Abstract
Purpose
Accurate transcriptional sequencing (RNA-seq) from formalin-fixation and paraffin-embedding (FFPE) tumor samples presents an important challenge for translational research and diagnostic development. In addition, there are now several different protocols to prepare a sequencing library from total RNA. We evaluated the accuracy of RNA-seq data generated from FFPE samples in terms of expression profiling.
Methods
We designed a biospecimen study to directly compare gene expression results from different protocols to prepare libraries for RNA-seq from human breast cancer tissues, with randomization to fresh-frozen (FF) or FFPE conditions. The protocols were compared using multiple computational methods to assess alignment of reads to reference genome, and the uniformity and continuity of coverage; as well as the variance and correlation, of overall gene expression and patterns of measuring coding sequence, phenotypic patterns of gene expression, and measurements from representative multigene signatures.
Results
The principal determinant of variance in gene expression was use of exon capture probes, followed by the conditions of preservation (FF versus FFPE), and phenotypic differences between breast cancers. One protocol, with RNase H-based rRNA depletion, exhibited least variability of gene expression measurements, strongest correlation between FF and FFPE samples, and was generally representative of the transcriptome from standard FF RNA-seq protocols.
Conclusion
Method of RNA-seq library preparation from FFPE samples had marked effect on the accuracy of gene expression measurement compared to matched FF samples. Nevertheless, some protocols produced highly concordant expression data from FFPE RNA-seq data, compared to RNA-seq results from matched frozen samples.
Keywords: Formalin-fixation and paraffin-embedding tissue, Gene expression, Library preparation, Breast cancer tissue, Coding region enrichment, RNA sequencing
INTRODUCTION
While it is generally best to identify gene expression biomarkers from cancer tissues using the highest quality of ribonucleic acids (RNA) purified from fresh frozen (FF) samples, any subsequent development toward diagnostic testing will require translation for use with formalin-fixed paraffin-embedded (FFPE) tissue samples. However, the variably fragmented and chemically modified RNA derived from FFPE samples presents a challenge for accurate measurement of gene-expression [1, 2].
In a different context, there is great interest to perform transcriptome sequencing (RNA-seq) for biomarker discovery research using large cohorts of precious archival FFPE samples from completed clinical trials. However, an unfavorable signal-to-noise ratio from FFPE samples could reduce the accuracy of biomarker discovery. Therefore, it is essential to select a protocol for FFPE RNA-seq libraries that yields data that is comparable with a “gold standard” result from FF samples. But there is more than one standard protocol for RNA-seq of high-quality RNA from FF tumor samples.
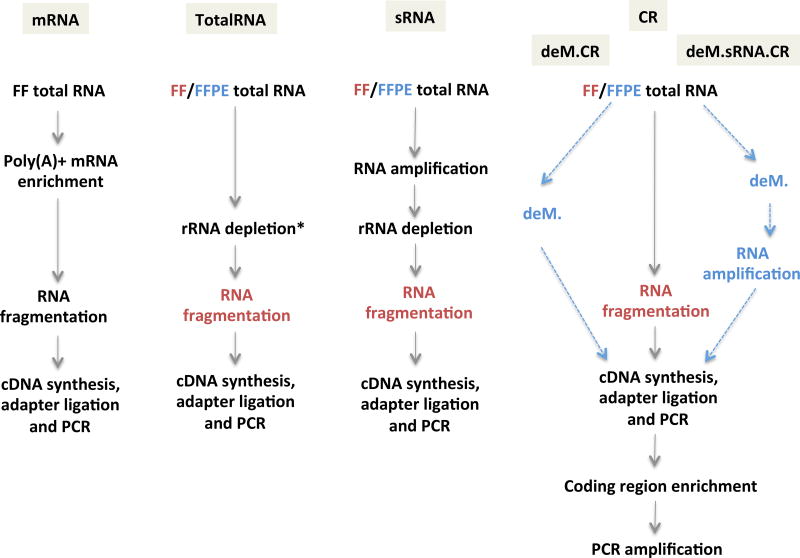
Different approaches to generating libraries for RNA-seq include: 1) selection of messenger RNA by targeting the poly(A) 3’ tail (mRNA protocol), 2) depletion of more abundant ribosomal RNA (rRNA depletion) using bead-based method (I.TotalRNA protocol) or enzymic method (K.TotalRNA protocol), and 3) exon capture probes for known coding region sequence (CDS) from an RNA-seq library prepared (CR protocol).
