Abstract
Background:
The central vein sign (CVS) is a promising magnetic resonance imaging (MRI) diagnostic biomarker for multiple sclerosis (MS). Recent studies demonstrate that patients with MS have higher proportions of white matter lesions with CVS compared to people with diseases that mimic MS on MRI. However, the clinical application of CVS as a biomarker is limited by inter-rater differences in the adjudication of CVS, as well as the time burden required for the determination of CVS for each lesion in a patient’s full MRI scan.
Methods:
We present an automated technique for the detection of CVS in white matter lesions.The method derives a CVS probability,πij for each lesion, as well as a patient-level CVS biomarker, ψi. The method is probabilistic in nature, allows for site-specific lesion segmentation methods, and is potentially robust to inter-site variability. The proposed algorithm was tested on imaging acquired at the University of Vermont in 16 participants who have MS, and 15 participants who do not.
Results:
Using the proposed automated technique, MS participants were found to have significantly higher values of ψ than non-MS participants ( = 0.55, sd = 0.18; = 0.31, sd = 0.12; p < 0.001). The algorithm was also found to show strong discriminative ability between MS and non-MS patients, with an area-under-the-curve of 0.88.
Conclusion:
The current study presents the first fully automated method for detecting CVS in white matter lesions and demonstrates promising performance in a sample of MS and non-MS cases.
1. Introduction
Multiple sclerosis (MS) is an inflammatory demyelinating disease of the central nervous system that is characterized by lesions in the brain and spinal cord. Currently, assessment of magnetic resonance imaging (MRI) factors heavily in the diagnosis of MS, with much importance placed on the distribution (dissemination in space) and time course of lesions (dissemination in time)1 in patients presenting with clinical symptoms typical for MS. However, current imaging-based diagnostic criteria favor sensitivity over specificity, making misdiagnosis of MS relatively common2,3. This is especially true among disorders that demonstrate white matter lesions similar to those found in MS4,5.
As a means for distinguishing MS lesions from white matter abnormalities arising from other diseases, the identification of a vein traversing the center of a lesion has been proposed as a diagnostic tool since inflammatory demyelination in the MS white matter is perivenular6,7. The potential for this marker to be used in the diagnosis of MS has been advanced by recent developments in MRI pulse sequences, which have enabled detailed imaging of veins in the brain8–10. Using these sequences, researchers have provided strong evidence that higher proportions of MS lesions show central vein sign (CVS) compared to lesions resulting from other disease processes commonly mistaken for MS7,11–16. This finding has been demonstrated for neuromyelitis optica spectrum disorder (NMOSD), systemic autoimmune diseases (SAD), cerebral small vessel disease (CSVD), Susac’s syndrome, and migraine. While further replication in a prospective setting is still necessary, a high proportion of brain MRI lesions demonstrating CVS appears to have potential as a biomarker with high specificity for MS.
Unfortunately, important barriers limit the feasibility of clinical application of CVS. Two such limitations are the presence of intra- and inter-rater variability in the subjective assessment of CVS, and the time required to adjudicate CVS in every MRI lesion per patient. Recent studies have attempted to mitigate the time burden associated with CVS assessment by limiting the number of lesions that are examined13,17. However, these techniques have the potential to increase variability, and have generally not been as successful as the evaluation of the proportion of CVS in all MRI lesions per patient7,18. Importantly, in studies that adjudicate all lesions per patient, optimal proportion cutoffs have differed across study sites and disease comparisons7,13,18. This variability highlights the need for thorough comparison and optimization of these cutoffs across samples and diseases, yet the same issues of rater subjectivity and temporal burden make this type of research difficult. As such, the current study introduces an algorithm for the automatic determination of CVS in white matter lesions, and presents a fully automated patient-level diagnostic biomarker. In this paper, we describe the CVS detection pipeline, present statistical measures of judgment accuracy, and discuss the implications and next steps for this line of research.
