Summary
Dickeya dadantii 3937 secretes pectate lyases (Pels) to degrade the plant cell wall. Previously, we have demonstrated that EGcpB and EcpC function as cyclic-di-GMP (c-di-GMP) specific phosphodiesterases (PDEs) to positively regulate Pel production. However, the diguanylate cyclase (DGC) responsible for the synthesis of c-di-GMP and dichotomously regulation of Pel has remained a mystery. Here, we identified GcpA is the dominant DGC to negatively regulate Pel production by specifically repressing pelD gene expression. Quantitative real-time PCR (qRT-PCR) assays revealed that the expression levels of histone-like nucleoid-structuring protein encoding gene hns and the post-transcriptional regulator encoding genes rsmA and rsmB were significantly affected by GcpA. Deletion of hns or rsmB in the gcpAD418A site-directed mutant restored its Pel production and pelD expression, demonstrating that H-NS and RsmB contribute to GcpA-dependent regulation of Pel in D. dadantii. In addition, RsmB expression was subject to positive regulation by H-NS. Thus, we propose a novel pathway consisting of GcpA-H-NS-RsmB-RsmA-pelD that controls Pel production in D. dadantii. Furthermore, we showed that H-NS and RsmB are responsible for the GcpA-dependent regulation of motility and T3SS gene expression, respectively. Among the two PDEs involved in the regulation of Pels, only EGcpB regulates pelD expression through the same pathway as GcpA.
Keywords: soft-rot pathogen, pectate lyase, c-di-GMP, virulence, type III secretion system, motility
Introduction
Dickeya dadantii 3937 is an Enterobacterium that causes soft-rot disease in a wide range of economically important crops (Ma et al., 2007, Czajkowski et al., 2011). Pectate lyases, which degrade the plant cell wall, are one of the major virulence factors that contribute to the pathogenesis of D. dadantii (Collmer & Keen, 1986). The production of Pel is controlled by a sophisticated regulatory mechanism that includes modifications of DNA topology, quorum sensing, and other regulatory systems associated with bacterial physiological status or environmental stimuli (Hugouvieux-Cotte-Pattat et al., 1996, Charkowski et al., 2012, Reverchon & Nasser, 2013). During the early stages of infection, several transcriptional repressors, such as FIS, H-NS, KdgR, PecS, PecT, Fur, and the PhoP/Q two-component regulatory system, negatively regulate the expression of pel genes in response to initially encountering the oxidative and acidic environments in the plant intercellular spaces (Reverchon et al., 1991, Franza et al., 2002, Llama-Palacios et al., 2005, Ouafa et al., 2012, Hérault et al., 2014). In addition, the post-transcriptional regulator RsmA/RsmB inhibits Pel via an unknown mechanism (Yang et al., 2008, Wu et al., 2014). RsmA facilitates specific mRNA degradation while RsmB is an untranslated regulatory RNA that binds to RsmA and neutralizes its effect on target gene expression (Liu et al., 1997). After adaptation to the intracellular spaces, D. dadantii secretes a massive amount of Pel into the plant apoplast following the inactivation of the previously mentioned Pel repressors, and the activation of Pel inducers, which include the GacS/A two-component system, MfbR, CRP, and Vfm quorum sensing system (Reverchon et al., 2010, Franza et al., 2002, Reverchon & Nasser, 2013, Charkowski et al., 2012, Yang et al., 2008, Hugouvieux-Cotte-Pattat et al., 1996, Nasser et al., 2013).
Bis-(3’–5’)-cyclic dimeric guanosine monophosphate (c-di-GMP) is a common bacterial second messenger found in most major bacterial phyla (Römling et al., 2013). It was first discovered as an allosteric activator for cellulose synthase in Gluconacetobacter xylinus (Ross et al., 1987). It is now established that c-di-GMP is involved in the regulation of many cellular activities including biofilm formation, motility, cell cycle, antibiotic production, and virulence (Cotter & Stibitz, 2007, Tamayo et al., 2007, Hengge, 2009, Jenal et al., 2017). The synthesis and hydrolysis of c-di-GMP are catalyzed by diguanylate cyclase (DGC) and c-di-GMP-specific phosphodiesterase (PDE) enzymes, respectively. DGC activity is associated with the GGDEF domain, which converts two molecules of guanosine-5’-triphosphate (GTP) to one molecule of c-di-GMP (Paul et al., 2004, Whiteley & Lee, 2015). PDE activity is associated with either an EAL or HD-GYP domain, which degrade c-di-GMP to 5’-phosphoguanylyl-(3’–5’)-guanosine (pGpG) or two molecules of guanosine monophosphate (GMP) (Tamayo et al., 2005, Schmidt et al., 2005, Ryan et al., 2006). The sophisticated c-di-GMP-mediated signaling network includes transcriptional, post-transcriptional, and post-translational regulation. The regulatory function of c-di-GMP is exerted through the binding of c-di-GMP to a variety of cellular effectors, such as PilZ domain proteins, transcription factors, enzymatically inactive GGDEF, EAL or HD-GYP domain proteins and RNA riboswitches (Römling et al., 2013, Ryan et al., 2012, Jenal et al., 2017, Orr et al., 2016).
It has been revealed that GGDEF and EAL domain proteins are abundantly present in many Gram-negative bacteria. For example, Escherichia coli K-12 contains 29 genes and Vibrio cholerae contains 53 (Povolotsky & Hengge, 2012, Waters et al., 2008). In D. dadantii, we found 12 gcp (GGDEF-domain-containing protein), 4 ecp (EAL-domain-containing protein) and 2 egcp (EAL-GGDEF-domains-containing protein) genes in the genome using Pfam program (pfam.xfam.org) (Fig. S1). Our previous studies demonstrated that two c-di-GMP specific PDEs, EGcpB and EcpC, positively regulate swimming motility, Pel production and T3SS gene expression, while negatively regulating biofilm formation (Yi et al., 2010, Yuan et al., 2015). EGcpA, a homologue of E. coli CsrD, negatively regulates Pel production and T3SS gene expression by modulating the expression of RsmB (Wu et al., 2014). Nevertheless, the function of other Gcp and Ecp proteins in D. dadantii, and the molecular mechanism of c-di-GMP signaling in the regulation of diverse virulence factors remain unclear.
In the present study, we first analyzed the regulatory roles of eighteen GGDEF and/or EAL domain proteins on Pel production. GcpA was identified to be the major DGC that negatively regulated Pel production by repressing the expression of pelD gene. We then demonstrated that GcpA regulates pelD through the H-NS-RsmB-RsmA pathway. Although EGcpB and EcpC are the two major PDEs that up-regulate Pel production, it appeared that only EGcpB positively regulated pelD gene expression through the same regulatory pathway as GcpA. Furthermore, we demonstrated that GcpA was involved in the regulation of swimming motility and T3SS gene expression through diverse mechanisms that were independent from its regulation on Pel. Together, our studies defined a comprehensive signaling network that links c-di-GMP signaling and multiple virulence factors in D. dadantii.
Results
The GGDEF-domain protein GcpA negatively regulates Pel production in D. dadantii
GGDEF and EAL domains are responsible for the enzymatic activities of DGCs and PDEs, respectively. In D. dadantii, eighteen genes were found to encode proteins that contain putative GGDEF and/or EAL domains at their C-terminal regions (Fig. S1A), implying a complicated c-di-GMP signaling network exists for regulating diverse cell behaviors. Our results showed that four proteins, GcpA, GcpD, GcpF, and EGcpB, contained two types of sensory domains, GAF (cGMP phosphodiesterase, Adenyl cyclase, FhlA domain) and PAS (Per/Arnt/Sim), at their N-terminus (Fig. S1A). Nine proteins including GcpB, GcpC, GcpG, GcpH, GcpJ, GcpK, GcpL, EGcpA, and EcpD, contained one or multiple N-terminal transmembrane domains (Fig. S1A). Amino acid sequence alignments between the known GGDEF and EAL domains from Caulobacter crescentus, Vibrio cholerae, Pseudomonas aeruginosa, and those from D. dadantii revealed that most of the GGDEF domains in D. dadantii contained an active-site (A-site) that is involved in GTP binding (Römling et al., 2013). Eight GGDEF domains from GcpA-H were annotated with an inhibition-site or I-site (RxxD motif), a secondary c-di-GMP binding site that represses the cyclase activity of DGC enzymes (Christen et al., 2006) (Fig. S1B).
