from the nonparametric Wilcoxon rank-sum test are shown. For each subtype, black dots indicate means and black bars indicate standard errors.
These observations in established cell lines prompted us to evaluate these pathways in tumor samples from patients with breast cancer. We performed the same analyses on 1,182 patients with breast cancer from TCGA, on the basis of RNAseq (Fig 1D),5 or a subset of 597 patients on the basis of microarray (Fig 1E), as well as 1,809 curated microarray gene expression profiles (Fig 1F).14 In each comparison, we found expression of the XBP1 gene signature was significantly higher in ER+ or non-TNBC than in ER− or TNBC tumor samples, respectively. In contrast, expression of the ATF4/CHOP gene signatures was consistently higher in ER− or TNBC than in ER+ or non-TNBC tumor samples, respectively. These observations are consistent with the results from our breast cancer cell line analysis with augmented significance as a result of much larger sample sizes in a real clinical setting.
To evaluate the association between higher XBP1 activity and ER+ status in a different manner, we checked the correlation of XBP1 activity with the ER pathway in patients with breast cancer from TCGA. Overall, we identified a strong correlation between the expression of XBP1, which is also a transcriptional target of activated XBP1 itself,4 and ER pathway genes (ie, ESR1, GATA3, and FOXA1; Fig 2A and 2B).This relationship was also significant when we correlated the average expression of the XBP1 gene signature with the average ER pathway gene expression (Fig 2C and 2D). These data are consistent with a previous report of XBP1 playing a crucial role in 17-β-estradiolinduced growth of ER+ breast cancer cells.15
Fig 2.
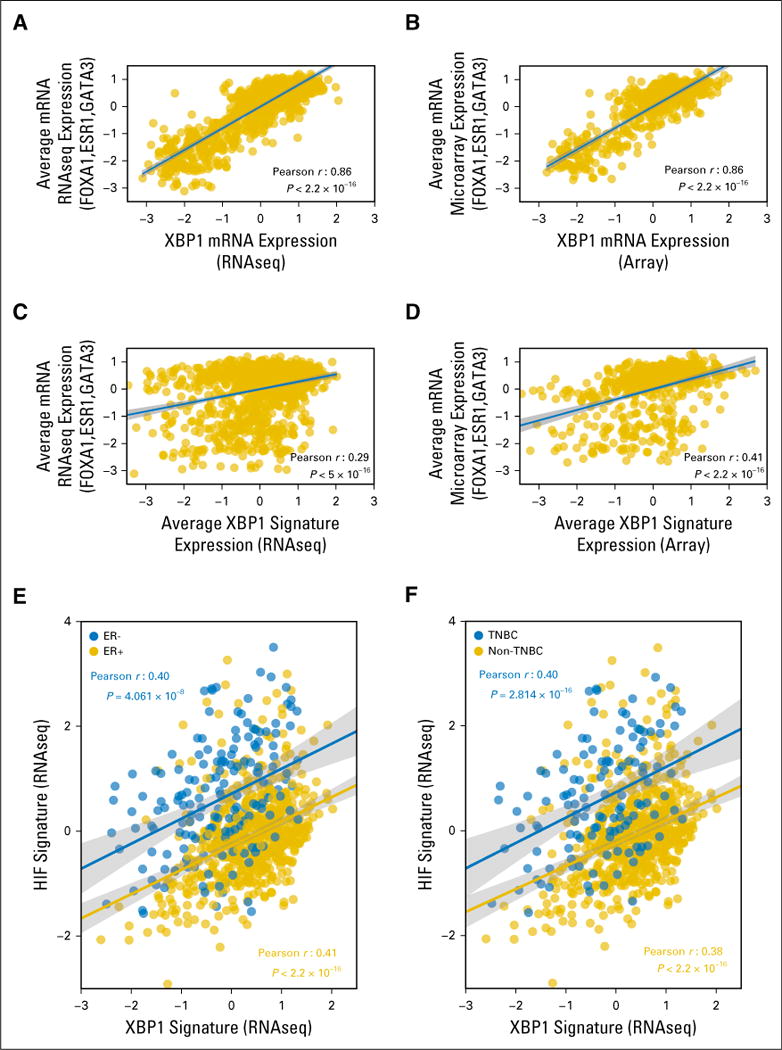
Correlation of XBP1 mRNA expression and the XBP1 gene signature with estrogen receptor (ER) pathway gene expression and correlation of the XBP1 and HIF1α gene signatures in the The Cancer Genome Atlas (TCGA) breast cancer data set. Correlation analysis of expression of the XBP1 gene and the average of three ER pathway genes (ESR1, FOXA1, and GATA3) using (A) RNAseq or (B) microarray data from 1,182 patients with breast cancer in TCGA breast cancer database and the same correlation analysis of the average expression of the XBP1 gene signature and the three ER pathway genes on the basis of (C) RNAseqor (D) microarray data in the TCGA database. (E,F) Correlation of the average expression of the XBP1 and HIF1α gene signatures in the groups of patients with breast cancer, on the basis of the RNAseq data in the TCGA database: (E) ER+ (n = 600 patients, gold) and ER− (n = 179 patients, blue); and (F) triple-negative breast cancer (TNBC; n= 125 patients, blue) and non-TNBC (n=647 patients, gold). The Pearson coefficients of correlation as well as the P values are shown in (A–D) black or the (E, F) corresponding colors. The fitted lines and the 95% CIs (shades) are also shown. Only patients with known status for ER/progesterone receptor/HER2 defined experimentally were used for analysis.