2. Methods
2.1 CVS detection algorithm
To adjudicate CVS for each lesion in a given participant, the several steps are carried out. We first present the overall summary and then address each step, with associated rationale, in detail below. To perform the algorithm, a T1-weighted volume (T1), T2-weighted FLAIR volume (T2-FLAIR), and T2*-weighted segmented echo-planar imaging volume (T2*-EPI) are required. 1) A map of the veins present in the T2*-EPI volume is created using a process referred to as vesselness filtering, and the vein map is rigidly registered to the T1 volume. 2) White matter lesions are segmented using the T1 and T2-FLAIR volumes. 3) Clear lesion boundaries are then determined using a process that removes ambiguous boundary voxels. 4) Periventricular lesions are removed from candidacy, per guidelines given by the North American Imaging in Multiple Sclerosis (NAIMS) cooperative19. 5) A permutation procedure is carried out to determine whether identified veins occur in the center of a given lesion to a greater degree than would be expected by chance. This yields a probability of CVS for each lesion j in patient i’s scan, denoted πij . Lesion-level CVS probabilities are then averaged to obtain a patient-level CVS biomarker, denoted ψi . 6) To account for scan motion, lesions’ contributions to the average can be weighted by the noise in their T2*-EPI intensities. Figure 1 demonstrates the algorithm’s steps on an example lesion. Importantly, while figures are necessarily presented in 2D space, all methods undertaken for this procedure are conducted in 3D volumetric space, and simultaneously consider all three planes of the image.
Figure 1:
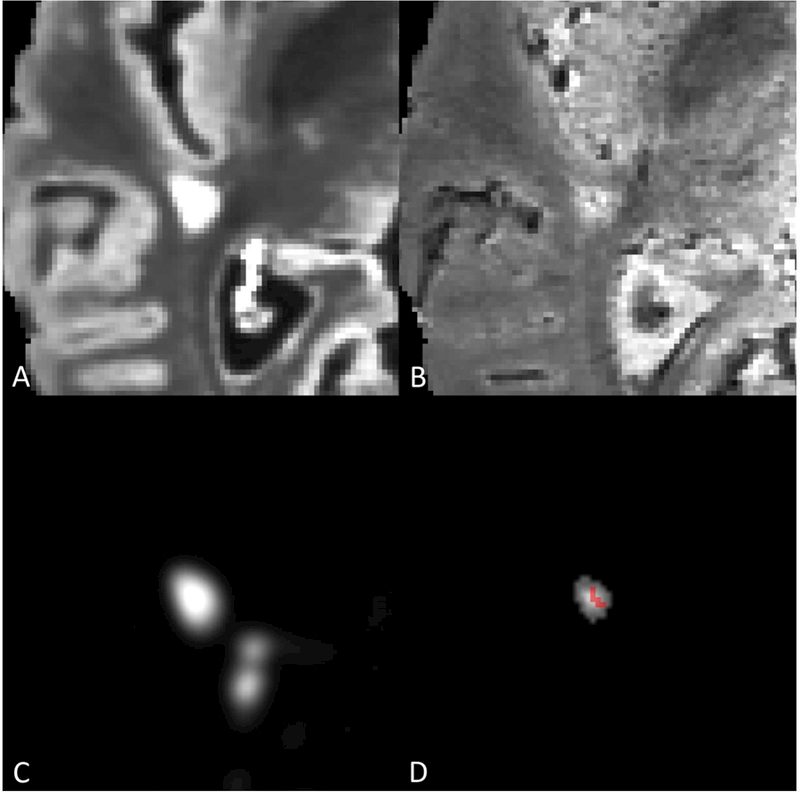
A) Axial slice of lesion on T2-FLAIR volume. B) Axial slice of lesion on T2*-EPI volume. C) MIMoSA lesion probability map. D) Distance-to-lesion-boundary mask with vesselness filter overlay. Lesion-level CVS probability following permutation was 0.975.