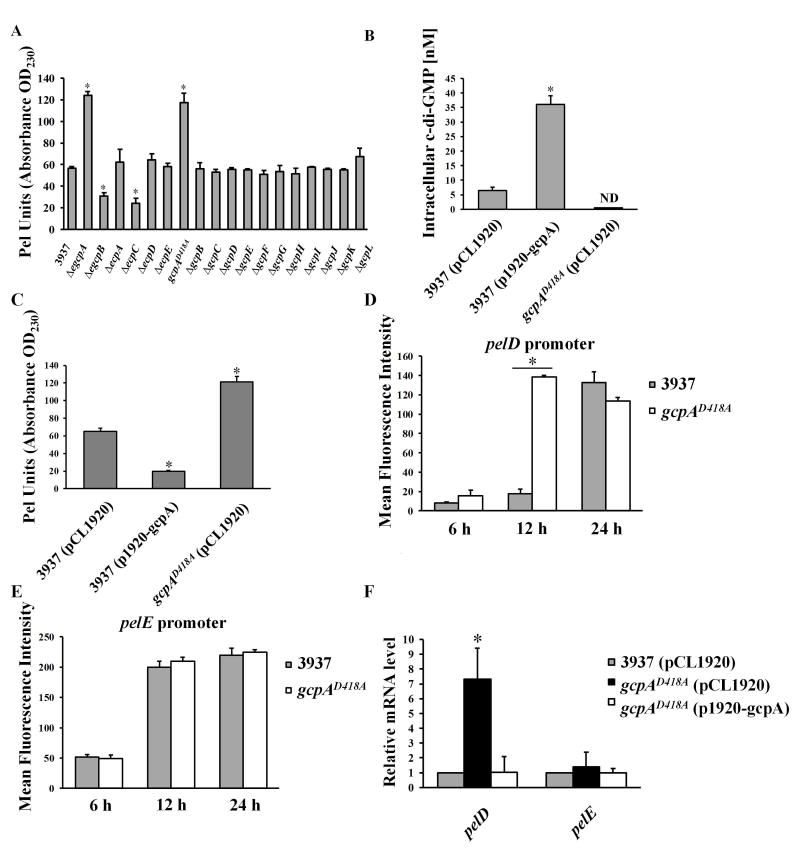
To fully investigate the network of c-di-GMP signaling in D. dadantii involved in Pel regulation, the impacts of each Gcp and Ecp protein on Pel were investigated. Nine gcp and ecp gene deletion mutants were newly constructed (Table S1). However, several attempts to delete gcpA were not successful, suggesting that gcpA might be essential for the viability of D. dadantii. We then disrupted the predicted A-site motif (SGDEF) in the GGDEF domain of GcpA by replacing the essential aspartic acid residue to alanine (SGAEF), resulting in a gcpAD418A site-directed mutant. This mutant was not defective for growth when compared with the wild-type strain (data not shown). Therefore, together with eight mutants that were previously constructed, we expanded the mutant library to cover each individual gcp, ecp or egcp gene (Table S1). The Pel activity was measured in the eighteen mutants and the wild-type strain. As previously reported, deletion of egcpB and ecpC showed reduced Pel activities while ΔegcpA showed enhanced Pel activity relative to the wild-type strain (Yi et al., 2010, Wu et al., 2014) (Fig. 1A). No difference in Pel activity was observed in ΔecpA, ΔecpD, or ΔecpE (Fig. 1A). Interestingly, among the twelve gcp gene deletion mutants, only gcpAD418A exhibited increased Pel activity compared with the wild-type strain (Fig. 1A). Complementation assays confirmed that in trans expression of gcpA drastically reduced Pel activity in gcpAD418A (Fig. S2). These findings suggested that GcpA negatively regulates Pel production in D. dadantii.
Fig. 1.
GcpA synthesizes c-di-GMP to negatively regulate Pel production and pelD gene expression in D. dadantii. (A) Pel production of wild-type D. dadantii and GGDEF and/or EAL deletion mutant strains cultured in MM+0.1% PGA for 12 h at 28°C. (B) Measurement of intracellular c-di-GMP and (C) Pel production in wild-type D. dadantii harboring empty vector pCL1920, wild type harboring pCL1920-gcpA and gcpAD418A harboring pCL1920-gcpA strains. The pelD (D) and pelE (E) promoter activities were measured in the parental strain D. dadantii and gcpAD418A. Cells cultured in MM+0.1% PGA were harvested at 6, 12 and 24 h respectively to measure the mean fluorescence intensity (MFI) by flow cytometry. (F) Quantitative RT-PCR (qRT-PCR) analysis of mRNA levels of pelD and pelE in D. dadantii strains. The data represent expression levels of each gene relative to that in the wild type, which was mathematically designated as 1. rplU gene was used as an endogenous control for the calculation. All results are from one representative experiment. Three independent experiments were performed and three replicates were used for each experiment. Error bars indicate standard errors of the means. ND represents not detectable. Asterisks indicate statistically significant differences of the means (P<0.05 by Student’s t test).
The DGC activity of GcpA is essential for its regulation on Pel production
Since mutation of the GcpA A-site enhanced the production of Pel and a functional A-site is required for the GGDEF domain activity, we hypothesized that GcpA is an active DGC and it regulates Pel production through c-di-GMP signaling. To test this, the intracellular concentrations of c-di-GMP were compared in the wild-type strain, gcpAD418A, and wild-type expressing gcpA, using ultra performance liquid chromatography coupled with tandem mass spectrometry (UPLC-MS-MS) approach. The results showed that the c-di-GMP concentration in the strain over-expressing gcpA was about 6-fold higher than the same strain carrying the empty vector (Fig. 1B). Alternatively, the c-di-GMP concentration was below detection in gcpAD418A (Fig. 1B). These data confirmed that GcpA is an active DGC. We also observed overexpression of gcpA drastically reduced the Pel activity in the wild-type strain (Fig. 1C). Altogether, these results supported the conclusion that GcpA synthesizes c-di-GMP in D. dadantii to negatively regulate the production of Pel.
Expression of pelD is enhanced in gcpAD418A A-site mutant
To reveal the underlying mechanism of GcpA to inhibit Pel production, we analyzed the effects of GcpA on two major Pel genes, pelD and pelE. In the presence of polygalacturonate (PGA), the promoter activities of pelD and pelE were induced by 22- and 4.5-fold at 24 h, respectively (Fig. S3), suggesting that these pelpromoter-GFP transcriptional fusions are sensitive to the addition of pectin catabolic products. Next, the promoter activities of pelD and pelE were determined in wild-type and gcpAD418A strains cultured in the presence of PGA for 6, 12 and 24 h, respectively. At 12 h, a significant increase of the pelD promoter activity was observed in the gcpAD418A mutant compared with the wild-type strain (Fig. 1D). Interestingly, no significant change was observed for the pelE promoter activity (Fig. 1E). To confirm the negative effect of GcpA on pelD, the mRNA of pelD and pelE were measured by quantitative real-time PCR (qRT-PCR) in wild-type, gcpAD418A, and the complemented strain containing p1920-gcpA plasmid. Consistent with the change in the pelD promoter activity, pelD transcript levels increased by 7-fold in gcpAD418A relative to the wild-type strain (Fig. 1F). The elevated mRNA level of pelD in gcpAD418A was restored to wild-type levels by the complementation plasmid p1920-gcpA (Fig. 1F). In contrast, pelE transcript levels were not altered in the tested strains (Fig. 1F). Thus, these results suggested that GcpA negatively regulates Pel production by repressing the expression of pelD in D. dadantii.
Expression of Pel regulators in the gcpAD418A mutant
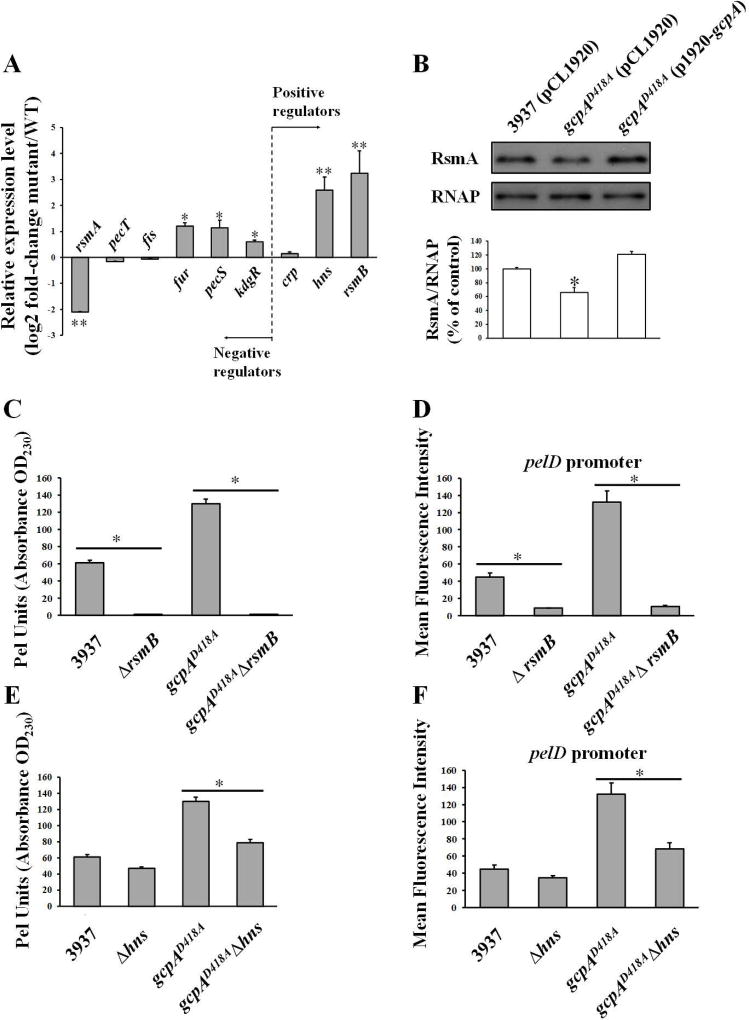
To assess whether the regulation of Pel by GcpA is through any known Pel regulators, we determined the expression of six negative regulators and three positive regulators in wild-type and gcpAD418A strains using qRT-PCR assays (Fig. 2A). Mutation of gcpA resulted in increased RNA levels of rsmB, hns, fur, pecS and kdgR, and decreased RNA levels of rsmA. No significant difference was observed for pecT, fis and crp. Given that Fur, PecS and KdgR are the pel gene repressors, we reasoned that the enhanced Pel activity in gcpAD418A was not due to increased expression of these genes. Since hns, rsmA, and rsmB showed the highest fold changes by the qRT-PCR analysis, we asked whether there were any changes at the level of promoter activity. These promoters were examined using transcriptional fusions to a GFP reporter in wild-type and gcpAD418A strains. None of the promoter activities were significantly influenced in the gcpAD418A mutant (Fig. S4), suggesting that GcpA regulates expression of these genes at the post-transcriptional level. A western blot assay confirmed that the protein levels of RsmA were reduced in the gcpAD418A mutant (Fig. 2B). Altogether, these results implied that the control of Pel production by GcpA might rely on its positive regulation of RsmA and negative regulation of RsmB and H-NS at the post-transcriptional level.
Fig. 2.
RsmB and H-NS play important roles in the GcpA-dependent Pel regulation. (A) Quantitative RT-PCR analysis of RNA levels of rsmA, pecT, fis, fur, pecS, kdgR, crp, hns and rsmB in D. dadantii wild type and gcpAD418A. The mutant/WT ratio for each gene expression was calculated as described in the Experimental Procedures. (B) Western blot analysis of RsmA protein in D. dadantii strains. Pel production (C and E) and pelD promoter activities (D and F) were tested in D. dadantii strains. Values are a representative of three independent experiments. Three replicates were used in each experiment. Error bars indicate standard errors of the means. Asterisks indicate statistically significant differences of the means (*P< 0.05 or **P<0.01 by Student’s t-test).
