Abstract
The study objective was to determine the prevalence of Staphylococcus aureus colonisation in the nares and oropharynx of healthy persons, and identify any risk factors associated with such S. aureus colonisation. 263 participants (177 adults and 86 minors) comprising 95 families were enrolled in a year-long prospective cohort study from one urban and one rural county in eastern Iowa, USA, through local newspaper advertisements and email lists and through the Keokuk Rural Health Study. Potential risk factors including demographic factors, medical history, farming, and healthcare exposure were assessed. Among the participants, 25.4% of adults and 36.1% minors carried S. aureus in their nares and 37.9% of adults carried it in their oropharynx. The overall prevalence was 44.1% among adults and 36.1% for minors. Having at least one positive environmental site for S. aureus in the family home was associated with colonisation (prevalence ratio: 1.34, 95% CI: 1.07–1.66). The sensitivity of the oropharyngeal cultures was greater than that of the nares cultures (86.1% compared to 58.2% respectively). In conclusion, the nares and oropharynx are both important colonisation sites for healthy community members and the presence of S. aureus in the home environment is associated with an increased probability of colonisation.
Keywords: S. aureus, MRSA, Epidemiology, Community
INTRODUCTION
Staphylococcus aureus colonises approximately 30% of the population [1] with the anterior nares historically considered the most frequent carriage site [2, 3]. Colonised persons carry the bacterium asymptomatically and may transmit it to susceptible persons either directly or through contact with fomites. While such colonisation often does not harm the host, it increases the risk of infections in both community and hospital settings [4].
S. aureus is a common cause of infections in hospitals and in the community. In 2014, the Active Bacterial Core surveillance program reported that methicillin-resistant S. aureus (MRSA) caused 72,444 invasive infections in the USA, 16,522 of which were community onset and although overall MRSA infections decreased from 2013 to 2014 by 5.36%, community-associated infections increased by 1.57%[5]. It has been suggested understanding the patient’s environment, household transmission dynamics, and colonisation among persons living in the community are key to reducing community-associated infections [6].
While many prior studies have assessed S. aureus colonisation in healthcare settings[7, 8], few have addressed colonisation in the community[9] and more rarely among persons in rural areas[10] who make up 20% of the United States population[11]. Prevalence estimates for S. aureus oropharyngeal colonisation in healthy community populations has ranged from 7.7% to 56.7% and the proportion carrying S. aureus in their oropharynx, but not in their nares, has ranged from 6% to 22.7% [12–16]. Moreover, there is a paucity of data on differences in S. aureus colonisation prevalence based upon population density owing to the lack of studies designed specifically to look for these differences.
To address this knowledge gap, we assessed the prevalence of S. aureus carriage in one rural and one urban population in Iowa to better define colonisation prevalence and associated risk factors in the community setting. Specifically, we recorded the prevalence of nasal and oropharyngeal carriage, risk factors for colonisation, and applied molecular epidemiological tools to characterise S. aureus isolates recovered from participants and their household environment.
MATERIALS AND METHODS
Study design and sample collection
Families from Johnson and Keokuk Counties, Iowa that met the study inclusion criteria were enrolled in a prospective cohort study between October 6, 2011 and January 4, 2012. These counties were chosen to reflect an urban (Iowa City, within Johnson County) and a rural (Keokuk County) population in Iowa [11]. This paper presents the baseline data and cohort description for all participants in the cohort.
Participants were recruited, either through local newspaper advertisements and email lists, and/or for rural families through their existing participation in the Keokuk County Rural Health Study [17]. Eligibility of participants was assessed and a scheduled visit was arranged for family enrollment. Participants in Keokuk County were contacted initially by letter and later by phone to determine eligibility and schedule the enrollment visit. Trained staff enrolled eligible families in their homes, administered questionnaires, collected environmental samples, and trained adult participants to swab their nares and oropharynx.
Study inclusion criteria were: must be able to provide consent, assent, or have parents willing to provide consent; speak English; be willing to complete the enrollment questionnaire as well as the weekly follow-up questionnaires. The University of Iowa institutional review board approved the study protocols. After consenting, all participants completed questionnaires providing detailed information on demographic factors, medical history, f arming exposure, healthcare exposure, and other possible risk factors. Farming exposure for minors, designated as “worked with livestock”, refers to work performed for pay as well as work performed as a part of family responsibilities and chores.
Isolation and Identification of S. aureus
For self-swabbing, participants were instructed to insert the swab into their nostril so the bottom part of the cotton was just inside the anterior naris, circle the swab three times and then switch nostrils and repeat. To sample their oropharynx, participants were instructed to swab the roof of their mouth beyond the soft palate, but not far enough back to cause a gag reflex. Additionally, parents were trained to swab the nares of participating children in their household. Samples were collected using BBL CultureSwabs with Liquid Stuart Medium (Becton, Dickinson & Co., Sparks MD, USA) and transported on ice directly to the Center for Emerging Infectious Diseases (CEID).