Vesselness filtering
Vein maps in the brain are created in order to later determine presence or absence of veins in each lesion. To do this, the Frangi vesselness filter20 is applied to the unregistered T2*- EPI volume (for the application to data, this study used the Convert3D toolbox), producing a map of scores ≥0, with scores of 0 implying no vesselness qualities. The Frangi filter is a vessel enhancement algorithm based on the Hessian matrix at each voxel, in which the second-order structure of the image is obtained through convolution with derivatives of Gaussian kernels. The scores are calculated using the eigenvalues of the Hessian matrix, specifically picking up on tubular structures that are darker (or lighter, depending on the implementation) than their surroundings. After being obtained in the unregistered T2*-EPI space, these “vesselness” maps are then rigidly registered to the T1 space.
Lesion segmentation
To determine the location and shape of white matter lesions, automatic lesion segmentation is performed on co-registered T1 and T2-FLAIR volumes. For the application to data, this study used the Method for Inter-Modal Segmentation Analysis (MIMoSA) model21 in the R statistical environment22. The lesion segmentation algorithm produces a map containing the probabilities that each voxel is part of a lesion. For the results presented in this paper, a threshold of 0.30 is applied to this probability map in order to create a binary lesion mask. The threshold of 0.30 was chosen because previous work has found it to be a conservative cutoff that can limit the amount of false positive lesion tissue23,24. Following the definition for a CVS+ lesion given by the NAIMS cooperative19, lesions detected by the MIMoSA model that are smaller than 3 mm in any plane are removed from candidacy.
Lesion boundary determination
Thresholding of the lesion probability map often results in pathologically distinct lesions being connected by ambiguous boundary voxels. For these lesions to be properly assessed for CVS, the proposed algorithm addresses this pseudo-confluence through a recently described technique that removes voxels that are connecting pathologically distinct lesions24. The technique works by finding regions in which the texture of the lesion probability map resembles the center of a lesion. Therefore, the centers that it produces are maintained and used for investigating CVS for the remainder of this algorithm. Further detail on the implementation of this method can be found in the original publication24.
As the NAIMS guidelines call for the exclusion of confluent lesions, in some cases of true confluence this step may represent a deviation from those recommendations. Yet it is important to note that many lesions that would be judged as discrete by expert raters are often merged by automated segmentation methods25. This can result in drastic and unrealistic degrees of pseudo-confluence in automated lesion masks, sometimes resulting in 50 or more distinct lesions being merged into fewer than 10 lesion components24. Thus, relying on automated determinations of confluence in automated lesion masks would likely result in the exclusion of many or most eligible lesions.
Periventricular lesion exclusion
The density and branching nature of veins near the ventricles makes assessment for CVS difficult in periventricular lesions, especially in cases where more than one distinct vein traverses the lesion. For this reason, the NAIMS Cooperative recommends excluding lesions with more than one vein or with branching veins19. The proposed algorithm addresses this consideration by excluding periventricular lesions from consideration, as periventricular lesions typically contain multiple veins. This exclusion is done by performing tissue-class segmentation on the T1 volumes (for the application to data, this study used the FAST algorithm in the FSL26), expanding the cerebrospinal fluid (CSF) region of the brain by 3mm, and eliminating lesions from the lesion-center mask that overlap with the expanded CSF region. The choice of a 3mm expansion was made based on visual inspection of randomly selected T2*EPI volumes, for which 3mm appeared to include most of the branching vein structure discussed in the consensus statement, without removing too much of the deep white matter. Notably, although this technique excludes periventricular lesions, it does not exclude other lesions that may have multiple veins. This represents a second deviation from the NAIMS recommendations, which could potentially be addressed by future advances in methods for segmenting and counting distinct veins.
CVS permutation procedure
In lesions that contain central veins, one would expect above-average coherence between voxels’ centrality within the lesion and their vesselness score. The proposed permutation procedure takes advantage of that expectation to examine the degree to which a lesion’s most vein-like voxels are more concentrated in the lesion’s center than one might expect to observe by chance. First, a vein-center coherence score for lesion j in patient i’s scan, Cij, is calculated by summing over the products of each voxel’s distance-to-nearest-lesion-boundary (i.e., centrality) score, dijv, and its Frangi vesselness score, fijv. The coherence formula is given by,
where V is the set of all voxels in lesion j. Thus, higher values of this score indicate that the highest vesselness values within the lesion tended to occur in the same voxels as the highest centrality values.