GcpA regulates Pel through the Rsm system
A significant increase of rsmB and decrease of rsmA in gcpAD418A strongly suggested that the Rsm system may play an important role in the GcpA-dependent signaling pathway for Pel regulation. To investigate this hypothesis, we constructed a rsmB deletion in the wild-type and gcpAD418A strains, respectively, and tested the Pel related phenotypes. We found that deletion of rsmB resulted in severe defects in Pel production and pelD promoter activities in both backgrounds (Fig. 2C and 2D). These results were in agreement with previous studies showing that RsmB is a positive regulator for the Pel (Yang et al., 2008). More importantly, deletion of rsmB in gcpAD418A (gcpAD418AΔrsmB) drastically reduced both its Pel production and pelD promoter activities to a level that is similar to that of the ΔrsmB mutant (Fig. 2C and 2D). This result suggested that RsmB plays a predominate role in controlling Pel in D. dadantii, and that the repression of Pel production by GcpA is possibly through the regulation of RsmB. Considering that RsmB functions mainly by titrating the effect of RNA-binding protein RsmA, we speculated that over-expression of rsmA might result in similar phenotypes. To test this hypothesis, we constructed a plasmid to express rsmA in trans and transformed it into the gcpAD418A mutant. As expected, over-expression of rsmA reduced the Pel production and pelD promoter activity to near-wild-type levels in the gcpAD418A mutant (Fig. S5). Therefore, we concluded that the RsmA/RsmB system plays an important role in the c-di-GMP signaling pathway mediated by GcpA to regulate Pel activity.
H-NS is involved in the GcpA-Rsm pathway
It has been previously shown that H-NS positively modulates Pel synthesis in D. dadantii (Nasser & Reverchon, 2002). Interestingly, we did not observe a significant reduction of Pel production or pelD promoter transcription when hns was deleted in the wild-type strain (Fig. 2E and 2F). However, when hns was deleted in the gcpAD418A mutant, Pel production and pelD promoter activity were dramatically reduced (Fig. 2E and 2F). These results, together with the observation that expression of hns was elevated in the gcpAD418A mutant (Fig. 2A), suggested that GcpA also represses Pel through H-NS.
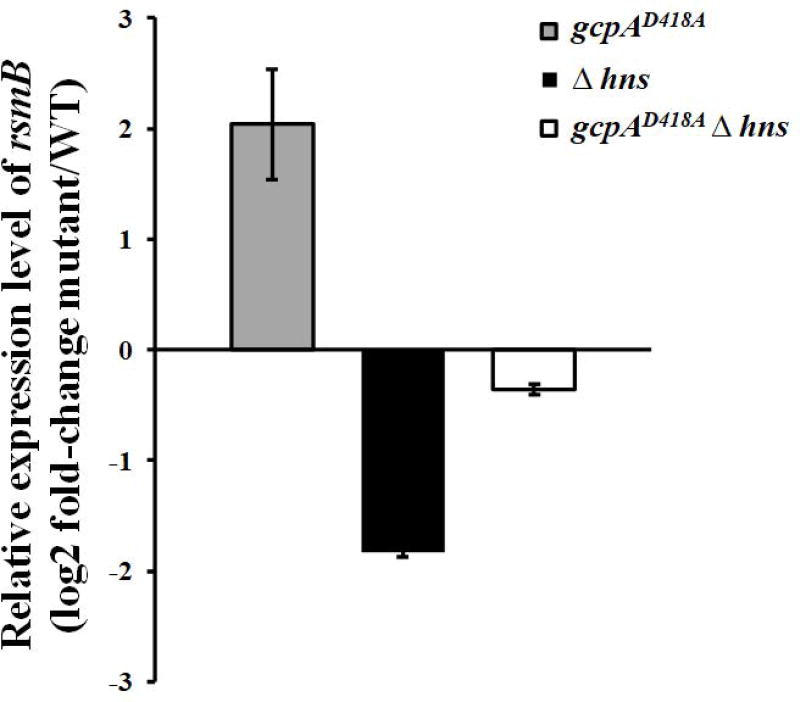
As we showed that both H-NS and the Rsm system were involved in the GcpA-dependent Pel regulation, we hypothesized that there might be a genetic link between these two regulators. To address this hypothesis, the RNA levels of rsmB were determined in wild-type, gcpAD418A, Δhns, and a gcpAD418AΔhns double mutant using qRT-PCR. In contrast to the approximate 4-fold increase of rsmB in the gcpAD418A mutant, the relative level of rsmB was decreased by about 4-fold in the Δhns mutant (Fig. 3). More importantly, rsmB expression was recovered to near-wild-type levels in the gcpAD418AΔhns double mutant. The difference of rsmB RNA levels in the different strains was further confirmed by northern blot analysis (data not shown). Together, our data supported that GcpA regulates Pel through the GcpA-H-NS-RsmB-RsmA-pelD pathway.
Fig. 3.
H-NS is involved in the GcpA-dependent regulation on rsmB. rsmB RNA levels were examined in D. dadantii using qRT-PCR. The mutant/wild-type ratio for rsmB gene expression was calculated as described in the Experimental Procedures. One representative experiment was chosen, and three independent experiments were performed. Error bars indicate standard errors of the means. Asterisks indicate statistically significant differences of the means (P<0.05 by Student’s t test).
EGcpB and EcpC differentially affect Pel production
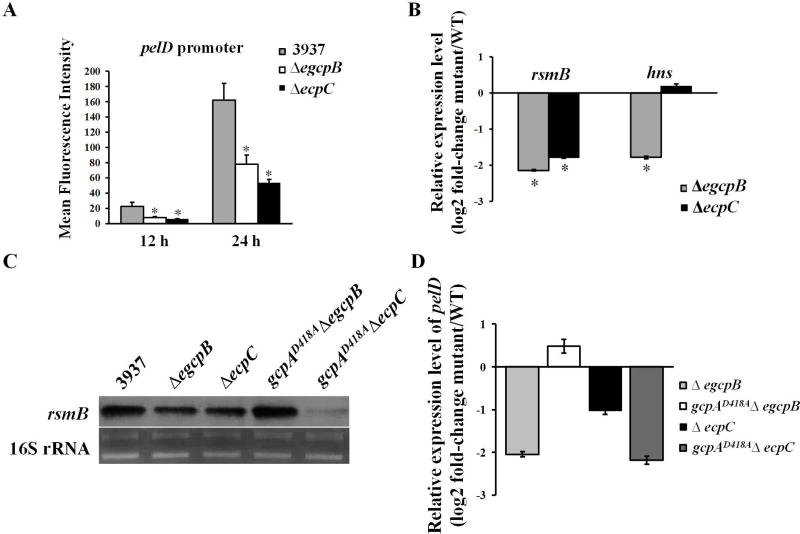
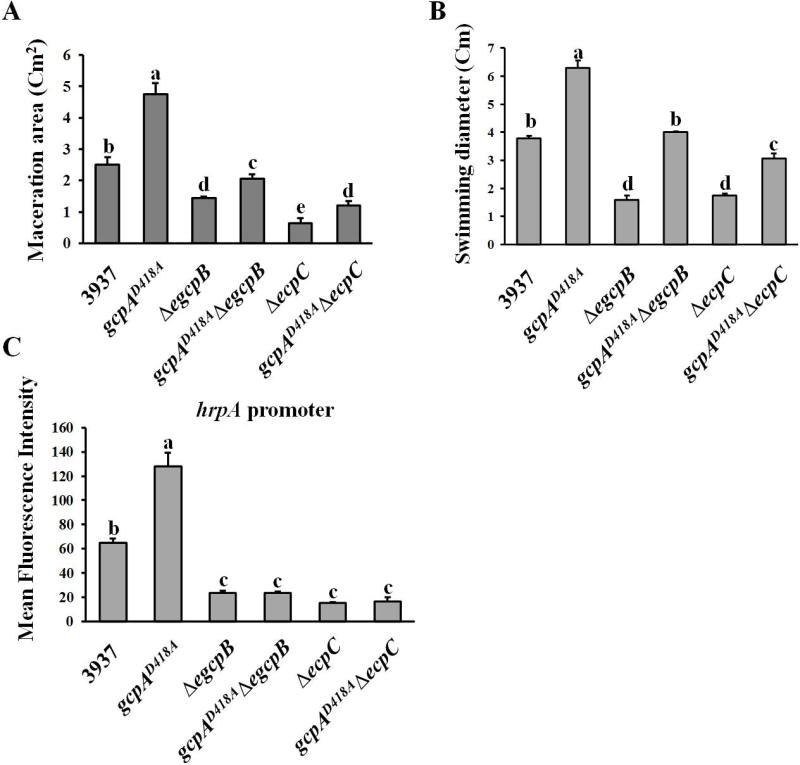
Previously, we have shown that the PDEs EGcpB and EcpC increase Pel production in D. dadantii by reducing c-di-GMP, which is in contrast to GcpA which synthesizes c-di-GMP to inhibit Pel production (Yi et al., 2010, Yuan et al., 2015). Therefore, we asked whether these two PDEs control Pel through the same regulatory pathways as GcpA. To address this question, we first examined the promoter activities of pelD in ΔegcpB and ΔecpC. As expected, deletion of either egcpB or ecpC in the wild-type strain strongly reduced the expression of pelD (Fig. 4A). Next, we compared the RNA levels of hns and rsmB in ΔegcpB and ΔecpC mutants to the wild-type strain using qRT-PCR. Interestingly, although a strong reduction in rsmB transcripts was detected in both ΔegcpB and ΔecpC, the hns transcript levels were only reduced in ΔegcpB but not in ΔecpC (Fig. 4B). These data suggested that EGcpB might regulate Pel through the H-NS-RsmB-RsmA-pelD pathway while EcpC might regulate rsmB expression through a different mechanism. To further investigate their genetic interactions, the egcpB or ecpC gene was deleted in gcpAD418A resulting in the double mutants, gcpAD418AΔegcpB and gcpAD418AΔecpC. We then detected rsmB transcripts in the wild-type strain and the single and double mutants by northern blots. Consistent with the qRT-PCR results, both ΔegcpB and ΔecpC mutants showed reduced levels of rsmB (Fig. 4C). Interestingly, rsmB expression was recovered to a near-wild-type level in the gcpAD418AΔegcpB double mutant, while it appeared to be even more reduced in the gcpAD418AΔecpC double mutant than the ΔecpC mutant. Moreover, the expression levels of pelD in various mutants were detected by qRT-PCR assays. The results showed that expression of pelD was down-regulated in both ΔegcpB and ΔecpC (Fig. 4D), which is consistent with the reduced rsmB levels in these mutants. As expected, pelD expression was recovered to near-wild-type levels in the gcpAD418AΔegcpB double mutant, while it remained low in gcpAD418AΔecpC (Fig. 4D). In summary, our results suggested that GcpA and EGcpB inversely regulate pelD gene expression through the same regulatory pathway while EcpC utilizes a different mechanism.
