Abstract
Protein tagging is a powerful tool for performing comprehensive analyses of the biological functions of a protein of interest owing to the existence of a wide variety of tags. It becomes indispensable in some cases, such as in tracking protein dynamics in a live cell or adding a peptide epitope due to the lack of optimal antibodies. However, efficiently integrating an array of tags into the gene of interest remains a challenge. Traditional DNA recombinant technology based on type II restriction endonucleases renders protein tagging tedious and inefficient as well as the introduction of an unwanted junction sequence. In our attempt to tag Thrombospondin type 1 domain-containing 1 (THSD1) that we identified as the first intracranial aneurysm gene (Santiago-Sim et al., 2016), we developed a novel precision tagging technique by combinational use of type II and IIS restriction endonucleases (Xu et al., 2017), which generates a seamless clone with high efficiency. Here, we describe a protocol that not only provides a generalized strategy for any gene of interest but also takes its application of 11 different tags in THSD1 as a step-by-step example.
Keywords: DNA cloning, Protein tagging, Type IIS restriction enzyme, Non-palindromic, THSD1
Background
Versatile tags with different features serve as a set of tools to dissect protein function molecularly. Various tags such as Green Fluorescence Protein (GFP) tag and its derivatives, tandem affinity purification tags, such as FLAG-HA or ProtA-CBP, have revolutionized the biological research over the years. Some newly developed chemical tags, such as SNAP or CLIP, allow conditional labeling of the protein of interest in a time-controlled fashion (Bodor et al., 2012). However, a method to incorporate as many different tags as possible into a gene of interest efficiently has been poorly developed.
Traditional DNA recombination utilizes type II restriction endonucleases that recognize palindromic sequences. For example, EcoRI recognizes 5′-GAATTC and cleaves inside to make a 3′-AATT sticky end. When a protein needs different tags, it is a tedious and inefficient process that may also lead to the introduction of unwanted junction sequences due to the existence of a restriction recognition sequence. To improve the cloning efficiency, gateway technology takes advantage of another enzyme called integrase, which allows for site-specific recombination (Esposito et al., 2009). However, as patented technology, it requires that many essential reagents be purchased from designated resources. Also, the recognition sequence for integrase imposes a longer unwanted junction sequence between the tag and the protein of interest.
In our recent study to add 11 different tags to the N-terminus of THSD1, a single-span transmembrane protein responsible for cerebral aneurysm pathogenesis (Santiago-Sim et al., 2016), we developed a new cloning strategy by combinational use of type II and type IIS restriction endonucleases (Xu et al., 2017). Unlike type II, type IIS restriction endonucleases recognize non-palindromic sequences and cleave the DNA outside of their recognition site. For example, BsaI recognizes 5′-GGTCTC and cleaves the DNA a nucleotide downstream, resulting in a 5′ overhang 4 nucleotides long, thus making a custom sticky end that matches the gene of interest. Therefore, we can generate a seamless clone by completely eliminating the unwanted junction sequences (Xu et al., 2017). Even more importantly, using type II and type IIS restriction endonucleases in combination makes our method highly compatible with the traditional cloning system that is still widely used by many research labs. In addition, unlike gateway technology, precision tagging does not require designated destination vectors. Since each lab may favor a different set of destination vectors and tags for its gene of interest, our protocol affords researchers great flexibility in making their personalized tagging choices.
Materials and Reagents
PCR tubes (Corning, Axygen®, catalog number: PCR-02-C)
Pipette tips
Razor blade (Fisher Scientific, catalog number: 12-640)
Microcentrifuge tubes, 1.5 ml (Fisher Scientific, catalog number: 05-408-129)
Petri dish, 100 mm (Fisher Scientific, catalog number: FB0875713)
DH5α competent cells (Thermo Fisher Scientific, Invitrogen™, catalog number: 18265017)
- Plasmids containing different tags:
- pBabe-GFP (Addgene, catalog number: 10668)
- mCherry-Talin-H-18 (Addgene, catalog number: 62749)
- Dendra2-Vinculin-N-21 (Addgene, catalog number: 57749)
- pSNAPf-C1 (Addgene, catalog number: 58186)
- miniSOG-Zyxin-6 (Addgene, catalog number: 57781)
- Type II restriction endonucleases such as:
- EcoRI-HF (New England Biolabs, catalog number: R3101S)
- BamHI-HF (New England Biolabs, catalog number: R3136S)
- SalI-HF (New England Biolabs, catalog number: R3138S)
- Bsu36I (New England Biolabs, catalog number: R0524S)
- Type IIS restriction endonucleases such as:
- BsaI-HF (New England Biolabs, catalog number: R3535S)
- BbsI-HF (New England Biolabs, catalog number: R3539S)
- BsmBI (New England Biolabs, catalog number: R0580S)
LB broth (Fisher Scientific, catalog number: BP1427-500)
LB agar (Fisher Scientific, catalog number: BP1425-500)
Ampicillin sodium salt (Fisher Scientific, catalog number: BP1760-5)
Calf intestinal alkaline phosphatase (CIP) (New England Biolabs, catalog number: M0290S)
T4 polynucleotide kinase (New England Biolabs, catalog number: M0201S)
-
pBS-KSII-4B was modified from pBS-KSII (accession number: X52327) by synonymously destroying BsaI site (Xu et al., 2017)
Note: It is available upon request and will be deposited to Addgene soon.