Data generated from the popular mRNA protocol using FF tissue samples (FF.mRNA library) are highly concordant with microarray data in tumor gene expression signature study [3]. But this protocol is not appropriate for degraded mRNAs from FFPE samples [4]. On the other hand, total RNA library protocols do not restrict enrichment to poly(A)+ tailed mRNA, allowing less biased quantification of isoform abundance [4–6].
Corresponding protocols for RNA-seq from FFPE tumor samples include an adaptation of the mRNA protocol that combines random and poly(A) primers (sRNA protocol) optimized for gene expression microarrays (SensationPlus kit, Affymetrix, CA); or are unchanged for the I.TotalRNA, K.TotalRNA and CR protocols (Figure 1). Total RNA protocols have achieved Pearson correlations with FF counterparts of >0.9 [4, 6, 7]. Exon capture using the CR protocol has potential for stronger correlation, but involves selected coverage [8]. Finally, since pre-treatment heat and methyl saturation have been claimed to reduce methylol adducts on FFPE RNA [9], we evaluated pre-analytical demethylation (deM) of total RNA prior to library preparation using the CR and sRNA protocols (Figure 1).
Figure 1.
Workflows of RNA-seq library preparation. The red color indicates steps only applied to FF samples, while the blue indicates steps only applied to FFPE samples. The grey shaded boxes contain the names for each protocol. The * indicates different rRNA depletion methods that result in two different TotalRNA protocols, that is, RiboZero for I.TotalRNA and RNase H for K.TotalRNA protocol. Abbreviations: mRNA, messenger RNA targeting protocol; sRNA, sense RNA protocol; CR, coding region capture protocol; deM, demodification protocol; PCR, polymerase chain reaction.
Consequently, this study was designed to directly compare the results from RNA-seq library protocols between optimally matched sample pairs (FF and FFPE) from representative breast cancers, in order to address three scenarios in translational research: 1) biomarker discovery from FF samples phase with intention to translate for FFPE samples in future studies for validation and diagnostic development, 2) biomarker discovery from FFPE samples that is intended to be representative had high quality FF samples been available, and 3) translation of existing biomarkers, developed using a different method (such as microarrays or RNA-seq using mRNA protocol), for use with RNA-seq data from FFPE samples.
METHODS
Tumor tissue samples
Fresh tumor tissue, was collected at intraoperative pathology evaluation, diced into pieces of 1–2mm diameter, stirred, and randomly assigned to: 1) RNAlater solution later stored at −80°C freezer (FF); or 2) 10% neutral buffered formalin and paraffin-embedded as a FFPE tissue block [10]. Phenotypically, the nine breast cancers were defined by pathologic status of hormone receptors (HR) and HER2 receptor as: HR+/HER2− in five, HR+/HER2+ in one, and triple receptor-negative (TN) in three. RNA was purified from FF samples using the RNeasy Mini Kit (Qiagen, Valencia, CA), and FFPE samples from 10µm sections using High Pure FFPE RNA Isolation Kit (Roche, Indianapolis, IN). A DNase-I treatment step was included in both.
Construction of RNA-seq library and sequencing
Full details of all methods for library construction and sequencing of RNA samples are in the Supplementary Methods. An overview diagram of the different RNA-seq library protocols is shown in Figure 1, and details of the number of libraries prepared, starting RNA requirement, cost and duration to perform each protocol are summarized in Supplementary Table 1.
The mRNA protocol (FF only) used oligo-dT beads for poly(A)+ mRNA enrichment, followed by standard procedures of TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA).
The I.TotalRNA protocol used Ribo-Zero™ Magnetic Gold Kit to deplete ribosomal RNA (rRNA) from total RNA, followed by library preparation using the Truseq Stranded Total RNA Sample Prep Kit (Illumina, San Diego, CA).
