Abstract
Radiation therapy is the primary intervention for nearly half of the patients with localized advanced prostate cancer and standard of care for recurrent disease following surgery. The development of radiation resistant disease is an obstacle for nearly 30–50% of patients undergoing radiotherapy. A better understanding of mechanisms that lead to radiation resistance could aid in the development of sensitizing agents to improve outcome. Here, we identified a radiation-resistance pathway mediated by CD105, downstream of BMP and TGF-β signaling. Antagonizing CD105-dependent BMP signaling with a partially humanized monoclonal antibody, TRC105, resulted in a significant reduction in clonogenicity when combined with irradiation. In trying to better understand the mechanism for the radio-sensitization, we found that radiation-induced CD105/BMP signaling was sufficient and necessary for the upregulation of sirtuin1 (SIRT1) in contributing to p53 stabilization and PGC-1α activation. Combining TRC105 with irradiation delayed DNA damage repair compared to irradiation alone. However, in the absence of p53 function, combining TRC105 and radiation resulted in no reduction in clonogenicity compared to radiation alone, despite similar reduction of DNA damage repair observed in p53-intact cells. This suggested DNA damage repair was not the sole determinant of CD105 radio-resistance. As cancer cells undergo an energy deficit following irradiation, due to the demands of DNA and organelle repair, we examined SIRT1’s role on p53 and PGC-1α with respect to glycolysis and mitochondrial biogenesis, respectively. Consequently, blocking the CD105-SIRT1 axis was found to deplete the ATP stores of irradiated cells and cause G2 cell cycle arrest. Xenograft models supported these findings that combining TRC105 with irradiation significantly reduces tumor size over irradiation alone (p value = 10−9). We identified a novel synthetic lethality strategy of combining radiation and CD105 targeting to address the DNA repair and metabolic addiction induced by irradiation in p53-functional prostate cancers.
Keywords: radiation sensitization, DNA damage repair, mitochondrial biogenesis, prostate cancer
Introduction
Prostate cancer is the second leading cause of cancer mortality in men. The standard of care for localized prostate cancer is radiotherapy or surgical resection. Radiation is also used as an adjuvant therapy following surgery, salvage therapy after biochemical recurrence, and for palliation in the setting of distant metastasis. Up to 30% of localized prostate cancer patients treated with definitive radiation therapy develop recurrent radio-resistant disease and the most common anatomic site of recurrence is within the prostate itself, even in patients at high risk of metastasis (1–3). Further, 50% of patients that undergo salvage radiation therapy after biochemical recurrence will have disease progression (4). Although dose-escalation improves biochemical control, toxicity remains a significant obstacle in optimizing local control (5, 6). Accordingly, sensitizing agents are needed to improve tumor eradication and minimize toxicity to normal structures. With the rational that targeting mechanisms of radio-resistance can yield durable sensitization, we identified a novel pathway affecting both DNA repair and energy demands manifested by irradiation of prostate cancer cells.
Endoglin (CD105), a type III transforming growth factor beta/bone morphogenic protein (TGF-β/BMP) co-receptor, recognized as a marker of proliferating endothelia, is upregulated in several cancers including prostate cancer (7). CD105 behaves like a switch on the cell surface to inhibit TGF-β signaling and promote BMP signaling. Therefore, silencing or knocking out CD105 results in the gain of TGF-β-mediated Smad 2/3 signaling and loss of Smad1/5 signaling associated with BMP activity (8). CD105 expression on various cancers has correlated with progression, metastasis, aggressiveness, and evasion to conventional therapeutics (9–12). Various DNA repair genes were found to be downregulated by CD105 silencing, thereby sensitizing ovarian cancer to DNA a damaging agent, cisplatin (13). However, these studies did not distinguish between the CD105 effects on TGF-β and BMP signaling on DNA damage repair. Significant data are reported for the use of specific TGF-β inhibition in radiation sensitizing breast cancer and glioblastoma (14, 15). However, limited information is known about the role of BMP signaling in response to radiation. In this study, we use TRC105, a partially humanized monoclonal antibody that blocks the CD105/BMP signaling complex. Importantly, as TRC105 does not affect the CD105/TGF-β signaling axis, the role of CD105/BMP signaling on radiation responsiveness was tested (16). Based on our finding that CD105 was elevated by irradiation, we hypothesized targeting CD105, using TRC105, could sensitize prostate cancer to irradiation. Of note, numerous phase I trials have shown TRC105 to be well tolerated, but it has had limited therapeutic efficacy for prostate cancer as a single agent (17, 18).
Probing CD105/BMP regulation of DNA repair genes led us to identify sirtuin 1 (SIRT1), a NAD+ dependent histone deacetylase, as a BMP regulated target. SIRT1 activation is observed in prostate cancer and in response to irradiation (19). In the context of cancer, SIRT1 has been studied primarily for its role in DNA damage response. Outside of cancer biology, SIRT1 de-regulation is associated with metabolic, neurodegenerative, and cardiovascular diseases (20–22). SIRT1 has both tumor suppressor and oncogenic properties (23). Apart from histones, SIRT1 regulates p53 and peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC-1α) (22, 24, 25). SIRT1-mediated deacetylation contributes to p53 de-stabilization. Accordingly, blocking SIRT1 in prostate cancer is reported to stabilize p53 leading to the inhibition of glycolysis (26). Further, SIRT1 potentiates PGC-1α transcriptional activity in promoting mitochondrial biogenesis and oxidative phosphorylation (21). With the rational that, irradiation elevates the energy needs of a cell to enable DNA and organelle repair for cell recovery (27), targeting metabolic pathways could mediate radiation resistance. We tested the role CD105 has on the acute effects on DNA damage repair as well as its chronic energy needs downstream of a new target, SIRT1, in the context of irradiation.
Results
CD105 expression in prostate cancer upon radiation
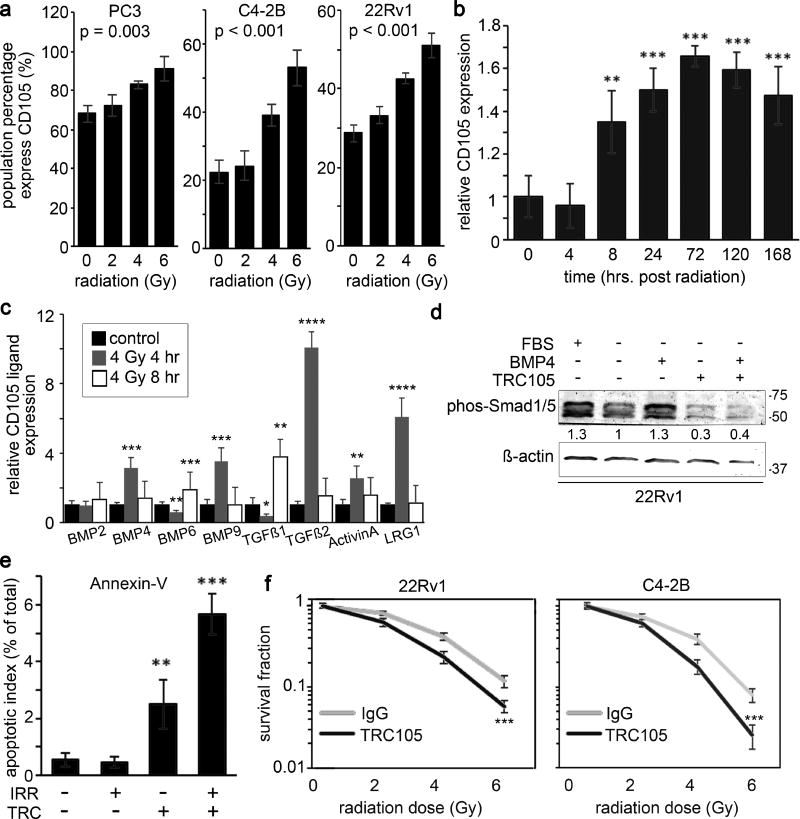
CD105 is implicated in resistance to therapy in several cancers including ovarian, gastric, and breast cancer (13, 28, 29). FACS analysis revealed that prostate cancer cell lines, PC3, C4-2B, and 22Rv1, upregulate cell surface CD105 expression when exposed to irradiation (Figure 1a). Expression of cell surface CD105 was both radiation dose and time dependent (Figure 1b). While 2 Gy radiation did not significantly upregulate CD105 expression, doses of 4 Gy and 6 Gy significantly increased CD105 for all three cell lines. Further, CD105 expression in 22Rv1 showed a significant elevation by 8 hours after 4 Gy radiation that persisted for at least one week. As CD105 can facilitate signaling by interacting with a number of different ligands, we next tested for the expression of a panel of TGFβ/BMP ligands post radiation. We found a significant elevation of BMP4, BMP6, BMP9, TGFβ1, TGFβ2, Activin A, and LRG1 by irradiation (Figure 1c).
