Abstract
ISG15 is a ubiquitin-like modifier that is expressed in response to type 1 interferon signaling (IFN-α/β) and plays a role in antiviral responses. The core E1, E2, and E3 enzymes for ISG15 are Ube1L, UbcH8, and Herc5, respectively, and these are all also induced at the transcriptional level by IFN-α/β. We recently showed that Herc5 associates with polysomes and modifies target proteins in a cotranslational manner. Here, we describe the expression of the core conjugating enzymes in human cells, the detection of ISG15 conjugates, and the methods for fractionation of Herc5 with polysomes.
Keywords: Interferon, ISG15, Herc5, UbcH8, Ube1L, Polysomes
1. Introduction
ISG15 is a 17 kDa ubiquitin-like modifier that is rapidly induced by type 1 interferon signaling. It was one of the first interferon-stimulated gene (ISG) products identified (1) and the first ubiquitin-like modifier, after ubiquitin, to be discovered (2, 3). As an interferon-induced protein, ISG15 was long presumed to function in the innate immune response to infectious agents, but it is only in recent years that it has been shown to have antiviral activity against a broad range of virus types, including Influenza, Ebola, Sindbis, Herpes, and HIV (4–7). Several virus types also appear to have evolved mechanisms for blocking the antiviral effects of ISG15. The Influenza NS1B protein and the Vaccinia E3 protein bind noncovalently to ISG15 and block conjugation (8, 9), and the SARS and Crimean Congo Hemmorhagic Fever virus encodes a protease that functions as a deconjugating enzyme for ISG15 (10, 11).
ISG15 is roughly twice the size of ubiquitin and resembles a translational dimer of ubiquitin, with two ubiquitin-like domains, each approximately 30% identical to ubiquitin. The X-ray crystal structure of free ISG15 has been solved, and there appears to be little or no stable interactions between the two domains (12). ISG15 is the only Ubl where the last six residues of the processed form of ISG15 are identical to those of ubiquitin (LRLRGG). While there may be no further significance to this C-terminal identity, this may be the basis of confusion over the identity of specific deconjugating enzymes for ISG15 (13–15). Interestingly, ISG15 was also reported to be a secreted protein with cytokine activities (16, 17); however, the relationship between this function and its intracellular function as a protein modifier is unknown.
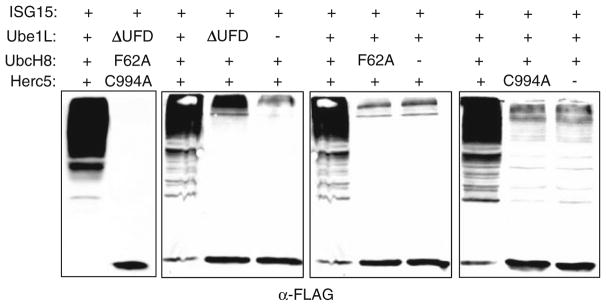
There is a single major E1, E2, and E3 enzyme for ISG15 in human cells, and all of these are induced at the transcriptional level by type 1 interferons. Ube1L is the E1 enzyme (8), and the E2 enzyme is UbcH8/Ube2L6 (18–20). UbcH8 has been widely reported to function with ubiquitin (21–27), but determination of kinetic constants of Ube1L and Ube1 for UbcH8 and UbcH7 suggested that UbcH8 is unlikely to function as a ubiquitin E2 in vivo (18). The major E3 for ISG15 is human Herc5, a HECT domain ligase with N-terminal RCC1 repeats (28, 29). Herc5 siRNA knockdown leads to a dramatic decrease in overall ISG15 conjugation activity, affecting conjugation to all apparent cellular target proteins (29). Herc5 is the only human HECT E3 known to be a ligase for anything other than ubiquitin. Co-expression of ISG15, Ube1L, UbcH8, and Herc5 in non-interferon stimulated cells reconstitutes robust ISG15 conjugation to the broad spectrum of target proteins (Fig. 1), consistent with the notion that these enzymes represent the core interferon-induced components of the ISGylation system (28–30). Plasmids expressing mutant forms of each of these components can be used as negative controls when examining ISGylation (Fig. 1). The C-terminal ubiquitin fold domain (UFD) of Ube1L is required for interaction with UbcH8 and deletion of this domain (Ube1LΔUFD) results in the inability of Ube1L to transfer ISG15 to UbcH8. F62 of UbcH8 is a conserved residue among E2 proteins that has been shown to be critical for the E2/E3 interaction, and mutation of this residue (UbcH8 F62A) prevents transfer of ISG15 from UbcH8 to Herc5. Mutation of the active site cysteine of Herc5 (Herc5 C994A) results in a catalytically inactive Herc5 incapable of supporting ISG15 conjugation. Co-transfection of ISG15 with the mutant forms of its conjugating enzymes prevents ISGylation of target proteins; however, a low level of ISGylation may be observed when using a mutant form of only one of the enzymes or leaving out one of the enzymes (Fig. 1). This is due to a basal level of each of the conjugating enzymes and varies depending on the cell line used in the experiment (29).