To test the reported coregulation of target gene expression by activated XBP1 and HIF1α4 in a large cohort of patients with breast cancer, we performed similar correlation analysis using the TCGA dataset. Overall, we found a strong correlation between the XBP1 and HIF1α gene signatures in patients either ER+ or ER−, and with non-TNBC or TNBC, who were from TCGA (Fig 2E and 2F). This result confirms the molecular-level interaction between the two pathways with large-scale gene-expression data from patients and suggests the XBP1 and HIF1α pathways are highly correlated in breast cancer regardless of the subtypes.
To assess the influence of the UPR and HIF1α pathways on survival of patients with breast cancer and extend the analysis of the previous study4 to larger patient cohorts, we performed survival analyses of the 1,809-patient data set, using the same high-low expression cutoff (58th percentile) as previously reported.4 Surprisingly, the XBP1 signature was not associated with worse RFS in the whole population of patients with breast cancer (Fig 3A, left). However, when the patient population was further stratified into TNBC versus non-TNBC, ER+ versus ER−, or HER2+ versus HER2−, higher expression of the XBP1 signature was associated with worse RFS in TNBC (Fig 3B, left), ER− (Fig 3C, left), or HER2+ (Fig 3D, left) patients, but not in patients positive for ER (Fig 3C, left) or negative for HER2 (Fig 3D, left). A similar association was also observed in patients with non-TNBC (P = .045; Fig 3B). Interestingly, the ATF4/CHOP signature was not associated with RFS (Fig 3A, middle), even after stratification into either TNBC/non-TNBC, ER+/− or HER2+/− status (Fig 3B–3D, middle). In contrast, the HIF1α signature was significantly associated with RFS regardless of how the patients were stratified (Fig 3A–3D, right).
Fig 3.
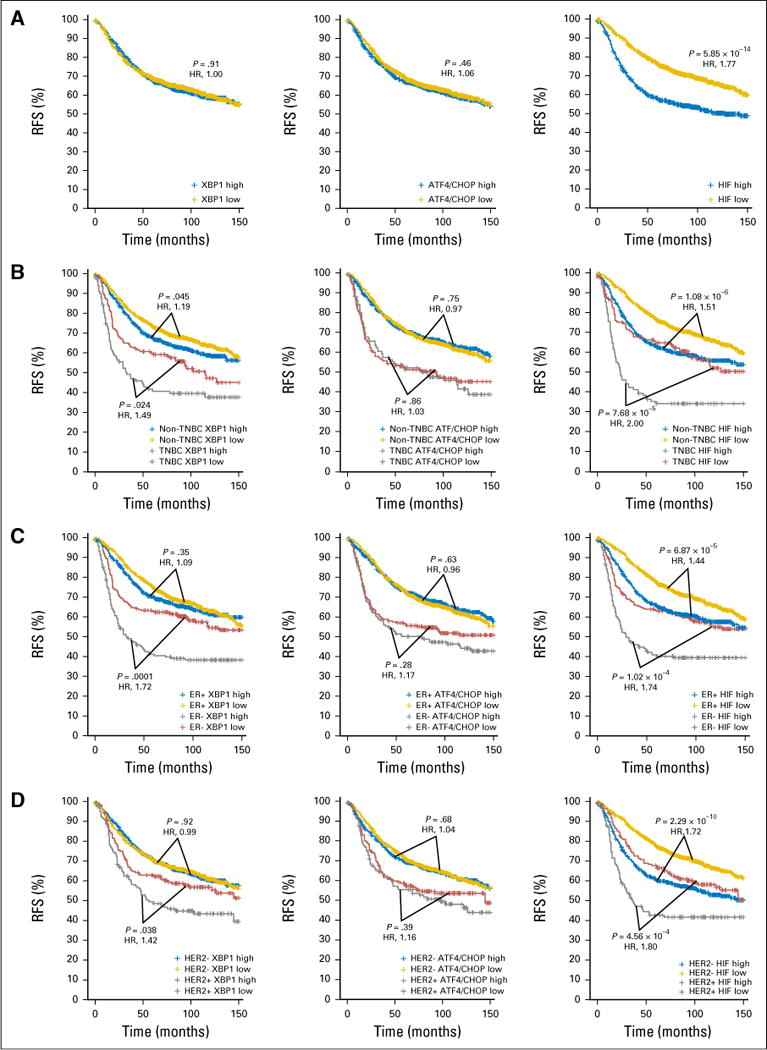
Kaplan-Meier graphs showing relapse-free survival (RFS) of the different subtypes of patients with breast cancer stratified by the XBP1, ATF4/CHOP, or HIF1α gene signatures. Kaplan-Meier graphs of RFS of (A) the total of 1,640 patients, (B) 225 patients with triple-negative breast cancer (TNBC) and 1,415 with non-TNBC, (C) 1,286 who are estrogen receptor (ER)+ and 354 who are ER−, or (D) 270 who are HER2+ and 1,370 who are HER2− with RFS ≤ 150 months separated by high and low (with the 58th percentile cutoff used previously4) levels of the average expression of the XBP1 (left column), ATF4/CHOP (middle column), or HIF1α (right column) gene signature. P values are after Benjamini-Hochberg correction from log-rank (Mantel-Cox) test and the hazard ratios (HRs; higher expression compared with lower expression) are shown in red.
