Abstract
It has been recently reported that the population of Fusobacterium, particularly Fusobacterium nucleatum (Fn), is overrepresented in colorectal cancers and adenomas. The promoting effects of Fn infection on adenoma and/or carcinoma formation have been shown in ApcMin/+mice. Characteristics of Fn-associated CRC were identified through studies using human CRC cohorts, and include right-sided colon location, CpG island methylation phenotype-high (CIMP-H), high level of microsatellite instability (MSI-H), and poor patient prognosis. A subset of Fn-associated CRC exhibits a low level of microsatellite instability (MSI-L) and elevated microsatellite alterations in selected tetra-nucleotide repeats (EMAST) induced by translocation of MSH3 from the nucleus to the cytoplasm in response to oxidative DNA damage or inflammatory signals. The association between CIMP/MSI-H and Fn-infection can be explained by the role of the mismatch repair (MMR) protein complex formed between MSH2 and MSH6 (MutSα) to repair aberrant bases generated by ROS to form 7,8-dihydro-8-oxo-guanine (8-oxoG). Clustered 8-oxoGs formed at CpG-rich regions including promoters by ROS is refractory to base excision repair (BER). Under these conditions, MutSα initiates repair in cooperation with DNA methyltransferases (DNMTs) and the polycomb repressive complex 4 (PRC4). DNMTs at damaged sites methylate CpG islands to repress transcription of target genes and promote repair reactions. Thus, continuous generation of ROS through chronic Fn infection may initiate 1) CIMP-positive adenoma and carcinoma in an MSH2/MSH6-dependent manner, and/or 2) MSI-L/EMAST CRC in an MSH3-dependent manner. The poor prognosis of Fn-associated CRC can be explained by Fn-induced immune-evasion and/or chemo-resistance.
Keywords: microsatellite instability (MSI), elevated microsatellite alterations at selected tetranucleotide repeats (EMAST), colorectal cancer (CRC), DNA mismatch repair (MMR), inflammation, MSH3, MSH6, MSH2, Fusobacterium nucleatum (Fn), CpG island methylate phenotype (CIMP)
Introduction
Microbiota imbalances in the colon and rectum are associated with an inflammatory microenvironment and promotion of colorectal cancer (CRC) (1). Several bacterial organisms including Fusobacterium nucleatum (Fn), Enterotoxigenic Bacteroides fragilis (ETBF), and colibactin-producing Escherichia col. (E. coli) are epidemiologically associated with CRC, and have been found to be enriched in CRC (1,2). Dejea et al showed that CRCs in the ascending colon and hepatic flexures but not on the left side of the colon are covered with invasive bacterial films (3). The enrichment of Fn was found in colorectal adenomas relative to non-adenomas or surrounding normal tissues (4,5,6). Recent studies further demonstrated that the CRCs with a high load of Fn infection is associated with MSI-H and CIMP-H CRC, and is associated with a right-side location and poor prognosis (7,8). There is also a sub-group of non-MSI-H CRC that exhibit microsatellite alterations in response to inflammatory tumor-microenvironment (9,10). For this review, we have compiled articles that deal specifically with Fn infection and CRC. Based upon data in these articles, we discuss possible mechanisms for genetic and epigenetic change induced by Fn and the possible role of Fn in the prognosis of CRCs.
Discovery of Fusobacterium in colorectal neoplasms
Fn is a one of 14 species belonging to the genus Fusobacterium. It is gram-negative, anaerobic, and is associated with various infections including periodontitis, otitis media, Lemierre’s disease, amniotic fluid infections and inflammatory bowel disease (11,12). Fn is a common resident in the human gut mucosa and is a one bacteria that colonizes CRC tumors more frequently than adjacent normal mucosa (11,13). Clear evidence for the abundance of Fn in CRC tumor tissues compared with nearby normal tissues was first reported by two studies in 2012 (14,15). Kostic et al used a whole genome sequence approach where they identified microbial DNA sequences by subtracting human DNA sequences from total DNA reads. They found an enrichment of genus Fusobacterium in most CRC tissues (8/9 cases). They further amplified microbial 16S ribosomal-DNA and sequenced from 95 tumor/normal CRC pairs, and also performed fluorescence in situ hybridization to show that Fusobacterium is enriched in tumor tissues. They identified Fn as the most enriched species among Fusobacterium species in CRC (14). Castellarin et al used an RNA-seq approach where human transcript sequences were subtracted from total RNA reads in 11 paired CRC tumor/normal tissues. They found enrichment of Fn in tumors (> 2-fold) as compared with matching normal tissues (9/11 cases). They developed a quantitative PCR assay to determine Fn-specific copy numbers using the nusG gene of Fn, and measured Fn copies in genomic DNA from 99 CRC tumor/normal tissues. Again, Fn was enriched in tumors compared to normal tissues. They also showed that Fn isolated from one CRC patient was invasive to cultured colon cancer cells. Finally, they showed that a high level of Fn in CRC was associated with lymph node metastasis (15). Later, an enrichment of Fn in CRC tumor tissues was found in cohorts from Japan (6), Europe (16) and China (17), indicating that this phenomenon is universal among human populations.
McCoy et al first described the association between Fusobacterium and colorectal adenoma (4). They compared the abundance of Fusobacterium in normal rectal mucosa isolated from subjects with adenoma (48 cases) and from subjects without adenoma (67 cases), and found that rectal mucosa from adenoma patients were significantly enriched in Fusobacterium compared to mucosa from subjects without adenoma (4). In contrast to McCoy’s study, Kostic et al directly quantified Fusobacterium spp. loads in adenomas and matched adjacent normal tissues from the same patient. They found significant enrichment of the Fusobacterium species in adenoma tissues (P<0.004) (5). Flanagan et al quantified Fn loads in 52 colorectal adenomas and nearby normal mucosa, and found that there was a trend but no significant association between the Fn level and adenoma state. However, they found a significantly higher Fn load in high-grade dysplasia and in CRC, thus suggesting that the Fn level was associated with progression of adenoma to carcinoma (16). Although a higher level of Fn load was detected in adenoma and carcinoma tissues compared to adjacent normal tissues, carcinoma tissues contained more Fn than adenoma tissues (16). Similarly, Yu et al observed a gradual increase in the Fn load during the transition from adenoma to carcinoma (18). Taken together, these results suggest that Fn infection may be involved in progression from adenoma to carcinoma in some CRC cases.