The K.TotalRNA protocol used an RNase H-based method to deplete rRNA from total RNA, followed by library preparation using KAPA Stranded RNA-Seq Kit with RiboErase (Kapa Biosystems, Wilmington, MA).
The sRNA protocol used SensationPlus™ Amplification Kit (Affymetrix, Santa Clara, CA) with oligo-dT and random primers designed for whole-transcriptome amplification, then the sense RNA (sRNA) was synthesized by in vitro transcription and used as the template for the I.TotalRNA protocol described above.
The Coding-Region (CR) protocol was performed using Truseq Access RNAseq kit (Illumina, San Diego, CA), using random primers. Next, sequencing adapters were ligated to the resulting cDNA followed by the first PCR (15 cycles). The coding regions in those libraries were enriched using capture probes and amplified by PCR.
The de-modification (deM) protocol used heat in an amine-rich solution (70°C for 30 min in 1× TE buffer containing 20µM NH4Cl, pH7.0) [9, 11]. Starting with de-modified RNA, we tested two additional FFPE library preparation methods: FFPE.deM.CR, FFPE.deM.sRNA.CR.
Libraries were randomly assigned to a lane (4 per lane) and paired-end sequenced with Illumina Hi-Seq 2000 Sequencing System. We generated 100 base-paired reads for sample C and 50 base-paired reads for the other eight samples for the FF.mRNA and FFPE.sRNA protocols. All remaining libraries had 75 base-paired reads. For the mRNA and sRNA protocols, the libraries were prepared with two technical replicates to test reproducibility. No technical replicates could share the same sequencing lane.
RNA-seq data analysis
Full details of all data analysis methods have been provided in the Supplementary Methods. An overview diagram of the analysis plan is shown in Supplementary Figure 1. Briefly, the different protocols for FF and FFPE samples were compared with respect to metrics as follows: mapping rates of RNA-seq reads (exonic, intronic, intergenic), read coverage uniformity and continuity, principal component analysis and hierarchical clustering analysis on expression levels, pairwise comparison per gene over the coding sequence, of all genes and of selected breast cancer gene expression signatures that had previously been developed from RT-PCR or microarray data from FF samples.
RESULTS
RNA extracted from FFPE samples was severely degraded, with RNA integrity number (RIN) of 1.2–2.2, versus 6.7–9.3 from FF samples (Supplementary Table 2). All libraries generated >49 million raw reads (mean= 113 million, sd= 27 million).
Post-alignment statistics
The mapping rates from FFPE samples differed from FF samples when using libraries from mRNA and TotalRNA protocols as follows: fewer exonic (overall mean difference= 0.335, P < 10−14), more intronic (overall mean difference= 0.309, P < 10−15), and comparable for intergenic sequence reads (Supplementary Figure 2, 3). The RNAseq data generated from the sRNA protocol had significantly lower concordant pair alignment rate as compared to those from non-sRNA protocols (p-value<0.001, Supplementary Figure 2). Using the CR protocol, mapping was highly concordant between FF and FFPE and the exonic mapping rate increases compared to using non-CR methods (Supplementary Figure 2, 3, 4). Overall, The number of genes with read coverage (TPM > 0.1) was slightly higher in FFPE samples than in FF samples for both non-CR and CR protocols (Supplementary Figure 5) [12].
Uniformity and continuity of read coverage of transcripts
Uniformity of read coverage was measured by the mean coefficient of variation (CV), and continuity of coverage as the percentage of gaps without read coverage, across the top 1000 highly expressed transcripts (Supplementary Figures 6–8). FFPE.I.TotalRNA and FFPE.K.TotalRNA libraries demonstrated the most uniform and continuous coverage among protocols for FFPE samples, and were equivalent to protocols for FF samples. In contrast, the CR protocol produced non-uniform coverage, with high percentage of gaps, in both FF and FFPE libraries. The FFPE.sRNA protocol introduced modest non-uniformity.