To determine the degree to which this score deviates from chance in cases where there is no biological correspondence between vesselness and location within lesions, a lesion-specific null distribution of coherence scores is created using 1000 random permutations. For each permutation, p, the vesselness scores of the voxels in lesion j are randomly resampled without replacement, yielding a randomly ordered set of values, . A null coherence score is then calculated using the formula,
This permutation procedure is performed 1000 times, resulting in a sample of 1000 null coherence scores. The lesion-level CVS probability, πij, is then calculated as proportion of chance (null-distributed) CVS scores that are smaller than the observed score, given by,
To obtain a subject-level CVS biomarker, ψi, these probabilities are averaged over all lesions observed in patient i. The formula for ψi, is given by,
where NL is the number of candidate lesions in patient i’s scan. The biomarker, ψi can be interpreted similarly to the proportion of the patient’s lesions that demonstrate CVS.
Optional noise weighting
When taking the average of the CVS probabilities for a patient’s lesions, some lesions may have more reliable estimates than others. A more stable biomarker can potentially be obtained by weighting each lesion’s contribution to the biomarker by the amount of noise in the lesion’s voxels on the T2*-EPI volume. To estimate the level of noise in a lesion, a “noiseless” T2*-EPI is first constructed by performing anisotropic diffusion on the original scan27. This procedure results in a smoothed volume that maintains tissue boundaries and other image gradients. Then, for voxels in the lesion, the difference is taken between the original T2*-EPI and the smoothed T2*-EPI. A noise value is finally calculated by dividing the sum of the squared voxel differences by the total number of voxels in the lesion. For lesion j, this value is defined as,
where Iv and are voxel v’s intensity and smoothed intensity, respectively. The desired reliability weight is simply the inverse of this noise value,
A weighted subject-level biomarker, , is then calculated by summing the products of the lesions’ CVS probabilities with their weights, and dividing by the sum of the weights. The weighted biomarker is given by,
2.2 Implementation and software
To accompany this article, code for the central vein detection algorithm has been made freely available online (https://github.com/jdwor/cvs). One file, centralveins_full.R, contains code to run all preprocessing and analysis steps described in the previous section. This file serves to increase understanding of all steps used in this study and to provide a straightforward tool that can be applied to raw images. A second file, centralveins_simple.R, contains code to be run directly on a probability map and a vein map. This file serves to improve implementations across different sites and scanners, for which researchers and clinicians may have preferred pipelines for preprocessing and lesion segmentation. Following preprocessing and structure segmentation, the centralveins_simple function was found to take an average of 17.7 minutes (sd = 9.1), and was roughly broken down as a 10 minute baseline with an additional 20 seconds per lesion, when run without parallelization. Finally, a third file, helperfunctions.R, provides additional functions used within the previous two files.
2.3 Validation
Data
For this study, data were analyzed for 40 research participants recruited from the University of Vermont neurology clinic as part of a study aiming to improve diagnostic specificity for MS17. Participants were between 20 and 67 years of age, and 37 were female. 10 had MS and no comorbidities known to produce MRI white matter abnormalities; 10 had MS and comorbidities known to produce MRI white matter abnormalities; 10 had migraine with MRI white matter abnormalities and no other white matter comorbidities; and 10 were previously incorrectly diagnosed with MS and had MRI white matter abnormalities and a variety of diagnoses (Table 1).
Table 1.