Mouse Model of Fn-associated Intestinal Tumorigenesis
To evaluate Fn infection in intestinal tumorigenesis, Kostic et al used C57BL/6 ApcMin/+ mice which develop intestinal tumors due to an inactive mutation in one copy of the Apc gene (5). Oral introduction of Fn (13 of 15 mice) but not of control Streptococcus spp. (2 of 12 mice) or tryptic soy broth (1 of 20 mice) significantly increased the number of colonic tumors in ApcMin/+ mice. Kostic et al also replicated observations found in human tissues, that of enrichment of Fn in tumor tissues compared to surrounding normal mucosa and invasiveness of Fn to tumor tissues.
Yu et al used both C57BL/6 and C57BL/6 ApcMin/+ mice models (18). In the C57BL/6 model, mice were treated with a chemical carcinogen, 1,2-ddimethylhydrazine (DMH), followed by introduction of Fn or control bacteria (E. coli). Introduction of Fn increased the number of DMH-induced aberrant crypt foci (ACF) and tumors compared to introduction by E. coli. Similar to Kostic’s results, introduction of Fn increased the number of tumors in C57BL/6 ApcMin/+ mice (18). Similarly, Yang et al showed that introduction of Fn into C57BL/6 ApcMin/+ mice increased the number and size of tumors, and shortened overall survival compared with the non-treated controls (19). Although these three studies suggest that Fn infection may promote loss of a wild-type copy of Apc in ApcMin/+ colon cells to form adenoma, they do not give evidence that Fn infection may be involved in adenoma-to-carcinoma transition in the colon and rectum. It would be interesting to see the genetic and epigenetic landscapes of the Fn-induced tumors.
In contrast to the three studies discussed above, Tomkovich et al did not see any tumor-enhancing effects of Fn on ApcMin/+ mice (20). They used germ-free (GF) or GF-derived, specific-pathogen-free (SPF) 129/SvEv ApcMin/+ mice while other studies used SPF C57BL/6 ApcMin/+. Thus, the mouse genetic background and/or different microbial exposures before and/or after Fn infection may change the susceptibility of ApcMin/+ mice to Fn. There was also a difference in the strain of Fn used by Tomkovich’s study as compared to others. These discrepancies highlight the possibility that 1) there may be individual genetic and/or epigenetic backgrounds that are susceptible for Fn associated tumorigenesis; and/or 2) there may be a select group of Fn that are a component of diverse microbiota associated with colorectal tumorigenesis (20,21).
Characteristics of Fusobacterium-associated CRCs
Following the discovery of the association between Fusobacterium infection and CRC, it was recognized that the infection is significantly associated with two sub-groups of CRCs. The first group of CRCs exhibit a high level of CpG island methylation phenotype (CIMP-H) and/or a high level of microsatellite instability (MSI-H), and are located on the right side of the colon (6,7,8). The second group of CRCs exhibit a low level of microsatellite instability (MSI-L) and/or elevated microsatellite alterations in selected tetra-nucleotide repeats (EMAST), and are also located on the right side of the colon (unpublished data).
CIMP and MSI-H CRCs
Tahara et al found that 74% of tumor tissues from 149 CRC cases were tested positive for infection by the Fusobacterium species, including Fn (52.3%;78/149). Among infected CRCs, 14 (9.4%) and 8 (5.4%) cases were heavily infected with Fusobacterium spp. or Fn respectively. They further showed that the Fusobacterium-high CRCs were enriched in MSI-H (P=0.018), CIMP)-positive (P=0.001), hMLH1 methylation-positive (P=0.0028), p53 wild-type (P=0.015), CDH7/8 mutant CRCs (P=0.002) and located on the right-side (7). Ito et al also showed that 56% of tumor tissues from 511 cases of CRC were positive for Fn infection. They divided Fn-infected groups into high and low based on the median number of Fn bacteria present and found that Fn-high was associated with tumor tissues rather than premalignant tissues (P=0.0001), larger tumor size (P=0.0005) and CIMP-high (P=0.0013) (6). Mima et al also showed that a high load of Fn in CRC tissues was associated with MSI-H, CIMP, and BRAF mutation by univariate analysis but not by multivariate analysis adjusted for MSI-H (8).
The human cancer genome exhibits aberrant DNA methylation (22,23). Increased methylation of promoter region CpG islands is prominent in tumor DNA and is associated with transcriptional inactivation of tumor-suppressor genes. Toyota et al first systematically examined CpG islands that were differentially methylated between tumor and normal genomic DNA from CRC patients (24). They found a group of the CpG sites specifically associated with the cancer state in a sub-set of CRC and gave this phenotype the name CpG island methylation phenotype (CIMP). Thus, CRC can be divided into CIMP-positive (CIMP+) and CIMP-negative (CIMP-). They further demonstrated that (1) CIMP is seen in a sub-group of adenoma; (2) CRC with MSI-H due to promoter hypermethylation of hMLH1 (25) is a sub-group of CIMP+ CRC; (3) CDKN2A and THBS1 are frequently methylated in CIMP+ CRC; and (4) CIMP+ CRC is associated with proximal site (24). Later, it was recognized that CIMP is not a dichotomous trait but rather continuous and can be divided into CIMP+, CIMP-intermediate, and CIMP-. Sánchez-Vega et al analyzed methylation data from The Cancer Genome Atlas (TCGA) CRC cohort (274 cases: reference 26) using CIMP+, CIMP-intermediate, and CIMP- categories, and confirmed that a large portion of CIMP+ exhibit hypermethylation of hMLH1 (MSI-H), and are associated with the right side of the colon. They also showed that hypermethylation of MGMT is frequent in CIMP+ CRC, and is associated with FBXW7, APC and KRAS mutations (27).
Microsatellites, or simple sequence repeats, are composed of 1–6 repeated nucleotides. Microsatellite instability (MSI) is defined as continuous length changes in simple DNA repeats within microsatellite loci. MSI-H is caused by deficiencies in mismatch repair (MMR) genes including MSH2, MLH1, MSH6 and PMS2 (28). As mentioned above, the MSI-H exhibited in 10~15% of sporadic CRC cases is due to transcriptional down-regulation of MLH1 expression through promoter hyper-methylation (25).