To evaluate whether using gene signature expression cutoffs at the extremes would reveal stronger association, we applied quartile (the highest 25% v the lowest 25%) cutoff and performed the same survival analysis. The ATF4/CHOP signature became significantly associated with RFS in the whole population (Data Supplement) and the XBP1 signature became significantly associated with RFS in patients who were HER2− (Data Supplement). However, the positive association between the XBP1 signature and RFS in the patients with non-TNBC using the 58th percentile cutoff became negative when using the quartile cutoff, which likely was due to the reduced number of patients in the new categories (Data Supplement). Nevertheless, the major conclusions of the current study remain unchanged.
DISCUSSION
In summary, on the basis of a comprehensive analysis on large cohorts of patients with breast cancer, our findings confirm the initial observations from Chen et al4 that XBP1 activity was associated with a worse prognosis in TNBC, but not ER+ patients, and extend this relationship to patients who are ER− (Fig 3C, left). We also found that XBP1 activity was associated with a worse prognosis in patients who are HER2+, but not in those who are HER2−. This may partially explain why the XBP1-RFS association is stronger in patients who are ER− than in those with TNBC (Fig 3B and 3C, left), because some of the patients who are ER−/HER2+ are included in the patients who are ER− but not in the patients with TNBC. However, through analyzing large-scale gene expression profiles, we identified that average expression of the same XBP1 gene signature reported by Chen et al4 was higher in ER+ or non-TNBC than ER− or TNBC cell lines or patients, respectively (Fig 1C–1F), to correct the initial conclusion by Chen et al.4 These data are consistent with other previous studies. For example, FOXA1, GATA3, and XBP1 were found to associate with ESR1 expression in breast cancers16–18 and XBP1 was previously recognized as an estrogen-regulated gene.15,19 Interestingly, activity of another UPR branch represented by expression of the ATF4/CHOP gene signature was higher in ER− or TNBC cell lines or patients instead (Fig 1C–1F). These findings were significant regardless of the technical platform (RNAseq or microarray) or source of the data collection. Our findings suggest that, albeit with higher XBP1 activities, patients who are ER+, and other patients with non-TNBC (excluding HER2+), may not benefit from therapeutic strategies targeting XBP1 activity. In contrast, patients with TNBC, and those who are ER− and HER2+, may be the major target populations for anti-XBP1 therapy. The design and aims of the current study are summarized in the schematic shown in the Data Supplement.