Fig. 1.
Co-transfection of ISG15 with mutant forms of the ISG15 conjugating enzymes. HEK293T cells were transfected with plasmids expressing the indicated wild-type or mutant proteins. The plasmid expressing ISG15 contains an N-terminal 3×FLAG tag and the plasmid expressing Herc5 contains an N-terminal TAP tag. Cell extracts were prepared 24 h post-transfection and analyzed by immunoblotting with an antibody against the FLAG tag.
Several proteomics studies, together, identified approximately 300 cellular proteins that are targeted for ISGylation (28, 30–33). This raised the question of how a single ligase, Herc5, could recognize such a diverse set of proteins that shared no apparent distinguishing features. In addressing this problem, we discovered that the range of substrates went far beyond those identified by proteomics, and that nearly all proteins tested, including foreign proteins (bacterial, yeast, viral) could be ISGylated, provided that they were being synthesized in the same window of time that the conjugation system was active (34). The ISGylation of newly synthesized proteins suggested that modification might occur cotrans-lationally. We discovered that endogenously expressed Herc5 (as well as HA-tagged Herc5) cofractionated with polysomes. In addition, a mutant lacking the N-terminal RCC repeats and incapable of supporting ISGylation of target proteins did not cofractionate with polysomes. With the discovery that Herc5 fractionates with polysomes, we proposed that Herc5 modifies proteins stochastically in a cotranslational manner as they emerge from the ribosome. An implication of this model is that, in interferon-stimulated cells, newly synthesized viral proteins may be the key biologically relevant targets of this system, while modification of cellular proteins may be largely collateral (34).
Some of the outstanding mechanistic questions concerning ISGylation are (1) what factors determine the sites and efficiency of modification? (2) how is Herc5 tethered to the ribosome and how does it engage nascent polypeptides? and (3) what are the biochemical consequences of ISGylation on target proteins? In this chapter, we describe methods for generating and detection of ISG15 conjugates in either interferon-stimulated or transfected cells and methods for cofractionating Herc5 with polysomes.
2. Materials
2.1. Cell Culture, Transfections, and Immunoblotting
Human HeLa and HEK293T cells are maintained in DMEM supplementedwith10%sterilefetalbovineserum(DMEM+FBS) and penicillin–streptomycin solution (Cellgro).
Interferon-β (Betaseron, Bayer HealthCare Pharmaceuticals).
Lipofectamine 2000 (Invitrogen).
Plasmids for ISG15 conjugation: pc3 × FLAG-ISG15 (33), pcDNA Ube1L (33), pcDNA UbcH8 (33), and pcTAP Herc5 (29).
ControlplasmidsforISG15conjugation:pcDNAUbe1LΔUFD (18), pcDNA UbcH8 F62A (29), and pcTAP Herc5 C994A (29) (see Note 1).
Plasmids for Herc5 co-fractionation with polysomes: pcHA Herc5 and pcHA Herc5ΔRCC (34).
NP40 Lysis buffer: 0.1 M Tris–HCl, pH 7.9, 0.1 M NaCl, 1% Nonidet P-40, 1 mM DTT, 100 μM phenylmethylsulfonyl fluoride, 4 μM leupeptin, and 0.3 μM aprotinin.