Three-inch by four-inch disposable duster cloths (Swiffer® Sweeper™ Dry Pad; Procter & Gamble, Cincinatti, OH, USA) were used to collect samples from six commonly touched sites (kitchen sink handle, oven knobs, refrigerator door handle, primary television remote, main bathroom light switch, and main bathroom toilet flush lever). Each site was sampled using a new, sterile cloth. Surfaces were wiped in all directions for one minute. The cloth was placed into a dry, sterile bag and placed on ice for transport to the CEID.
Swabs were inoculated into 5mL of Baird-Parker broth and incubated for 24 hours at 35°C. Environmental sample cloths were homogenised for one minute in 25mL of 1.0% peptone after which 5mL was removed and inoculated into 5mL of 2× Baird-Parker broth and incubated for 24 hours. All samples were plated onto Baird-Parker agar and BBL CHROMagar MRSA II (Becton, Dickinson & Co., La Jolla CA, USA) and following incubation at 35°C for 48 hours, presumptive S. aureus colonies were streaked onto Columbia CNA agar with 5% sheep blood (Becton, Dickinson & Co) and incubated for 24 hours.
S. aureus was confirmed with the catalase, coagulase, and Pastorex Staph Plus rapid latex agglutination tests (Bio-Rad, Redmond, Washington, USA) and all isolates were subjected to molecular testing.
Molecular testing
Bacterial genomic DNA was extracted with the Promega Wizard Genomic DNA purification kit (Promega Corporation, Madison WI, USA). Multiplex polymerase chain reaction (PCR) was used to assess the presence of the mecA gene, the S. aureus specific nuc gene, and 16S rRNA gene to confirm the identity of S. aureus [18]. PCR was also used to detect the Panton-Valentine leukocidin (PVL) [19] and to amplify the spa gene [20] for strain typing using the Ridom StaphType software (Ridom GmbH, Germany). Isolates unable to be typed after three attempts were labeled as non-typable (NT) [21]. The Based Upon Repeat Pattern (BURP) algorithm was used to identify and characterise genetic clusters [22]. All molecular procedures included known positive (USA300 SF8300 strain) and negative (sterile water) controls.
As carriage of the same S. aureus strain by multiple members of the same family artificially inflates the strain prevalence, we adjusted for duplicate strains within-family colonisation, by including only one isolate of a specific strain per family when calculating spa type prevalence as well as for mecA and PVL genes. Isolates were considered to be the same strain if they had the same spa type, mecA and PVL profiles.
Statistical analysis
Data were analysed using SAS version 9.3 and R version 2.15.3. The demographics of participants were compared using the Student’s T-Test (age), the Chi-Squared Test (gender), and the Fisher’s Exact Test (race). Random effects logistic regression was utilised to model the effect of possible risk factors on the probability of colonisation while accounting for correlation within families and counties[23]. Risk factors were modeled separately for adults and minors as minors submitted only nasal swabs. The adult model was adjusted for age while the minor’s model was adjusted for age, gender, and interaction between age and gender. Conditional prevalence ratios (PRs) were estimated from the random effects logistic regression model [23]. All associations were considered significant at ≤ 0.05.
Preliminary models run using proc glimmix with both family and county random effects indicated that county did not have an effect after the risk factors and the random effect for family were included (data not shown). The only random effect utilised within the analyses presented here was a family random effect. For each risk factor, proc glimmix was used to obtain starting values for the final model which used proc nlmixed, the latter allowed for the estimation of prevalence ratios (PR) rather than odds ratios as PRs have been shown to be truer estimates of the risk for non-rare outcomes [23]. PRs were estimated at the midpoint age for adults (43 years), and for minors were estimated for females at the midpoint age (10 years). Risk factors were assessed as dichotomous or categorical with the exception of house size, number of children, and environmental contamination, which were all modeled as discrete numeric variables. Individuals with a missing data point were removed from the specific analyses for that data point.
RESULTS
A total of 263 participants were enrolled (177 adults, 86 minors), comprising 95 families. The average age of adults was 43.2 years (range: 22.2–67.7 years) and 9.9 years (0.4–18.2 years) for minors. Adult participants were significantly older in the rural county than in the urban county (P= <0.001). One hundred and forty-four (54.7%) participants were female; three (2 adults, 1 minor) participants did not indicate gender. Most participants considered themselves White (92.7% of adults, 89.5% of minors), with 1.7% of adults and 5.8% of minors considering themselves Black/African American, and 5.6% of adults and 4.7% of minors indicating “other” (American Indian/Alaska native, Asian, Hispanic or Latino, or Native Hawaiian or other Pacific Islander) as their race. Gender and race did not differ significantly by county for adults and the distributions of age, gender, and race did not differ significantly for minors. Additional demographic characteristics are provided in Table S1.
S. aureus was detected on 147 (33.4%) of the 440 swabs collected. Of the adult participants, 11 (6.2%) carried S. aureus only in the nares, 33 (18.6%) only in the oropharynx, and 35 (19.8%) in both sites; across both sites, the prevalence of carriage among adults was 44.6% (Table 1). Minors provided nares samples only and 36.1% carried S. aureus. The prevalence of nasal carriage in adults was 23.1% in the urban county and 27.9% in the rural county; for minors, it was 40.8% in the urban county and 29.7% in the rural county. Oropharyngeal carriage prevalence was 47.3% and 27.9% in the urban and rural county, respectively. Overall, the sensitivity of nares cultures was 58.2% (46/79) and oropharyngeal cultures was 86.1% (68/79) (Table 2). In the urban county, oropharynx cultures proved more sensitive than nares cultures, but cultures of both sites were equally sensitive in the rural county.