Pre-analytical sources of variance
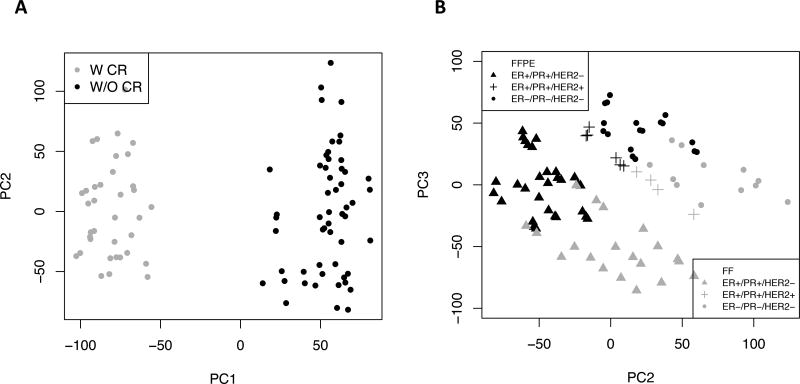
In RNA-seq studies, the variance across samples usually grows with the mean of gene expression (also known as heteroscedasticity), and this can be problematic for correctly uncovering the underlying pattern in data using techniques such as distance-based clustering [13]. We therefore applied the variance-stabilizing transformation method to approximate the independence between variance and mean (Supplementary Figure 9). Principal component analysis (PCA) of expression of a total of 20,381 CR protocol targeted poly(A)+ genes for all libraries showed that the 26.5% of total variation captured by the first principal component was due to use of exon capture probes (CR protocol), and 20.6% from the second and third components combined - effects of FFPE and biological differences (Figure 2). Hierarchical clustering results, with high confidence (average bootstrap probability= 0.93), showed that the major tumor phenotypes (HR+ vs. HR−) and the source tumor, clustered together with FFPE samples (Supplementary Figure 10).
Figure 2.
Scatter plot of the first three principal components for CPM-normalized and variance stabilizing transformed counts of 20,381 CR-targeted poly(A)+ genes. Each point corresponds to one of 90 libraries. A) the gray color indicates samples prepared with CR and the black for those without CR treatment. A 26.5% of total variation comes from CR treatment. B) the gray color indicates FF samples and the black for FFPE samples. The symbol shape indicates the different biological group. The biological differences and FFPE effects are captured, which accounts for 20.6% of total variation. Abbreviations: PC, principal component; W CR, with coding region capture; W/O CR, without coding region capture.
Technical replicates
Technical replicates (both FF.mRNA and FFPE.sRNA protocols in all 9 tumors) were highly correlated (Spearman rho ≥ 0.992) for all samples after normalization by total count and transformation to log2 count per million (CPM) (Supplementary Figure 11).
Protocols that target mRNA or deplete rRNA
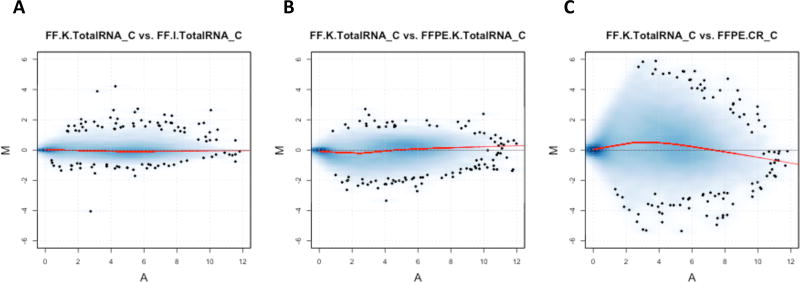
Figure 3 illustrates, for one tumor (C), MA plots of gene expression for pairs of libraries. Comparing both TotalRNA protocols with FF samples, differences were centered around zero, with small variation across different mean expression levels (Figure 3A). Comparing FF and FFPE samples using the same TotalRNA protocol, the log ratio values were still centered around zero at different mean expression levels (Figure 3B). However, comparing the FFPE.CR protocol to the FF reference, the log ratio values deviated from zero at both low and high expression levels (Figure 3C). The same patterns were observed for all other tumor samples (Supplementary Figure 12–14). These observations suggest that the TotalRNA protocols produced high-quality FFPE RNA-seq data that was comparable to the FF RNA-seq data.