Demographics of the study sample
| MS (n=10) | |
|---|---|
| Age | 44 (16) |
| Sex | 9/10 Female |
| Disease duration | 9 (7) |
| Disease subtype | 10/10 RRMS |
| MS with comorbidities (n=10) | |
| Age | 43 (9) |
| Sex | 9/10 Female |
| Disease duration | 9 (6) |
| Disease subtype | 10/10 RRMS |
| Migraine (n=10) | |
| Age | 47 (13) |
| Sex | 10/10 Female |
| Misdiagnosed with MS (n=10) | |
| Age | 53 (7) |
| Sex | 9/10 Female |
Whole-brain 3D T2-FLAIR, T1, and T2*-EPI28 volumes were acquired in a 3 tesla (T) Philips dStream MRI scanner with a 32-channel head coil. FLAIR and T1 volumes were obtained with 1-mm isotropic resolution, and T2*-EPI volumes were obtained with 0.55-mm isotropic resolution. N4 bias correction29 was performed on all images, and the T2-FLAIR volume for each participant was interpolated to a voxel size of 1 mm3 and rigidly co-registered to the T1 volume. Extracerebral voxels were removed from the T1 volume using a skull-stripping procedure30, and the brain mask was applied to the T2-FLAIR volume.
Motion exclusion criteria
Since head motion might occur during the T2*-EPI scan, potentially producing uninterpretable images, each participant’s T2*-EPI scan was manually rated for motion in the relevant white matter regions. Scans were scored from 1 to 5, where 1 indicated “perfect, no artifacts and excellent signal-to-noise,” 2 indicated “only one minor artifact that does not obscure any vessels in supratentorial white matter,” 3 indicated “more than one artifact that do not obscure any vessels in supratentorial white matter,” 4 indicated “more than one artifact that do obscure some vessels in supratentorial white matter,” and 5 indicated “severe artifacts or bad signal-to-noise that do obscure most vessels in supratentorial white matter.” It was decided a priori that scans that were rated 5 would be removed for the primary analysis, as scans with that degree of motion may be unusable in clinical practice as well.
Performance assessment
As CVS shows great promise as a diagnostic biomarker, the performance of this algorithm in distinguishing between MS and non-MS cases is of primary interest. To determine whether the automated biomarkers, ψi and , replicate the findings from previous work that the distribution of manually adjudicated central vein proportion differs between MS and its mimics, t-tests were used to compare the automated CVS values for MS and non-MS cases. To determine the diagnostic utility of ψi and , the area under the curve (AUC) values of the receiver operating characteristic (ROC) curves were estimated. The presence of a difference in performance between of ψi and was tested with DeLong’s test for comparing the areas under correlated ROC curves31, using the pROC package in the R statistical environment22,32. Sensitivity and specificity were calculated using the 40% cutoff7, under which inflammatory demyelination is diagnosed if 40% or more of white matter lesions exhibit CVS, as well as the more recently proposed 50% cutoff18. Additionally, locally optimal cutoffs were determined, and their sensitivity and specificity values were compared to those obtained using established cutoffs.
Finally, these cutoffs were compared to the performance of proportion cutoffs applied to manual determinations of CVS in previous research7,13,18, as well as the performance of three recently proposed clinical decision rules that do not require the assessment of the full set of lesions in a scan. The first such rule, referred to as the rule of 613, states that inflammatory demyelination is diagnosed if there are more than six lesions with CVS, or if more than half of lesions show CVS. The second and third, referred to as select315 and select3*17, state that inflammatory demyelination is diagnosed if CVS is found in at least 2 of 3 lesions pre-selected on T2-FLAIR and FLAIR*9 imaging, respectively.
3. Results
Following manual ratings of scan noise due to motion, 9 participants were excluded and 31 remained for the primary analysis. Of the remaining 31 participants, 16 had MS and 15 did not. Automated CVS detection was performed on these 31 participants, using the algorithms and software packages described in the previous section. Two-sample t-tests were run to determine whether the automated CVS scores differed between the 16 MS and 15 non-MS cases. In both the unweighted (MMS = 0.56, SDMS = 0.17; Mnon-MS = 0.37, SDnon-MS = 0.12; p < 0.01) and weighted (MMS = 0.55, SDMS = 0.18; Mnon-MS = 0.31, SDMS = 0.12; p < 0.001) variants of the algorithm, the within-patient average CVS probabilities were higher in patients with MS compared to patients without MS. See Figure 2 for breakdowns across all four groups.