pCMV5 (accession number: AF239249, from lab stock and available upon request) and pBabe-puro (Addgene, catalog number: 1764)
Ultrapure water (Thermo Fisher Scientific, Invitrogen™, catalog number: 10977015)
Phusion high-fidelity DNA polymerase (New England Biolabs, catalog number: M0530S)
Custom DNA Oligonucleotides (Integrated DNA Technology)
1× Low-EDTA TE buffer pH 8.0 (Quality Biological, catalog number: 351-324-721)
6× Gel loading dye, purple (New England Biolabs, catalog number: B7024S)
Agarose (Thermo Fisher Scientific, Thermo Scientific™, catalog number: 17850)
GeneRuler 1 kb DNA ladder (Thermo Fisher Scientific, Thermo Scientific™, catalog number: SM0313)
50× TAE (Bio-Rad Laboratories, catalog number: 1610773)
UltraPure Ethidium Bromide (Thermo Fisher Scientific, Invitrogen™, catalog number: 15585011)
QIAquick gel extraction (QIAGEN, catalog number: 28704)
QIAquick PCR purification kit (QIAGEN, catalog number: 28104)
QIAprep spin miniprep kit (QIAGEN, catalog number: 27106)
Overlapping PCR (Recipe 1)
In vitro digestion by restriction endonucleases (Recipe 2)
Linearization of a vector by restriction endonucleases and dephosphorylation by calf intestinal alkaline phosphatase (CIP) (Recipe 3)
In vitro DNA ligation (Recipe 4)
DNA transformation (Recipe 5)
In vitro DNA oligonucleotide annealing (Recipe 6)
In vitro T4 polynucleotide kinase phosphorylation (Recipe 7)
Equipment
Pipettes
Thermo Cycler (Applied Biosystems) (or any other convention PCR device)
DNA electrophoresis system (Takara Bio, model: Mupid-exU)
3UV lamp (Fisher Scientific)
Water bath (VWR International), set at 37 °C for enzymatic digestion and 42 °C for DNA transformation into DH5α competent cells
Heat block (VWR International), set at 55 °C for agarose gel dissolution
Air incubator (VWR International), set at 37 °C for bacteria growth on LB plates
Microcentrifuge (Eppendorf, model: 5424)
Orbital shaker (Forma Scientific), set at 37 °C for bacteria growth in LB medium
ChemiDoc MP imaging system (Bio-Rad Laboratories, model: ChemiDoc™ MP)
Software
DNA sequence analysis software: Word, Vector NTI, DNAstar, or other
Procedure
A. Choosing the appropriate combination of type II and type IIS restriction endonucleases
Choose the gene of interest (GOI). For example, we recently focused on adding different tags to THSD1, a single-span transmembrane protein.
Determine the tags to be inserted. We applied 11 different tags to THSD1 (Table 1).
Find type II restriction enzymes that are absent for the GOI and the tags to be inserted. For simplicity, we term them as ‘non-cutters’ hereafter. For human THSD1 (accession number: NM_018676), some non-cutters are available such as BamHI, ClaI, EcoRI, EcoRV, NheI, SalI, XbaI, XhoI, etc. Since the sequences of the tags are custom designed, alternative codon usage can be potentially used to remove the restriction site.
Design the appropriate combination of the identified non-cutters based on pBS-KSII-4B and your destination vector. pBS-KSII-4B is a modified vector for precision tagging (Xu et al., 2017) and it shares the identical multiple cloning site with the widely-used cloning vector pBS-KSII (accession number: X52327). To express tagged THSD1 in mammalian cells, we chose two different destination vectors including pCMV5 and pBabe-puro for transient or stable protein expression, respectively. Accordingly, BamHI, SalI and EcoRI were selected since BamHI/SalI can transfer the insert into pCMV5 while BamHI/EcoRI does the same for pBabe-puro.
Subclone the GOI into pBS-KSII-4B vector using selected non-cutters (Figures 1A to 1B). We termed this new construct as ‘entry clone’ and labeled it as pBS-KSII-4B-GOI. Using regular PCR, we added a BamHI site at the N-terminus, and a sequential SalI-EcoRI site at the C-terminus of THSD1 in pBS-KSII-4B. Therefore, tagged THSD1 in pBS-KSII-4B can work for both pCMV5 and pBabe-puro. If more destination vectors are needed, we may consider readjusting their sequence order of multiple cloning sites to be compatible with pBS-KSII-4B.