Figure 3.
MA-plot of 20,381 CR targeted poly(A)+ genes for tumor sample C when using FF.K.TotalRNA sample C library as the reference. A) MA plot for tumor C between FF.K.TotalRNA and FF.I.TotalRNA; B) MA plot for tumor C between FF.K.TotalRNA and FFPE.K.TotalRNA; C) MA plot for tumor C between FF.K.TotalRNA and FFPE.CR. M is the log2-transformed expression of a gene from first library divided by that from the second library, while the A is the mean log2-transformed expression of the gene. The red curve indicates the lowess smoother fitted to the data. Abbreviation: MA-plot, difference in measurements as log ratio (M) versus mean average expression (A) of each gene.
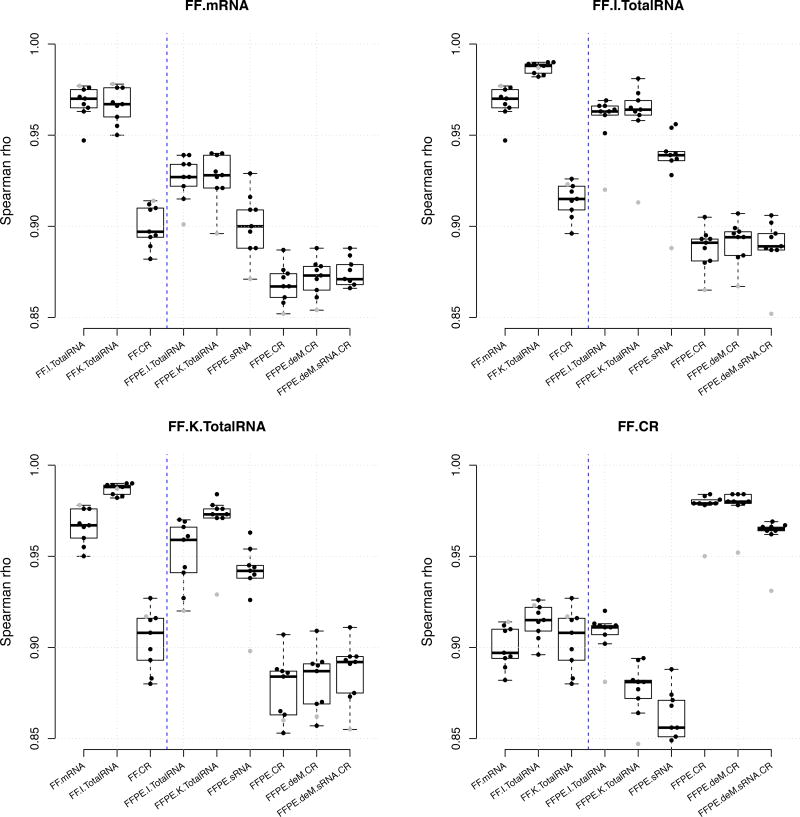
The FF.K.TotalRNA and FFPE.K.TotalRNA libraries had highly correlated transcripts per million (TPM) measure, with median rank correlation 0.973. This was significantly higher than for FF.K.TotalRNA with FF.CR (mean difference = 0.066, P < 10−6), or any other FFPE protocol (least mean difference = 0.019, P = 0.031) (Figure 4). Results were similar using CPM and FPKM measures (Supplementary Figure 15–16). The FFPE.K.TotalRNA also had the highest median rank correlation with FF.mRNA and FF.I.TotalRNA, in spite of normalization methods used (Figure 4, Supplementary Figure 15–16, Supplementary Table 3).
Figure 4.
Summary of between-protocol correlation coefficients based on TPM. The main title of each figure is the reference protocol used for comparison. Each dot is the Spearman rho estimate calculated between the reference library and the library showing on the × axis. Each box summarizes the Spearman rho estimates from nine breast tumor samples. The gray dot indicates the tumor sample N.
