Abstract
We have developed a protocol to purify RNA from DSS (Dextran Sulfate Sodium)-treated mouse tissues. This method, which prevents downstream inhibition of q-RT-PCR observed in DSS-treated tissues, relies on successive precipitations with lithium chloride.
Keywords: Dextran sodium sulfate, Colitis, Lithium chloride, RNA, q-RT-PCR
[Background]
Dextran Sulfate Sodium (DSS) is very commonly used in laboratories to induce colitis in rodents. Specifically, it mimics the clinical and histological features of human Inflammatory Bowel Disease (IBD) with Ulcerative Colitis (UC) characteristics. DSS is diluted in the drinking water and penetrates tissues. We have observed that contamination of RNA extracts with DSS prevented successful subsequent amplification processes from the colon and small intestine, but also blood and other tissues obtained from DSS-treated animals. We had previously shown that the presence of DSS in the samples inhibited reverse transcription and polymerase chain reaction amplification (Viennois et al., 2013). This inhibitory effect was observed in a dose depended manner by Kerr et al. and they suggested a poly-A-purification based technique to remove DSS from total RNA extract (Kerr et al., 2012). We hereby propose another efficient and economical method for purifying total RNA extracts from DSS traces based on lithium chloride (LiCl) precipitations. This method has been extensively used in our laboratory as well as in others (Chassaing et al., 2012, Li et al., 2016); however, no attempt has been taken to document the procedure in detail. Therefore, we provide a detailed description of LiCl purification procedure of total RNA primarily isolated from DSS-treated murine tissue with another method (Trizol, Spin column-based nucleic acid purification…).
Materials and Reagents
Pipette tips (0.1-10 μl, 1-200 μl, 100-1,000 μl)
Eppendorf Safe-Lock Tubes, 1.5 ml (Eppendorf, catalog number: 022363204)
8 M lithium chloride (LiCl) (SIGMA Lithium Chloride Solution, 8 M Solution, Sigma-Aldrich, catalog number: L7026-100ML, 090M8728)
RT-PCR Grade Water (Thermo Fisher Scientific, Ambion™, catalog number: AM9935)
Pure 100% ethanol (Decon Labs, catalog number: 2716)
3 M sodium acetate, pH 5.2 (see Recipes)
70% ethanol (see Recipes)
Equipment
Pipettes 0.5-10 μl, 10-100 μl and 100-1,000 μl (Eppendorf, model: Research® plus, Variable Adjustable Volume Pipettes)
Refrigerated centrifuge (Thermo Fisher Scientific, Thermo Scientific™, model: Sorvall™ Legend™ Micro 21R, or equivalent)
Multi-Mode Microplate Reader (BioTek Instruments, model: BioTek™ Synergy™ 2)
−20 °C freezer
Procedure
RNAs are in solution in RNase-free water after previous isolation with any method (Trizol, Spin column-based nucleic acid purification).
Add 0.1 volume of 8 M LiCl solution to 1 volume of RNA solution (i.e., 5 μl of 8 M LiCl for 50 μl RNA solution).
Mix well by pipetting up and down and incubate on ice for 2 h.
After the incubation, centrifuge at 14,000 x g for 30 min, at 4 °C.
Discard the supernatant and dissolve the pellet (that might be almost invisible depending on the initial quantity of RNA) in 200 μl RNase-free water.
Repeat steps from 1 to 5. Briefly, perform a second precipitation with 0.1 volume of LiCl (20 μl), mix by pipetting and incubate the solution for 2 h on ice, centrifuge and dissolve the pellet in 200 μl of water.
Add 0.1 volume (i.e., 20 μl) of 3 M sodium acetate (pH = 5.2) and 2.0 volumes (i.e., 400 μl) of −20 °C prechilled 100% ethanol to 1 volume of RNA solution. Incubate at −20 °C for 30 min.
Centrifuge at 14,000 × g for 30 min at 4 °C.
Discard the supernatant and wash the pellet with 100 μl of −20 °C prechilled 70% ethanol.
Centrifuge at 14,000 × g for 10 min at 4 °C.
-
Remove the supernatant carefully with a pipet without disturbing the pellet which might be invisible at this step.
Note: The supernatant should be removed carefully. The expected location of the pellet (which is determined by the position of the tube in the centrifuge) must be considered for pipetting up the supernatant.
Let the pellet air dry at room temperature for 5-10 min.
Dissolve the pellet in RNase free water in a volume smaller or equal to the initial volume of RNA solution in Step 1.
Determine RNA yield by measuring the absorbance peak at 260 nm with a multi-mode microplate reader. The absorbance ratio at 260/280 nm is also determined to evaluate the purity of RNA with an acceptable ratio from 2 to 2.2.
RNAs from DSS-treated tissues can now be used for cDNA synthesis and qPCR according to standard protocols.