Table 1.
Prevalence of S. aureus and MRSA isolated from participants by county
| MSSAa | MRSAb | Total S. aureus | |
|---|---|---|---|
| Adults | |||
| Urban county | 39/90 (43%) | 7/90 (7%) | 46/90 (51%) |
| Rural county | 31/87 (35%) | 2/87 (2%) | 33/87 (37%) |
| Total | 70/177 (40%) | 9/177 (5%) | 79/177 (45%) |
| Minors | |||
| Urban county | 19/49 (38%) | 1/49 (2%) | 20/49 (40%) |
| Rural county | 11/37 (29%) | 0/37 (0%) | 11/37 (29%) |
| Total | 30/86 (34%) | 1/86 (1%) | 31/86 (36%) |
Note: The numerator for adults was the number of participants with S. aureus isolated from the nares, the oropharynx, or both. The numerator for minors was the number of participants with S. aureus isolated from the nares. The denominator is the total number of participants by county for adults and minors.
MSSA = methicillin-susceptible S. aureus
MRSA = methicillin-resistant S. aureus
Table 2.
Sensitivity of screening the nares and the oropharynx to identify adult individuals who are colonised with S. aureus
| Urban county (n=46) | Rural county (n=33) | Total (n=79) | |
|---|---|---|---|
| Nares | 21/46 (45%) | 25/33 (75%) | 46/79 (58%) |
| Oropharynx | 43/46 (93%) | 25/33 (75%) | 68/79 (86%) |
Note: The denominator is the total number of adults colonised with S. aureus in their nares, their oropharynx, or both. The numerator is the number of adults identified by culture screening of the specific anatomical site.
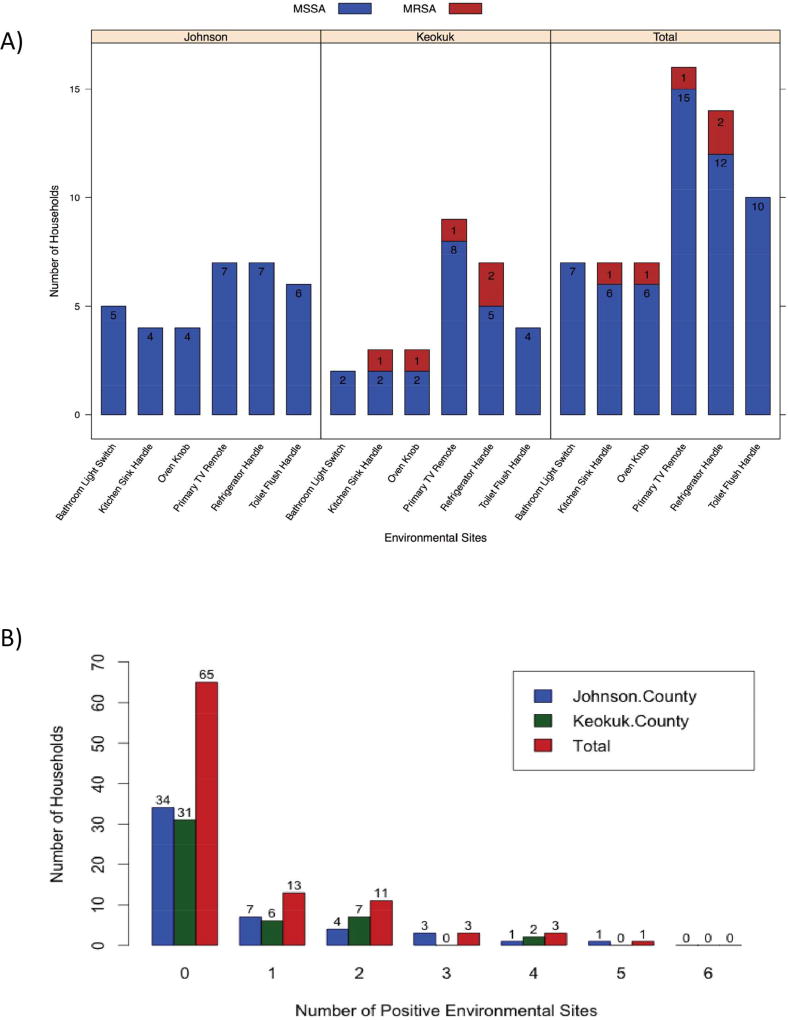
Six environmental sites that were most likely to be contacted by all members of the household in each home were screened for contamination (Figure 1) and S. aureus was not isolated from any of these sites in the majority (65/95, 68.4%) of households; 13 of 95 (13.7%) households had a single positive site. The most frequent positive sites were the primary TV remote (16/95, 16.8%) and the refrigerator handle (14/95, 14.7%), and at least one of the six sampled sites in each household was found positive at least once in both counties. MRSA was isolated from three (3/46, 6.5%) rural households (two houses with two positive sites and one house with a single positive site), but MRSA was not recovered from the environment in urban households.