Figure 1. Radiation induced CD105 expression in prostate cancer cells support radio-resistance.
a. Cell surface CD105 expression was measured in cell lines at 72 hours following a dose range of irradiation (0, 2, 4, or 6 Gy). b. The durability of cell surface CD105 expression in 22Rv1 was determined 0, 0.5, 4, 8, 24, 48, 72, 120, and 168 hours following 4 Gy irradiation. CD105 cell surface expression fold change was normalized to levels expressed prior to irradiation. c. The mRNA expression of CD105 ligands was measured at 0, 4, and 8 hours post radiation by rtPCR. Expression was normalized to GAPDH and to the 0 hour timepoint. d. Western blot for phosphorylated-Smad1/5 was measured in 22Rv1 cells in the presence or absence of serum starvation and treatment with 50 ng/ml BMP4 or TRC105. β-actin expression served as the loading control. Molecular Weight (kDa) is indicated. e. Annexin-V expression was measured in 22Rv1 cells by FACS analysis 5 days following 4 Gy irradiation and treatment of IgG or TRC105. f. Clonogenic assay was measured 10 days following irradiation of 22Rv1 and C4-2B cells in a dose range of 0 to 6 Gy in the presence of IgG or TRC105. Data are reported as a mean +/− S.D. of 3 independent experiments (**p < 0.01, ***p < 0.001 compared to control, unless otherwise indicated).
Next, we sought to identify the role of CD105/BMP signaling in prostate cancer radiation response by blocking BMP-dependent CD105 signaling using TRC105. To confirm the ability of TRC105 in modulating BMP signaling, we analyzed phosphorylation of SMAD1/5 and the expression of ID1, a BMP target gene, in 22Rv1 stimulated with BMP4 under serum free conditions (Figure 1d, Supplemental Figure 1). A known BMP antagonist, noggin, was used to confirm BMP dependent regulation of ID1 expression by TRC105. Importantly, TRC105 did not affect TGF-β dependent expression of COL1A1, while the TGF-β inhibitor LY-364947 effectively inhibited TGF-β induction of COL1A1 (Supplemental Figure 1). Combining TRC105 with radiation significantly increased apoptosis as measured by cell surface Annexin-V expression, compared to radiation alone (p value < 0.01, Figure 1e). To determine if CD105 confers radio-resistance, clonogenic survival assays were performed comparing IgG or TRC105 treated 22Rv1 and C4-2B cell lines with increasing doses of radiation (Figure 1f). In both these cell lines, treatment with TRC105 sensitized prostate cancer cells to radiation (p value < 0.001). Together, radiation-induced CD105 seemed to regulate prostate epithelial cell death and clonogenicity.
Radiation induced BMP mediates SIRT1-dependent DNA damage repair
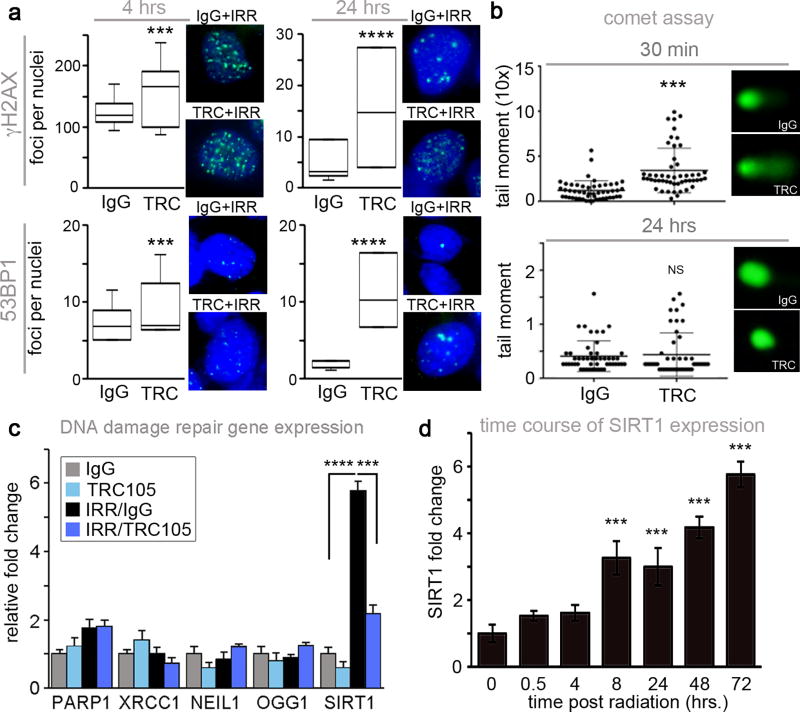
The upregulation of CD105 by irradiation and its potential consequence on cell death suggested that CD105 may be involved in the DNA damage response. To test if impaired of DNA damage repair is the mechanism by which TRC105 conferred radio-sensitivity, γ-H2AX and p53 binding protein (53BP1) foci quantitated following 4 Gy radiation. Combined, irradiation and TRC105 treatment resulted in a significant elevation in γ-H2AX and 53BP1 foci 4 to 24 hours post irradiation, compared to irradiation and IgG control (Figure 2a). However, by 48 hours post irradiation, there were no significant differences in DNA double stranded breaks between the experimental groups (data not shown). The alkaline comet assay provided a measure of single stranded DNA breaks induced by irradiation in the presence and absence of TRC105. There was a significant increase in tail moment of TRC105 treated cells 30 minutes following irradiation compared to radiation plus IgG (p value < 0.001), but there was no difference between the two groups after 24 hours (Figure 2b). Antagonizing BMP signaling downstream of CD105 by TRC105 administration impaired the repair of both double and single stranded DNA damage mediated by irradiation, to suggest CD105 as a target for radiation sensitivity.
Figure 2. TRC105 induces transient DNA damage and repression of SIRT1 induction.
22Rv1 were pre-treated with IgG or TRC105 24 hours prior to irradiation with 4 Gy. a. γ-H2AX or 53BP1 were immunolocalized at 4, and 24hours post-irradiation. Foci per nuclei were quantified (n = 100). Representative images are shown for γ-H2AX (green), and 53BP1 (green) foci, nuclear counterstained with DAPI (blue). b. Comet assay was performed 30 minutes and 24 hours following irradiation. The tail moment was quantified (n = 50). c. The mRNA expression of DNA damage repair genes were measured by rtPCR 72 hours post-irradiation. d. SIRT1 mRNA expression was measured in a time course 0–72 hours following 4 Gy irradiation of 22Rv1. SIRT1 mRNA expression was normalized to GAPDH and to untreated. Data are reported as a mean +/− S.D. of 3 independent experiments (***p < 0.001, ****p < 0.0001, NS – not significant, compared to control).
To better understand the observed longer term effects of radiation in the presence of TRC105, we examined the expression of DNA repair genes following 4 Gy irradiation. As expected, double stranded DNA repair genes (PARP1, XRCC1) and the base excision repair genes (NEIL1, OGG1) were back to baseline levels 72 hours following irradiation as most DNA damage had been repaired (Figure 2c). Interestingly, SIRT1, a critical DNA damage repair component with de-acetylase activity was found to be elevated approximately six-fold by irradiation over control (p value < 0.0001) and nearly restored to control levels by TRC105 72 hours after irradiation (Figure 2c). Similar SIRT1 mRNA expression patterns were observed with C4-2B cells (Supplemental Figure 2a). SIRT1 was found to be significantly upregulated in a time-dependent and radiation dose-dependent manner in both 22Rv1 and C4-2B cells at the mRNA and protein levels (Figure 2d and Supplemental Figure 2b–e). Unlike the other DNA damage repair genes that are acutely active following irradiation, SIRT1 is chronically expressed in irradiated prostate cancer cells.