Figure 2:
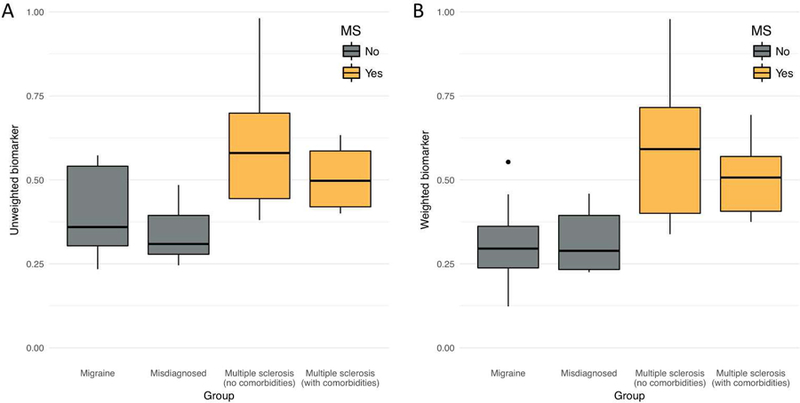
Boxplots of patient-level central vein sign (CVS) biomarker score, by diagnostic group. The score can be interpreted as the proportion of lesions that are CVS+ according to the method described in this paper. Groups shaded gold do not carry an MS diagnosis, whereas groups shaded gray do. A) Boxplots for the unweighted biomarker. B) Boxplots for the noise-weighted biomarker. Points outside of the boxplots represent outliers within their respective groups.
To determine the diagnostic utility of the automated biomarkers, ψi and , ROC curves were estimated, and their AUCs were calculated. For the unweighted case, ψi yielded an AUC of 0.84 (Figure 3A). Based on the 40% rule, applying a cutoff of 0.40 to ψi yielded a sensitivity of 0.94 and a specificity of 0.67. Based on the 50% rule, applying a cutoff of 0.50 to this biomarker yielded a sensitivity of 0.56 and a specificity of 0.80. Three locally optimal cutoffs appear to occur at 0.38, at which sensitivity was 1.00 and specificity was 0.67, at 0.44, at which sensitivity was 0.75 and specificity was 0.73, and at 0.50, at which sensitivity was 0.56 and specificity was 0.80 (Table 2).
Figure 3:
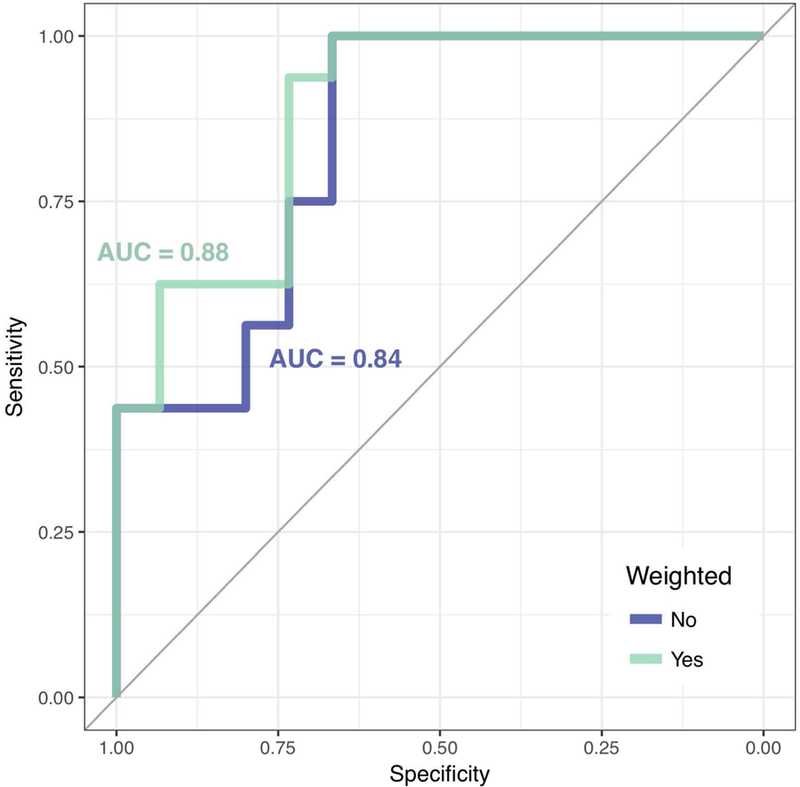
Receiver operating characteristic (ROC) curves of MS diagnosis based on patient-level automated CVS biomarker scores. The ROC curve for the unweighted biomarker is shaded blue, and the ROC curve for the weighted biomarker is shaded green. The AUC values for both curves are displayed in their respective colors.