Recently, we have shown that doxorubicin blocks XBP1 activation through the inhibition of IRE1α RNase activity.9 These data suggest that patients with TNBC, who are ER−, and possibly those who are HER2+, may benefit the most from anthracycline-based therapies. We also identified 3,6-DMAD as a novel IRE1α/XBP1 inhibitor that inhibits both the RNase activity and oligomerization of IRE1α.11To test the hypothesis that TNBC cells may respond better to XBP1 inhibition-based therapy, we compared the sensitivity of a panel of TNBC cell lines to doxorubicin or 3,6-DMAD with that of a panel of ER+ breast cancer cell lines. We found that the TNBC cell lines were consistently more sensitive to doxorubicin or 3,6-DMAD treatment than the ER+ cell lines (Fig 4), which supports our hypothesis that TNBC cells should be more sensitive to XBP1 inhibition. Thus, our results have significant therapeutic implications for targeting the IRE1α-XBP1 branch of the UPR in breast cancer treatment.
Fig 4.
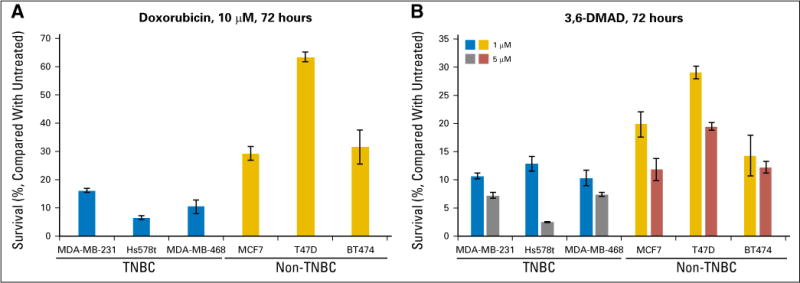
Triple-negative breast cancer (TNBC) cells are more sensitive to XBP1 inhibitors than estrogen receptor (ER)+ breast cancer cells. A panel of three TNBC cell lines (MDA-MB-231, Hs578t, and MDA-MB-468) and three ER+ breast cancer cell lines (MCF7, T47D, and BT474) were treated with (A) 10 μM doxorubicin or (B) 1 or 5 μM 3,6-DMAD, and cell viability was measured by XTT assay after 72 hours. XTT measurements were normalized to 0 μM (untreated), which was set to be 100% viable. Error bars represent standard deviation from four replicate wells.
DMAD, dimethyl acetylenedicarboxylate.
Supplementary Material
Acknowledgments
Funded by the National Institutes of Health (Grant No. 1R01CA161585 and P01CA067166 to Q.T.L. and A.C.K.; Grant No. 1U19AI109662, U19AI057229, U54I117925, and U01AI089859 to P.K.).
Footnotes
AUTHOR CONTRIBUTIONS
Conception and design: Dadi Jiang, Purvesh Khatri, Albert C. Koong
Administrative support: Maximilian Diehn, Albert C. Koong
Financial support: Albert C. Koong
Collection and assembly of data: Dadi Jiang
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or po.ascopubs.org/site/ifc.
Dadi Jiang
Consulting or Advisory Role: Capella Biosciences
Brandon Turner
No relationship to disclose
Jie Song
No relationship to disclose
Ruijiang Li
No relationship to disclose
Maximilian Diehn
Stock and Other Ownership Interests: Quanticel Pharma, CiberMed
Consulting or Advisory Role: Quanticel Pharma, Roche
Research Funding: Varian Medical Systems
Patents, Royalties, Other Intellectual Property: Patent filings on ctDNA detection assigned to Stanford University (Inst), patent filings on tumor treatment resistance mechanisms assigned to Stanford University (Inst)
Travel, Accommodations, Expenses: Roche, Varian Medical Systems
Quynh-Thu Le
Stock and Other Ownership Interests: Aldea
Research Funding: Amgen (Inst), RedHill (Inst)
Travel, Accommodations, Expenses: Merck, Bristol-Myers Squibb, Pfizer, Varian Medical Systems
Purvesh Khatri
Stock and Other Ownership Interests: Inflammatix
Consulting or Advisory Role: Inflammatix
Patents, Royalties, Other Intellectual Property: Inflammatix has licensed infectious diseases diagnosis signatures of which I am listed as a co-inventor.
Albert C. Koong
No relationship to disclose
Contributor Information
Dadi Jiang, Stanford University School of Medicine, Stanford, CA.