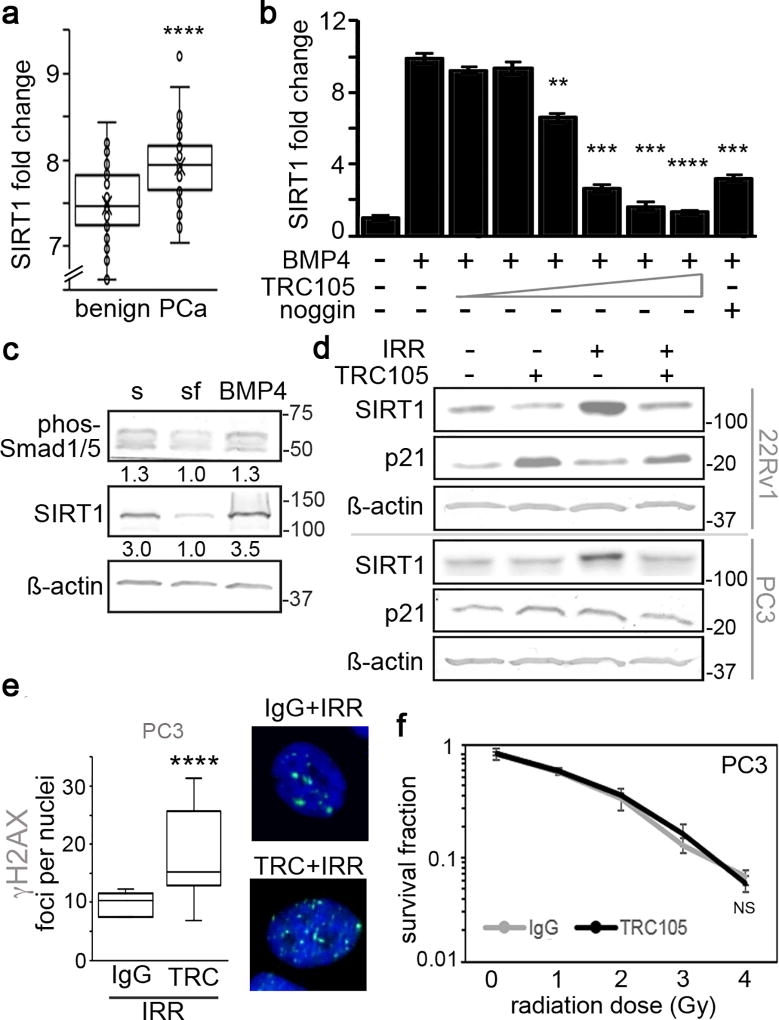
The role of SIRT1 in tumors has long been contentious as it has been shown to act as both a tumor suppressor as well as tumor promoter (23). Therefore, we sought to compare SIRT1 levels in patient samples to determine its role in prostate cancer. Using R2-Genomics analysis, we compared SIRT1 expression in patient samples from the German Cancer Research Center and National Center of Tumor Diseases Affymetrix GeneChip exon array dataset with benign tissue (n = 48) and prostate cancer tissue (n = 47) (30). The comparison validated SIRT1 expression was significantly upregulated in prostate cancer samples (p value < 0.0001, Figure 3a). Since blocking CD105/BMP signaling with TRC105 resulted in limiting radiation-induced SIRT1 expression, we investigated the role of BMP4 on SIRT1 expression. We found antagonizing CD105 with TRC105 effectively blocked BMP4-dependent induction of SIRT1 in a TRC105 dose dependent manner, similar to that mediated by noggin (Figure 3b). Strikingly, the BMP4-induced SIRT1 protein expression in serum-starved 22Rv1 associated with phosphorylated SMAD1/5 (Figure 3c), suggesting a role for CD105 in the regulation of SIRT1 via canonical BMP signaling.
Figure 3. Radiation induces BMP-mediated SIRT1 expression.
a. Fold change of SIRT1 mRNA in benign prostate and prostate cancer patients, obtained from R2-Genomics analysis is expressed (n = 95). b. SIRT1 mRNA expression was measured in 22Rv1 under serum free conditions with 50 ng/ml BMP4, in the context of increasing doses of TRC105 (0.05, 0.1, 0.5, 1, 5, or 10 µg/ml) or with 50 ng/ml noggin. SIRT1 mRNA expression was normalized to GAPDH and to serum treated control. c. Western blot for phosphorylated Smad1/5, SIRT1 and β-actin expression was measured in 22Rv1 cells following serum starvation and treatment with 50 ng/ml BMP4. Densitometric quantitation is indicated under each band. d. SIRT1 and p21 protein expression was measured 72 hours after irradiating (4 Gy) 22Rv1 and PC3 cells in the presence of IgG or TRC105 prior to irradiation. e. γ-H2AX (green) was immunolocalized in PC3 cells following treatment with IgG, TRC105, and 4 Gy irradation at 4 hours. Cells were nuclear counterstained with DAPI (blue). Foci per nuclei were quantified (n = 100). (****p<0.0001 as compared to control). f. Clonogenic survival assay was performed on p53 null PC3 cells at indicated doses of radiation. No significant (NS) radiation sensitization was had with TRC105 compared to IgG control. mRNA expression are reported as a mean +/− S.D. (**p < 0.01, ***p < 0.001, ****p < 0.0001, compared to control).
SIRT1 expression is a known result of radiation treatment. We found that CD105/BMP signaling is necessary and sufficient for SIRT1 expression downstream of radiation treatment, as TRC105 limited radiation-induced SIRT1 protein expression (Figure 3d). Interestingly both 22Rv1 and p53-null, PC3 cells similarly induced SIRT1 in a CD105 dependent manner. However, the lack of p53 in PC3 disabled p21 induction by TRC105 treatment, observed in 22Rv1 cells. This suggested CD105/BMP inhibition of p21 was p53-dependent. However, SIRT1 is known to de-stabilize p53 by de-acetylating p53-K382. Thus, to test SIRT1 function downstream of CD105, we measured p53 regulation by immuno-precipitation and immunoblotting for acetylated p53-K382 following treatment with either TRC105 or nicotinamide, an inhibitor of SIRT1 activity, in the context of radiation. As expected, radiation alone resulted in increased total-p53 expression compared to control. Inhibiting SIRT1 expression with TRC105 resulted in elevated p53 acetylation and total p53 expression, compared to radiation alone (Supplemental Figure 3a). Irradiation-induced CD105 mediated SIRT1 expression and function as revealed by suppressed p53 expression.
The apparent SIRT1 regulation of p53 by CD105/BMP signaling prompted us to test the efficacy of TRC105-mediated radiation sensitization in PC3 cells. Yet, in PC3 cells, TRC105 caused significantly elevated DNA double stranded breaks following irradiation, compared to IgG control (Figure 3e). However, when we knocked down p53 in 22Rv1 (siP53), TRC105 was not found to further γ-H2AX foci numbers significantly compared to scrambled siRNA or IgG controls (Supplemental Figure 3b, c). Accordingly, in clonogenic assays treating PC3 with TRC105 at increasing doses of radiation did not provide radiation sensitization over IgG control (Figure 3f). PC3 cells have previously been described as unresponsive to SIRT1 inhibitors (24). While loss-of-function p53 mutations are rare in prostate cancer, 50–75% of pancreatic cancers have p53 mutations (31, 32). We therefore tested two p53 mutant pancreatic cancer cell lines, MIAPACA-2 and HPAF-II, for radiation responsiveness in the context of TRC105 treatment. In validating the findings with PC3 cells, neither MIAPACA-2 nor HPAF-II were sensitized to radiation by CD105 antagonism (Supplemental Figure 3d, e). This suggested the novel CD105-SIRT1 signaling axis requires p53 for radiation responsiveness.