Protocol with subsequent exon capture
Subsequent exon capture (CR protocol) resulted in a median rank correlation of 0.980 between FF and FFPE, but the FF.CR had much lower correlation with non-CR libraries (least mean difference = 0.063, P < 10−9 using TPM) (Figure 4, Supplementary Table 3). Generally, the CR protocol tended to overly enrich the highly expressed genes, and was more likely to not capture low expressed genes (Figure 3C, Supplementary Figure 17–19). This was not improved by prior de-modification (deM) of methyol adducts from FFPE tissue-derived RNA using heat and amines, or the sRNA protocol (Figure 4, Supplementary Table 3). Although both approaches appeared to slightly increase concordance of expression, neither was statistically significant.
Further investigating these protocol-induced biases, we calculated the number of genes that would be considered as “differentially expressed” or “false positives” compared to each reference FF standard protocol (Supplementary Figure 20 and 21). Fewer FP genes would suggest fewer artifacts introduced by a protocol. FFPE.K.TotalRNA RNA-seq data had the fewest genes with significantly differential expression at various p-value thresholds and using different data normalization methods. In contrast, FF.CR was the most biased method, compared to FF.mRNA, with 84.2% of all genes significantly differentially expressed at an adjusted p-value cutoff of 0.01.
Pattern dissimilarity in measurement of coding sequence
We used a pattern dissimilarity score to measure the differences in expression patterns of coding DNA sequences (CDS) between library protocols, allowing direct comparison of non-CR and CR protocols. A smaller value of the score indicates higher similarity between a protocol and a FF reference. The distributions of dissimilarity scores across all genes were similar within each protocol, but varied across protocols (Supplementary Figure 22). FFPE.K.TotalRNA had the lowest mean dissimilarity score when using FF non-CR libraries as the reference (Supplementary Figure 23 and Supplementary Table 4).
Gene expression patterns associated with tumor phenotype
We analyzed differential expression (DE) of genes comparing HR+/HER2− and TN breast cancers within each protocol. Overall, the normalized data were distributed around zero relative log expression, and were clustered by tumor phenotypes in the first two principal components. The p-value from DE analysis followed the ideal uniform distribution for non-DE genes, with a spike close to zero for the DE genes (Supplementary Figure 24). Receiver operating characteristic (ROC) curves represented the sensitivity and specificity of the DE analyses using each FF reference as the gold standard. FFPE.K.TotalRNA achieved high and stable area under the curve (AUC) (0.921 – 0.933) at different cutoffs set for each FF gold standard, even after the strongest DE genes in the gold standards had been filtered out (Supplementary Figure 25–28, Supplementary Table 5). The best agreement between FFPE protocols and each FF standards was as follow: FFPE.sRNA with FF.mRNA, FFPE.K.TotalRNA with both FF.I.TotalRNA and FF.K.TotalRNA, and FFPE.CR with FF.CR (Supplementary Table 5).
Representative gene signatures of prognosis
We compared five published breast cancer gene expression signatures: recurrence score (Oncotype DX), PAM50, sensitivity to endocrine therapy (SET) index, mammaprint and PI3-kinase index (PI3K) [14–19]. Those were compared between three FFPE protocols (I.TotalRNA, K.TotalRNA and sRNA) and three FF protocols (mRNA, I.totalRNA and K.TotalRNA). Best correlations using FFPE protocols with FF.mRNA (range 0.911 – 0.934) were not as strong as with FF.I.TotalRNA (range 0.952 – 0.975) or FF.K.TotalRNA (range 0.956 – 0.986) protocols (Supplementary Table 6). The FFPE.K.TotalRNA protocol had the highest observed Spearman correlation coefficient in 13 of these 15 comparisons.