Fig. 4.
EGcpB and EcpC positively regulate pelD gene expression via different pathways. (A) The promoter activity of pelD was examined in D. dadantii. (B) RNA levels of rsmB and hns were examined using qRT-PCR. The mutant/WT ratio of each gene was calculated as described in the Experimental Procedures. (C) Northern blot analysis of rsmB mRNA in D. dadantii strains. (D) Relative mRNA levels of pelD in mutant strains to that in the wild-type strain. Values are a representative of three independent experiments. Three replicates were used in each experiment. Error bars indicate standard errors of the means. Asterisks indicate statistically significant differences of the means (P<0.05 by Student’s t test).
GcpA negatively regulates the virulence, swimming motility, and T3SS gene expression of D. dadantii
Since mutation of gcpA showed an opposite effect on Pel production as mutation of either egcpB or ecpC, and the virulence of ΔegcpB and ΔecpC on host plants were reduced, we further determined whether the virulence of gcpAD418A was altered. As shown in Fig. 5A, gcpAD418A exhibited a two-fold increase in maceration areas relative to the wild-type strain in the leaves of host plant Chinese cabbage (Brassica campestris). Furthermore, in comparison with the reduced maceration areas caused by ΔegcpB and ΔecpC, double mutants of both gcpAD418AΔegcpB and gcpAD418AΔecpC partially restored the virulence phenotype, but not to the extent of the wild-type strain (Fig. 5A). This result was noteworthy for gcpAD418AΔecpC since its pelD expression was not restored at all (Fig. 4D), indicating that GcpA might regulate other virulence factors besides Pel in D. dadantii.
Fig. 5.
Effects of GcpA on swimming motility, T3SS gene expression and virulence. (A) Bacterial cells of D. dadantii were inoculated in the leaves of Chinese cabbage (Brassica campestris). The maceration areas were measured 16 h post-inoculation. The swimming motility (B), T3SS gene hrpA promoter activity (C) were examined. Values are a representative of three independent experiments. Three replicates were used in each experiment. Error bars indicate standard errors of the means. Different lowercase letters above the bar indicate statistically significant differences between treatments (Fisher’s LSD, P<0.05).
Thus, we further tested the swimming motility and expression of the T3SS gene, hrpA, in the gcpAD418A mutant, both of which are virulence factors previously shown to be regulated by c-di-GMP signaling in D. dadantii (Yi et al., 2010). The results showed that the swimming motility and hrpA promoter activity were significantly enhanced in the gcpAD418A mutant compared with the wild-type strain (Fig. 5B and 5C). More interestingly, mutation of gcpA in the ΔegcpB mutant fully recovered its swimming motility (Fig. 5B) but not its hrpA promoter activity (Fig. 5C). The gcpAD418AΔecpC mutant exhibited low hrpA promoter activity that was equivalent to the ΔecpC mutant (Fig. 5C), while its swimming motility was partially restored from ΔecpC (Fig. 5B). These results could explain the partial restoration of virulence in the gcpAD418AΔegcpB and gcpAD418AΔecpC double mutants, suggesting that all three virulence factors including Pel production, swimming motility and T3SS are essential for the full virulence of D. dadantii in host plants.
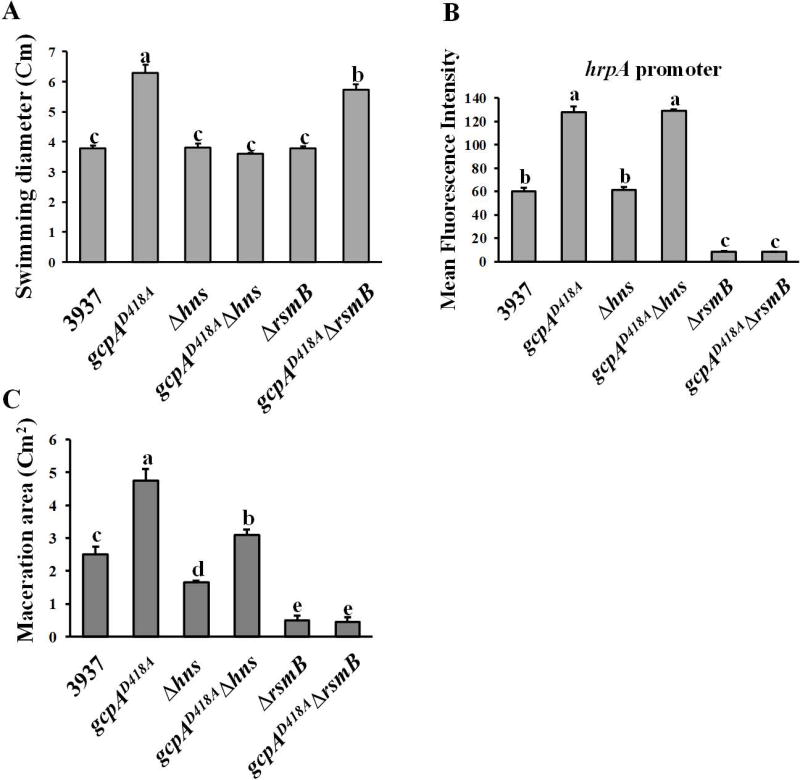
H-NS and Rsm played different roles in GcpA-dependent regulation of swimming motility and T3SS
Since we proposed a novel regulatory pathway in which GcpA controls Pel production via H-NS-RsmB-RsmA, we asked whether the same regulatory pathway was involved in the GcpA-dependent regulation of swimming motility and T3SS gene expression. As shown in Fig. 6A, Δhns and ΔrsmB mutants exhibited wild-type levels of swimming motility, however, deletion of hns but not rsmB in the gcpAD418A mutant fully recovered its swimming motility to the wild-type level. This result, together with the observation that GcpA negatively regulated H-NS (Fig. 2A), suggested that H-NS is essential for GcpA to control swimming motility. Next, the hrpA promoter activity was determined, and the results indicated that H-NS is not involved in the regulation of T3SS in both the D. dadantii wild-type and gcpAD418A strains (Fig. 6B). Deletion of rsmB drastically decreased the promoter activity of hrpA relative to the wild-type, and deletion of rsmB in gcpAD418A reduced its hrpA promoter activity to the ΔrsmB level (Fig. 6B). To investigate how H-NS and RsmB contribute to virulence through various GcpA-regulated virulence factors, virulence assay was performed in the host plant, B. campestris. The maceration ability of Δhns was reduced compared with that of the wild-type strain. In addition, deletion of hns in gcpAD418A significantly reduced its maceration ability to nearly wild-type levels (Fig. 6C). On the other hand, despite a hyper swimming motility phenotype observed in the gcpAD418AΔrsmB double mutant (Fig. 6A), deletion of rsmB in either the wild-type strain or the gcpAD418A mutant resulted in a non-pathogenic phenotype (Fig. 6C). Together, we concluded that the molecular mechanisms of GcpA to control various virulence factors are diverse; H-NS and RsmB are essential for the GcpA-dependent regulations of swimming motility and T3SS, respectively. However, the H-NS-RsmB-RsmA pathway was not a major component modulating these two virulence factors in a GcpA-dependent manner.
Fig. 6.
Effects of H-NS and RsmB on swimming motility, T3SS gene expression, and virulence. The swimming motility (A), T3SS gene hrpA promoter activity (B) and maceration on the leaves of Chinese cabbage (C) were examined. Values are a representative of three independent experiments. Three replicates were used in each experiment. Error bars indicate standard errors of the means. Different lowercase letters above the bar indicate statistically significant differences between treatments (Fisher’s LSD, P<0.05).
Discussion
D. dadantii produces Pels to degrade the plant cell wall, and production of this virulence factor is negatively regulated by c-di-GMP (Yi et al., 2010, Yuan et al., 2015). In this study, we propose a unique regulatory model that connects c-di-GMP regulation of bacterial virulence to both the global transcriptional and post-transcriptional regulatory systems in D. dadantii. To our knowledge, this is the first report implicating the H-NS-Rsm systems in the c-di-GMP signaling network for the negative regulation of pelD.
We confirmed GcpA which contains conserved A- and I-sites is a genuine DGC (Fig. 1B). More importantly, our findings clearly demonstrated that GcpA relies on its DGC activity to regulate Pel (Fig. 1B and 1C). Moreover, the unsuccessful attempt to delete gcpA suggests that GcpA might be essential for bacterial viability and may play additional roles other than its DGC activity. Interestingly, no other Gcp or Ecp proteins were shown to affect Pel production, suggesting that there might be specificity in different c-di-GMP signaling pathways.