Figure 1.
Prevalence of S. aureus environmental contamination.
(A) Prevalence at 6 sites in participant households stratified by county and presence of the mecA gene, which is indicative of methicillin-resistant S. aureus.
(B) Total number of environmental sites contaminated by S. aureus for each household by county.
After adjusting for age, there was a significant positive relationship between S. aureus colonisation among adults and the number of positive environmental sites in the participants’ homes (Table 3). The largest PR for the number of positive environmental sites was observed for participants living in homes with one positive environmental site compared with participants in homes with no positive environmental sites (PR: 1.34, CI: 1.07–1.66, P= 0.0095). The PRs decreased, but remained significant, for all other comparisons. Borderline positive associations of S. aureus colonisation were evident with race (PR: 1.64, CI: 0.98–2.74, P= 0.062), having a family member hospitalised in the past three months (PR: 1.82, CI: 0.96–3.47, P= 0.069), and having a heart condition (PR: 1.61, CI: 0.96–2.71, P= 0.07); likewise a borderline negative relationship of colonisation was found with a family member having a skin or soft tissue infection (PR: 0.36, CI: 0.11–1.18, P= 0.09). After adjustment for within-family colonisation, a significant positive relationship was observed between S. aureus colonisation and female and male minors with asthma, the number of children in a household, and the number of positive environmental sites (Table 3). Other factors not reaching statistical significance are included in Tables S1 and S2.
Table 3.
Bivariable analysis assessing the association between colonisation with S. aureus in the nares and/or oropharynx and participant demographics and risk factors
| Risk Factor | Number Positive (%) |
Total (N) | PRa | 95% CI | p-value |
|---|---|---|---|---|---|
| Adults (n=174) | |||||
| Age | --- | 177 | 0.98 | 0.97 –1.002 | 0.082 |
| Number of S. aureus positive environmental sites in the home | |||||
| Zero | 45 (36.8) | 122 | -ref- | -ref- | -ref- |
| One | 11 (47.8) | 23 | 1.34 | 1.07–1.66 | 0.009 |
| Two | 12 (66.7) | 18 | 1.26 | 1.10–1.43 | <0.001 |
| Three | 4 (80.0) | 5 | 1.18 | 1.11–1.26 | <0.001 |
| Four | 3 (75.0) | 4 | 1.12 | 1.09–1.16 | <0.001 |
| Five | 1 (50.0) | 2 | 1.08 | 1.03–1.13 | <0.001 |
| Minors (n=83) | |||||
| Number of Children | |||||
| One | 0 (0) | 1 | -ref- | -ref- | -ref- |
| Two | 3 (15) | 19 | 1.68 | 0.83–3.41 | 0.150 |
| Three | 13 (35) | 37 | 1.54 | 0.90–2.64 | 0.113 |
| Four | 13 (59) | 22 | 1.39 | 1.01–1.91 | 0.041 |
| Five | 0 (0) | 4 | 1.25 | 1.07–1.46 | 0.004 |
| Number of S. aureus positive environmental sites in the home | |||||
| Zero | 12 (22) | 53 | -ref- | -ref- | -ref- |
| One | 4 (44) | 9 | 1.68 | 1.07–2.64 | 0.025 |
| Two | 4 (57) | 7 | 1.53 | 1.08–2.18 | 0.018 |
| Three | 2 (50) | 4 | 1.38 | 1.10–1.72 | 0.005 |
| Four | 6 (85) | 7 | 1.24 | 1.09–1.41 | <0.001 |
| Five | 2 (66) | 3 | 1.14 | 1.04–1.25 | 0.006 |
| Asthma | |||||
| No | 21 (30) | 70 | -ref- | -ref- | -ref- |
| Yes | 8 (66) | 12 | 2.18 | 1.15–4.15 | 0.017 |
| NA | 1(100) | 1 | NA | NA | NA |
PR = prevalence ratio. For adults, PRs were adjusted for age and estimated at age=43.
For minors, PRs were adjusted for the age, gender, and age by gender interaction. Estimates for PR are calculated at the values of gender=Female and age=10. For all discrete numeric variables, PR estimates are calculated as each discrete value over the previous discrete value (i.e. Prevalence ratio for number of positive environmental sites =2 was calculated as prevalence given positive environmental sites =2/ prevalence given positive environmental sites =1). For dichotomous variables, PR estimates are estimated for prevalence given yes over prevalence given no.