Antibodies: Anti-ISG15 (Santa Cruz Biotechnology; sc-50366), Anti-Herc5 (Enzo Life Sciences; BML-PW0920), Anti-Flag M2 (Sigma; F3165), and Anti-HA (Covance; MMS-101P).
1× protein loading buffer: 62 mM Tris–HCl, pH 6.8, 10% glycerol, 2% SDS, 100 mM DTT, 0.001% bromphenol blue.
2.2. Sucrose Gradient Fractionation
10 mg/mL Cycloheximide (CHX). Make a 10 mL stock and store at 4°C.
0.22 μm Millipore Express PLUS Membrane Steritop (Millipore).
10× Polysome Buffer (10× PB): 100 mM Tris–HCl, pH 7.4, 100 mM NaCl, 30 mM MgCl2. Sterile filter 500 mL of 10× buffer and store at room temperature.
Sucrose for linear sucrose gradients (7, 27.5, and 47%): In a 1 L beaker, add sucrose (35 g for 7%, 137.5 g for 27.5%, and 235 g for 47%) to 100 mL water plus 50 mL 10× PB. Alternate solution between stirplate and 65°C water bath until sucrose is dissolved. Bring volume to 500 mL, stir, and sterile filter. Store at 4°C.
65% Sucrose for fractionation of gradient: In a 1 L beaker, slowly add 325 g sucrose to 200 mL water. Alternate solution between stirplate and 65°C water bath until sucrose is dissolved. Bring volume to 500 mL, stir, and sterile filter. Store at 4°C.
100 mm Tissue culture dishes.
50 mL Centrifuge tubes with screw caps.
5 and 10 mL Plastic disposable serological pipettes, individually wrapped, sterile.
Pipette filler.
1× Polysome Lysis Buffer: 0.5% (v/v) Triton X-100, 200 μg/mL Heparin, and 10× PB. Prepare just before use and keep on ice.
DMEM with 10% FBS and 100 μg/mL cycloheximide (DMEM + FBS + CHX). Prepare just before use and use immediately.
Phosphate-buffered saline with 50 μg/mL cycloheximide (PBS + CHX). Prepare just before use and keep on ice.
15 mL Centrifuge tubes with screw caps.
Polyallomer centrifuge tubes, 14 mm × 89 mm.
Ultracentrifuge and Beckman SW41Ti rotor.
10% (v/v) Trichloroacetic acid (TCA).
3. Methods
3.1. Production and Detection of ISG15 Conjugates in Interferon-β-Stimulated Cells
HeLa cells are a convenient and widely used cell line that is interferon-responsive. To detect IFN-β-induced ISGylation, incubate one 35 mm culture dish of subconfluent HeLa cells with 1,000 U of IFN-β in 1 mL media for 24–48 h (see Note 2).
Lyse cells in 200 μL NP40 lysis buffer for 20 min on ice. Centrifuge for 10 min at 16,300 × g at 4°C.
Separate 30 μg of total protein by SDS-PAGE, transfer protein to a nitrocellulose membrane, and probe with anti-ISG15 to detect unconjugated and conjugated ISG15 (see Note 3).
3.2. Production and Detection of ISG15 Conjugation in Noninterferon-Stimulated Cells
Reconstitution of broad ISGylation without IFN-β treatment requires co-transfection of plasmids expressing ISG15, Ube1L, UbcH8, and Herc5.
HEK293T cells are easy to grow and to transfect and therefore work well for reconstitution of ISGylation by co-transfection of plasmids expressing the ISG15 pathway components.
HEK293T cells should be 70–80% confluent at the time of transfection. For transfection of one 35 mm culture dish, incubate 0.25 μg pcDNA Ube1L, 0.25 μg pcDNA UbcH8, 0.5 μg pcTAP Herc5, and 0.5 μg pc3 × FLAG-ISG15 with 3 μL Lipofectamine 2000 (2 μL transfection reagent/1 μg DNA). Follow the Lipofectamine 2000 product instructions.
Substitution with a plasmid expressing a mutant form of any one of the ISG15 conjugation components can be used as a control: pcDNA Ube1LΔUFD, pcDNA UbcH8 F62A, or pcTAP Herc5 C994A.