PelD and PelE, which share high homology (89% similarity in amino acid sequence), are the most important Pels for the virulence of D. dadantii (Boccara et al., 1988). Expressions of pelD and pelE are differentially regulated in both plant tissues and media (Tardy et al., 1997, Hugouvieux-Cotte-Pattat et al., 1992, Masclaux et al., 1996). Here, we observed that the expression of pelD was much more significantly induced (22-fold) than pelE (4.5-fold) when PGA was supplied in minimal medium (MM), probably because the basal expression level of pelE is higher than pelD (Fig. S3). These results are in agreement with the previous studies, which indicate that the high basal level of pelE expression is essential to initiate rapid pectin degradation, while the highly induced expression of pelD is necessary for the maximum production of Pels during the infection in plant (Robert-Baudouy et al., 2000, Ouafa et al., 2012). Furthermore, the transcription of pelD was monitored along with the growth in MM supplemented with PGA, and the results showed that the expression of pelD gene was only highly induced at the mid-log phase of growth, but not in the early log phase of growth (Fig. S6). This result suggested that the transcription of the pelD gene is growth-phase dependent. Interestingly, we observed that the expression of pelD at both transcriptional and post-transcriptional levels was significantly enhanced in gcpAD418A A-site mutant relative to the wild-type at 12 h, which corresponded to early log phase, while the expression levels of pelE were not altered (Fig. 1D, 1E and 1F). A recent study demonstrated that the transcriptional start site shift after gene duplication might be one of the reasons for the different expression patterns of pelD and pelE (Duprey et al., 2016). However, the upstream regulatory mechanism remains unclear. Our results implied that the GcpA-mediated c-di-GMP signaling pathway is involved in the mechanism of differentially expression of pelD and pelE.
The post-transcriptional regulatory system, RsmA/RsmB has been shown to regulate several virulence factors including Pel in soft-rot pathogens D. dadantii and Pectobacterium carotovorum, but how RsmA/RsmB regulates pel gene expression remains unclear (Yang et al., 2008, Chatterjee et al., 1995, Mukherjee et al., 1996). Deletion of rsmA was lethal to D. dadantii 3937, unlike previous results studying P. carotovorum. Therefore, we were not able to generate a rsmA mutant to examine its direct effect on Pel production. Instead, we observed that over-expression of rsmA significantly repressed the pelD expression and Pel production (data not shown). Nevertheless, our results demonstrated that GcpA regulates the expression of RsmB and RsmA (Fig. 2). This regulation was further determined to be essential for the GcpA-dependent Pel regulation since either deletion of rsmB or overexpression of rsmA in the gcpAD418A mutant drastically reduced its pelD promoter activity and Pel production (Fig. 2C, 2D and S5). In P. aeruginosa, the RsmY and RsmZ sRNAs sequester the mRNA-binding protein RsmA in a mechanism similar to the RsmA/RsmB system (Lapouge et al., 2008). Several studies revealed that different DGCs negatively regulate RsmA activity through RsmY or RsmZ to control biofilm formation (Colley et al., 2016, Moscoso et al., 2014, Valentini et al., 2016). Thus, our findings strongly suggest that the regulation of c-di-GMP signaling on the Rsm system might be common in different bacterial species.
H-NS is a nucleoid-associated protein that functions as a global transcriptional regulator in many Gram-negative bacteria (Falconi et al., 1998, Yu & DiRita, 2002, Castang et al., 2008). In D. dadantii, it has been shown that H-NS positively regulates swimming motility and Pel production, but negatively regulates exopolysaccharides (EPS) synthesis (Nasser et al., 2001). Our findings demonstrated that GcpA negatively regulates hns expression at the post-transcriptional level (Fig. 2A and S4). Strikingly, deletion of hns in the gcpAD418A mutant not only restored its pelD promoter activity and Pel production (Fig. 2E and 2F), but also restored rsmB RNA to the wild-type level (Fig. 3). Taken together, we propose a regulatory pathway in which GcpA represses pelD gene expression via H-NS-RsmB-RsmA. Besides directly functioning as a repressor of pelD gene expression, H-NS is also known to be a positive regulator for Pel synthesis due to its negative impacts on PecT production (Nasser & Reverchon, 2002). Since we did not observe significant changes of pecT expression in the gcpAD418A mutant (Fig. 2A), this regulatory pathway might not be related to PecT.
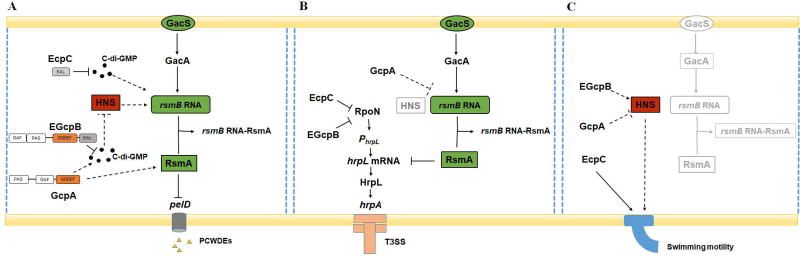
It is of interest to note that bacteria use multiple GGDEF and/or EAL domain proteins to regulate the same cellular behaviors in a sophisticated manner (Lindenberg et al., 2013, Valentini et al., 2016). Our findings demonstrated that the Rsm system plays an essential role in the c-di-GMP signaling and regulation of Pel production in D. dadantii. Although EGcpB, EcpC, and GcpA were shown to modulate the expression of rsmB at a post-transcriptional level (Fig. 2A and 4C), our results hinted that their regulatory mechanisms might be different. GcpA and EGcpB may respond to similar environmental signals via their GAF and PAS sensory domains (Fig. S1A), and modulate the same c-di-GMP pool to control pelD gene expression through the H-NS-RsmB-RsmA pathway (Fig. 7A). In Acetobacter xylinum, Qi and colleagues reported that the PAS domain of the DGC AxDGC2 enhances its cyclase activity by binding to the flavin adenine dinucleotide (FAD) cofactor under redox conditions (Qi et al., 2009). Similarly, oxygen levels may play a role in modulating c-di-GMP metabolism in D. dadantii. EcpC, the sole-EAL domain protein, is very likely to modulate a different c-di-GMP pool that directly targets RsmB bypassing H-NS. This convergence and divergence in c-di-GMP signaling is also supported by the regulation of swimming motility and T3SS gene expression in D. dadantii (Fig. 7B and 7C). We showed here that GcpA and EGcpB inversely modulate swimming motility through H-NS, which is different from EcpC. It is worth noting that our data indicated that the regulatory mechanism of GcpA on T3SS gene expression might be different from EGcpB and EcpC, which previous reports had indicated that these two PDEs positively regulate RpoN at the post-transcriptional level to control the T3SS master regulator HrpL. Indeed, we observed that only the transcript of hrpL not rpoN was increased in gcpAD418A A-site mutant compared with the wild-type strain (Fig. S7). Finally, our virulence assay further confirmed that swimming motility, T3SS gene expression, and Pel production are essential for D. dadantii to express full virulence in plants. Nevertheless, the environmental signals triggering the c-di-GMP-dependent regulation, the expression patterns and localizations of different DGCs and PDEs, and the c-di-GMP effectors that contribute to the signaling specificity on diverse cellular behaviors remain to be determined.
Fig. 7.
Working model for the c-di-GMP signaling pathway in D. dadantii.
(A) The regulation of c-di-GMP signaling on Pel production in D. dadantii is complex, involves several Gcp and Ecp proteins and takes place at both transcriptional and post-transcriptional levels. Rsm system is a central component in the c-di-GMP-related Pel regulation. GcpA and EGcpB modulate the same c-di-GMP pool to control pelD gene expression through H-NS-rsmB-RsmA pathway. EcpC modulates a different c-di-GMP pool that directly targets RsmB bypassing H-NS. GcpA positively regulates RsmA at post-transcriptional level. (B) The regulation of GcpA on T3SS gene expression is dependent on its impact on RsmB, which controls the expression of hrpL at post-transcriptional level. This regulation is different from EGcpB and EcpC that regulate hrpL at transcriptional level via RpoN-hrpL pathway. (C) GcpA and EGcpB regulate swimming motility through H-NS, while EcpC is different from them. ⊥represents negative control; →represents positive control. The dotted lines indicate regulatory mechanisms identified in this study.
Overall, this multilevel regulation of c-di-GMP signaling on diverse virulence factors assures an accurate control of the virulence process for the infection of D. dadantii. It provides evidence on the complexity and specificity of c-di-GMP signaling in bacteria and sheds light on the understanding of D. dadantii infection strategies under various environmental conditions or during different infection status.
Experimental Procedures
Bacterial strains, plasmids, primers, and media
The bacterial strains and plasmids used in this study are listed in Table S1 (see Supporting Information). Dickeya dadantii strains were grown in Luria-Bertani (LB) medium (1% tryptone, 0.5% yeast extract, and 1% NaCl), mannitol-glutamic acid (MG) medium (1% mannitol, 0.2% glutamic acid, 0.05% potassium phosphate monobasic, 0.02% NaCl, and 0.02% MgSO4) or M9 minimal medium (MM) supplemented with 0.1% polygalacturonic acid (PGA) at 28°C (Yang et al., 2007). Escherichia coli strains were grown in LB at 37°C. Antibiotics were added to the media at the following concentrations: ampicillin (100 µg/ml), kanamycin (50 µg/ml), chloramphenicol (20 µg/ml), and spectinomycin (100 µg/ml). The D. dadantii 3937 genome sequence was retrieved from a systematic annotation package for community analysis of genomes (ASAP) (https://asap.ahabs.wisc.edu/asap/home.php). Primers used for cloning and qPCR in this study are listed in Table S2 (see Supporting Information).
Mutant construction and complementation
The GGDEF and/or EAL domain encoding genes, hns, rsmB, and pecT were deleted from the genome by allelic exchange mutagenesis (Yang et al., 2002). In brief, upstream and downstream fragments flanking each target gene were amplified by polymerase chain reaction (PCR) with specific primers (Table S2). The kanamycin cassette was amplified from the pKD4 plasmid (Datsenko & Wanner, 2000) and was cloned between two flanking regions using three-way cross-over PCR. The PCR construct was inserted into the suicide plasmid pWM91, and the resulting plasmid was transformed into D. dadantii 3937 by conjugation using E. coli strain S17-1 λ-pir. Recombinants that grew on kanamycin medium, were plated on 10% sucrose plate to select strains with chromosomal deletions. Cells that were resistant to sucrose due to the loss of SacB-mediated toxicity were then plated on an ampicillin plate, and the ampicillin sensitive cells were confirmed by PCR using outside primers. Mutations were confirmed by sequencing.