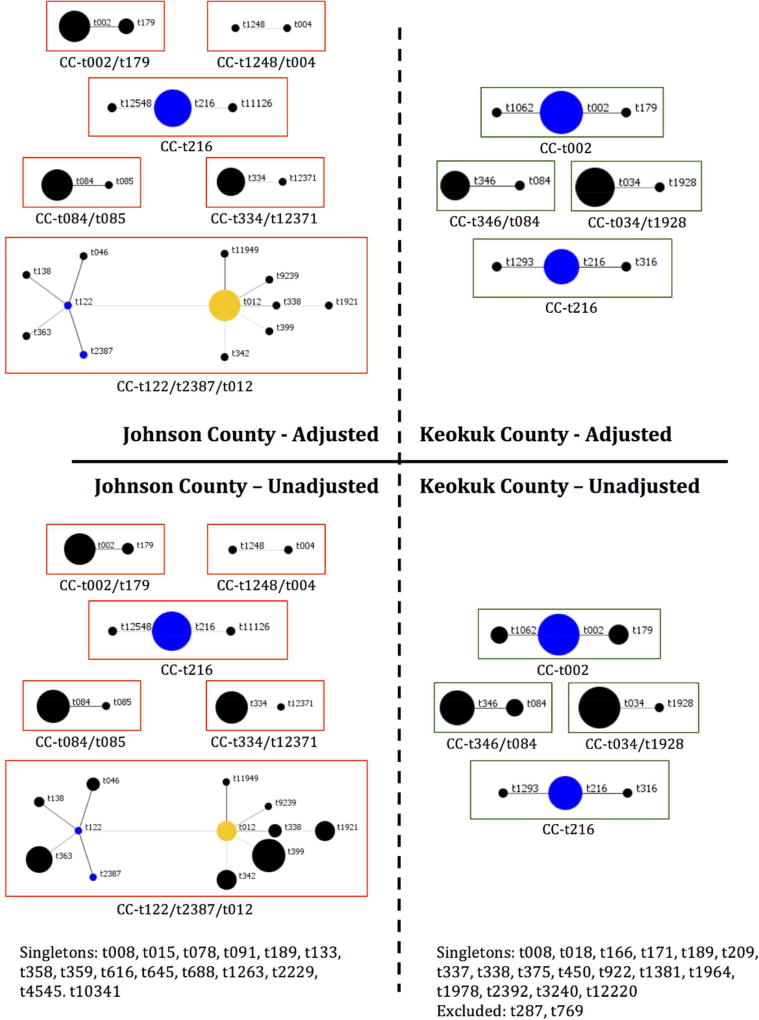
Of the 147 S. aureus isolates, there were 57 unique spa types, t he most common being t002 (n=21), t008 (n=21), t034 (n=11), and t216 (n=10). After adjusting, the number of unique isolates was reduced to 98 with t002 (n=11), t008 (n=8), and t216 (n=8) the most prevalent. Six clusters of spa types were identified among the isolates from the urban county compared with four clusters among rural (Figure 2), indicating there is a the large diversity of circulating spa types in both locations. BURP analysis identified 28 spa types with a genetic cost greater than four from any clustered spa type, indicating these spa types were not likely related to any cluster and thus were excluded from Figure 2 [22]. Also, two spa types were excluded as they had fewer than 5 repeat sequences. The genetic relatedness of all spa types by county are shown using a minimum spanning tree (Figure S1), where the genetic distance between isolates is represented by the thickness of connecting lines, and the proportion of isolates for each spa type is represented by circle size. There were no observable differences between the two counties.
Figure 2.
BURP analysis of S. aureus spa typing by county.
Unadjusted proportions represent all 147 total isolates; adjusted proportions control for familial clustering of clonal isolates. Each circle represents a single spa type and size of the circle is proportionate to the relative number of isolates of that spa type. Blue circles indicate the putative founder, yellow circles are secondary putative founders.
Ten (11.6%) and two (3.3%) isolates from the urban and rural counties, respectively, carried the mecA gene (Table 4) and after adjustment for within-family colonisation, this was reduced to seven isolates (12.5%) from the urban county and one (2.4%) from the rural county (Figure S1b). The PVL gene was not identified in urban isolates but was present in seven (11.5%) rural county isolates. After adjustment for within-family colonization, five (11.9%) isolates from the rural county had the PVL gene.
Table 4.
Molecular characteristics of S. aureus isolates from participants
| Unadjusted mecA | Adjusted mecA | Unadjusted PVLa | Adjusted PVLa | |
|---|---|---|---|---|
| Urban county | 10/86 (11%) | 7/56 (12.5%) | 0/86 (0%) | 0/56 (0%) |
| Rural county | 2/61 (3%) | 1/42 (2%) | 7/61 (11%) | 5/42 (11%) |
| Total | 12/147 (8%) | 8/98 (8%) | 7/147 (4.8%) | 5/98 (5%) |
Note: Unadjusted values include all S. aureus isolates from participants broken down my county. Adjusted values include only a single copy of isolates with the same molecular profile (same spa type, and presence/absence of mecA and PVL) to account for familial clustering and the presence of clonal isolates in the nares and oropharynx of adult participants.