Determine the position of the tag of interest. In our case, we inserted the tag right after the signal peptide of THSD1 (Figure 2A). In general, tags can be placed before the N-terminal or after the C-terminal of a full-length protein. For a transmembrane protein, tags have to be inserted after the signal peptide if the N-terminal tagging is needed, since they will be cleaved upon entering the lumen of the endoplasmic reticulum. In particular, THSD1 is a type I transmembrane protein, and its N-terminal region will be located outside the cytoplasmic membrane. To avoid any potential interference with THSD1 intracellular signaling transduction, we decided to tag its extracellular domain corresponding to its N-terminal region.
Find a single-cut type II restriction endonuclease that is close to the position of the tag of interest. Henceforth, we will refer to this as a ‘unique cutter’.
Check whether the unique cutter is absent for both the tags and pBS-KSII-4B. For THSD1, we chose Bsu36I that is 455 bp away from the tag position (Figure 2A).
Note that four type IIS restriction endonucleases, including BsaI, BbsI, BsmBI, and BfuAI, are absent for pBS-KSII-4B. All of them cleave DNA outside its non-palindromic recognition sequences, and more product information is available on the New England Biolabs’ website.
Check which type IIS restriction endonuclease is absent for the GOI or the tag. For example, BsaI is absent for THSD1, and different type IIS enzymes may be selected for different tags. In our case, we applied 11 tags (Table 1) and used BsaI, BbsI, and BsmBI, accordingly.
Choose a type IIS DNA cassette. Four type IIS restriction endonuclease-mediated DNA cassettes containing detailed nucleotide information are shown in Figure 3. For THSD1, we chose a BsaI DNA cassette.
Record the DNA sequence after inserting the selected type IIS DNA cassette into the tag position of the GOI. Word, Vector NTI, DNAstar, or other DNA sequence analysis software is helpful. We named the sequence that contains the type IIS DNA cassette from one end to the site of unique cutter in the GOI as ‘template fragment’.
Table 1. Information of 11 tags used for THSD1.
Information on the amino acids of the tags smaller than 150 bp is shown here, and the addgene number for each tag larger than 150 bp is referenced.
| HA-tag | YPYDVPDYA |
| Myc-tag | EQKLISEEDL |
| FLAG-tag | DYKDDDDK |
| V5-tag | GKPIPNPLLGLDST |
| Avitag | GLNDIFEAQKIEWHE |
| Strep-tag II | WSHPQFEK |
| EGFP | Addgene:10668 |
| mCherry | Addgene: 62749 |
| Dendra2 | Addgene: 57749 |
| SNAPf | Addgene: 58186 |
| MiniSog | Addgene: 57781 |
Figure 1. Overview of the workflow for precision tagging by combinational use of type II and type IIS restriction endonucleases.
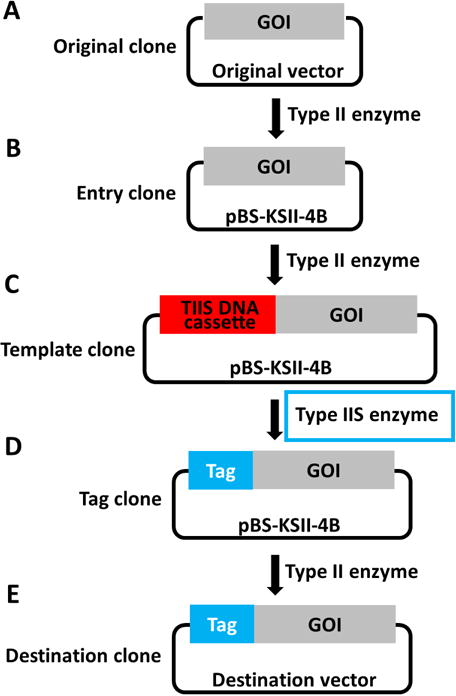
A–E. GOI in different stages is specified in the precision tagging workflow. Type IIS DNA cassette in the GOI is highlighted in the red box. Type IIS restriction endonucleases are used between (C) and (D) and emphasized in blue frame. GOI is the abbreviation of ‘Gene of Interest’.
Figure 2. Schematic of making a template clone from an entry clone of THSD1.