PGC-1α and cellular energy production are regulated by CD105/BMP
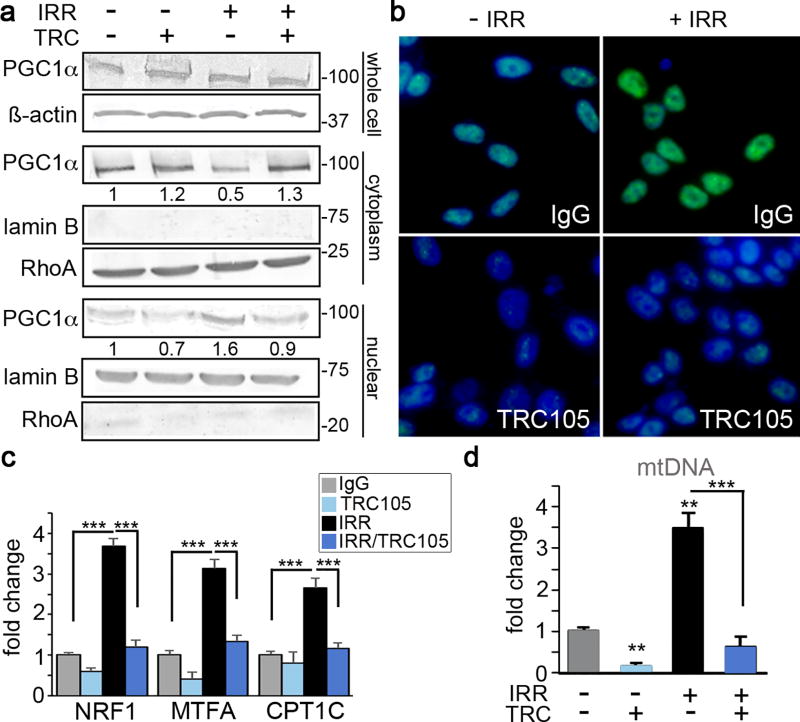
Cell recovery from radiation-induced damage requires large amounts of energy. Further, radio-resistant cancer cells have been shown to induce mitochondrial content, and mitochondrial DNA (mtDNA) accumulation in response to radiation (33). Therefore, we reasoned that targeting cellular metabolism may play a role in the radiation sensitization seen with TRC105 and its inhibition of radiation-induced SIRT1 upregulation. We tested another downstream function of SIRT1, the activation of PGC-1α, a transcription factor involved in mitochondrial biogenesis. Activation and nuclear localization of PGC-1α requires de-acetylation by SIRT1 (21). The treatment of 22Rv1 cells with 4 Gy radiation in the presence of IgG or TRC105 had no effect on PGC-1α expression, by Western blotting of the whole cell lysate (Figure 4a). However, closer examination of sub-cellular localization through organelle fractionation demonstrated PGC-1α depletion from the cytoplasmic fraction and accumulation in the nuclear fraction in the context of radiation. Blocking CD105 prevented radiation-induced nuclear translocation of PGC-1α. Immunofluorescent localization corroborated these same findings (Figure 4b). PGC-1α target genes involved in oxidative stress, mitochondrial biogenesis, and fatty acid oxidation: NRF1, MTFA, and CPT1C, respectively were significantly elevated by radiation (p value < 0.001, Figure 4c). The same genes were significantly downregulated by the added treatment with TRC105 in both 22Rv1 and C4-2B cells (Figure 4c and Supplemental Figure 4a). Consequently, mitochondrial DNA content was significantly elevated by irradiation (p value < 0.0001), to be restored to control levels by antagonizing CD105 (Figure 4d). The evaluation of specific mitochondrial electron transport chain proteins showed TRC105 treatment downregulated complex I-NDUF88 and complex IV-MTCO1 (Supplemental Figure 4b). To further validate the importance of PGC-1α in TRC105 mediated radiation sensitization, we knocked down PGC-1α in 22Rv1 and measured γ-H2AX (Supplemental Figure 4c). Silencing of PGC-1α, resulted in a significant increase in radiation-induced γ-H2AX foci per nuclei, indicating mitochondrial biogenesis was necessary for DNA damage repair. Together, we found that CD105 regulation of SIRT1 expression affected both DNA damage and maintenance of mitochondrial integrity through PGC-1α in the context of irradiation.
Figure 4. PGC-1α and mitochondrial biogenesis are regulated by CD105/BMP.
22Rv1 cells were incubated with IgG or TRC105 with or without 4 Gy irradiation. All measurements were made 72 hours post-irradiation. a. Western blot for whole cell lysate, nuclear and cytoplasmic fractions were independently analyzed for PGC-1α expression. Loading controls included β-actin (whole cell), lamin B (nuclear marker), and Rho A (cytoplasm marker). Molecular weights (kDa) of the ladder are indicated. b. Immunofluorescent localization of PGC-1α (green) was visualized with DAPI (blue) nuclear counterstain. c. The mRNA expression of PGC-1α target genes, NRF1, MTFA, and CPT1C were measured. MRNA expression was normalized to GAPDH and untreated. d. Mitochondrial DNA (mtDNA) was measured from total DNA extracts and normalized to nuclear DNA and to untreated. Data are reported as means +/− S.D. of 3 independent experiments. (*p < 0.05, **p < 0.01, ***p < 0.001, compared to control unless otherwise indicated).
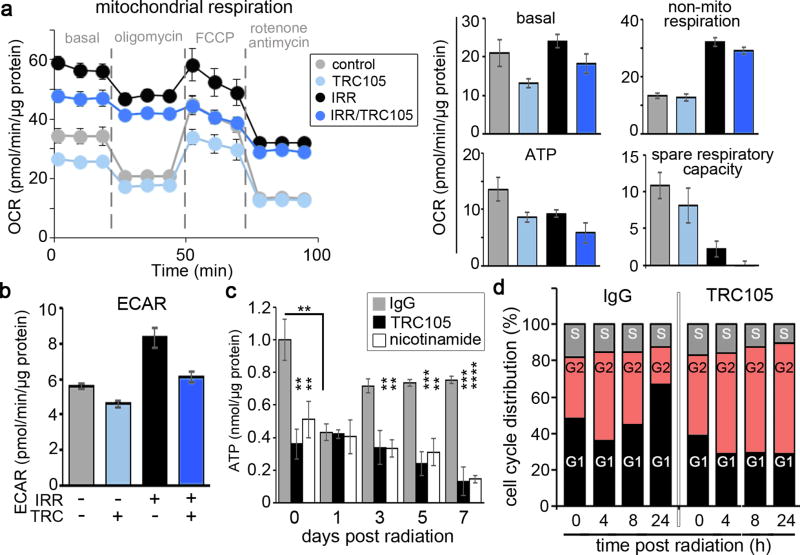
Since PGC-1α was crucial for DNA damage repair, we studied the functionality of the mitochondria after radiation and TRC105 treatment through the measurement of oxygen consumption rates as an indicator of oxidative phosphorylation activity. Radiation treatment elevated non-mitochondrial respiration compared to cells not irradiated, regardless of TRC105 treatment (Figure 5a). However, when comparing only mitochondrial respiration, the basal oxygen consumption of irradiated to non-irradiated cells were similar. Not surprisingly, radiation-mediated mitochondrial damage manifested in decreased ATP production and a depletion of spare respiratory capacity. Antagonizing CD105 in the context of radiation resulted in a decrease in basal oxidative phosphorylation, further decrease in ATP production, and spare respiratory capacity compared to radiation alone. Mitochondrial ATP production downregulated by irradiation was found to increase reliance on glycolysis, as measured by extracellular acidification rate, in 22Rv1 cells (Figure 5b). But, further addition of TRC105 inhibited glycolysis in 22Rv1 cells.
Figure 5. Metabolic changes induced by CD105 antagonism.
a, b. Cells were analyzed for mitochondrial activity 168 hours following 4 Gy irradiation in the presence of IgG or TRC105. a. Basal respiration, non-mitochondrial respiration, mitochondrial ATP, spare respiratory capacity, and b. extracellular acidification rate were quantitated. Data are reported as mean +/− S.D. of a representative experiment (n = 5) of 3 independent experiments. c. Total cellular ATP was measured 0, 24, 72, 120, and 168 hours following 4 Gy irradiation in the context of IgG, TRC105 or nicotinamide treatment. Data are reported as mean +/− S.D. of 3 independent experiments. (**p < 0.01, ***p < 0.001, ****p < 0.0001). d. Cell cycle analysis was performed on 22Rv1 at 0, 4, 8, and 24 hours post-irradiation in the presence of IgG or TRC105 (n=3) in 3 independent experiments.