To construct the site-specific point mutation in the GGDEF motif of GcpA, single nucleotide substitution was performed using the QuikChange XL Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA). Briefly, a primer set, gcpA-D418A-1 and gcpA-D418A-2 (Table S2), was used to generate gcpAD418A, in which the SGDEF motif was changed to SGAEF. Substitution was confirmed by sequencing. The gcpAD418A fragment was then amplified using primer set, gcpA-A-SacI and gcpA-B (Table S2), and cloned upstream to the kanamycin cassette followed by the downstream fragment flanking gcpA using three-way cross-over PCR. The construct was inserted into pWM91, and the resulting plasmid was transferred into D. dadantii by conjugation using E. coli strain S17-1 λ-pir. The above described allelic exchange mutagenesis was conducted to replace wild-type gcpA with gcpAD418A. Mutation was confirmed by sequencing using outside primers.
To construct double mutants, rsmB, hns, egcpB, and ecpC, were allelic exchanged in a gcpAD418A unmarked mutant strain, respectively. In brief, the pFLP2 plasmid encoding FLP (flipase) recombinase enzyme was transferred into the gcpAD418A::Km strain by conjugation using E. coli S17-1 λ-pir. Two FLP recombinase target (FRT) sites flanking the kanamycin cassette allowed for flipase mediated excision of Km. Transconjugants that were sensitive to Km and high-concentration of sucrose, were then confirmed using outside primers and sequencing. To generate complemented strains, the promoter and ORF regions of target genes were amplified and cloned into the low-copy-number plasmid pCL1920 (Table S1). The resulting plasmids were then confirmed by sequencing and electroporated into mutant strains.
Swimming motility assay
Swimming motility was examined by inoculating 10 µl of overnight bacterial cultures (OD600=1.0) onto the center of MG plates containing 0.2% agar. The inoculated plates were incubated at 28°C for 16 h. The diameter of the radial growth was measured (Antúnez-Lamas et al., 2009).
Pel activity assay
Extracellular Pel activity was measured by spectrometry as previously described (Matsumoto et al., 2003). Bacterial cells were cultured in MM media supplemented with 0.1% PGA at 28°C for 16 h. 1 ml bacterial cultures were then centrifuged at 15,000 rpm for 2 min, and the supernatant was collected. 10 µl of supernatant was added to 990 µl of the reaction buffer (0.05% PGA, 0.1 M Tris-HCl [pH 8.5], and 0.1 mM CaCl2, prewarmed to 30°C). Pel activity was monitored at A230 for 3 min and calculated based on one unit of Pel activity being equal to an increase of 1 × 10−3 OD230 in 1 min.
GFP reporter plasmid construction and flow cytometry assay
To generate the reporter plasmids pAT-pelE, pAT-rsmA, and pAT-hns, the promoter regions of each gene were PCR amplified and cloned into the promoter probe vector pPROBE-AT, which contains a ribosomal binding site upstream of the gfp gene (Miller et al., 2000, Leveau & Lindow, 2001). The reporter plasmids pAT-pelD, pAT-hrpA and pAT-rsmB was constructed previously following the same procedure (Li et al., 2015, Peng et al., 2006, Yang et al., 2007). Promoter activity was monitored by measuring GFP intensity through flow cytometry (BD Biosciences, San Jose, CA) as previously described (Peng et al., 2006). Briefly, bacterial cells with the reporter plasmid were grown in LB media overnight and inoculated 1:100 into MM media with or without 0.1% PGA. Samples were collected at 6 h, 12 h and 24 h, respectively, and promoter activity was quantified by detecting GFP intensity using flow cytometry.
Determination of the intracellular c-di-GMP concentration
Intracellular c-di-GMP concentrations were determined by using ultra performance liquid chromatography coupled with tandem mass spectrometry (UPLC-MS-MS) using a method that has been previously described (Massie et al., 2012). Briefly, overnight bacterial cultures were inoculated 1:1000 into 50 ml LB media in a flask. After the OD600 of the bacterial culture reached about 0.8, corresponding to mid- to late-exponential growth, all cells were centrifuged in 50 ml polystyrene centrifuge tubes for 30 min at 4,000 rpm. The supernatant was then removed, and the pellet was re-suspended in 1.5 ml extraction buffer (40% acetonitrile-40% methanol in 0.1 N formic acid). To lyse the cells and release intracellular c-di-GMP, cells re-suspended in extraction buffer were dried by speed-vac, resuspended in 100 µL of HPLC grade water, centrifuged for 5 min at 21,000×g in a tabletop centrifuge to pellet insoluble debris, filtered through a Titan syringe filter (PVDF, 0.45 µm, 4 mm), and analyzed by UPLC-MS-MS.
Western blot analysis
D. dadantii cells were grown in MM broth supplemented with 0.1% PGA at 28°C for 12 h during the exponential growth phase and 1 ml samples were taken. Cells were then resuspended in phosphate-buffered saline (PBS) buffer and lysed by sonication. The protein in crude lysates was quantified using the Bradford protein assay (Bio-Rad). Samples were boiled before loading onto 12% sodium dodecyl sulfate polyacrylamide gels. Proteins were then transferred onto a polyvinylidene fluoride membrane (Millipore). Blots were washed with PBS containing 0.05% Tween-20 and probed with an anti-RsmA antibody (Proteintech, Rosemont, IL). Anti-RNA polymerase monoclonal antibody (Neoclone) was used as a control. The resulting blots were incubated for 1 min in enhanced chemiluminescence reagent (GE Healthcare) and detected using O-MAT X-ray film.
Northern blot analysis
To measure the RNA levels of rsmB in D. dadantii strains, bacterial cells grown in MM supplemented with 0.1% PGA for 12 h were harvested and total RNA was isolated using TRI reagent (Sigma-Aldrich, St Louis, MO). The residual DNA was removed with a Turbo DNA-free DNase kit (Invitrogen, Ausin, TX). Northern blot analysis was performed using biotin-labelled probe and a biotin detection system (BrightStar Psolaren-Biotin and Bright Star BioDetect, Ambion). 16S rRNA was used as an internal control.
qRT-PCR analysis
The mRNA levels of pelD, pelE, rsmA, rsmB, pecT, pecS, fis, fur, kdgR, crp, hns, hrpL and rpoN were measured by qRT-PCR. Briefly, bacterial cells cultured in MM broth supplemented with 0.1% PGA for 12 h were harvested and total RNA was extracted using PureLink RNA Mini Kit (Ambion, Carlsbad, CA) according to the manufacturer’s instruction. On-column DNase treatment (Invitrogen, Carlsbad, CA) was performed. cDNA was synthesized using the QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany). The cDNA level of different samples was quantified by real-time PCR using a PowerUp SYBR Green Master Mix (Life Technologies, Carlsbad, CA). The relative levels of gene expression were determined by using the 2−∆∆CT method (Livak & Schmittgen, 2001), with the rplU gene as the internal control (Mah et al., 2003). Three technical replicates were used each time.
Virulence assay
The local leaf maceration assay was performed using the leaves of Chinese cabbage (B. campestris) as described (Yuan et al., 2015). In brief, 10 µl of bacterial suspension at 107 CFU ml−1 were inoculated into the wounds punched with a sterile pipette on the leaves. Five leaves were used for each strain. Inoculated Chinese cabbage leaves were kept in growth chamber at 28°C with 100% relative humidity for 16 h before pictures were taken. To evaluate disease symptoms, APS ASSESS 1.0 software (Image Analysis Software for Plant Disease Quantification) was used to determine the leaf maceration areas.
Statistical analysis
Means and standard deviations of experimental results were calculated using Excel and the statistical analysis was performed using a two-tailed student’s t-test (Microsoft, Redmond, WA) or Fisher’s Lease Significant Difference (LSD) test using DPS data processing system (http://www.dpsw.cn/dps_eng).
Supplementary Material
Fig. S1 Analysis of GGDEF and/or EAL domain proteins in D. dadantii. (A) Summary of GGDEF and/or EAL domain proteins. Proteins were shown with the encoded gene names and protein length. Protein domains were predicted by the simplified modular architecture research tool (SMART). (B) Amino acid sequence alignment for the GGDEF domains in D. dadantii. PleD and WspR are two active DGCs from Caulobacter crescentus and Pseudomonas aeruginosa, respectively. Inhibition site RxxD motif (I-site) and enzymatic activity site GGDEF motif (A-site) are marked. (C) Amino acid sequence alignment for the EAL domains in D. dadantii. VC1086 is an active PDE from Vibrio cholerae. The arrow indicates glutamate residue in the EAL motif. “*” means that the residues are identical in all sequences in the alignment, “:” means that conserved substitutions have been observed, “.” Means that semi-conserved substitutions are observed.
Fig. S2. Complementation of gcpAD418A on Pel production. Values are a representative of three independent experiments. Three replicates were used in each experiment. Error bars indicate standard errors of the means. Asterisks indicate statistically significant differences of the means (P<0.05 by Student’s t test).
Fig. S3 PGA induces the transcription of pelD and pelE genes. (A–B) The promoter activities of pelD and pelE in D. dadantii were examined at 12 and 24 h, respectively. The experiments were repeated three independent times with similar results. The figure represents results from one experiment which includes three technical replicates. Error bars indicate standard errors of the means. Asterisks indicate statistically significant differences of the means (P<0.05 by Student’s t test).
Fig. S4 Promoter activities of rsmA, rsmB and hns are not affected in gcpAD418A. (A–C) The promoter activities of rsmA, rsmB and hns in D. dadantii strains. One representative experiment was chosen, and three independent experiments were performed. Assays were performed as described in the Experimental Procedures. Error bars indicate standard errors of the means.
Fig. S5 Expression of rsmA restored Pel in gcpAD418A A-site mutant. (A–B) Pel production and pelD promoter activity were measured in D. dadantii. Three independent experiments were performed with three replicates in each experiment. Values are from one representative experiment. Error bars indicate standard errors of the means. Asterisks indicate statistically significant differences of the means (P < 0.05 by Student’s t-test).