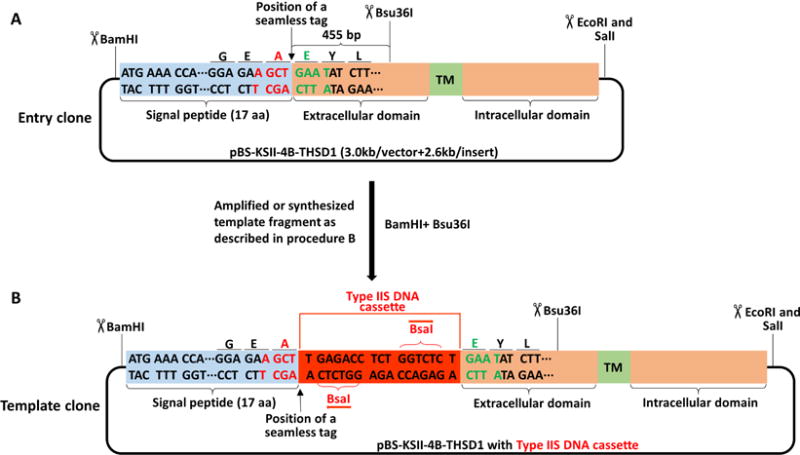
A–B. The signal peptide of THSD1 is highlighted in the light blue box while the rest in the orange box containing a green box with the labeled transmembrane domain (TM). BamHI and Bsu36I were selected to replace the original fragment in (A) with the template fragment containing BsaI DNA cassette (highlighted in red box) in (B).
Figure 3. Four kinds of type IIS DNA cassettes for pBS-KSII-4B.
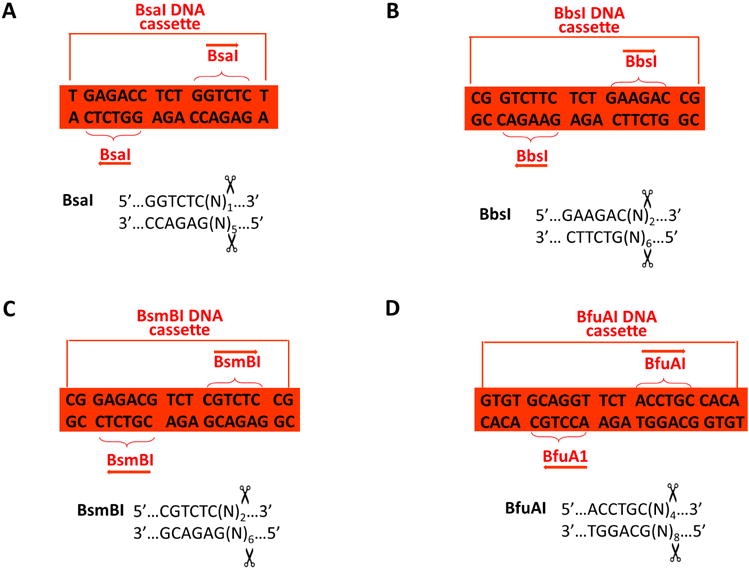
(A) BsaI (B) BsmB1 (C) BbsI (D) BfuA1 recognized sequences are indicated in the braces. The cutting positions for them are indicated at both ends of the DNA cassette or the bottom panel.
B. Construction of a template clone by type II restriction endonucleases
- Perform overlapping PCR to make a template fragment. For THSD1, we performed the following experiment as indicated in Figure 4 and Recipe 1 using Thermo Cycler.
- Order two oligos (BamHI-THSD1-F and Bsu36I-THSD1-R) that match both ends of the template fragment, with additional type II restriction enzymes such as BamHI or Bsu36I.
- Two internal overlapping oligos contain 18–24 nt of the left (light blue box) and right side (orange box) of the type IIS DNA cassette (red box), which is in total around 54–68 nt (BsaI-OL-F and BsaI-OL-R). The sequence information for above oligos is listed in Table 2.
Run the PCR product on a gel using the Mupid-exU DNA electrophoresis system, and check the size of amplified NT and CT fragment followed by gel purification (QIAquick gel extraction kit). Purify the DNA band of interest using QIAquick gel extraction kit (melt the gel in a 55 °C heat block). Next, perform overlapping PCR by mixing NT and CT fragment as indicated in Recipe 1 and gel purify the template fragment of correct size in the same way.
Alternatively, order the template fragment from Integrated DNA Technology (IDT) or other companies that provide similar services involving large DNA fragment synthesis. We ordered it with additional BamHI at the N-terminus, Bsu36I at the C-terminus, and BsaI DNA cassette right after the signal peptide of THSD1. Use the type II restriction endonucleases to digest the template fragment obtained either from overlapping PCR or the commercial gene synthesis. For THSD1, digest both fragments with BamHI and Bsu36I for 2 h at 37 °C using a water bath (Recipe 2 for enzymatic digestion).
Linearize pBS-KSII-4B-GOI with the same restriction endonucleases that are used for the template fragment, followed by calf intestinal phosphatase treatment (Recipe 3 for vector linearization). For example, we linearized pBS-KSII-4B-THSD1 with BamHI and Bsu36I.
Ligate the digested template fragment with linearized pBS-KSII-4B-GOI (Recipe 4 for DNA ligation). Accordingly, we joined the template fragment with the above linearized pBS-KSII-4B-THSD1 (Figure 2).
Transform DH5α competent cells with the ligation product and spread them on LB agar plates containing 100 μg/ml ampicillin (Recipe 5 for DNA transformation).