DISCUSSION
Overall, FFPE RNA-seq data reliably captured transcriptional profiles and differences in tumor phenotype-based expression in breast cancer samples, just not quite as well as FF RNA-seq data. Principal component analyses demonstrated the following order of variables influencing gene expression measurements from RNA-sequencing: i) whether the library preparation protocol used exon capture for coding region (CR); ii) whether the samples was from FF tissue or FFPE tissue; and the biological phenotype of the breast cancer based on hormone receptors and HER2 receptor status (Figure 2). Generally, we observed small differences in performance between non-CR protocols. However, even small differences can have important effects on large-scale genomic data for biomarker discovery, validation or subsequent diagnostic development. Nevertheless, we identified one protocol, FFPE.K.TotalRNA, with consistently good transcript coverage uniformity and continuity; most concordant expression; and least differential expression when compared to the different non-CR protocols with fresh tissue. This protocol utilized RNase H-based rRNA depletion method and outperformed another similar TotalRNA-seq method, which used RiboZero to remove rRNA. It had a reasonable requirement of total RNA input (100ng) for FFPE biopsy samples.
The first translational research scenario that we posed, in the Background section, considered the best pairing of protocols that would enable discovery using FF samples with intention to later translate for use with FFPE samples. Overall, we favor the K.TotalRNA as consistently best, or close to best performance with FFPE protocols, when compared to FF.mRNA, FF.I.TotalRNA, or FF.K.TotalRNA as reference FF protocols. This interpretation was supported by the quality of read coverage, pattern of coding sequence expression, translation of overall or phenotype-related gene expression profiles and prognostic signatures.
The CR protocols yielded concordant results, but very different from all other (non-CR) protocols. So a CR protocol used for discovery (FF) would preclude other protocols for later translation to FFPE samples (Figures 2, 4). Also, changes to the population of exon capture probes within a commercial kit over time could be a potential risk to this approach.
The most generalizable results for discovery research from FFPE samples were obtained using the Total.RNA protocols without exon capture. Although similar, the FFPE.K.TotalRNA protocol produced slightly stronger results than the FFPE.I.TotalRNA protocol. So for our second scenario, we prefer the K.TotalRNA protocol for best representation of the transcriptome in FFPE samples utilized for discovery research – aiming to represent the transcriptional information that FF samples would have provided.
Our third translational research scenario involves the translation of an existing gene expression signature that was previously developed using a different method (e.g. microarray) or a particular RNA-seq protocol. Again, the FFPE.K.TotalRNA protocol had the best performance for total transcriptional profile, coding sequence, phenotypic discrimination, and for specific gene expression signatures.
The formalin fixation process is known to cause cross-linkage between nucleic acids and proteins, and mono-methyl addition to the RNA bases [2]. Although we tested a method of chemical de-modification of total RNA, our results showed negligible effect and argue against the incorporation of this method for RNA-seq of FFPE samples (Figure 4). However, we did not test the performance of potential protocols combining de-modification with sRNA alone or TotalRNA methods, due to limited tumor sample total RNAs.
The inclusion of random and dT primers, and the T7 promoter region (sRNA protocol) to simulate the FF.mRNA protocol produced good concordance overall, but introduced a high number of non-concordant mapped reads, non-uniformity and discontinuity of read coverage across the transcriptome.
Limitations to our study include small sample size (although cancers were selected to represent biologic diversity), optimally short time to fixation of tissues and possibly as a result a modest degree of degradation of FFPE samples (DV200 ranges from 65% to 85%), optimal amount of input RNA used for non-CR protocols (at least 100ng) and lack of generalizability (single institution conditions of tissue processing). Also, the effects of long-term storage of FFPE samples could not be tested – but would be expected from a completed clinical trial. Also, several of the cases had prolonged storage of cut FFPE sections (at 4°C) until RNA purification. This could have compromised the FFPE library protocols for this comparison, but can also be viewed as stress-testing the FFPE-derived RNA. Notwithstanding these limitations, we believe that the results from this study will be helpful to translational researchers as they consider how to obtain accurate gene expression by applying RNA-seq methods to FFPE tumor samples.
Supplementary Material
Acknowledgments
JL and WW were supported by the Cancer Prevention Research Institute of Texas through grant number RP130090. WW was also supported by the U.S. National Cancer Institute through grant numbers 1R01CA174206, 1R01 CA183793 and P30 CA016672. TPS was supported by National Health and Medical Research Council Program Grant 1054618. The tissue processing, library preparation, and sequencing was funded by a research grant from the Breast Cancer Research Foundation to WFS.