Glycolysis and oxidative phosphorylation are crucial for energy production in the form of ATP. We found a significant depletion of cellular ATP stores within 1 day of radiation treatment (p value < 0.01), that seemed to be restored to levels close to control by 3 days in 22Rv1 cells (Figure 5c). When SIRT1 expression was inhibited by CD105 antagonism or its function with nicotinamide, cellular ATP stores were significantly lower than the non-irradiated control and further depleted by irradiation. The reliance of intact p53 for TRC105 radio-sensitization suggested inhibition of glycolysis by p53 is critical to radio-sensitization (34, 35). To independently test the consequence of ATP derived from oxidative phosphorylation on cell proliferation, 22Rv1, siP53 22Rv1,and p53-null PC3 cells were treated with oligomycin, an ATP synthase inhibitor. 22Rv1 proliferation limited by irradiation was further downregulated by oligomycin (p value < 0.01, Supplemental Figure 5). In contrast, the PC3 cells, had reduced cell counts with irradiation by about 50%, but were insensitive to inhibition of mitochondrial ATP synthesis. Silencing of p53 resulted in reduced susceptibility of 22Rv1 to oligomycin treatment than scrambled siRNA. Silencing of p53 was not as robust as PC3 response to oligomycin, possibly due to incomplete silencing or long term metabolic adaptation of p53 loss in PC3. Hence, the p53 response is important for radiation-induced maintenance of energy homeostasis and cell division.
The impact of radiation on the cell cycle is well described as causing a G2 cell cycle arrest followed by cell cycle redistribution. In view of the fact that both p53-dependent p21 activity and mitochondrial dysregulation can similarly impact G2 cell cycle arrest (36–38), we found that irradiating 22Rv1 cells, in the presence of IgG, caused an accumulation of cells in G2 phase by 4 hours, to then recover to control levels by 8 hours (Figure 5d). Interestingly, TRC105 alone expanded the G2 cell population. The combination of radiation and TRC105 treatment resulted in G2 cell cycle arrest that did not resolve by 24 hours. Therefore, radiation-induced CD105 signaling helps restore metabolic activity chronically to enable the G2/M cell cycle transition.
Antagonizing CD105 confers radio-sensitivity in vivo
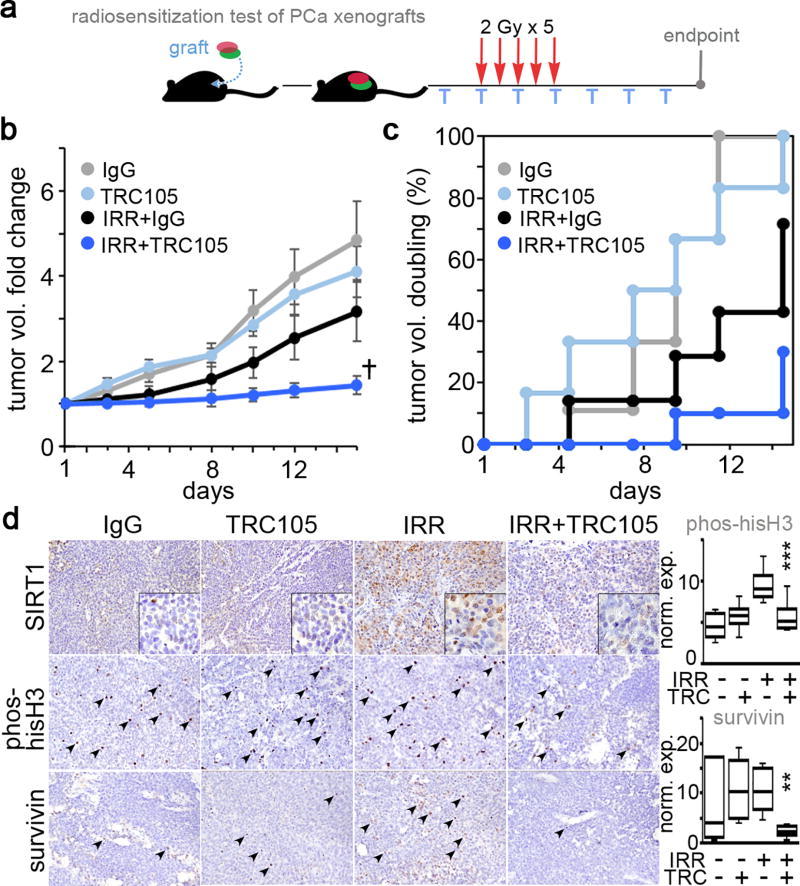
Lastly, we tried to determine the role of CD105 on radio-resistance using a 22Rv1 xenograft model. Mice engrafted with 22Rv1 were given one dose of IgG or TRC105 72 hours prior to irradiation when the tumor reached 0.8 cm2. The tumors were irradiated (2 Gy) for 5-consecutive days and TRC105 was administered 3 times a week for the duration of the treatment schedule (Figure 6a). We found TRC105 alone did not influence tumor volume compared to the control IgG treated group (Figure 6b). The tumor volumes for the irradiated IgG group was significantly lower a week after irradiation compared to control, but by 2 weeks this group was not significantly different from the non-irradiated groups. Conversely, the combination of radiation and TRC105 treated tumor volume was dramatically lower than the other three experimental groups (repeated measure ANOVA p value = 1×10−9, and F-statistic of 11.4). The tumor doubling time was appreciably reduced by combining TRC105 with irradiation compared to either treatment alone. Immunohistochemical staining of the tumors showed a radiation-induced increase in SIRT1, abrogated by the treatment with TRC105 (Figure 6c). The mitotic index measurement by phosphorylated-histone H3 quantitation indicated a significant down regulation by the combination of TRC105 and irradiation (p value = 0.0002). Concomitantly, the expression of survivin, an anti-apoptotic protein, was also markedly decreased in irradiated tumors treated with TRC105 (p value = 0.002). Thus, mitigating radiation elevated CD105-induced SIRT1 by TRC105 is an effective radiation sensitizer for p53-intact prostate cancer.
Figure 6. Antagonizing CD105 confers radio-sensitivity in vivo.
a. Tumor volumes were longitudinally measured. When tumor average volume reached 80 mm3 mice were treated with IgG or TRC105 (T) in the context of radiation (2 Gy for 5 days). Tumors were harvested 15 days after the first dose of radiation (n = 6). b. Tumor volume fold change was normalized to the first dose of radiation (†p = 1×10−9). Each treatment was compared for doubling of tumor volume as a function of time as depicted in the cumulative incidence plot. d. Histochemical localization of SIRT1, phosphorylated-histone H3, and survivin was performed on paraffin embedded tumor tissues. Phosphorylated-histone H3 and survivin expression was quantitated as a measure of total cells per field per tumor. (**p < 0.01, ***p < 0.001).
Discussion
Our work demonstrates the role of CD105 upregulation in response to irradiation. Most of what is known about CD105 signaling has been elucidated from studying endothelial cells and vascular diseases characterized by CD105 mutations such as hereditary hemorrhagic telangiectasia and preeclampsia. In endothelia, CD105 expression is tightly regulated by hypoxia inducible factor-1 alpha (HIF-1α) (39). Since radiation induces HIF-1α, an increase in CD105 expression with radiation treatment was anticipated (40). Instead of performing studies where CD105 is silenced or knocked out, as others have done in the context of DNA damaging chemotherapy studies (13), we deliberately used a neutralizing antibody that inhibited the CD105/BMP signaling axis without affecting TGF-β signaling. In breast cancer, radiation causes an increase in serum TGF-β levels and inhibiting tumor TGF-β signaling can sensitize to radiation (15, 41, 42). The mechanism of TGF-β inhibition associated radiation sensitization is primarily associated with impaired DNA damage repair (42). Thus, knocking out CD105 would achieve BMP signaling inhibition, but would activate TGF-β signaling – not a desired outcome. We found inhibiting BMP signaling through TRC105 or noggin could inhibit SIRT1 expression. We identified a new role for CD105 in mediating metabolic adaptations to stress caused by radiation through the regulation of a novel CD105/BMP target, SIRT1 (Figure 7). SIRT1 inhibitors have been effective in sensitizing a variety of cancer cell lines to DNA damaging agents, including radiation. However, the mechanism by which SIRT1 inhibitors, such as nicotinamide, sensitize cancers to therapy has largely been attributed to SIRT1’s role in DNA damage repair. We show that suppressing SIRT1 expression by antagonizing CD105/BMP signaling leads to increased DNA damage with radiation acutely (Figure 2), exacerbated by severe depletion of energy chronically (Figure 5).