Incubate the plates at 37 °C air incubator overnight.
Pick up several colonies on each plate and inoculate them separately into a clean tube containing 2–5 ml LB medium with 100 μg/ml ampicillin.
Grow the bacteria in an orbital shaker with a shaking speed of 220 rpm at 37 °C overnight.
Extract the plasmids from the overnight cultures using QIAprep spin miniprep kit.
Confirm the correct clones by restriction analysis and Sanger sequencing.
Figure 4. Schematic of making a template fragment of THSD1 from overlapping PCR.
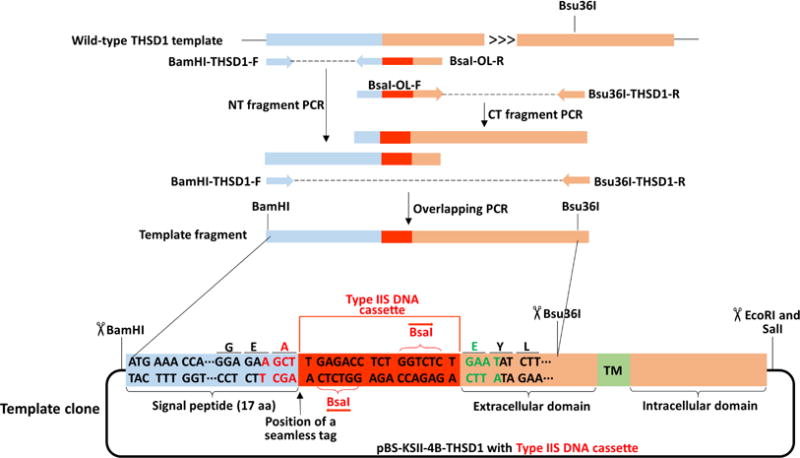
Detailed information for the oligos is provided in Table 2. BamHI was added to the forward primer for NT-fragment PCR. Overlapping PCR contains part of a signal peptide (highlighted in the light blue box), BsaI DNA cassette (selected from Figure 2, highlighted in the red box), and part of THSD1 coding sequence (highlighted in the orange box). Bsu36I is in the middle of the reserve primer for CT fragment PCR. The detailed information of the final PCR product is indicated in the region of the template clone between two black lines.
Table 2. Information of overlapping PCR.
BsaI DNA cassette is highlighted in red letters. BamHI and Bsu36I cutting sites are italicized. Nucleotides are colored according to Figure. OL is short for ‘over-lapping’.
| BamHI-THSDl-F: | |
| GGTggatccACC | |
| Bsal-OL-R | |
|
| |
| Bsal-OL-F | |
|
| |
| Bsu36l-THSD1-R: | |
|
|
C. Construction of a tag clone by type IIS restriction endonucleases
Define the tag sequence using DNA analysis software, such as Word or DNAstar.
For the tags that are smaller than 150 bp, such as HA, Myc or FLAG, order two annealing oligos as indicated in Figure 5A. So far, a 200 bp long oligo is the maximum length that can be provided commercially, and that is why 150 bp was chosen as an arbitrary cutoff. It still leaves extra spaces for adding gene-specific DNA sequences at both ends.
-
For the tags that are larger than 150 bp, such as GFP, miniSOG, or SNAPf, order two amplifying oligos with type IIS restriction endonuclease sequences at both ends as indicated in Figure 5B. To amplify these tags, perform PCRs under the same condition for each tag by following a standard protocol:
Template DNA 1 ng Amplifying Oligo-F (50 μM) 1 μl Amplifying Oligo-R (50 μM) 1 μl 10× HF buffer 5 μl dNTP (25 μM) 1 μl Phusion high-fidelity DNA polymerase 0.5 μl H2O added to total 50 μl Initial denaturation: 98 °C 30 sec; 28 cycles: 98 °C 10 sec, 60 °C 15 sec, 72 °C 1 min; final extension: 72 °C 5 min.
Following Step C2, oligos are annealed and phosphorylated by T4 polynucleotide kinase.
Following Step C3, amplified PCR products are digested by type IIS enzymes for 2 h at 37 °C (Recipe 2).
Linearize the template clone of pBS-KSII-4B-GOI containing type IIS DNA cassette by the predesigned type IIS restriction endonucleases. For example, pBS-KSII-4B-THSD1 containing BsaI cassette is digested by BsaI for 2 h at 37 °C, and dephosphorylated by calf intestinal alkaline phosphatase (Recipe 2 and Figures 6A–6B).
Ligate the tag fragments with two sticky ends that are generated at Step C4 or Step C5, into the linearized template clone from Step C6 (Figures 6B–6C).
Transform DH5α competent cells with the ligation product and spread them on LB agar plates containing 100 μg/ml ampicillin (Recipe 5 for DNA transformation).
Grow the plates at 37 °C air incubator overnight.