Brandon Turner, Stanford University School of Medicine, Stanford, CA.
Jie Song, Stanford University School of Medicine, Stanford, CA.
Ruijiang Li, Stanford University School of Medicine, Stanford, CA.
Maximilian Diehn, Stanford University School of Medicine, Stanford, CA.
Quynh-Thu Le, Stanford University School of Medicine, Stanford, CA.
Purvesh Khatri, Stanford Center for Biomedical Informatics Research, Stanford University, Stanford, CA.
Albert C. Koong, Stanford University School of Medicine, Stanford, CA
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 3.Davies MP, Barraclough DL, Stewart C, et al. Expression and splicing of the unfolded protein response gene XBP-1 are significantly associated with clinical outcome of endocrine-treated breast cancer. Int J Cancer. 2008;123:85–88. doi: 10.1002/ijc.23479. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Iliopoulos D, Zhang Q, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature. 2014;508:103–107. doi: 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 7.Ding L, Yan J, Zhu J, et al. Ligand-independent activation of estrogen receptor alpha by XBP-1. Nucleic Acids Res. 2003;31:5266–5274. doi: 10.1093/nar/gkg731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finlin BS, Gau CL, Murphy GA, et al. RERG is a novel ras-related, estrogen-regulated and growth-inhibitory gene in breast cancer. J Biol Chem. 2001;276:42259–42267. doi: 10.1074/jbc.M105888200. [DOI] [PubMed] [Google Scholar]
- 9.Jiang D, Lynch C, Medeiros BC, et al. Identification of doxorubicin as an inhibitor of the IRE1α-XBP1 axis of the unfolded protein response. Sci Rep. 2016;6:33353. doi: 10.1038/srep33353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang D, Niwa M, Koong AC. Targeting the IRE1α-XBP1 branch of the unfolded protein response in human diseases. Semin Cancer Biol. 2015;33:48–56. doi: 10.1016/j.semcancer.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang D, Tam AB, Alagappan M, et al. Acridine derivatives as inhibitors of the IRE1α-XBP1 pathway are cytotoxic to human multiple myeloma. Mol Cancer Ther. 2016;15:2055–2065. doi: 10.1158/1535-7163.MCT-15-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han J, Back SH, Hur J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15:481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 15.Sengupta S, Sharma CG, Jordan VC. Estrogen regulation of X-box binding protein-1 and its role in estrogen induced growth of breast and endometrial cancer cells. Horm Mol Biol Clin Investig. 2010;2:235–243. doi: 10.1515/HMBCI.2010.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacroix M, Leclercq G. About GATA3, HNF3A, and XBP1, three genes co-expressed with the oestrogen receptor-alpha gene (ESR1) in breast cancer. Mol Cell Endocrinol. 2004;219:1–7. doi: 10.1016/j.mce.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 17.Tozlu S, Girault I, Vacher S, et al. Identification of novel genes that co-cluster with estrogen receptor alpha in breast tumor biopsy specimens, using a large-scale real-time reverse transcription-PCR approach. Endocr Relat Cancer. 2006;13:1109–1120. doi: 10.1677/erc.1.01120. [DOI] [PubMed] [Google Scholar]
- 18.Andres SA, Wittliff JL. Relationships of ESR1 and XBP1 expression in human breast carcinoma and stromal cells isolated by laser capture microdissection compared to intact breast cancer tissue. Endocrine. 2011;40:212–221. doi: 10.1007/s12020-011-9522-x. [DOI] [PubMed] [Google Scholar]
- 19.Wang DY, Fulthorpe R, Liss SN, et al. Identification of estrogen-responsive genes by complementary deoxyribonucleic acid microarray and characterization of a novel early estrogen-induced gene: EEIG1. Mol Endocrinol. 2004;18:402–411. doi: 10.1210/me.2003-0202. [DOI] [PubMed] [Google Scholar]
- 20.Cline MS, Craft B, Swatloski T, et al. Exploring TCGA pan-cancer data at the UCSC Cancer Genomics browser. Sci Rep. 3(2652):2013. doi: 10.1038/srep02652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karn T, Metzler D, Ruckhäberle E, et al. Data-driven derivation of cutoffs from a pool of 3,030 Affymetrix arrays to stratify distinct clinical types of breast cancer. Breast Cancer Res Treat. 2010;120:567–579. doi: 10.1007/s10549-009-0416-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