PVL = Panton-Valentine leukocidin
DISCUSSION
The overall prevalence of S. aureus colonisation was higher (44.1%) than most prior community-based studies [9] and of hospitalised patients[7, 8], whereas, the nasal carriage rate (25.4%) was similar to that reported by others[1, 7–9]. Our higher overall carriage rate is likely due to the addition of oropharyngeal swabs as carriage at this site was markedly more frequent than the nares in adults (37.9% and 25.4%, respectively). This observation supports the statement by Mertz et al. that “throat carriage may indeed be more common among healthy individuals than among individuals who are exposed to the health care system”[16]. Moreover, our results corroborate those of Hamdan-Partida et al. [15] who found 46.5% of healthy workers in a Mexican community had S. aureus oropharyngeal carriage while 37.1% were nasal carriers [15]; these prevalences compare favourably with our finding of 47.3% and 29.3% for nasopharyngeal and nasal carriage, respectively. The difference in nasal carriage between the studies is possibly due to their inclusion of a greater proportion of study participants being minors [15], which generally have higher S. aureus carriage rates [24] compared with working age adults [9], as well as an urban population. The similarity to the urban county – and not the rural county – may be due to differences in population density, family size, and environmental exposures between the two counties, and is an association worthy of further study to elucidate this complex relationship. Two other studies have also found lower colonisation prevalence among persons in the community with reported rates for oropharyngeal carriage of 17.4% [13] and 10.8% [14], and 17.5% and 21.2% for nasal carriage [13,14]. Of note, the estimates of oropharyngeal colonisation among persons in the community provided by our study and these other studies are equal to or greater than the colonisation prevalences of 5.8% to 30% frequently observed in healthcare settings [25–29]. Additionally, the positive association between S. aureus colonisation and prevalence or exacerbation of asthma in our study is consistent with a number of recent publications [30–32], and warrants follow up to further interrogate this potential risk factor and elucidate the mechanism for asthma development in the presence of S. aureus colonisation
PVL prevalence was low (5.1%) after adjusting for within-family colonisation. This differs markedly from Wardyn et al. [13] who reported a PVL prevalence of 31% among S. aureus isolates colonising rural Iowans. The adjusted their estimates if a participant was colonised in both the nares and the oropharynx, but did not account for strains shared by family members, as was addressed in the current study. In contrast to 11.9% of isolates from the rural county that were PVL positive, none were found in the urban county. Similar to the population studied by Wardyn et al [13], Keokuk County is rural, but our adjusted prevalence was still substantially lower than reported by them.
Oropharyngeal cultures identified a substantially higher percentage of S. aureus carriers than nasal cultures (86.1% vs. 58.2%). This may help explain why Bradley [34] found greater than 40% of decolonised persons who were treated with intranasal antimicrobials became recolonised with their initial strains [33]. Other investigators have found the oropharynx to be difficult to decolonise with topical antimicrobials applied directly to the oropharynx [34] or with indirect intranasal antimicrobial application [35]. This high recolonisation rate and the importance of the oropharynx as a colonization site in healthy persons suggest that investigators evaluating the efficacy of decolonisation protocols should assess recolonisation rates of both the nares and oropharynx.
Three of the 109 (2.8%) colonised persons, each from different families, harboured a strain of spa type t034, which has been associated with livestock exposure, particularly swine[36]. All four participants were from the rural county, two were actively farming and one had previously farmed and had an actively farming family member; the fourth individual had no discernible livestock exposure.
The two households with MRSA environmental contamination had at least one family member colonised with an isolate of the same spa type but lacking the mecA gene. This finding is surprising as it suggests the mecA gene could be lost when an isolate moves from the environment to humans [37–39]. Alternate hypotheses are: these isolates are not truly clonal and have the same spa type by chance; or the strain acquired mecA in the environment following human shed. Extended storage at −20°C is unlikely to cause loss of the mecA gene [40] as samples from humans and the environment were stored together and the number of freeze-thaw cycles did not differ.
Environmental contamination has been postulated to be an important mode of S. aureus transmission, albeit less important than direct person-to-person transmission. Importantly, Alam et al. found the household to be a hotspot for transmission of MRSA isolates causing infections but did not discuss the possibility of environmental contamination as a fomite [41], thus we evaluated environmental contamination to gain insights into the role the environment plays in colonisation. In our population, having at least one positive environmental site had the strongest association with colonisation for both adults and minors, indicating it may play a greater role in transmission than previously thought. Our study did not elucidate the direction of transmission (i.e. environment to person or person to environment) or whether there is a dose-response of increased environmental contamination associated with increased colonisation prevalence, due to the cross-sectional nature of this study and the small number of individuals in houses with greater than one contaminated environmental site, respectively. A contaminated environment may also be a proxy for other risk factors, such as hygiene standards, that we did not assess directly. Additional research is needed to further characterise this association to identify which sites within a household are most commonly contaminated and/or associated with colonisation. Further research is also needed to assess whether fomites, aerosolisation, o r settled dust aid the transmission of S. aureus in homes [42].
A potential limitation of this study is the use of different recruitment approaches for each county. It is possible these strategies led to selection bias, however, as individuals are unlikely to know if they are colonised with S. aureus it is unlikely that such differences had any impact on an individual’s decision to enroll in the study. Additionally, participants were asked if anyone in their household had had a skin and soft tissue infection or S. aureus or MRSA infection in the past three months. The proportion of individuals responding yes was not statistically different between the two counties, reducing the possibility that participants were motivated by S. aureus colonization status or knowledge.