Pick up several colonies on each plate and inoculate them separately into a clean tube containing 2–5 ml LB medium with 100 μg/ml ampicillin.
Grow the bacteria in an orbital shaker with a shaking speed of 220 rpm at 37 °C overnight.
Extract the plasmids from the overnight cultures using QIAprep spin miniprep kit.
Confirm the correct clones by restriction analysis and Sanger sequencing.
Figure 5. Generation of the tag fragment with custom sticky ends.
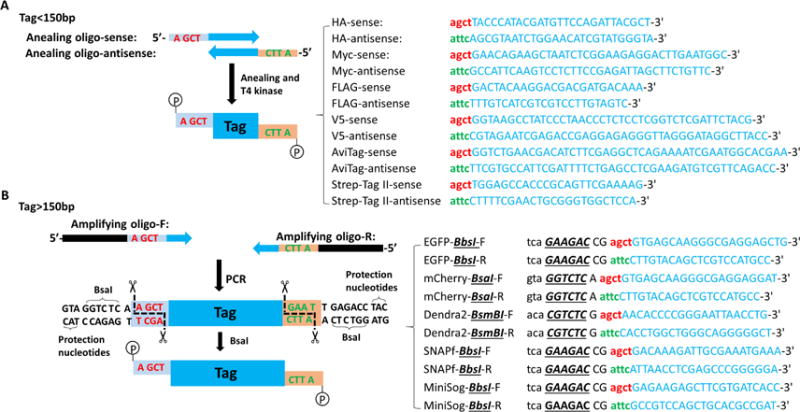
A. The sense annealing oligo (annealing oligo-S) is composed of 5′-AGCT (letters in red) and tag-specific sequence (letters in blue); the anti-sense annealing oligo (annealing oligo-AS) is composed of 5′-ATTC (letters in green) and tag-specific sequence (letters in blue), as shown in the right table. B. Both amplifying oligos contain a BsaI recognition site (letters in black), THSD1-specific sticky end (letters in red or green), and tag-specific sequences (letters in blue), as detailed on the right table.
Figure 6. Schematic of making a tag clone from a template clone of THSD1.
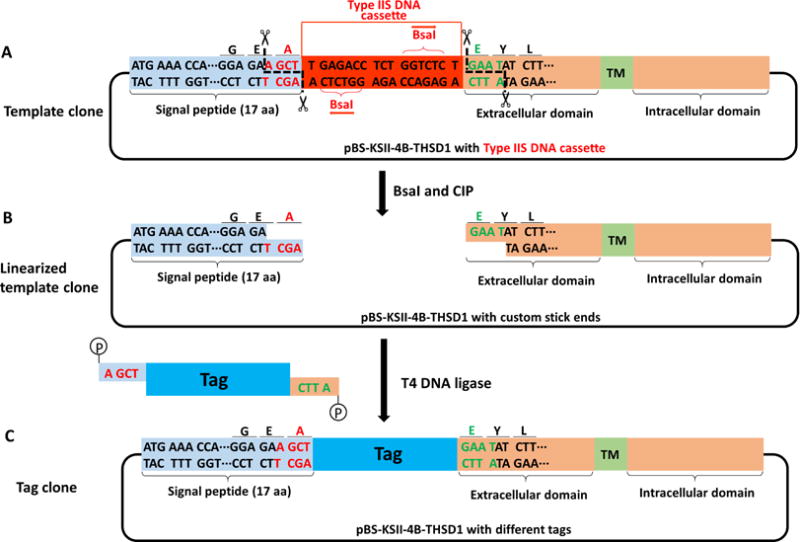
A–B. The template clone contains two THSD1-specific sticky ends (letters in red or green). C. All above tags containing a THSD1-specific sticky end and 5′-phosphorylation are joined with the linearized template clone by T4 DNA ligase, thus generating a seamless tag clone.
D. Construction of a destination clone by type II restriction endonucleases
Linearize your destination vector with the preselected type II restriction enzymes from Procedure A. For instance, we linearized pCMV5 with BamHI and SalI, or pBabe-puro with BamHI and EcoRI (Recipe 3).
Cut your tagged GOI from previous tag clones in Procedure C with the same enzymes, followed by running agarose gel electrophoresis and extracting DNA band of correct size from the gel. For pBS-KSII-4B-THSD1 with different tags, we cut tagged THSD1 by either BamH/SalI or BamHI/EcoRI, and recovered the DNA band of correct size by gel purification (QIAquick gel extraction kit).
Ligate the tagged GOI with the linearized destination vector by compatible sticky ends. In Figure 6, how tagged THSD1 are transferred from the tag clone into the destination clone like pCMV5 are shown (Figure 7).
Transform DH5α competent cells with the ligation product and spread them on LB agar plates containing 100 μg/ml ampicillin (Recipe 5 for DNA transformation).
Incubate the plates at 37 °C air incubator overnight.