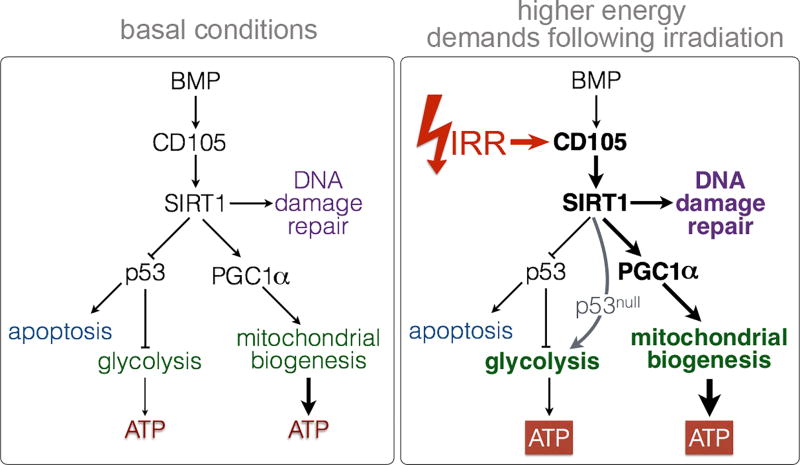
Figure 7. A Schematic depiction of the mechanism of CD105/BMP signaling in conferring radiation resistance.
Radiation of prostate cancer results in upregulation of cell surface CD105 expression. The ensuing CD105/BMP signaling is essential and necessary for SIRT1 expression and downstream activity. SIRT1 facilitates DNA damage repair, destabilizes p53, and activates of PGC-1α in response to radiation. Consequently, CD105 can regulate glycolysis and mitochondrial biogenesis to meet the higher energy demands necessary for survival following irradiation. The loss of functional p53 enables glycolysis as a source of ATP generation and survival.
The induction of CD105 by radiation in the tumor epithelia is exploited by the administration of TRC105. As a tool to interrogate the mechanism of action, TRC105 demonstrated that effective radiation sensitizers do not necessary need to solely act as DNA damage effectors. In addition to the capacity of the BMP-CD105-SIRT1 signaling axis to support DNA repair downstream of p53, we demonstrated it conferred radio-resistance through the induction of mitochondrial biogenesis through PGC-1α activity. TRC105, as a single agent, has little impact on cell proliferation despite its inhibitory effects on mitochondrial biogenesis and glycolysis, as the energy needs of a non-irradiated cells are minimal. However, the added ATP requirement to repair radiation-induced damage makes mitochondrial biogenesis obligatory following irradiation. Prostate cancer cells respond to irradiation by increasing SIRT1 expression (Figure 2) to transiently increase glycolysis, by stabilizing p53 (Figure 3), and mitochondrial biogenesis, by potentiating PGC-1α activity (Figure 4), to sustain immediate and long term energy requirements (Figure 5) (20, 34). Antagonizing CD105 with TRC105 acts by acutely limiting DNA damage repair and chronically preventing recovery by depleting their energy stores, thereby limiting prostate cancer expansion. PARP1 inhibitors have been effective radio-sensitizers for prostate and other cancers by furthering DNA damage accumulation. Unlike for PARP1 inhibitors, intact p53 function was necessary for TRC105 mediated radio-sensitization. TRC105 exploits the metabolic addiction induced by radiation in p53-intact cells. Addressing the CD105-mediated radiation resistance mechanism with TRC105 can serve as a synthetic lethal strategy for patients on radio-therapy.
Material and Methods
Cell lines and culture
CWR22Rv1 (22Rv1), PC3, C4-2B, MIAPACA-2, and HPAF-II cells were purchased (ATCC). Cell lines were cultured in RPMI-1640 medium in 10% FBS. For counting cells for proliferation, 25,000 cells per 24-well were treated with oligomycin 1 hour before radiation. Cells were collected and counted using a hemocytometer 72 hours post-treatment using 5 wells per treatment. The Gammacell 40 Exactor (Best Theratronics, Ottawa, CA) was used for irradiation at indicated doses. 22Rv1 were silenced using pooled target specific 19–25 nt siRNAs by transfecting with either control siRNA-A (sc-37007, Santa Cruz Biotechnology), PGC-1α siRNA (sc-38884, Santa Cruz Biotechnology) or P53 siRNA (sc-29435, Santa Cruz Biotechnology). Cells were transfected using lipofectamine 3000 Reagent (ThermoFisher) according to manufacturer’s protocol.
Reagents
TRC105 was provided from TRACON Pharmaceuticals, Inc. Cells were treated with TRC105 at a concentration of 1 µg/mL, unless noted otherwise. BMP4 (PHC9534, Gibco) and noggin (120–10C, Peprotech) were used at 50 ng/ml. TGF-β was used at 5 ng/ml and LY-364947 was used at 10 µM. Nicotinamide was used at 200 µg/ml. Oligomycin (495455, EMD Milipore,) was used at 1, 1.5, 2, and 2.5 µM concentrations for proliferation assays.
Clonogenic survival assay
Cells were seeded 2 hours prior to irradiation and treated with either IgG or TRC105. Cells were grown for 7–10 days to allow for colony formation and then fixed and stained with crystal violet in methanol. Colonies were delineated as greater than 50 cells. Survival Fraction was calculated as the ratio of the number of colonies formed to the number of colonies seeded times the plating efficiency (43).
Immunofluorescence
Cells grown on coverslips were fixed with 4% paraformaldehyde at room temperature followed by PBS rinses. Cells were permeabilized and blocked with 0.1% Triton X-100 and 1% BSA in PBS for 1 hour at room temperature, followed by incubation with γ-H2AX (05-636, EMD-Millipore), 53BP1 (SC-22760, Santa Cruz Biotechnology), or PGC-1α (SC-13067, Santa Cruz Biotechnology) antibodies at 4°C. Alexa 488 anti-mouse and Alexa 488 anti-rabbit (Life Technologies) secondary antibodies were used at room temperature. Coverslips were mounted with Vectashield Hardset Antifade Mounting Medium with DAPI (H-1400, Vector Laboratories). Images were taken with Olympus FSX-100 and quantitated as foci per nuclei using ImageJ.
Alkaline Comet assay
Cells were collected at indicated time points and re-suspended in low melting point agarose provided by Cell Biolabs's COMET Assay kit (STA-351, Cell Biolabs). The assay was run per manufactures protocol. Images were taken using an Olympus FSX-100 microscope and quantitated using the OpenComet plugin for ImageJ.
FACS analysis
FACS experiments were performed with anti-human CD105-APC (17-1057-41, e-Biosciences) and anti-human Annexin-V-PE (BDB556422, BD Biosciences). Cell cycle was analyzed as previously reported(44). All events were acquired on a BD Accuri C6 Plus flow cytometer and analyzed by FlowJo software v10.2.
Protein analysis
Whole lysate western blots were probed for the following antibodies phos-SMAD1/5 (9516, Cell Signaling Technologies), SIRT1 (9475, Cell Signaling Technologies), p21 (4060, Cell Signaling Technologies), PGC-1α (ST1202, EMD Millipore), Total OXPHOS Rodent (ab110413,Abcam), PGC-1α (ST1202, EMD Millipore), lamin B (sc-6217, Santa Cruz Biotechnology), Rho A (sc-418, Santa Cruz Biotechnology), p53 (sc-126, Santa Cruz Biotechnology), K382 acetyl-p53 (2525, Cell Signaling Technologies), and β-actin (sc-47778, Santa Cruz Biotechnology). The NE-PER Nuclear and Cytoplasmic Extraction Reagent Kit (PI-78833, Thermo Scientific) was used according to protocol. To enrich for p53, samples were immunoprecipitated using p53 N-term-Trap (pta-20, Chromotek) according to manufacter’s protocol with addition of 200 µg/ml of nicotinamide to lysis and wash buffers.
qRT-PCR
RNA was extracted using the RNeasy mini kit (74106, Qiagen Inc.) according to the manufacturer’s protocol. Reverse transcription and quantitative real-time PCR data was calculated by ΔΔCt method and represented relative to 18s rRNA expression. Mitochondrial DNA was quantified as previously described using MTCO2 expression normalized to genomic ACTB expression(45). (Refer to Supplemental Table 1 for primer sequences).