Fig. S6 pelD gene transcription is growth-phase dependent. The promoter activity of pelD gene was determined throughout growth in MM medium supplemented with 0.1% PGA at 28°C. The experiments were repeated two independent times with similar results. Three replicates were used for each experiment. Error bars indicate standard errors of the means. Open square represents OD600 values. Filled triangle represents mean fluorescence intensity.
Fig. S7 Impact of GcpA on the expression of hrpL and rpoN. Relative mRNA levels of hrpL and rpoN in gcpAD418A to that in the wild type, which was mathematically designated as 1. All results are from one representative experiment. Three independent experiments were performed and three replicates were used for each experiment. Error bars indicate standard errors of the means. Asterisks indicate statistically significant differences of the means (P < 0.05 by Student’s t-test).
Table. S1 Strains and plasmids used in this study.
Table. S2 Primers used in this study.
Acknowledgments
Xiaochen Yuan was supported by the Postdoctoral Workstation of Jiangsu Academy of Agricultural Sciences. This work was funded by United States Department of Agriculture-National Institute of Food and Agriculture-Agriculture and Food Research Initiative-Exploratory Research program (2016-67030-24856) and Research Growth Initiative of the University of Wisconsin-Milwaukee awarded to Ching-Hong Yang; National Key Research and Development program of China (2017YFC200604) awarded to Fengquan Liu; China Scholarship Council (201503250007) awarded to Fang Tian; National Science Foundation of China (31370160; 31671990) awarded to Chenyang He; National Institutes of Health grants GM109259 awarded to Christopher M. Waters.
References
- Antúnez-Lamas M, Cabrera-Ordonez E, Lopez-Solanilla E, Raposo R, Trelles-Salazar O, Rodríguez-Moreno A, et al. Role of motility and chemotaxis in the pathogenesis of Dickeya dadantii 3937 (ex Erwinia chrysanthemi 3937) Microbiology. 2009;155:434–442. doi: 10.1099/mic.0.022244-0. [DOI] [PubMed] [Google Scholar]
- Boccara M, Diolez A, Rouve M, Kotoujansky A. The role of individual pectate lyases of Erwinia chrysanthemi strain 3937 in pathogenicity on saintpaulia plants. Physiological and Molecular Plant Pathology. 1988;33:95–104. [Google Scholar]
- Castang S, McManus HR, Turner KH, Dove SL. H-NS family members function coordinately in an opportunistic pathogen. Proceedings of the National Academy of Sciences. 2008;105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charkowski A, Blanco C, Condemine G, Expert D, Franza T, Hayes C, et al. The role of secretion systems and small molecules in soft-rot Enterobacteriaceae pathogenicity. Annual Review of Phytopathology. 2012;50:425–449. doi: 10.1146/annurev-phyto-081211-173013. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Cui Y, Liu Y, Dumenyo CK, Chatterjee AK. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-L-homoserine lactone. Applied and Environmental Microbiology. 1995;61:1959–1967. doi: 10.1128/aem.61.5.1959-1967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, et al. Allosteric control of cyclic di-GMP signaling. Journal of Biological Chemistry. 2006;281:32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- Colley B, Dederer V, Carnell M, Kjelleberg S, Rice SA, Klebensberger J. SiaA/D interconnects c-di-GMP and RsmA signaling to coordinate cellular aggregation of Pseudomonas aeruginosa in response to environmental conditions. Frontiers in Microbiology. 2016;7 doi: 10.3389/fmicb.2016.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collmer A, Keen NT. The role of pectic enzymes in plant pathogenesis. Annual Review of Phytopathology. 1986;24:383–409. [Google Scholar]
- Cotter PA, Stibitz S. c-di-GMP-mediated regulation of virulence and biofilm formation. Current Opinion in Microbiology. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Czajkowski R, Perombelon MC, van Veen JA, van der Wolf JM. Control of blackleg and tuber soft rot of potato caused by Pectobacterium and Dickeya species: a review. Plant Pathology. 2011;60:999–1013. [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprey A, Nasser W, Léonard S, Brochier-Armanet C, Reverchon S. Transcriptional start site turnover in the evolution of bacterial paralogous genes–the pelE-pelD virulence genes in Dickeya. The FEBS Journal. 2016;283:4192–4207. doi: 10.1111/febs.13921. [DOI] [PubMed] [Google Scholar]
- Falconi M, Colonna B, Prosseda G, Micheli G, Gualerzi CO. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. The EMBO Journal. 1998;17:7033–7043. doi: 10.1093/emboj/17.23.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franza T, Michaud-Soret I, Piquerel P, Expert D. Coupling of iron assimilation and pectinolysis in Erwinia chrysanthemi 3937. Molecular Plant-Microbe Interactions. 2002;15:1181–1191. doi: 10.1094/MPMI.2002.15.11.1181. [DOI] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nature Reviews Microbiology. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Hérault E, Reverchon S, Nasser W. Role of the LysR-type transcriptional regulator PecT and DNA supercoiling in the thermoregulation of pel genes, the major virulence factors in Dickeya dadantii. Environmental Microbiology. 2014;16:734–745. doi: 10.1111/1462-2920.12198. [DOI] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. Regulation of pectinolysis in Erwinia chrysanthemi. Annual Reviews in Microbiology. 1996;50:213–257. doi: 10.1146/annurev.micro.50.1.213. [DOI] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N, Dominguez H, Robert-Baudouy J. Environmental conditions affect transcription of the pectinase genes of Erwinia chrysanthemi 3937. Journal of bacteriology. 1992;174:7807–7818. doi: 10.1128/jb.174.23.7807-7818.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Reinders A, Lori C. Cyclic di-GMP: second messenger extraordinaire. Nature Reviews Microbiology. 2017;15:271–284. doi: 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- Lapouge K, Schubert M, Allain FHT, Haas D. Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Molecular Microbiology. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- Leveau JH, Lindow SE. Predictive and interpretive simulation of green fluorescent protein expression in reporter bacteria. Journal of Bacteriology. 2001;183:6752–6762. doi: 10.1128/JB.183.23.6752-6762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hutchins W, Wu X, Liang C, Zhang C, Yuan X, et al. Derivative of plant phenolic compound inhibits the type III secretion system of Dickeya dadantii via HrpX/HrpY two-component signal transduction and Rsm systems. Molecular Plant Pathology. 2015;16:150–163. doi: 10.1111/mpp.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. The EAL domain protein YciR acts as a trigger enzyme in ac-di-GMP signalling cascade in E. coli biofilm control. The EMBO journal. 2013;32:2001–2014. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MY, Gui G, Wei B, Preston JF, Oakford L, Yüksel Ü, et al. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. Journal of Biological Chemistry. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Llama-Palacios A, López-Solanilla E, Rodríguez-Palenzuela P. Role of the PhoP-PhoQ system in the virulence of Erwinia chrysanthemi strain 3937: involvement in sensitivity to plant antimicrobial peptides, survival at acid pH, and regulation of pectolytic enzymes. Journal of Bacteriology. 2005;187:2157–2162. doi: 10.1128/JB.187.6.2157-2162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Hibbing ME, Kim H-S, Reedy RM, Yedidia I, Breuer J, et al. Host range and molecular phylogenies of the soft rot enterobacterial genera Pectobacterium and Dickeya. Phytopathology. 2007;97:1150–1163. doi: 10.1094/PHYTO-97-9-1150. [DOI] [PubMed] [Google Scholar]
- Mah T-F, Pitts B, Pellock B, Walker GC, Stewart PS, O'toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- Masclaux C, Hugouvieux-Cotte-Pattat N, Expert D. Iron is a triggering factor for differential expression of Erwinia chrysanthemi strain 3937 pectate lyases in pathogenesis of African violets. Molecular Plant-Microbe Interactions. 1996;9:198–205. [Google Scholar]
- Massie JP, Reynolds EL, Koestler BJ, Cong J-P, Agostoni M, Waters CM. Quantification of high-specificity cyclic diguanylate signaling. Proceedings of the National Academy of Sciences. 2012;109:12746–12751. doi: 10.1073/pnas.1115663109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H, Muroi H, Umehara M, Yoshitake Y, Tsuyumu S. Peh production, flagellum synthesis, and virulence reduced in Erwinia carotovora subsp. carotovora by mutation in a homologue of cytR. Molecular Plant-Microbe Interactions. 2003;16:389–397. doi: 10.1094/MPMI.2003.16.5.389. [DOI] [PubMed] [Google Scholar]
- Miller WG, Leveau JH, Lindow SE. Improved gfp and inaZ broad-host-range promoter-probe vectors. Molecular Plant-Microbe Interactions. 2000;13:1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- Moscoso JA, Jaeger T, Valentini M, Hui K, Jenal U, Filloux A. The diguanylate cyclase SadC is a central player in Gac/Rsm-mediated biofilm formation in Pseudomonas aeruginosa. Journal of Bacteriology. 2014;196:4081–4088. doi: 10.1128/JB.01850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Cui Y, Liu Y, Dumenyo CK, Chatterjee AK. Global regulation in Erwinia species by Erwinia carotovora rsmA a homologue of Escherichia coli csrA: repression of secondary metabolites, pathogenicity and hypersensitive reaction. Microbiology. 1996;142:427–434. doi: 10.1099/13500872-142-2-427. [DOI] [PubMed] [Google Scholar]
- Nasser W, Dorel C, Wawrzyniak J, Van Gijsegem F, Groleau MC, Déziel E, et al. Vfm a new quorum sensing system controls the virulence of Dickeya dadantii. Environmental Microbiology. 2013;15:865–880. doi: 10.1111/1462-2920.12049. [DOI] [PubMed] [Google Scholar]
- Nasser W, Faelen M, Hugouvieux-Cotte-Pattat N, Reverchon S. Role of the nucleoid-associated protein H-NS in the synthesis of virulence factors in the phytopathogenic bacterium Erwinia chrysanthemi. Molecular Plant-Microbe Interactions. 2001;14:10–20. doi: 10.1094/MPMI.2001.14.1.10. [DOI] [PubMed] [Google Scholar]