Pick up several colonies on each plate and inoculate them separately into a clean tube containing 2–5 ml LB medium with 100 μg/ml ampicillin.
Grow the bacteria in an orbital shaker with a shaking speed of 220 rpm at 37 °C overnight.
Extract the plasmids from the overnight cultures using QIAprep spin miniprep kit.
Confirm the correct clones by restriction analysis and Sanger sequencing.
Figure 7. Schematic of making a destination clone from a tag clone of THSD1.
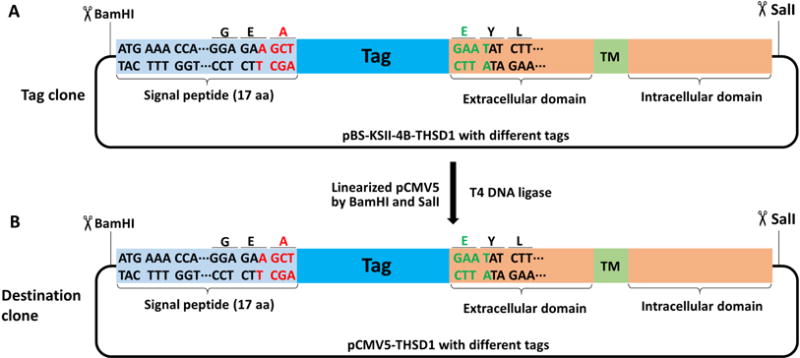
BamHI and SalI were selected to transfer the tagged THSD1 in pBS-KSII-4B to pCMV5 vector using traditional cloning method.
Data analysis
All clones at different stages (Figure 1) can be characterized by restriction endonucleases and followed by agarose gel electrophoresis. Gel images are recorded in the ChemiDoc imaging system. The shuttling vector pBS-KSII-4B does not contain BsaI and other three type IIS enzymes, such as BbsI, BsmBI, and BfuA1, are naturally absent (Figure 8).
Figure 8. Features of pBS-KSII-4B.
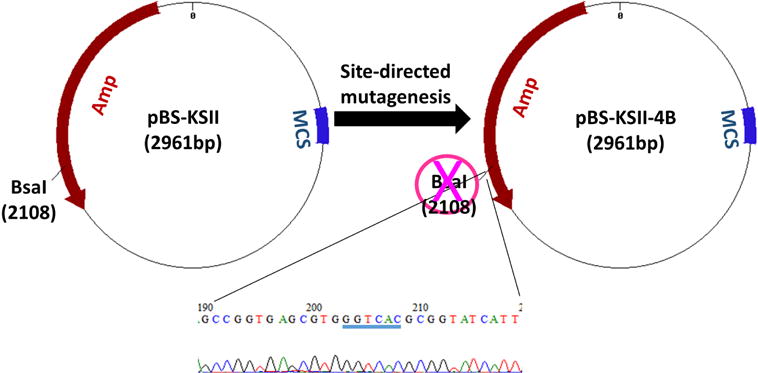
BsaI-recognized ‘GGTCTC’ was mutated to ‘GGTCAC’ (highlighted by blue underline), which synonymously destroyed BsaI site.
For further verification of different clones, DNA needs to be sent out for local sequencing service. Softwares such as Vector NTI or DNAstar is helpful for DNA sequence analyses.
Tagged GOI in destination vectors can be functionally evaluated. For example, THSD1 with different tags can be confirmed by immunoblotting against overexpressed HEK293T cells.
Notes
-
Unsuccessful PCR amplification of template
Unsuccessful PCR includes two scenarios: lack of amplified fragment of correct size, or weak detection of target band in addition to multiple non-specific fragments. For the former situation, make sure that PCR reaction system is still working such as the expiration date of the enzyme, buffer or dNTP, which can be assessed by using other primers and template that has been successfully amplified previously. Double check the new primers to see if they match the template in the right orientation. On occasions, repetitive sequences in the primers that form special secondary structures will interfere with effective PCR amplification, which can be improved by adding 5 M betaine into the reaction system (final concentration of betaine is 3 M). For the latter issue such as detection of weak amplification of target band co-existing with other non-specific fragments, make sure that the Tm values of each pair of primers are close. If not, adjust them simply by changing the length of primers. Besides, increasing the annealing temperature will help increase PCR specificity in general.
-
Only empty vector produced by ligation
It may be caused by incomplete digestion of a DNA fragment of interest. Reducing the amount of DNA fragment, increasing the units of digestive enzymes, or maximizing their activities using a compatible buffer may help. New England Biolabs has produced many new versions of the enzymes that cut the same DNA sequence and have 100% activity in different buffers in comparison with the old versions. Therefore, these new enzymes also called high-fidelity enzymes provide more options on buffer selections when maximized activities of double enzymatic digestion are needed.