CRCs have been divided into several subgroups based on their genetic and epigenetic landscapes (26), and their transcriptional (29) or proteomic similarities or differences (30), and on their immunological landscapes (31). In 2012, genetic and epigenetic landscapes of sporadic CRC identified through massive DNA/RNA sequencing were reported by the TCGA Consortium (26). One of the major findings was that there are 2 types of CRC that differ in in respect to the frequency of somatic gene mutations. Sixteen percent of CRC showed a hyper-mutated genotype where three-quarters exhibited MSI-H due to MLH1 silencing by promoter-methylation, and one-quarter had somatic mutations in the mismatch-repair (MMR) gene and polymerase ε (POLE) (26).
The transcriptional landscapes in CRC can be divided into 4 distinctive sub-groups: 1) the consensus molecular subtype 1 (CMS1) (14%), that is enriched in hyper-mutated and MSI-H CRC and characterized by gene expression with strong immune activation; 2) CMS2 (37%), marked with WNT and MYC signaling activation; 3) CMS3 (13%) with metabolic dysregulation; and 4) CMS4 (23%), marked with prominent transforming growth factor-β activation, stromal invasion and angiogenesis. CRCs with mixed features (13%) also exist and may represent a transitional phenotype (29). Although most of MSI-H CRC are grouped into CMS1, some of them are grouped into CMS3 and CMS4 (29). In proteomic sub-grouping, there are 5 distinctive groups within the TCGA CRC cohort. Proteomic sub-type B and C are enriched in MSI-H/CIMP+, and hyper-mutated CRC. These observations indicate that MSI-H/CIMP+ CRCs are heterogeneous in terms of m-RNA and protein expression (29,30). Angelova et al analyzed TCGA CRC data to identify a dominant immune cell subpopulation associated with a particular molecular subtype of CRCs. They found that tumor-infiltrating lymphocytes (TILs) from MSI-H CRC tumors are enriched by lymphocytes with anti-tumor functions including central and effector memory CD4+/CD8+ cells and natural killer T cells, compared to TILs from MSS CRC (31).
MSI-L/EMAST CRCs
MSI in CRS was defined in 1998 at an international workshop meeting sponsored by the National Cancer Institute (28). In this meeting, MSI-H, low-frequency of MSI (MSI-L), and microsatellite stable (MSS) CRCs were defined using a specific panel of microsatellite markers. Although MSI-H is caused by the functional loss of MMR proteins, the etiology of MSI-L and the distinction between MSI-L and MSS CRC remained unclear until recently (10,28). Another type of microsatellite alteration, termed EMAST, where insertion/deletion mutations in the loci with tetra-nucleotide but not with mono- and/or dinucleotide repeats, were recognized as a component of CRC (32,33). MSI-L and EMAST have been observed in many human cancers (10,32). Haugen et al examined the frequency of EMAST in CRC, its relationship to MSI-L and its possible causes (33). They found that EMAST is frequent in sporadic cases of non-MSI-H CRC (~50%) and is associated with decreased nuclear MSH3 expression in tumor cells. Using isogenic MSH3-proficient and –deficient cell lines, they also showed that EMAST and MSI-L in non-MSI-H CRC cells are caused by loss of MSH3 (33). The frequent incidence of EMAST in CRCs was confirmed by two other studies (34,35). The fact that EMAST is due to the loss of MSH3 was also proven by other studies using tissue cultured human cells (36,37). Later, Adam et al found patients with bi-allelic inactivation mutations at the MSH3 locus who suffered from colorectal adenoma polyposis syndrome. The adenoma polyps from these patients exhibited MSI at loci with dinucleotide repeats and EMAST loci but not at loci with mononucleotide repeats (38). These results strongly suggest that MSI-L/EMAST is caused by loss of the MSH3 function. The biochemical basis for MSI-L/EMAST formation through loss of MSH3 has been described elsewhere (10).
Immunohistochemical staining using anti-MSH3 showed that nuclear MSH3 is completely lost in adenoma polyps with bi-allelic germline MSH3 mutations (38) while localized loss of nuclear MSH3 is detected in sporadic CRCs exhibiting MSI-L/EMAST (33,35). These results suggest that loss of MSH3 expression in sporadic MSI-L/EMAST CRC may be due to an somatic epigenetic event. The frequency of MSH3 somatic mutations in CRC is about 6.6% (26) which does not explain the high incidence of MSI-L/EMAST (~50%) in CRC.
Lee et al first showed evidence that inflammation may be linked to MSI-L/EMAST in CRC (35). They found that EMAST CRC is enriched in CD8+ T cells in the tumor microenvironment compared to non-EMAST CRC. They also found that EMAST is significantly high in ulcerated tumors. These results suggest that some immunological and inflammatory responses are active in EMAST CRC. Later, Tseng-Rogenski et al demonstrated that in several cancer cell lines (37,39) inflammatory factors including oxidative stress (hydrogen peroxide), interleukin-6 (IL6) and prostaglandin E2 (PGE2) induce displacement of MSH3 from the nucleus to the cytoplasm, whereas the other MMR proteins do not displace. Repeated treatment of microsatellite stable colon cancer cell lines with IL6 induced EMAST. These studies convincingly showed that some inflammatory factors induce EMAST through loss of MSH3 in the nucleus.
Evidence that the inflammatory micro-environment induces MSI-L had been found in regenerated colon tissues from ulcerated colitis (UC) patients. Brentnall et al showed for the first time the presence of MSI-L but not MSI-H in colon tissues from UC patients (40). Ozaki et al also examined crypts from UC –derived CRC, UC-derived hyperplasia and UC-regenerated colons, and tested them for the presence of microsatellite instability. They again detected MSI-L but not MSI-H in some crypts but not in stroma cells regardless of whether they were from cancer or non-cancer tissues (41). Recently, we have shown that regenerated colon epithelial cells and tumors from UC patients show a high frequency of MSH3 displacement from the nucleus to the cytoplasm and of MSI-L /EMAST. These results support the role of inflammation in displacement of MSH3, which induces MSI-L/EMAST in human tissues including cancers (42).