Data analysis
Agarose gel electrophoresis demonstrates that, before LiCl precipitation, housekeeping gene 36B4 cDNA amplification products are obtained from non-DSS-treated mice as indicated by a clear band while amplification is inhibited in DSS-treated mice as indicated by the smear (Figure 1). After LiCl precipitation, cDNA obtained from non-DSS and DSS treated samples were both successfully amplified, as attested by a distinct band (Figure 1).
Figure 1. LiCl purification allows amplification of cDNA from DSS-treated tissues.
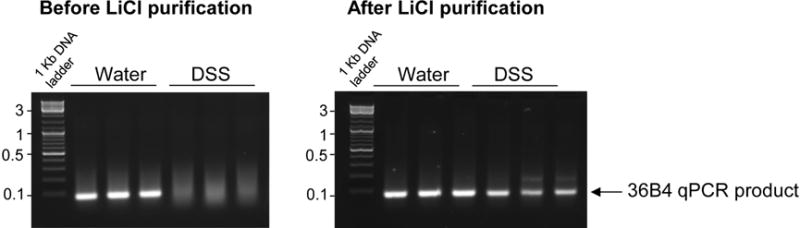
Total RNAs were extracted from colonic tissues obtained from mice treated with DSS diluted at 3% in the drinking water or water controls. After cDNA synthesis, qPCR for the housekeeping gene 36B4 was performed and the product of amplification was visualized by electrophoresis. The presence of a smear indicates that the amplification of 36B4 cDNA was inhibited in DSS-treated mice before LiCl purification. After purification of the DSS- and non-DSS RNA samples with LiCl, clear bands indicate that 36B4 cDNA was successfully amplified from both DSS- and non-DSS samples.
Notes
This protocol might be associated with a decrease of RNA yield. The volume of re-suspension in Step 13 might be adjusted according to the initial volume and should be the same or smaller (if the RNA should be at a similar or higher concentration than before the LiCl purification).
Due to the partial loss of RNA that might occur during the successive precipitations, the protocol should be performed on RNA samples with an initial concentration of no less than 200-300 ng/μl. The best results are obtained on samples from 500 ng/μl initial RNA concentration and above.
Within the same experiment, all groups of samples, whether they were obtained from mice administered with DSS or from the water control group, should be equally subjected to the LiCl purification process.
Recipes
-
3 M sodium acetate, pH 5.2 (for 100 ml)
24.61 g of sodium acetate
100 ml of Milli-Q® ultra-pure water
Adjust pH to 5.2 with HCl
-
70% ethanol
70% of 100% ethanol
30% of Milli-Q® ultra-pure water
Acknowledgments
EV is a recipient of the Career Development Award from the Crohn’s and Colitis Foundation. DM is a recipient of a Research Scientist Award from the Department of Veteran Affairs. This work was supported by grants from the National Institutes of Health of Diabetes and Digestive and Kidney (DK116306, DK107739; DK071594 to DM). We thank Samantha Spencer for proofreading the manuscript. This protocol was adapted from published work by Cathala et al. (1983) and Viennois et al. (2013). The authors declare no conflicts of interest within this work.
References
- Cathala G, Savouret JF, Mendez B, West BL, Karin M, Martial JA, Baxter JD. A method for isolation of intact, translationally active ribonucleic acid. DNA. 1983;2(4):329–35. doi: 10.1089/dna.1983.2.329. [DOI] [PubMed] [Google Scholar]
- Chassaing B, Srinivasan G, Delgado MA, Young AN, Gewirtz AT, Vijay-Kumar M. Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS One. 2012;7(9):e44328. doi: 10.1371/journal.pone.0044328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr TA, Ciorba MA, Matsumoto H, Davis VR, Luo J, Kennedy S, Xie Y, Shaker A, Dieckgraefe BK, Davidson NO. Dextran sodium sulfate inhibition of real-time polymerase chain reaction amplification: a poly-A purification solution. Inflamm Bowel Dis. 2012;18(2):344–348. doi: 10.1002/ibd.21763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Xiao HT, Hu DD, Fatima S, Lin CY, Mu HX, Lee NP, Bian ZX. Berberine ameliorates chronic relapsing dextran sulfate sodium-induced colitis in C57BL/6 mice by suppressing Th17 responses. Pharmacol Res. 2016;110:227–239. doi: 10.1016/j.phrs.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Viennois E, Chen F, Laroui H, Baker MT, Merlin D. Dextran sodium sulfate inhibits the activities of both polymerase and reverse transcriptase: lithium chloride purification, a rapid and efficient technique to purify RNA. BMC Res Notes. 2013;6:360. doi: 10.1186/1756-0500-6-360. [DOI] [PMC free article] [PubMed] [Google Scholar]