- Nasser W, Reverchon S. H-NS-dependent activation of pectate lyases synthesis in the phytopathogenic bacterium Erwinia chrysanthemi is mediated by the PecT repressor. Molecular Microbiology. 2002;43:733–748. doi: 10.1046/j.1365-2958.2002.02782.x. [DOI] [PubMed] [Google Scholar]
- Orr MW, Galperin MY, Lee VT. Sustained sensing as an emerging principle in second messenger signaling systems. Current Opinion in Microbiology. 2016;34:119–126. doi: 10.1016/j.mib.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouafa Z-A, Reverchon S, Lautier T, Muskhelishvili G, Nasser W. The nucleoid-associated proteins H-NS and FIS modulate the DNA supercoiling response of the pel genes, the major virulence factors in the plant pathogen bacterium Dickeya dadantii. Nucleic Acids Research. 2012;40:4306–4319. doi: 10.1093/nar/gks014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, et al. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes & Development. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q, Yang S, Charkowski AO, Yap M-N, Steeber DA, Keen NT, et al. Population behavior analysis of dspE and pelD regulation in Erwinia chrysanthemi 3937. Molecular Plant-Microbe Interactions. 2006;19:451–457. doi: 10.1094/MPMI-19-0451. [DOI] [PubMed] [Google Scholar]
- Povolotsky TL, Hengge R. ‘Life-style’control networks in Escherichia coli: signaling by the second messenger c-di-GMP. Journal of Biotechnology. 2012;160:10–16. doi: 10.1016/j.jbiotec.2011.12.024. [DOI] [PubMed] [Google Scholar]
- Qi Y, Rao F, Luo Z, Liang Z-X. A Flavin Cofactor-Binding PAS Domain Regulates c-di-GMP Synthesis in Ax DGC2 from Acetobacter xylinum. Biochemistry. 2009;48:10275–10285. doi: 10.1021/bi901121w. [DOI] [PubMed] [Google Scholar]
- Reverchon S, Nasser W. Dickeya ecology, environment sensing and regulation of virulence programme. Environmental Microbiology Reports. 2013;5:622–636. doi: 10.1111/1758-2229.12073. [DOI] [PubMed] [Google Scholar]
- Reverchon S, Nasser W, Robert-Baudouy J. Characterization of kdgR, a gene of Erwinia chrysanthemi that regulates pectin degradation. Molecular Microbiology. 1991;5:2203–2216. doi: 10.1111/j.1365-2958.1991.tb02150.x. [DOI] [PubMed] [Google Scholar]
- Reverchon S, Van Gijsegem F, Effantin G, Zghidi-Abouzid O, Nasser W. Systematic targeted mutagenesis of the MarR/SlyA family members of Dickeya dadantii 3937 reveals a role for MfbR in the modulation of virulence gene expression in response to acidic pH. Molecular Microbiology. 2010;78:1018–1037. doi: 10.1111/j.1365-2958.2010.07388.x. [DOI] [PubMed] [Google Scholar]
- Robert-Baudouy J, Nasser W, Condemine G, Reverchon S, Shevchik VE, Hugouvieux-Cotte-Pattat N. Pectic enzymes of Erwinia chrysanthemi, regulation and role in pathogenesis. Plant-Microbe Interactions. 2000;5:221–268. [Google Scholar]
- Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiology and Molecular Biology Reviews. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He Y-W, et al. Cell–cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proceedings of the National Academy of Sciences. 2006;103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ryan RP, Tolker-Nielsen T, Dow JM. When the PilZ don’t work: effectors for cyclic di-GMP action in bacteria. Trends in Microbiology. 2012;20:235–242. doi: 10.1016/j.tim.2012.02.008. [DOI] [PubMed] [Google Scholar]
- Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. Journal of Bacteriology. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annual Review of Microbiology. 2007;61:131. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Tischler AD, Camilli A. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. Journal of Biological Chemistry. 2005;280:33324–33330. doi: 10.1074/jbc.M506500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardy F, Nasser W, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Comparative analysis of the five major Erwinia chrysanthemi pectate lyases: enzyme characteristics and potential inhibitors. Journal of Bacteriology. 1997;179:2503–2511. doi: 10.1128/jb.179.8.2503-2511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini M, Laventie B-J, Moscoso J, Jenal U, Filloux A. The Diguanylate Cyclase HsbD Intersects with the HptB Regulatory Cascade to Control Pseudomonas aeruginosa Biofilm and Motility. PLoS Genetics. 2016;12:e1006354. doi: 10.1371/journal.pgen.1006354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. Journal of Bacteriology. 2008;190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley CG, Lee D-J. Bacterial diguanylate cyclases: Structure, function and mechanism in exopolysaccharide biofilm development. Biotechnology Advances. 2015;33:124–141. doi: 10.1016/j.biotechadv.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Wu X, Zeng Q, Koestler BJ, Waters CM, Sundin GW, Hutchins W, et al. Deciphering the components that coordinately regulate virulence factors of the soft rot pathogen Dickeya dadantii. Molecular Plant-Microbe Interactions. 2014;27:1119–1131. doi: 10.1094/MPMI-01-14-0026-R. [DOI] [PubMed] [Google Scholar]
- Yang C-H, Gavilanes-Ruiz M, Okinaka Y, Vedel R, Berthuy I, Boccara M, et al. hrp genes of Erwinia chrysanthemi 3937 are important virulence factors. Molecular Plant-Microbe Interactions. 2002;15:472–480. doi: 10.1094/MPMI.2002.15.5.472. [DOI] [PubMed] [Google Scholar]
- Yang S, Peng Q, Zhang Q, Yi X, Choi CJ, Reedy RM, et al. Dynamic regulation of GacA in type III secretion, pectinase gene expression, pellicle formation, and pathogenicity of Dickeya dadantii (Erwinia chrysanthemi 3937) Molecular Plant-Microbe Interactions. 2008;21:133–142. doi: 10.1094/MPMI-21-1-0133. [DOI] [PubMed] [Google Scholar]
- Yang S, Zhang Q, Guo J, Charkowski AO, Glick BR, Ibekwe AM, et al. Global effect of indole-3-acetic acid biosynthesis on multiple virulence factors of Erwinia chrysanthemi 3937. Applied and Environmental Microbiology. 2007;73:1079–1088. doi: 10.1128/AEM.01770-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Yamazaki A, Biddle E, Zeng Q, Yang CH. Genetic analysis of two phosphodiesterases reveals cyclic diguanylate regulation of virulence factors in Dickeya dadantii. Molecular Microbiology. 2010;77:787–800. doi: 10.1111/j.1365-2958.2010.07246.x. [DOI] [PubMed] [Google Scholar]
- Yu RR, DiRita VJ. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Molecular Microbiology. 2002;43:119–134. doi: 10.1046/j.1365-2958.2002.02721.x. [DOI] [PubMed] [Google Scholar]
- Yuan X, Khokhani D, Wu X, Yang F, Biener G, Koestler BJ, et al. Cross-talk between a regulatory small RNA, cyclic-di-GMP signalling and flagellar regulator FlhDC for virulence and bacterial behaviours. Environmental Microbiology. 2015;17:4745–4763. doi: 10.1111/1462-2920.13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Analysis of GGDEF and/or EAL domain proteins in D. dadantii. (A) Summary of GGDEF and/or EAL domain proteins. Proteins were shown with the encoded gene names and protein length. Protein domains were predicted by the simplified modular architecture research tool (SMART). (B) Amino acid sequence alignment for the GGDEF domains in D. dadantii. PleD and WspR are two active DGCs from Caulobacter crescentus and Pseudomonas aeruginosa, respectively. Inhibition site RxxD motif (I-site) and enzymatic activity site GGDEF motif (A-site) are marked. (C) Amino acid sequence alignment for the EAL domains in D. dadantii. VC1086 is an active PDE from Vibrio cholerae. The arrow indicates glutamate residue in the EAL motif. “*” means that the residues are identical in all sequences in the alignment, “:” means that conserved substitutions have been observed, “.” Means that semi-conserved substitutions are observed.
Fig. S2. Complementation of gcpAD418A on Pel production. Values are a representative of three independent experiments. Three replicates were used in each experiment. Error bars indicate standard errors of the means. Asterisks indicate statistically significant differences of the means (P<0.05 by Student’s t test).
Fig. S3 PGA induces the transcription of pelD and pelE genes. (A–B) The promoter activities of pelD and pelE in D. dadantii were examined at 12 and 24 h, respectively. The experiments were repeated three independent times with similar results. The figure represents results from one experiment which includes three technical replicates. Error bars indicate standard errors of the means. Asterisks indicate statistically significant differences of the means (P<0.05 by Student’s t test).
Fig. S4 Promoter activities of rsmA, rsmB and hns are not affected in gcpAD418A. (A–C) The promoter activities of rsmA, rsmB and hns in D. dadantii strains. One representative experiment was chosen, and three independent experiments were performed. Assays were performed as described in the Experimental Procedures. Error bars indicate standard errors of the means.
Fig. S5 Expression of rsmA restored Pel in gcpAD418A A-site mutant. (A–B) Pel production and pelD promoter activity were measured in D. dadantii. Three independent experiments were performed with three replicates in each experiment. Values are from one representative experiment. Error bars indicate standard errors of the means. Asterisks indicate statistically significant differences of the means (P < 0.05 by Student’s t-test).
Fig. S6 pelD gene transcription is growth-phase dependent. The promoter activity of pelD gene was determined throughout growth in MM medium supplemented with 0.1% PGA at 28°C. The experiments were repeated two independent times with similar results. Three replicates were used for each experiment. Error bars indicate standard errors of the means. Open square represents OD600 values. Filled triangle represents mean fluorescence intensity.
Fig. S7 Impact of GcpA on the expression of hrpL and rpoN. Relative mRNA levels of hrpL and rpoN in gcpAD418A to that in the wild type, which was mathematically designated as 1. All results are from one representative experiment. Three independent experiments were performed and three replicates were used for each experiment. Error bars indicate standard errors of the means. Asterisks indicate statistically significant differences of the means (P < 0.05 by Student’s t-test).
Table. S1 Strains and plasmids used in this study.
Table. S2 Primers used in this study.