Our recent investigations not only confirmed that Fn is associated with MSI-H but also showed that Fn is associated with MSI-L/EMAST when compared to non-MSI-H, non-MSI-L/EMAST CRC (unpublished data). These findings raise the possibility that inflammatory factors generated in tumor-microenvironments in response to Fn infection, and/or Fn itself may induce translocation of MSH3 from the nucleus to the cytoplasm in colon tumor cells, resulting in MSI-L/EMAST.
Poor Prognosis for patients with Fn-associated CRCs
Increased loads of Fn are associated with advanced CRC stages (6,16–18) and with poor prognoses (19,43–46). Mima et al showed that colon cancer-specific death, but not overall survival is associated with high levels of tissue Fn (43). Yu et al looked at the relationship between CRC recurrence and microbiota alterations in the gut. They found that Fn is the most enriched bacteria species in tumors from recurrent CRC compared to tumors from non-recurrent CRC. Using additional cohorts, they showed that the higher load of Fn is an independent determinant and predictor of recurrence-free survival (RFS) (44). Yan et al also showed that a high level of Fn in stage III and IV CRC is associated with shorter cancer-specific survival and shorter RFS (45). They also observed that adjuvant chemotherapy is more effective in preventing recurrence in stage III patients with a low level of Fn compared to the those with a high level of Fn (45). Bullman et al showed that patients from TCGA cohorts who suffered from right-side colon cancer with a high load of Fn infection had poor overall survival compared with patients with a low load of Fn infection. Interestingly, they detected Fn infection not only in primary CRC but also in matching liver metastasis. They also demonstrated that antibiotic treatment not only reduced the Fusobacterium load but also inhibited growth of xenograft tumors from patients, suggesting that infection by Fusobacterium accelerates tumor growth in vivo. (46).
Possible explanation of why Fusobacterium infection is associated with CIMP CRC
CIMP has been detected in many human cancers but its causes are not clear (27). Several observations suggest that infection by micro-organisms and/or inflammation associated with micro-organism infection may induce CIMP in tumor genomes. For instance, in gastric cancers, there are two types of CIMP+ tumors: one is associated with Epstein-Barr virus (EBV) infection, and another is associated with MSI-H (hMLH1 promoter hypermethylation) (47). Infection of Helicobacter pylori in gastric cells also induces inflammation that causes aberrant DNA methylation (48). In fact, when human cell lines were co-cultured with Fn in vitro, expression of pro-inflammatory genes including TNF, IL8, and IL1β as well as ROS was induced (49,50). Kostic et al analyzed transcriptome sequences from 133 cases of CRC in the TCGA data set (26) to identify the genes associated with Fusobacterium species infections (5). They found that the expression of inflammatory response genes including IL1β, IL24, PTGS2 (COX-2), IL8, IL6 and TNF were enriched in Fusobacterium –infected CRCs. Importantly, these gene signatures were specific to Fusobacterium but not to other bacteria detected in CRC tissues including Bacteroides, Escherichia, Streptococcus and Propionobacterium (5). They further demonstrated that Fn-induced tumors in ApcMin/+ mice express elevated levels of mouse homologs of PTGS2, IL8, IL6 and TNF genes compared to tumors not induced by Fn. Finally, they showed that NF-κB, which drives the pro-inflammatory response, was activated in Fn-enriched CRC tissues (5). Similarly, Yang et al reported that Fn-infection activates Toll-like receptor 4 (TLR4)/NF-κB axis in cultured cells, and NF-κB is highly activated in Fn-enriched CRC tissues (19). These observations strongly suggest that the tumor-microenvironment of Fn-infected CRC is highly inflammatory.
One of the factors that would be constantly generated by chronic Fusobacterium infection and could be responsible for an aberrant DNA methylation is ROS (51). Although there are many cases where ROS is associated with DNA hypermethylation (48,52), until recently, there has been no direct evidence that oxidative DNA damage causes a genome-wide hyper-methylation of promoter CpG islands and hypo-methylation of CG sites at other parts of the genome. The aberrant base, 7,8-dihydro-8-oxo-guanine (8-oxoG) is the most abundant DNA modification generated by ROS. Therefore, 8-oxoG would be enriched at promoter CpG islands within the whole genome after ROS exposure. Base excision repair (BER) is primarily responsible for removing 8-oxoG; however clustered 8-oxoG lesions are known to be refractory to BER (53). Zlatanou et al first reported that MutSα, the heterodimer formed between the MMR proteins MSH2 and MSH6, recognizes and initiates removal of the clustered 8-oxoG in cooperation with mono-ubiquitinated PCNA and DNA polymerase eta (53). Independently, O’Hagan et al discovered that a large protein complex consisting of DNA methyltransferase I (DNMT1), DNMT3B, and a member of the polycomb repressive complex 4 (PRC4), linked to transcriptional silencing, tightly binds to a promoter CpG island of the expressed gene when the island is damaged by ROS. Binding of this complex leads to a reduction of gene expression and induces DNA methylation at the CpG islands of the target gene (54). Ding et al successively demonstrated when MutSα recognizes and repairs 8-oxoG, it also recruits DNMT1 to the damaged site (55). Taken together, these results support the idea that enriched 8-oxoG at the promoter CpG islands attracts MutSα, and at the same time, DNMTs and PRC4 are recruited to reduce the transcriptional activity of the target gene through promoter methylation during MutSα-directed repair. This imbalance in DNMTs/PRC4 localization between promoter and non-promoter CpG sites explains how genome-wide hyper-methylation of promoter CpG islands, CIMP, and hypo-methylation of CG sites in other parts of the genome, develop in response to ROS damage in the cancer genome. Finally, this mechanism has been proven by Maiuri et al using a mouse model of colitis-associated CRC (56). They showed that inflammation-induced tumors gained DNA hypermethylation at CpG islands in an MSH2-dependent manner (56). They used MinApcΔ716+/− and MSH2l/lVC/Min mice and inoculated mice with Bacteroides fragilis (ETBF) to generate colitis-associated CRC (57). They found that ETBF-induced tumors had more hypermethylated regions and fewer hypomethylated regions in their genome compared to ETBF-mock tumors. They observed a transient increase in the amount of 8-oxoG in colon epithelium cells from ETBF-infected mice and a synchronous reduction of gene expression from the loci that were hypermethylated in ETBF-induced tumors. They detected MSH2 interacting with DNMTs and components of PRC4 in ETBF-inflamed colon cells. Lastly, they demonstrated that loss of MSH2 reduces CpG island hypermethylation in ETBF-induced tumors (56).