Abbreviations
- FFPE
formalin-fixation and paraffin-embedding
- FF
fresh-frozen
- RNA-seq
RNA-sequencing
- CR
coding-region targeted
- CDS
coding DNA sequences
- deM
de-modification of the methyl adducts and methylene bridges
- PCR
polymerase chain reaction
- PCA
principal component analysis
- ROC
receiver operating characteristic curves
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
WW and WFS designed the research; CF performed experiments; JL, WW and TPS analyzed data; All authors interpreted the results; JL, WFS, and WW prepared the manuscript. All authors edited, read and approved the final manuscript.
References
- 1.Penland SK, Keku TO, Torrice C, et al. RNA expression analysis of formalin-fixed paraffin-embedded tumors. Lab Invest. 2007;87(4):383–91. doi: 10.1038/labinvest.3700529. [DOI] [PubMed] [Google Scholar]
- 2.Masuda N, Ohnishi T, Kawamoto S, et al. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27(22):4436–43. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fumagalli D, Blanchet-Cohen A, Brown D, et al. Transfer of clinically relevant gene expression signatures in breast cancer: from Affymetrix microarray to Illumina RNA-Sequencing technology. BMC Genomics. 2014;15:1008. doi: 10.1186/1471-2164-15-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao W, He X, Hoadley KA, et al. Comparison of RNA-Seq by poly (A) capture, ribosomal RNA depletion, and DNA microarray for expression profiling. BMC Genomics. 2014;15:419. doi: 10.1186/1471-2164-15-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matranga CB, Andersen KG, Winnicki S, et al. Enhanced methods for unbiased deep sequencing of Lassa and Ebola RNA viruses from clinical and biological samples. Genome Biol. 2014;15(11):519. doi: 10.1186/s13059-014-0519-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adiconis X, Borges-Rivera D, Satija R, et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat Methods. 2013;10(7):623–9. doi: 10.1038/nmeth.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norton N, Sun Z, Asmann YW, et al. Gene expression, single nucleotide variant and fusion transcript discovery in archival material from breast tumors. PLoS One. 2013;8(11):e81925. doi: 10.1371/journal.pone.0081925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Exon capture protocol. http://www.illumina.com/content/dam/illumina-marketing/documents/products/datasheets/datasheet-truseq-rna-access.pdf.
- 9.Method for optimized isolation of RNA from fixed tissue. 2009127350 A1. WO. http://www.google.com/patents/WO2009127350A1?cl=en.
- 10.Hatzis C, Sun H, Yao H, et al. Effects of Tissue Handling on RNA Integrity and Microarray Measurements From Resected Breast Cancers. Journal of the National Cancer Institute. 2011 doi: 10.1093/jnci/djr438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonin S, Stanta G. RNA Temperature Demodification. Chapter of Guidelines for Molecular Analysis in Archive Tissues. 2011:67–69. [Google Scholar]
- 12.Li P, Conley A, Zhang H, et al. Whole-Transcriptome profiling of formalin-fixed, paraffin-embedded renal cell carcinoma by RNA-seq. BMC Genomics. 2014;15:1087. doi: 10.1186/1471-2164-15-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zwiener I, Frisch B, Binder H. Transforming RNA-Seq data to improve the performance of prognostic gene signatures. PLoS One. 2014;9(1):e85150. doi: 10.1371/journal.pone.0085150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byron SA, Van Keuren-Jensen KR, Engelthaler DM, et al. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nature Reviews Genetics. 2016;17(5):257–271. doi: 10.1038/nrg.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. New England Journal of Medicine. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 16.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loi S, Haibe-Kains B, Majjaj S, et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(22):10208–10213. doi: 10.1073/pnas.0907011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Symmans WF, Hatzis C, Sotiriou C, et al. Genomic Index of Sensitivity to Endocrine Therapy for Breast Cancer. Journal of Clinical Oncology. 2010;28(27):4111–4119. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van't Veer LJ, Dai HY, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.