Oxygen consumption and acidification analysis
Respirometry was conducted on 22Rv1 cells using the Seahorse XFe24 Extracellular Flux Analyzer (Seahorse Biosciences) 7 days after radiation treatment for real-time measurements of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR, as a reporter of glycolysis). Cells were seeded in XF24 cell culture plates at a density of 100,000 cells/ well and assay was conducted 16 hours after. Prior to performing the assay, culture media was exchanged for Seahorse XF Base media (supplemented to 10mM Glucose, 1mM Pyruvate, and 1mM Glutamine, pH 7.4) and equilibrated for 1 hour at 37C in a non-CO2 incubator. Final concentration of inhibitors are as follow: 2µM Oligomycin, 1.5µM FCCP (carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone), 1µM antimycin A and 1µM rotenone (Sigma). Results were normalized to protein concentration determined by Pierce BCA Protein Assay Kit.
ATP assay
22Rv1 cells were collected at Days 0, 1, 3, 5, and 7 after radiation and pellets were frozen. ATP was quantified immediately after lysis of pellets using the ATP Determination Kit (A22066, Invitrogen) according to the manufacture protocol.
Xenograft model
22Rv1 (1×106) were suspended in 100 µl of saline with 50% rat-tail collagen and were implanted subcutaneously into the flank of 6 week old male athymic nude mice (Envigo, Indianapolis, IN). N=6 mice were used per a condition, based on previous subcutaneous tumor experiments. When average tumor volume reached 80mm3, the mice were placed into 4 groups (IgG alone, TRC105 alone, IgG with radiation, TRC105 with radiation) by randomization and the first dose of TRC105 or IgG was administered. Mice were treated with either IgG or TRC105 (50 µg) three times a week, unblinded. Tumor volume was recorded three times a week with digital calipers. No animals were excluded from analysis. All animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee at Cedars-Sinai Medical Center.
Immunohistochemistry
Paraffin embedded tissues (5 µm thick) were subjected to immunohistochemical staining as previously reported (46). Anti-phosphorylated histone H3 (PH-H3, 06-570, Millipore), anti-survivin (2808, Cell Signal Technologies), anti-SIRT1 (sc-74504, Santa Cruz Biotechnology) were incubated at 4°C overnight. Secondary antibody development was performed with Dako Cytomation EnVision+ mouse or rabbit labeled polymer kits (K4001 and K4003, Dako Cytomation) and visualized using 3,3’-diaminobenzidine tetrahydrochloride substrate (K3468, Dako Cytomation). Up to five fields per tissue (n=4) were quantitated with Fiji (ImageJ) using a custom written macro. Mitotic (PH-H3) index was calculated by taking the total number of positive (brown) nuclei divided by the total number of nuclei.
Statistical analysis
Student’s T-test was used to compare radiation alone to radiation with treatment. 2-way ANOVA was used to compare the effect of multiple treatment groups. The repeated measures analysis of variance (R-Anova) in MATLAB was used to calculate the p values for detecting tumor size differences over time. Results were expressed as individual data points or as the mean ± S.D. P values of less than 0.05 were considered statistically significant (* P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001).
Supplementary Material
Acknowledgments
This work was supported by grants from the Department of Defense (PC161016 to AM), Department of Veterans Affairs (I01BX0011040 to NAB), and the National Cancer Institute (NIH, CA108646 and CA098912 to NAB). The metabolic analysis was possible because of help from the Cedars-Sinai Medical Center Metabolism and Mitochondrial Research Core. We also thank Tracon Pharmaceuticals for providing the CD105 antagonist, TRC105.
Footnotes
Conflicts of Interest:
The authors declare no conflict of interest.
References
- 1.Moon DH, Efstathiou JA, Chen RC. What is the best way to radiate the prostate in 2016? Urol Oncol. 2017;35(2):59–68. doi: 10.1016/j.urolonc.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Rukstalis DB. Treatment options after failure of radiation therapy-a review. Rev Urol. 2002;4(Suppl 2):S12–7. [PMC free article] [PubMed] [Google Scholar]
- 3.Zumsteg ZS, Spratt DE, Romesser PB, Pei X, Zhang Z, Kollmeier M, et al. Anatomical Patterns of Recurrence Following Biochemical Relapse in the Dose Escalation Era of External Beam Radiotherapy for Prostate Cancer. The Journal of urology. 2015;194(6):1624–30. doi: 10.1016/j.juro.2015.06.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shipley WU, Seiferheld W, Lukka HR, Major PP, Heney NM, Grignon DJ, et al. Radiation with or without Antiandrogen Therapy in Recurrent Prostate Cancer. N Engl J Med. 2017;376(5):417–28. doi: 10.1056/NEJMoa1607529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris WJ, Tyldesley S, Rodda S, Halperin R, Pai H, McKenzie M, et al. Androgen Suppression Combined with Elective Nodal and Dose Escalated Radiation Therapy (the ASCENDE-RT Trial): An Analysis of Survival Endpoints for a Randomized Trial Comparing a Low-Dose-Rate Brachytherapy Boost to a Dose-Escalated External Beam Boost for High- and Intermediate-risk Prostate Cancer. Int J Radiat Oncol Biol Phys. 2017;98(2):275–85. doi: 10.1016/j.ijrobp.2016.11.026. [DOI] [PubMed] [Google Scholar]
- 6.Yamoah K, Showalter TN, Ohri N. Radiation Therapy Intensification for Solid Tumors: A Systematic Review of Randomized Trials. Int J Radiat Oncol Biol Phys. 2015;93(4):737–45. doi: 10.1016/j.ijrobp.2015.07.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonker L. TGF-beta & BMP receptors endoglin and ALK1: overview of their functional role and status as antiangiogenic targets. Microcirculation. 2014;21(2):93–103. doi: 10.1111/micc.12099. [DOI] [PubMed] [Google Scholar]
- 8.Lee NY, Ray B, How T, Blobe GC. Endoglin promotes transforming growth factor beta-mediated Smad 1/5/8 signaling and inhibits endothelial cell migration through its association with GIPC. J Biol Chem. 2008;283(47):32527–33. doi: 10.1074/jbc.M803059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miao ZF, Wu JH, Wang ZN, Zhao TT, Xu HM, Song YX, et al. Endoglin overexpression mediates gastric cancer peritoneal dissemination by inducing mesothelial cell senescence. Hum Pathol. 2016;51:114–23. doi: 10.1016/j.humpath.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 10.Pal K, Pletnev AA, Dutta SK, Wang E, Zhao R, Baral A, et al. Inhibition of endoglin-GIPC interaction inhibits pancreatic cancer cell growth. Mol Cancer Ther. 2014;13(10):2264–75. doi: 10.1158/1535-7163.MCT-14-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romero D, Terzic A, Conley BA, Craft CS, Jovanovic B, Bergan RC, et al. Endoglin phosphorylation by ALK2 contributes to the regulation of prostate cancer cell migration. Carcinogenesis. 2010;31(3):359–66. doi: 10.1093/carcin/bgp217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith SJ, Tilly H, Ward JH, Macarthur DC, Lowe J, Coyle B, et al. CD105 (Endoglin) exerts prognostic effects via its role in the microvascular niche of paediatric high grade glioma. Acta Neuropathol. 2012;124(1):99–110. doi: 10.1007/s00401-012-0952-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziebarth AJ, Nowsheen S, Steg AD, Shah MM, Katre AA, Dobbin ZC, et al. Endoglin (CD105) contributes to platinum resistance and is a target for tumor-specific therapy in epithelial ovarian cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(1):170–82. doi: 10.1158/1078-0432.CCR-12-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardee ME, Marciscano AE, Medina-Ramirez CM, Zagzag D, Narayana A, Lonning SM, et al. Resistance of glioblastoma-initiating cells to radiation mediated by the tumor microenvironment can be abolished by inhibiting transforming growth factor-beta. Cancer research. 