To examine ISGylation of a specific target protein, an epitope tagged target protein can be co-expressed with the ISG15 components. Modify the transfection in step 3 by including 0.5 μg of an HA-tagged target protein and increasing the Lipofectamine 2000 from 3 to 4 μL (see Note 4).
24–48 h Posttransfection, lyse cells in 200 μL NP40 lysis buffer for 20 min on ice. Centrifuge for 10 min at 16,300 × g at 4°C.
Separate 30 μg of total protein by SDS-PAGE, transfer protein to a nitrocellulose membrane, and probe with anti-FLAG to detect unconjugated and conjugated ISG15.
3.3. Co-fractionation of Herc5 with Polysomes
For detection of endogenous, interferon-induced Herc5: incubate HeLa cells in one 100 mm culture dish with 8,000 U of human IFN-β in 8 mL DMEM + FBS. For detection of exogenous, epitope-tagged Herc5: transfect one 100 mm culture dish of HEK293T cells with 2 μg DNA and 4 μL Lipofectamine 2000 according to product instructions. Begin harvest of cells no later than 24 h posttreatment (see Note 5).
2 h before harvesting cells, pour linear 7–47% (w/v) sucrose gradients containing 200 μg/mL cycloheximide. Label a 50 mL tube for each sucrose concentration (7, 27.5, and 47%) and add cyclo-heximide to each tube (total volume will depend on the number of gradients being made). Using a 10 mL sterile plastic pipette, transfer 7% sucrose to the 50 mL tube. The same pipette can be used to then transfer the 27.5% and 47% sucrose. Cap tubes, gently swirl to mix, and place on ice. Using a 5 mL sterile plastic pipette (one forw each sucrose concentration and pipette filler, layer sucrose in 13.2 mL polyallomer centrifuge tubes): first add 3.5 mL 47% sucrose, followed by 3.7 mL 27% sucrose, and end with 3.7 mL 7% sucrose (see Note 6). Parafilm each tube and gently lay horizontally at 4°C for a minimum of 1.5 h to achieve a linear gradient.
24 h Postinterferon treatment or transfection, replace the culture media with DMEM + FBS + CHX and incubate for 30 min.
Wash cells in ice-cold PBS + CHX and transfer to a 15 mL tube with screw cap. Centrifuge tube for 5 min at 162 × g (tabletop centrifuge) at 4°C.
Aspirate the PBS and resuspend pellet in 300 μL ice-cold 0.5% Triton X-100. Transfer lysate to a 1.5 mL eppendorf tube and lyse on ice for 10 min. Centrifuge lysates for 10 min at 16,300×g at 4°C.
Remove the 7–47% (w/v) sucrose gradients (now linear) from 4°C and balance tubes using 7% sucrose. Gently layer the clarified cell lysate on the gradient, balance tubes using 1× PB, and centrifuge at 222,000 × g for 90 min at 4°C in Beckmann SW41Ti rotor.
Remove samples from rotor buckets and keep on ice until fractions from each tube are collected.
Polysome profiles can be generated by monitoring the absorbance at 254 nm while collecting fractions (~1 mL each) using a density gradient fractionator.
To precipitate proteins from sucrose fractions, add 10% TCA (see Note 7). Incubate on ice for 20 min and centrifuge for 10 min at 16,300 × g at 4°C. Carefully aspirate the sucrose/ TCA and resuspend the pellet in 40 μL 1× protein loading buffer (see Note 8).
Precipitated proteins can be analyzed by SDS-PAGE and immunoblotting (see Note 9).
Footnotes
Using a mutant form of the E1, E2, or E3 enzymes (or leaving out the E1, E2, or E3 enzymes) for ISG15 may not completely abrogate ISGylation. Some cell lines contain a low basal level of these enzymes.
ISG15 conjugates do not accumulate to appreciable levels until ~18 h after IFN stimulation. This corresponds to the delayed induction of the E1, E2, and E3 enzymes (29).
Typically, ISG15 conjugates migrate between ~45 kDa and well over 200 kDa, whereas unconjugated ISG15 migrates at ~15 kDa. In order to detect both free and conjugated ISG15, a 12% (or higher) polyacrylamide gel must be used. In addition, using a semidry transfer apparatus increases detection of ISG15 conjugates.