Recipes
- Overlapping PCR
-
The below reaction is for N-terminal (NT) fragment PCR. Template DNA 1 ng
Oligo-F (50 μM) 1 μl Overlapping-R (50 μM) 1 μl 10× HF buffer 5 μl dNTP (25 μM) 1 μl Phusion high-fidelity DNA polymerase 0.5 μl H2O added to total 50 μl Note: The annealing temperature is 55–65 °C with 28 cycles. - For C-terminal (CT) fragment PCR, use oligo-R and overlapping-F primers
- Gel purify the amplified NT and CT fragment using QIAquick gel extraction kit
- Mix purified fragments in a ratio of 1:1 as a new template DNA and run overlapping PCR as follows:
Mixed template DNA around 100 ng 10× HF buffer 5 μl dNTP (25 μM) 1 μl Phusion high-fidelity DNA polymerase 0.5 μl H2O added to total 50 μl - Run 10 cycles with annealing temperature being 55 °C or higher. Continue to run extra 20 cycles after adding oligo-F (1 μl) and oligo-R (1 μl) to the reaction system
- Extract the fragment of correct size with gel purification
-
-
In vitro digestion by restriction endonucleases
DNA 1 μg 10× buffer 5 μl Restriction endonuclease #1 0.75 μl Restriction endonuclease #2 0.75 μl H2O added to total 50 μl Incubate at 37 °C for 2 h Note: Optimal temperature for digestion may vary for different enzymes such as 55 °C for BsmBI.
- Linearization of a vector by restriction endonucleases and dephosphorylation by calf intestinal alkaline phosphatase (CIP)
- Linearization of a vector: same as Recipe 2
- Vector de-phosphorylation: after a vector linearization, 1 μl CIP was added and incubated at 37 °C for 40 min
-
In vitro DNA ligation
DNA insert 12.5 μl Linearized vector 1 μl 10× T4 DNA ligase buffer 1.5 μl Room temperature, 1 h Note: Negative control is the ligation of linearized vector alone by replacing DNA insert with water.
- DNA transformation
- Ligation product is incubated with DH5α competent cells on ice for 30 min
- Heat shock at 42 °C for 1 min
- Cells are allowed to recover for 1 h at 37 °C with a shaking speed of 220 rpm prior to plating on LB agar with appropriate antibiotics
- In vitro DNA oligonucleotide annealing
- Each oligo was dissolved in 1× low EDTA TE buffer to 50 μM
- Mix equal volume of annealing oligos, and put them at 95 °C heat block for 5 min, and turn it off to cool slowly to room temperature
-
In vitro T4 polynucleotide kinase phosphorylation
Annealed oligos 1 μl 10× T4 DNA ligase buffer 1 μl T4 polynucleotide kinase 1 μl H2O added to total 10 μl Incubate at 37 °C for 1 h Store at −20 °C Note: For later DNA ligation, the phosphorylated oligos need to be diluted 1:500 before use.
Acknowledgments
We appreciate the critical proofreading of this manuscript by Dr. Joanna O’Leary. The project was supported by RO3NS087416 from the US National Institute of Health.
Footnotes
Conflict of interest statement: The authors declare no conflict of interest or competing interests.
References
- Bodor DL, Rodriguez MG, Moreno N, Jansen LE. Analysis of protein turnover by quantitative SNAP-based pulse-chase imaging. Curr Protoc Cell Biol. 2012 doi: 10.1002/0471143030.cb0808s55. Chapter 8: Unit8 8. [DOI] [PubMed] [Google Scholar]
- Esposito D, Garvey LA, Chakiath CS. Gateway cloning for protein expression. Methods Mol Biol. 2009;498:31–54. doi: 10.1007/978-1-59745-196-3_3. [DOI] [PubMed] [Google Scholar]
- Santiago-Sim T, Fang X, Hennessy ML, Nalbach SV, DePalma SR, Lee MS, Greenway SC, McDonough B, Hergenroeder GW, Patek KJ, Colosimo SM, Qualmann KJ, Hagan JP, Milewicz DM, MacRae CA, Dymecki SM, Seidman CE, Seidman JG, Kim DH. THSD1 (thrombospondin Type 1 domain containing protein 1) mutation in the pathogenesis of intracranial aneurysm and subarachnoid hemorrhage. Stroke. 2016;47(12):3005–3013. doi: 10.1161/STROKEAHA.116.014161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2(12):905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Xu Z, Rui YN, Balzeau J, Menezes MR, Niu A, Hagan JP, Kim DH. Highly efficient one-step scarless protein tagging by type IIS restriction endonuclease-mediated precision cloning. Biochem Biophys Res Commun. 2017;490(1):8–16. doi: 10.1016/j.bbrc.2017.05.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Song Y, Li Y, Chang J, Zhang H, An L. The tandem affinity purification method: an efficient system for protein complex purification and protein interaction identification. Protein Expr Purif. 2010;72(2):149–156. doi: 10.1016/j.pep.2010.04.009. [DOI] [PubMed] [Google Scholar]