2012;72(16):4119–29. doi: 10.1158/0008-5472.CAN-12-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biswas S, Guix M, Rinehart C, Dugger TC, Chytil A, Moses HL, et al. Inhibition of TGF-beta with neutralizing antibodies prevents radiation-induced acceleration of metastatic cancer progression. The Journal of clinical investigation. 2007;117(5):1305–13. doi: 10.1172/JCI30740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S, Pan CC, Bloodworth JC, Nixon AB, Theuer C, Hoyt DG, et al. Antibody-directed coupling of endoglin and MMP-14 is a key mechanism for endoglin shedding and deregulation of TGF-beta signaling. Oncogene. 2014;33(30):3970–9. doi: 10.1038/onc.2013.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosen LS, Hurwitz HI, Wong MK, Goldman J, Mendelson DS, Figg WD, et al. A phase I first-in-human study of TRC105 (Anti-Endoglin Antibody) in patients with advanced cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(17):4820–9. doi: 10.1158/1078-0432.CCR-12-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karzai FH, Apolo AB, Cao L, Madan RA, Adelberg DE, Parnes H, et al. A phase I study of TRC105 anti-endoglin (CD105) antibody in metastatic castration-resistant prostate cancer. BJU Int. 2015;116(4):546–55. doi: 10.1111/bju.12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung-Hynes B, Nihal M, Zhong W, Ahmad N. Role of sirtuin histone deacetylase SIRT1 in prostate cancer. A target for prostate cancer management via its inhibition? The Journal of biological chemistry. 2009;284(6):3823–32. doi: 10.1074/jbc.M807869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canto C, Auwerx J. Targeting sirtuin 1 to improve metabolism: all you need is NAD(+)? Pharmacol Rev. 2012;64(1):166–87. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20(2):98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14(4):312–23. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Z, Fang D. The Roles of SIRT1 in Cancer. Genes Cancer. 2013;4(3–4):97–104. doi: 10.1177/1947601912475079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Ahmad N, Liu X. Combining p53 stabilizers with metformin induces synergistic apoptosis through regulation of energy metabolism in castration-resistant prostate cancer. Cell cycle (Georgetown, Tex) 2016;15(6):840–9. doi: 10.1080/15384101.2016.1151582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munoz-Fontela C, Gonzalez D, Marcos-Villar L, Campagna M, Gallego P, Gonzalez-Santamaria J, et al. Acetylation is indispensable for p53 antiviral activity. Cell cycle (Georgetown, Tex) 2011;10(21):3701–5. doi: 10.4161/cc.10.21.17899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Shen L, Sun LQ. The regulation of radiosensitivity by p53 and its acetylation. Cancer Lett. 2015;363(2):108–18. doi: 10.1016/j.canlet.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Young VR. Energy metabolism and requirements in the cancer patient. Cancer research. 1977;37(7 Pt 2):2336–47. [PubMed] [Google Scholar]
- 28.Koyama Y, Okayama H, Kumamoto K, Saito K, Nakamura I, Ohki S, et al. Overexpression of endoglin (CD105) is associated with recurrence in radically resected gastric cancer. Exp Ther Med. 2010;1(4):627–33. doi: 10.3892/etm_00000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vo MN, Evans M, Leitzel K, Ali SM, Wilson M, Demers L, et al. Elevated plasma endoglin (CD105) predicts decreased response and survival in a metastatic breast cancer trial of hormone therapy. Breast Cancer Res Treat. 2010;119(3):767–71. doi: 10.1007/s10549-008-0261-5. [DOI] [PubMed] [Google Scholar]
- 30.Borno ST, Fischer A, Kerick M, Falth M, Laible M, Brase JC, et al. Genome-wide DNA methylation events in TMPRSS2-ERG fusion-negative prostate cancers implicate an EZH2-dependent mechanism with miR-26a hypermethylation. Cancer Discov. 2012;2(11):1024–35. doi: 10.1158/2159-8290.CD-12-0041. [DOI] [PubMed] [Google Scholar]
- 31.Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol. 2008;3:157–88. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winter JM, Maitra A, Yeo CJ. Genetics and pathology of pancreatic cancer. HPB (Oxford) 2006;8(5):324–36. doi: 10.1080/13651820600804203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamori T, Yasui H, Yamazumi M, Wada Y, Nakamura Y, Nakamura H, et al. Ionizing radiation induces mitochondrial reactive oxygen species production accompanied by upregulation of mitochondrial electron transport chain function and mitochondrial content under control of the cell cycle checkpoint. Free Radic Biol Med. 2012;53(2):260–70. doi: 10.1016/j.freeradbiomed.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 34.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107–20. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 35.Zhong J, Rajaram N, Brizel DM, Frees AE, Ramanujam N, Batinic-Haberle I, et al. Radiation induces aerobic glycolysis through reactive oxygen species. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2013;106(3):390–6. doi: 10.1016/j.radonc.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antico Arciuch VG, Elguero ME, Poderoso JJ, Carreras MC. Mitochondrial regulation of cell cycle and proliferation. Antioxid Redox Signal. 2012;16(10):1150–80. doi: 10.1089/ars.2011.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoeferlin LA, Oleinik NV, Krupenko NI, Krupenko SA. Activation of p21-Dependent G1/G2 Arrest in the Absence of DNA Damage as an Antiapoptotic Response to Metabolic Stress. Genes Cancer. 2011;2(9):889–99. doi: 10.1177/1947601911432495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deckbar D, Jeggo PA, Lobrich M. Understanding the limitations of radiation-induced cell cycle checkpoints. Crit Rev Biochem Mol Biol. 2011;46(4):271–83. doi: 10.3109/10409238.2011.575764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez-Elsner T, Botella LM, Velasco B, Langa C, Bernabeu C. Endoglin expression is regulated by transcriptional cooperation between the hypoxia and transforming growth factor-beta pathways. The Journal of biological chemistry. 2002;277(46):43799–808. doi: 10.1074/jbc.M207160200. [DOI] [PubMed] [Google Scholar]
- 40.Moeller BJ, Dewhirst MW. Raising the bar: how HIF-1 helps determine tumor radiosensitivity. Cell cycle (Georgetown, Tex) 2004;3(9):1107–10. [PubMed] [Google Scholar]
- 41.Bouquet F, Pal A, Pilones KA, Demaria S, Hann B, Akhurst RJ, et al. TGFbeta1 inhibition increases the radiosensitivity of breast cancer cells in vitro and promotes tumor control by radiation in vivo. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(21):6754–65. doi: 10.1158/1078-0432.CCR-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ewan KB, Henshall-Powell RL, Ravani SA, Pajares MJ, Arteaga C, Warters R, et al. Transforming growth factor-beta1 mediates cellular response to DNA damage in situ. Cancer research. 2002;62(20):5627–31. [PubMed] [Google Scholar]
- 43.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1(5):2315–9. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 44.Zumsteg ZS, Morse N, Krigsfeld G, Gupta G, Higginson DS, Lee NY, et al. Taselisib (GDC-0032), a Potent beta-Sparing Small Molecule Inhibitor of PI3K, Radiosensitizes Head and Neck Squamous Carcinomas Containing Activating PIK3CA Alterations. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22(8):2009–19. doi: 10.1158/1078-0432.CCR-15-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andres AM, Tucker KC, Thomas A, Taylor DJ, Sengstock D, Jahania SM, et al. Mitophagy and mitochondrial biogenesis in atrial tissue of patients undergoing heart surgery with cardiopulmonary bypass. JCI insight. 2017;2(4):e89303. doi: 10.1172/jci.insight.89303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Placencio VR, Sharif-Afshar AR, Li X, Huang H, Uwamariya C, Neilson EG, et al. Stromal transforming growth factor-beta signaling mediates prostatic response to androgen ablation by paracrine Wnt activity. Cancer research. 2008;68(12):4709–18. doi: 10.1158/0008-5472.CAN-07-6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.