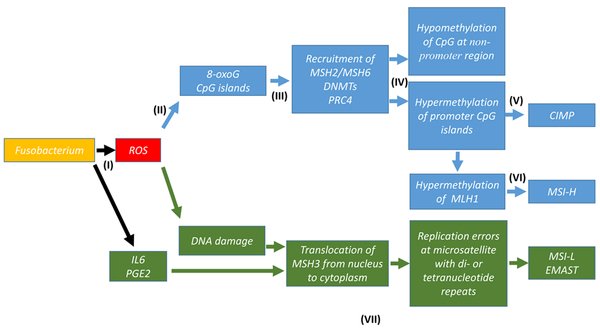
Although the ETBF-induced mouse CRC model highlights a casual role of ROS in the CIMP phenotype, whether Fn-infection also activates MSH2/MSH6-dependent hypemethylation of promoter CpG islands in CRC remains to be determined (Figure 1).
Fig. 1:
Hypothetical Pathways of Genetic and Epigenetic Alterations in CRC induced by Chronic Fusobacterium Infection
(I): Chronic infection of colon mucosa by Fusobacterium induces ROS and other pro-inflammatory factors including IL6 and PGE2 (references 5,50).
(II): ROS generates clustered 8-oxoG lesions at promoter CpG island.
(III): MSH2/MSH6, DNMT1, DMNT3B and PRC4 are recruited from whole genome and enriched at damaged promoter CpG islands.
(IV): non-promoter CpG sites become DNMT-poor, leading to hypomethylation.
(V): Recruited DNMT1 and DMNT3B methylate promote CpGs to enhance DNA repair by MSH2/MSH6, leading to hypermethylation of CpG islands (CIMP) (references 53–56).
(VI): hypermethylation of the hMLH1 promoter CpG island leads to MSH-H.
(VII): DNA damage (8-oxoG) or IL6/PGE2 induces translocation of MSH3 from nucleus to cytoplasm, leading to MSI-L/EMAST (references 37,39).
Blue arrows and boxes represent the pathway to CIMP/MSI-H triggered by Fn infection.
Green arrows and boxes represent the pathway to MSI-L/EMAST triggered by Fn infection.
Possible explanation of why Fusobacterium infection is associated with MSI-H CRC
Although the CpG island of the MLH1 promoter would be a one of many sites damaged by ROS and methylated by DNMTs/PRC4, resulting in MSI-H (Figure 1), it is not clear why inactivation of this locus has an advantage for CRC initiation triggered by inflammation. One possible explanation is that silencing MLH1 may aid the damaged cells in escaping apoptosis induced by oxidative stress. Hardman et al showed that MLH1-deficient cells are more resistant to the cytotoxic effects of hydrogen peroxide or tert-butyl hydroperoxide than are MLH1-proficient cells. They observed a lack of apoptotic events including increased mitochondrial permeability, release of cytochrome c and caspase 3 activation in MLH1-deficient cells after exposure to hydrogen peroxide (58). Yanamadala et al confirmed Harman’s results and showed that apoptosis induced by hydrogen peroxide is MLH1-dependent and associated with global inhibition of mRNA (59). Chen et al found that MLH1 is cleaved by caspase-3 during apoptosis activated by DNA damage and that this cleaved MLH1 product plays a role in the execution of apoptosis (60). Thus, for colonic epithelial cells continuously exposed to ROS by chronic infection of Fn, silencing MLH1 would be one way to avoid apoptosis.
Fusobacterium infection is associated with MSI-L/EMAST
As mentioned above, cumulative evidence supports the idea that the etiology of MSI-L/EMAST formation in cancer genome is the inflammation-induced loss of nuclear MSH3 in dividing cells (10,37,39). Therefore, it is reasonable to speculate that a sub-group of MSI-L/EMAST CRC may be associated with pathogenic infections of microbiota including Fusobacterium. We have examined a CRC cohort for the MSI-H, MSI-L/EMAST genotypes, and for the level of Fn in each CRC case. We have confirmed that Fn-positivity is associated with MSI-H compared to non-MSI-H. We have also found that Fn-positivity is significantly associated with MSI-L/EMAST as opposed to non-MSI-H, non-MSI-L/EMAST CRC (unpublished data). We are currently determining whether direct infection by Fn induces MSI-L/EMAST in tissue cultured cells and/or in mouse models (Figure 1).
Furthermore, it is possible that MSI-L/EMAST CRC also exhibits CIMP. In support of this assumption, a sub-set of CIMP CRC exhibits hypermethylation of MGMT, which is associated with MSI-L CRC (27,61).
Possible explanation of why Fusobacterium infection is associated with poor prognosis of CRC
Enhancement of Tumor Cell Growth and Survival
Yang et al demonstrated that Fn directly promotes CRC cell growth and survival in vitro and in vivo. Fn activates NF-κB through Toll-like receptor 4 signaling, resulting in up-regulation of microRNA-21 that could be a marker for poor clinical outcomes for CRC patients (19). Fn secretes adhesion and FadA through which attachment and invasion of Fn to a host cell occurs (62). Rubinstein et al showed that FadA binds to E-cadherin on the host cell, and activates β-catenin, resulting in oncogenic and inflammatory responses (63). They observed that 1) Fn and purified FadA stimulate cell growth in several CRC cell lines in an E-cadherin-dependent manner; 2) FadA binds to E-cadherin on CRC cells and promotes Fn attachment and invasion via E-cadherin; 3) the binding of FadA to E-cadherin initiates β–catenin nuclear translocation and increases transcription of the down-stream oncogenic pathway including T cell factors, Myc and cyclin D genes. Further internalization of the FadA/E-cadherin complex by endocytosis also activates inflammatory NF-κB signaling. Thus, FadA on the surface of Fn stimulates tumor growth through bacterial attachment but also triggers the inflammatory response through bacterial invasion. There are many other uncharacterized Fn proteins that may control host-pathogen interactions (64). Further studies are required to identify and characterize these proteins in relation to CRC carcinogenesis.