In conclusion, S. aureus was isolated from both the nares and oropharynx of healthy community members, with the oropharynx being the more common site of colonisation. We observed a higher prevalence of colonisation than previously reported and postulate that this was due to the addition of oropharyngeal swabs. Environmental contamination was the factor most strongly associated with S. aureus colonisation. Previous studies of S. aureus colonisation have focused on patients in healthcare settings or on other high-risk groups and thus may not be applicable to community members. Our findings have implications for efforts to decrease community-acquired S. aureus infections and, thus, warrant additional studies.
Supplementary Material
Acknowledgments
Funding
This project was supported by AFRI food safety grant no. #2011-67005-30337 from the USDA National Institute of Food and Agriculture (TCS).
The authors would like to thank Elizabeth Chrischilles and Kelley Donham for their invaluable guidance in the analysis and preparation of this manuscript.
References
- 1.Gorwitz RJ, et al. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. The Journal of infectious diseases. 2008;197(9):1226–1234. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- 2.Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clinical microbiology reviews. 1997;10(3):505–520. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nouwen JL, et al. Predicting the Staphylococcus aureus nasal carrier state: derivation and validation of a "culture rule". Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2004;39(6):806–811. doi: 10.1086/423376. [DOI] [PubMed] [Google Scholar]
- 4.Wertheim HF, et al. The role of nasal carriage in Staphylococcus aureus infections. The Lancet infectious diseases. 2005;5(12):751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. [Last accessed: Mar 1, 2016];Active Bacterial Core Surveillance Report, Emerging Infections Program Network, Methicillin-Resistant Staphylococcus aureus. 2014 In, 2014. ( http://www.cdc.gov/abcs/reports-findings/survreports/mrsa12.pdf)
- 6.Malani PN. National burden of invasive methicillin-resistant Staphylococcus aureus infection. JAMA : the journal of the American Medical Association. 2014;311(14):1438–1439. doi: 10.1001/jama.2014.1666. [DOI] [PubMed] [Google Scholar]
- 7.Lipsky BA, et al. Factors affecting staphylococcal colonization among NIDDM outpatients. Diabetes care. 1987;10(4):483–486. doi: 10.2337/diacare.10.4.483. [DOI] [PubMed] [Google Scholar]
- 8.Lu PL, et al. Methicillin-resistant Staphylococcus aureus carriage, infection and transmission in dialysis patients, healthcare workers and their family members. Nephrology, dialysis, transplantation : official publication of the European Dialysis and Transplant Association - European Renal Association. 2008;23(5):1659–1665. doi: 10.1093/ndt/gfm806. [DOI] [PubMed] [Google Scholar]
- 9.Malik S, et al. Prevalence of community-associated methicillin-resistant Staphylococcus aureus colonization outside the healthcare environment. Epidemiology and infection. 2009;137(9):1237–1241. doi: 10.1017/S0950268809002222. [DOI] [PubMed] [Google Scholar]
- 10.Polgreen PM, et al. Epidemiology of methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus in a rural state. Infection control and hospital epidemiology : the official journal of the Society of Hospital Epidemiologists of America. 2006;27(3):252–256. doi: 10.1086/501537. [DOI] [PubMed] [Google Scholar]
- 11.Bureau USC. [Last accessed: June 9, 2016];2010 Census Urban and Rural Classification and Urban Area Criteria. In, 2010. ( https://www.census.gov/geo/reference/ua/urban-rural-2010.html)
- 12.Miller LG, et al. Staphylococcus aureus colonization among household contacts of patients with skin infections: risk factors, strain discordance, and complex ecology. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2012;54(11):1523–1535. doi: 10.1093/cid/cis213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wardyn SE, Forshey BM, Smith TC. High prevalence of Panton-Valentine leukocidin among methicillin-sensitive Staphylococcus aureus colonization isolates in rural Iowa. Microbial drug resistance (Larchmont, NY) 2012;18(4):427–433. doi: 10.1089/mdr.2011.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith TC, et al. Molecular and epidemiologic predictors of Staphylococcus aureus colonization site in a population with limited nosocomial exposure. American journal of infection control. 2012;40(10):992–996. doi: 10.1016/j.ajic.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 15.Hamdan-Partida A, Sainz-Espunes T, Bustos-Martinez J. Characterization and persistence of Staphylococcus aureus strains isolated from the anterior nares and throats of healthy carriers in a Mexican community. Journal of clinical microbiology. 2010;48(5):1701–1705. doi: 10.1128/JCM.01929-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mertz D, et al. Throat swabs are necessary to reliably detect carriers of Staphylococcus aureus. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2007;45(4):475–477. doi: 10.1086/520016. [DOI] [PubMed] [Google Scholar]
- 17.Stromquist AM, et al. The Keokuk County Rural Health Study. Journal of Agromedicine. 1997;4(3–4):243–248. doi: 10.1080/10599240801887850. [DOI] [PubMed] [Google Scholar]