An HA-tag is preferable because it has no lysine residues that might serve as ISG15 acceptors. The FLAG tag has two lysines, and a larger protein tag, like the TAP tag, contains many. Further, the “empty” TAP tag (two copies of protein A, one copy of calmodulin binding protein) is itself ISGylated (34).
At the time of harvest, cells should be 80–90% confluent. As cells approach confluency, there is a reduction in polysomes and increase in monosomes (80S).
In order to maintain the interface between each sucrose concentration, hold the polyallomer tube at 45° angle as the sucrose is added. The tip of the pipette should be touching the tube while slowly adding the sucrose so that it runs down the inside of the tube.
The expression of endogenous Herc5 is much lower than transfected Herc5. TCA precipitate ~500 μL of each gradient fraction for endogenous Herc5 and ~150 μL of each fraction for transfected Herc5.
Any remaining TCA may alter the pH of your sample. One option is to wash the pellet with 500 μL acetone and centrifuge for 5 min at 16,300 × g at 4°C before resuspending in 1× protein loading buffer. Alternatively, NaOH can be added to each sample after resuspension of the pellet in 1× protein loading buffer.
To detect endogenous or transfected Herc5, use an 8% poly-acrylamide gel.
References
- 1.Farrell PJ, Broeze RJ, Lengyel P. Accumulation of an mRNA and protein in interferon-treated Ehrlich ascites tumour cells. Nature. 1979;279:523–525. doi: 10.1038/279523a0. [DOI] [PubMed] [Google Scholar]
- 2.Haas AL, Ahrens P, Bright PM, Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J Biol Chem. 1987;262:11315–11323. [PubMed] [Google Scholar]
- 3.Loeb KR, Haas AL. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J Biol Chem. 1992;267:7806–7813. [PubMed] [Google Scholar]
- 4.Lenschow DJ, Giannakopoulos NV, Gunn LJ, et al. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J Virol. 2005;79:13974–13983. doi: 10.1128/JVI.79.22.13974-13983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okumura A, Pitha PM, Harty RN. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci USA. 2008;105:3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okumura A, Lu G, Pitha-Rowe I, Pitha PM. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci USA. 2006;103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsiang TY, Zhao C, Krug RM. Interferon-induced ISG15 conjugation inhibits influenza A virus gene expression and replication in human cells. J Virol. 2009;83:5971–5977. doi: 10.1128/JVI.01667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan W, Krug RM. Influenza B virus NS1 protein inhibits conjugation of the inter-feron (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 2001;20:362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang YG, Yan XZ, Xie YY, et al. Different roles for two ubiquitin-like domains of ISG15 in protein modification. J Biol Chem. 2008;283:13370–13377. doi: 10.1074/jbc.M800162200. [DOI] [PubMed] [Google Scholar]
- 10.Lindner HA, Lytvyn V, Qi H, et al. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch Biochem Biophys. 2007;466:8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akutsu M, Ye Y, Virdee S, et al. Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains. Proc Natl Acad Sci USA. 2011;108:2228–2233. doi: 10.1073/pnas.1015287108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narasimhan J, Wang M, Fu Z, et al. Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J Biol Chem. 2005;280:27356–27365. doi: 10.1074/jbc.M502814200. [DOI] [PubMed] [Google Scholar]
- 13.Catic A, Fiebiger E, Korbel GA, et al. Screen for ISG15-crossreactive deubiquitinases. PLoS ONE. 2007;2:e679. doi: 10.1371/journal.pone.0000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knobeloch KP, Utermohlen O, Kisser A, et al. Reexamination of the role of ubiquitin-like modifier ISG15 in the phenotype of UBP43-deficient mice. Mol Cell Biol. 2005;25:11030–11034. doi: 10.1128/MCB.25.24.11030-11034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malakhov MP, Malakhova OA, Kim KI, et al. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277:9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 16.D’Cunha J, Ramanujam S, Wagner RJ, et al. In vitro and in vivo secretion of human ISG15, an IFN-induced immunomodulatory cytokine. J Immunol. 1996;157:4100–4108. [PubMed] [Google Scholar]