Immune evasion
CRC infected with a high load of Fn is associated with MSI-H and poor prognoses. However, it is widely accepted that the prognosis of MSI-H CRC is better than for non-MSI-H CRC. Popat et al showed that MSI-H CRC exhibits better overall survival than does non-MSI-H CRC through meta-analysis of 32 studies examining 7642 cases including 1277 MSI-H tumors (65). As mentioned above, the tumor-microenvironment of MSI-H CRC is enriched in lymphocytes with anti-tumor functions including CD4+/CD8+ T cells and natural killer (NK) cells. A high immunoscore based on the abundance of CD3+ and CD8+ T cells at the central and invasion fronts of tumor tissues is associated with improved prognosis, and the majority of MSI-H cases have this high immunoscore (31). The reason for the improved prognosis of MSI-H CRC compared to non-MSI-H CRC is the presence of an anti-tumor T cell population in the tumor-microenvironment, elicited by continuous generations of frameshift neo-antigens in hypermutated MSI-H CRC (66). However, Mlecnik et al showed that MSI-H CRC is heterogeneous and consists of at least 2 sub-groups; the major group exhibiting a positive prognosis with high T cell activity and the minor group exhibiting poor prognosis with reduced T cell activity (67,68). Therefore, it could be that CRC with a high Fn load is a sub-group of MSI-H CRC with reduced T cell activity. Compatible with this assumption, Mima et al showed that CRCs with high levels of Fn exhibited a reduced density of CD3+ T cells in their tumor-microenvironments (69).
Reduced T cell activity in the tumor-microenvironment can be achieved by cancer cells through loss of HLA expression and/or disabling the antigen-processing machinery (APM). In fact, alterations in the HLA complex containing B2M and APM occur more frequently in MSI-H CRCs (70–73). Thus, this immune evasion mechanism may partially explain the immunological heterogeneity seen in MSI-H CRC.
MSI-H tumors that have lost HLA or APM function are still targets of NK cells (74). Fn secretes several proteins involved in binding to other microorganisms or to host cells and in invasion. Fap2 was first identified as an apoptosis-inducing protein for human lymphocytes (75,76). Gur et al showed that infection of cancer cells with Fn protects cancer cells from cytotoxic attacks of natural killer (NK) and/or T cells via Fap2-TIGIT (T cell immunoglobulin and ITIM domain) interactions (77). The NK cell activity is controlled by inhibitory and activating NK receptors. TIGIT is a one of the inhibitory NK receptors, expressed in all NK cells. When its ligands (for instance, Fap2) bind to TIGIT, NK cells stop killing their target cancer cells. Coppenhagen-Glazer et al demonstrated that Fap2 co-aggregates other microorganisms and attaches to the mammalian cell surface by binding to galactose- and N-acetyl-d-galactosamine (Gal/GalNAc) (78). Abed et al showed evidence that enrichment of Fn in CRC tumor tissues, as opposed to normal tissues surrounding these tissues, may be due to selective binding of Fap2 to Gal/GalNAc, which is over expressed on the surface of tumor cells (78). Thus, cancer cells coated by Fap2 would be protected from an NK attack.
Chemo-resistance
Yu et al convincingly demonstrated that a higher load of Fn in CRC is associated with cancer recurrence after surgery. They reasoned that chemo-resistance is a major factor for recurrence and proved that Fn induces cancer cells’ chemo-resistance through activation of autophagy (44). We previously showed that stage II/III MSI-L/EMAST CRC exhibit a shorter RFS compared to MSI-H or non-MSI-H, non-MSI-L/EMAST CRC (79,80). Because MSI-L/EMAST CRCs are infected by Fn, it is tempting to speculate that a majority of Fn-associated CRCs that exhibit poor prognoses, especially shorter RFS, could be MSI-L/EMAST CRCs but not MSI-H CRCs. Considering that only 4% of stage IV CRCs are MSI-H (81), a majority of Fn-infected CRCs with poor prognoses could be MSI-L/EMAST. These possibilities are under intensive investigation.
Conclusion
It is still questionable whether Fn and/or other bacterial infections cause CRC. However, cumulative evidence shows that Fn infection affects the course of CRC carcinogenesis linked to DNA repair and genetic/epigenetic alterations. In conclusion, Fn-associated CRCs are located on the right side of the colon, are inflamed by ROS, and are associated with CIMP/MSI-H and MSI-L/EMAST (10,82,83). Fn-associated CRCs exhibit shorter RFS. Mechanistically, DNA damage caused by ROS triggers MSH2/MSH6-dependent repair that results in CIMP, and in MSI-H when MLH1 is silenced, while it also induces translocation of MSH3 from the nucleus to the cytoplasm, resulting in MSI-L/EMAST. Poor prognoses for patients with Fn-associated CRC could be explained by Fn-induced chemo-resistance and/or immune evasion and/or immune suppression. It would be worthwhile to conduct further studies, especially into the detection, prevention and treatment of Fusobacterium.
Acknowledgments:
This work was supported by the United States Public Health Service (DK067287 and CA206010) and the A. Alfred Taubman Medical Research Institute of the University of Michigan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest are disclosed.