- 18.Louie L, et al. Rapid detection of methicillin-resistant staphylococci from blood culture bottles by using a multiplex PCR assay. Journal of clinical microbiology. 2002;40(8):2786–2790. doi: 10.1128/JCM.40.8.2786-2790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lina G, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 1999;29(5):1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 20.Shopsin B, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. Journal of clinical microbiology. 1999;37(11):3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baum C, et al. Non-spa-typeable clinical Staphylococcus aureus strains are naturally occurring protein A mutants. Journal of clinical microbiology. 2009;47(11):3624–3629. doi: 10.1128/JCM.00941-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellmann A, et al. Based Upon Repeat Pattern (BURP): an algorithm to characterize the long-term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC microbiology. 2007;7:98. doi: 10.1186/1471-2180-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santos CA, et al. Estimating adjusted prevalence ratio in clustered cross-sectional epidemiological data. BMC medical research methodology. 2008;8:80. doi: 10.1186/1471-2288-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogaert D, et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. 2004;363(9424):1871–1872. doi: 10.1016/S0140-6736(04)16357-5. [DOI] [PubMed] [Google Scholar]
- 25.Bignardi GE, Lowes S. MRSA screening: throat swabs are better than nose swabs. The Journal of hospital infection. 2009;71(4):373–374. doi: 10.1016/j.jhin.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Bitterman Y, et al. Characterization of the best anatomical sites in screening for methicillin-resistant Staphylococcus aureus colonization. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2010;29(4):391–397. doi: 10.1007/s10096-009-0869-3. [DOI] [PubMed] [Google Scholar]
- 27.Collins J, et al. Review of a three-year meticillin-resistant Staphylococcus aureus screening programme. The Journal of hospital infection. 2011;78(2):81–85. doi: 10.1016/j.jhin.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 28.Harbarth S, et al. Is throat screening necessary to detect methicillin-resistant Staphylococcus aureus colonization in patients upon admission to an intensive care unit? Journal of clinical microbiology. 2007;45(3):1072–1073. doi: 10.1128/JCM.02121-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall C, Spelman D. Re: is throat screening necessary to detect methicillin-resistant Staphylococcus aureus colonization in patients upon admission to an intensive care unit? Journal of clinical microbiology. 2007;45(11):3855. doi: 10.1128/JCM.01176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludwig S, et al. Analysis of home dust for Staphylococcus aureus and staphylococcal enterotoxin genes using quantitative PCR. The Science of the total environment. 2017;581–582:750–755. doi: 10.1016/j.scitotenv.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis MF, et al. Staphylococcus aureus colonization is associated with wheeze and asthma among US children and young adults. The Journal of allergy and clinical immunology. 2015;135(3):811–813.e815. doi: 10.1016/j.jaci.2014.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis MF, et al. Effect of home exposure to Staphylococcus aureus on asthma in adolescents. The Journal of allergy and clinical immunology. 2018;141(1):402–405.e410. doi: 10.1016/j.jaci.2017.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradley SF. Eradication or Decolonization of Methicillin-Resistant Staphylococcus aureus Carriage: What Are We Doing and Why Are We Doing It? Clinical Infectious Diseases. 2007;44(2):186–189. doi: 10.1086/510395. [DOI] [PubMed] [Google Scholar]
- 34.Buehlmann M, et al. Highly effective regimen for decolonization of methicillin-resistant Staphylococcus aureus carriers. Infection control and hospital epidemiology : the official journal of the Society of Hospital Epidemiologists of America. 2008;29(6):510–516. doi: 10.1086/588201. [DOI] [PubMed] [Google Scholar]
- 35.Wertheim HF, et al. Effect of mupirocin treatment on nasal, pharyngeal, and perineal carriage of Staphylococcus aureus in healthy adults. Antimicrobial agents and chemotherapy. 2005;49(4):1465–1467. doi: 10.1128/AAC.49.4.1465-1467.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith TC, et al. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS One. 2009;4(1):e4258. doi: 10.1371/journal.pone.0004258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence C, et al. Consecutive isolation of homologous strains of methicillin-resistant and methicillin-susceptible Staphylococcus aureus from a hospitalized child. The Journal of hospital infection. 1996;33(1):49–53. doi: 10.1016/s0195-6701(96)90028-6. [DOI] [PubMed] [Google Scholar]
- 38.Inglis B, el-Adhami W, Stewart PR. Methicillin-sensitive and -resistant homologues of Staphylococcus aureus occur together among clinical isolates. The Journal of infectious diseases. 1993;167(2):323–328. doi: 10.1093/infdis/167.2.323. [DOI] [PubMed] [Google Scholar]
- 39.Wagenvoort JH, et al. Hospital outbreak of methicillin-resistant Staphylococcus aureus followed by an in vivo change to a mecA-negative mutant with loss of epidemicity. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2000;19(12):976–977. doi: 10.1007/s100960000404. [DOI] [PubMed] [Google Scholar]
- 40.van Griethuysen A, et al. Loss of the mecA gene during storage of methicillin-resistant Staphylococcus aureus strains. Journal of clinical microbiology. 2005;43(3):1361–1365. doi: 10.1128/JCM.43.3.1361-1365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alam MT, et al. Transmission and microevolution of USA300 MRSA in U.S. households: evidence from whole-genome sequencing. MBio. 2015;6(2):e00054. doi: 10.1128/mBio.00054-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis MF, et al. Household transmission of meticillin-resistant Staphylococcus aureus and other staphylococci. The Lancet infectious diseases. 2012;12(9):703–716. doi: 10.1016/S1473-3099(12)70156-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.