- 17.D’Cunha J, Knight E, Jr, Haas AL, et al. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc Natl Acad Sci USA. 1996;93:211–215. doi: 10.1073/pnas.93.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durfee LA, Kelley ML, Huibregtse JM. The basis for selective E1-E2 interactions in the ISG15 conjugation system. J Biol Chem. 2008;283:23895–23902. doi: 10.1074/jbc.M804069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim KI, Giannakopoulos NV, Virgin HW, Zhang DE. Interferon-Inducible Ubiquitin E2, Ubc8, Is a Conjugating Enzyme for Protein ISGylation. Mol Cell Biol. 2004;24:9592–9600. doi: 10.1128/MCB.24.21.9592-9600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao C, Beaudenon SL, Kelley ML, et al. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc Natl Acad Sci USA. 2004;101:7578–7582. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chin LS, Vavalle JP, Li L. Staring, a novel E3 ubiquitin-protein ligase that targets syntaxin 1 for degradation. J Biol Chem. 2002;277:35071–35079. doi: 10.1074/jbc.M203300200. [DOI] [PubMed] [Google Scholar]
- 22.Kumar S, Kao WH, Howley PM. Physical interaction between specific E2 and Hect E3 enzymes determines functional cooperativity. J Biol Chem. 1997;272:13548–13554. doi: 10.1074/jbc.272.21.13548. [DOI] [PubMed] [Google Scholar]
- 23.Moynihan TP, Ardley HC, Nuber U, et al. The ubiquitin-conjugating enzymes UbcH7 and UbcH8 interact with RING finger/IBR motif-containing domains of HHARI and H7-AP1. J Biol Chem. 1999;274:30963–30968. doi: 10.1074/jbc.274.43.30963. [DOI] [PubMed] [Google Scholar]
- 24.Niwa J, Ishigaki S, Doyu M, et al. A novel centrosomal ring-finger protein, dorfin, mediates ubiquitin ligase activity. Biochem Biophys Res Commun. 2001;281:706–713. doi: 10.1006/bbrc.2001.4414. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka K, Suzuki T, Chiba T, et al. Parkin is linked to the ubiquitin pathway. J Mol Med. 2001;79:482–494. doi: 10.1007/s001090100242. [DOI] [PubMed] [Google Scholar]
- 26.Urano T, Saito T, Tsukui T, et al. Efp targets 14-3-3 sigma for proteolysis and promotes breast tumour growth. Nature. 2002;417:871–875. doi: 10.1038/nature00826. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Gao J, Chung KK, et al. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci USA. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong JJ, Pung YF, Sze NS, Chin KC. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc Natl Acad Sci USA. 2006;103:10735–10740. doi: 10.1073/pnas.0600397103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dastur A, Beaudenon S, Kelley M, et al. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J Biol Chem. 2006;281:4334–4338. doi: 10.1074/jbc.M512830200. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi T, Inoue S, Yokosawa H. Identification and Herc5-mediated ISGylation of novel target proteins. Biochem Biophys Res Commun. 2006;348:473–477. doi: 10.1016/j.bbrc.2006.07.076. [DOI] [PubMed] [Google Scholar]
- 31.Giannakopoulos NV, Luo JK, Papov V, et al. Proteomic identification of proteins conjugated to ISG15 in mouse and human cells. Biochem Biophys Res Commun. 2005;336:496–506. doi: 10.1016/j.bbrc.2005.08.132. [DOI] [PubMed] [Google Scholar]
- 32.Malakhov MP, Kim KI, Malakhova OA, et al. High-throughput immunoblotting. Ubiquitin-like protein ISG15 modifies key regulators of signal transduction. J Biol Chem. 2003;278:16608–16613. doi: 10.1074/jbc.M208435200. [DOI] [PubMed] [Google Scholar]
- 33.Zhao C, Denison C, Huibregtse JM, et al. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc Natl Acad Sci USA. 2005;102:10200–10205. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durfee LA, Lyon N, Seo K, Huibregtse JM. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol Cell. 2010;38:722–732. doi: 10.1016/j.molcel.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]