References
- 1.Brennan CA, Garrett WS. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu Rev Microbiol. 2016. September;70:395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014. March;15(3):317–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dejea CM, Wick EC, Hechenbleikner EM, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A. 2014. December;111(51):18321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013. January;8(1): e53653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013. August;14(2):207–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, Igarashi H, Takahashi T, Tachibana M, Takahashi H, Yoshii S, Takenouchi T, Hasegawa T, Okita K, Hirata K, Maruyama R, Suzuki H, Imai K, Yamamoto H, Shinomura Y. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015. September;137(6):1258–68. [DOI] [PubMed] [Google Scholar]
- 7.Tahara T, Yamamoto E, Suzuki H, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014. March;74(5):1311–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mima K, Cao Y, Chan AT, et al. Fusobacterium nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin Transl Gastroenterol. 2016. November 3;7(11):e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carethers JM, Koi M, Tseng-Rogenski SS. EMAST is a Form of Microsatellite Instability That is Initiated by Inflammation and Modulates Colorectal Cancer Progression. Genes (Basel). 2015. March;6(2):185–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koi M, Tseng-Rogenski S, Carethers JM. Inflammation-associated Microsatellite Alterations: Mechanisms and Significance in the Prognosis of Patients with Colorectal Cancer. World J Gastrointest Oncol. 2017. (https://www.f6publishing.com/ArticleInPressDetail?id=36858). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strauss J, White A, Ambrose C, et al. Phenotypic and genotypic analyses of clinical Fusobacterium nucleatum and Fusobacterium periodonticum isolates from the human gut. Anaerobe. 2008. December;14(6):301–9. [DOI] [PubMed] [Google Scholar]
- 12.Strauss J, Kaplan GG, Beck PL, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011. September;17(9):1971–8. [DOI] [PubMed] [Google Scholar]
- 13.Marchesi JR, Dutilh BE, Hall N, et al. Towards the human colorectal cancer microbiome. PLoS One. 2011. ;6(5):e20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012. February;22(2):292–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012. February;22(2):299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flanagan L, Schmid J, Ebert M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014. August;33(8):1381–90 [DOI] [PubMed] [Google Scholar]
- 17.Li YY, Ge QX, Cao J, et al. Association of Fusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J Gastroenterol. 2016. March;22(11):3227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu YN, Yu TC, Zhao HJ, et al. Berberine may rescue Fusobacterium nucleatum-induced colorectal tumorigenesis by modulating the tumor microenvironment. Oncotarget. 2015. October;6(31):32013–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Weng W, Peng J, et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017. March;152(4):851–866. e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomkovich S, Yang Y, Winglee K, et al. Locoregional Effects of Microbiota in a Preclinical Model of Colon Carcinogenesis. Cancer Res. 2017. May;77(10):2620–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holt RA, Cochrane K. Tumor Potentiating Mechanisms of Fusobacterium nucleatum, A Multifaceted Microbe. Gastroenterology. 2017. March;152(4):694–696. [DOI] [PubMed] [Google Scholar]
- 22.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983. January;301(5895):89–92. [DOI] [PubMed] [Google Scholar]
- 23.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002. June;3(6):415–28. [DOI] [PubMed] [Google Scholar]
- 24.Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999. July;96(15):8681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998. June;95(12):6870–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sánchez-Vega F, Gotea V, Margolin G, et al. Pan-cancer stratification of solid human epithelial tumors and cancer cell lines reveals commonalities and tissue-specific features of the CpG island methylator phenotype. Epigenetics Chromatin. 2015. April;8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998. November;58(22):5248–57. [PubMed] [Google Scholar]
- 29.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015. November;21(11):1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, Davies SR, Wang S, Wang P, Kinsinger CR, Rivers RC, Rodriguez H, Townsend RR, Ellis MJ, Carr SA, Tabb DL, Coffey RJ, Slebos RJ, Liebler DC; NCI CPTAC. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014. September;513(7518):382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angelova M, Charoentong P, Hackl H, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015. March;16:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watson MM, Berg M, Søreide K. Prevalence and implications of elevated microsatellite alterations at selected tetranucleotides in cancer. Br J Cancer. 2014. August 26;111(5):823–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haugen AC, Goel A, Yamada K, et al. Genetic instability caused by loss of MutS homologue 3 in human colorectal cancer. Cancer Res. 2008. October 15;68(20):8465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamada K, Kanazawa S, Koike J, et al. Microsatellite instability at tetranucleotide repeats in sporadic colorectal cancer in Japan. Oncol Rep. 2010. February;23(2):551–61. [PMC free article] [PubMed] [Google Scholar]
- 35.Lee SY, Chung H, Devaraj B, et al. Microsatellite alterations at selected tetranucleotide repeats are associated with morphologies of colorectal neoplasias. Gastroenterology. 2010. November;139(5):1519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campregher C, Schmid G, Ferk F, et al. MSH3-deficiency initiates EMAST without oncogenic transformation of human colon epithelial cells. PLoS One. 2012;7(11):e50541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tseng-Rogenski SS, Chung H, Wilk MB, et al. Oxidative stress induces nuclear-to-cytosol shift of hMSH3, a potential mechanism for EMAST in colorectal cancer cells. PLoS One. 2012;7(11):e50616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adam R, Spier I, Zhao B, et al. Exome Sequencing Identifies Biallelic MSH3 Germline Mutations as a Recessive Subtype of Colorectal Adenomatous Polyposis. Am J Hum Genet. 2016. August 4;99(2):337–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tseng-Rogenski SS, Hamaya Y, Choi DY, et al. Interleukin 6 alters localization of hMSH3, leading to DNA mismatch repair defects in colorectal cancer cells. Gastroenterology. 2015. March;148(3):579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brentnall TA, Crispin DA, Bronner MP, et al. Microsatellite instability in nonneoplastic mucosa from patients with chronic ulcerative colitis. Cancer Res. 1996. March 15;56(6):1237–40. [PubMed] [Google Scholar]
- 41.Ozaki K, Nagasaka T, Notohara K, et al. Heterogeneous microsatellite instability observed within epithelium of ulcerative colitis. Int J Cancer. 2006. December;119(11):2513–9. [DOI] [PubMed] [Google Scholar]
- 42.Munakata K, Koi M, Leconte P, et al. Loss of MSH3 and Subsequent EMAST Determines the Pathological Significance of MSI-L in Ulcerative Colitis. Gastroenterology. 2016. April; 150(4): S962 [Google Scholar]
- 43.Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016. December;65(12):1973–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017. July;170(3):548–563. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan X, Liu L, Li H, et al. Clinical significance of Fusobacterium nucleatum, epithelial-mesenchymal transition, and cancer stem cell markers in stage III/IV colorectal cancer patients. Onco Targets Ther. 2017. October;10:5031–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bullman S, Pedamallu CS, Sicinska E, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017. November; eaal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014. September 11;513(7517):202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niwa T, Tsukamoto T, Toyoda T, et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010. February;70(4):1430–40. [DOI] [PubMed] [Google Scholar]
- 49.Dharmani P, Strauss J, Ambrose C, et al. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect Immun. 2011. July;79(7):2597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang B, Wang K, Jia YP, Zhu P, Fang Y, Zhang ZJ, Mao XH, Li Q, Zeng DZ. Fusobacterium nucleatum-Induced Impairment of Autophagic Flux Enhances the Expression of Proinflammatory Cytokines via ROS in Caco-2 Cells. PLoS One. 2016. Nov;11(11):e0165701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, Kanno S, Yamamoto I, Ishigami K, Igarashi H, Maruyama R, Imai K, Yamamoto H, Shinomura Y. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. 2016. January;22(2):557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niwa T, Ushijima T. Induction of epigenetic alterations by chronic inflammation and its significance on carcinogenesis. Adv Genet. 2010;71:41–56. [DOI] [PubMed] [Google Scholar]
- 53.Zlatanou A, Despras E, Braz-Petta T, Boubakour-Azzouz I, Pouvelle C, Stewart GS, Nakajima S, Yasui A, Ishchenko AA, Kannouche PL. The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol η in response to oxidative DNA damage in human cells. Mol Cell. 2011. August;43(4):649–62. [DOI] [PubMed] [Google Scholar]
- 54.O’Hagan HM, Wang W, Sen S, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011. November;20(5):606–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding N, Bonham EM, Hannon BE, Amick TR, Baylin SB, O’Hagan HM. Mismatch repair proteins recruit DNA methyltransferase 1 to sites of oxidative DNA damage. J Mol Cell Biol. 2016. June;8(3):244–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maiuri AR, Peng M, Sriramkumar S, et al. Mismatch Repair Proteins Initiate Epigenetic Alterations during Inflammation-Driven Tumorigenesis. Cancer Res. 2017;77(13):3467–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu S, Rhee KJ, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009. September;15(9):1016–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hardman RA, Afshari CA, Barrett JC. Involvement of mammalian MLH1 in the apoptotic response to peroxide-induced oxidative stress. Cancer Res. 2001. February;61(4):1392–7. [PubMed] [Google Scholar]
- 59.Yanamadala S, Ljungman M. Potential role of MLH1 in the induction of p53 and apoptosis by blocking transcription on damaged DNA templates. Mol Cancer Res. 2003. August;1(10):747–54. [PubMed] [Google Scholar]
- 60.Chen F, Arseven OK, Cryns VL. Proteolysis of the mismatch repair protein MLH1 by caspase-3 promotes DNA damage-induced apoptosis. J Biol Chem. 2004. June;279(26):27542–8. [DOI] [PubMed] [Google Scholar]
- 61.Whitehall VL, Walsh MD, Young J, et al. Methylation of O-6-methylguanine DNA methyltransferase characterizes a subset of colorectal cancer with low-level DNA microsatellite instability. Cancer Res. 2001. February;61(3):827–30. [PubMed] [Google Scholar]
- 62.Han YW, Ikegami A, Rajanna C, et al. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol. 2005. August;187(15):5330–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013. August;14(2):195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Casasanta MA, Yoo CC, Smith HB, et al. A chemical and biological toolbox for Type Vd secretion: Characterization of the phospholipase A1 autotransporter FplA from Fusobacterium nucleatum. J Biol Chem. 2017. October: jbc.M117.819144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005. January 20;23(3):609–18. [DOI] [PubMed] [Google Scholar]
- 66.Xiao Y, Freeman GJ. The microsatellite instable subset of colorectal cancer is a particularly good candidate for checkpoint blockade immunotherapy. Cancer Discov. 2015. January;5(1):16–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mlecnik B, Bindea G, Angell HK, et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity. 2016. March;44(3):698–711. [DOI] [PubMed] [Google Scholar]
- 68.Koi M, Carethers JM. The colorectal cancer immune microenvironment and approach to immunotherapies. Future Oncol. 2017. August;13(18):1633–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mima K, Sukawa Y, Nishihara R, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015. August;1(5):653–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bicknell DC, Rowan A, Bodmer WF. Beta 2-microglobulin gene mutations: a study of established colorectal cell lines and fresh tumors. Proc Natl Acad Sci U S A. 1994. May 24;91(11):4751–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kloor M, Michel S, von Knebel Doeberitz M. Immune evasion of microsatellite unstable colorectal cancers. Int J Cancer. 2010. September;127(5):1001–10. [DOI] [PubMed] [Google Scholar]
- 72.Kloor M, Becker C, Benner A, Woerner SM, Gebert J, Ferrone S, von Knebel Doeberitz M. Immunoselective pressure and human leukocyte antigen class I antigen machinery defects in microsatellite unstable colorectal cancers. Cancer Res. 2005. July;65(14):6418–24. [DOI] [PubMed] [Google Scholar]
- 73.Giannakis M, Mu XJ, Shukla SA, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016. Apr. pii: S2211–1247(16)30364–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Purdy AK, Campbell KS. Natural killer cells and cancer: regulation by the killer cell Ig-like receptors (KIR). Cancer Biol Ther. 2009. December;8(23):2211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaplan CW, Ma X, Paranjpe A, et al. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect Immun. 2010. November;78(11):4773–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015. February;42(2):344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Coppenhagen-Glazer S, Sol A, Abed J, et al. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect Immun. 2015. March;83(3):1104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abed J, Emgård JE, Zamir G, et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe. 2016. August;20(2):215–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garcia M, Choi C, Kim HR, et al. Association between recurrent metastasis from stage II and III primary colorectal tumors and moderate microsatellite instability. Gastroenterology. 2012. July;143(1):48–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koi M, Garcia M, Choi C, et al. Microsatellite Alterations With Allelic Loss at 9p24.2 Signify Less-Aggressive Colorectal Cancer Metastasis. Gastroenterology. 2016. April;150(4):944–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koopman M, Kortman GA, Mekenkamp L, et al. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer. 2009. January 27;100(2):266–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee S-Y, Miyai K, Han HS, Hwang D-Y, Seong MK, Chung H, Jung BH, Devaraj B, McGuire KL, Carethers JM. Microsatellite instability, EMAST, and morphology associations with T cell infiltration in colorectal neoplasia. Dig Dis Sci 2012;57:72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carethers JM, Koi M, Tseng-Rogenski S. EMAST is a form of microsatellite instability that is initiated by inflammation and modulates colorectal cancer progression. Genes 2015;6:185–205. [DOI] [PMC free article] [PubMed] [Google Scholar]