Table 2.
Diagnostic performance of weighted and unweighted biomarkers.
| Threshold | Sensitivity | Specificity | |
|---|---|---|---|
| Unweighted biomarker (ψi ) |
0.38 | 1.00 (0.75-1.00) | 0.67 (0.42-0.84) |
| 0.40 | 0.94 (0.70-1.00) | 0.67 (0.42-0.84) | |
| 0.44 | 0.75 (0.50-0.90) | 0.73 (0.47-0.89) | |
| 0.50 | 0.56 (0.25-0.65) | 0.80 (0.58-0.94) | |
| Weighted biomarker |
0.37 | 0.94 (0.70-1.00) | 0.73 (0.42-0.84) |
| 0.40 | 0.75 (0.50-0.90) | 0.73 (0.42-0.84) | |
| 0.46 | 0.63 (0.40-0.80) | 0.93 (0.68-1.00) | |
| 0.50 | 0.56 (0.35-0.75) | 0.93 (0.74-1.00) |
For the noise-weighted case, yielded an AUC of 0.88 (Figure 3B). Applying a cutoff of 0.40 to yielded a sensitivity of 0.75 and a specificity of 0.73. Applying a cutoff of 0.50 yielded a sensitivity of 0.56 and a specificity of 0.93. Two locally optimal cutoffs for appear to occur at 0.37, at which sensitivity was 0.94 and specificity was 0.73, and at 0.46, at which sensitivity was 0.63 and specificity was 0.93 (Table 2). Although the weighting appeared to produce marginally improved performance, no significant difference was found using DeLong’s test (Z = 0.77, p = 0.22). Robustness analysis on the full sample of 40 participants after reintroducing the motion-obscured scans showed AUC values of 0.77 and 0.81 for ψi and respectively.
Previous studies that utilized CVS proportions within patients’ full set of lesions obtained optimal sensitivity/specificity of 1.00/1.00 when comparing MS cases to undiagnosed non-MS cases7, patients with microangiopathic lesions13, and patients with inflammatory valculopathies18. Prior research on a subset of the current sample was unable to obtain perfect discrimination between MS and migraine patients when adjudicating CVS for all lesions15. Compared to cutoffs that utilized the full set of lesions, decision rules based on a subset of lesions were generally less discriminative between MS and non-MS participants. The rule of 6 did obtain sensitivity/specificity of 1.00/1.00 for distinguishing patients with MS and small-vessel ischemia13, yet in the current sample of MS, migraine, and misdiagnosed patients, the select3 procedure obtained sensitivity/specificity of 0.81/0.95, and the select3* procedure obtained sensitivity/specificity of 0.81/0.8317.
4. Discussion
Preliminary studies have proposed and validated CVS as a promising biomarker for differentiation MS from other diseases that cause MRI white matter abnormalities7,15,19. Yet concerns remain regarding the heavy temporal burden on manual adjudication of CVS, as well as the subjective differences that may arise in response to variation in adjudicators’ time constraints and intuition. This study sought to address these issues by introducing an algorithm for automated CVS detection that could in principle, following further validation, be applied in clinical practice.
In the primary analysis, the algorithm was tested on a cohort of 16 MS (eight with and eight without other white matter comorbidities) and 15 non-MS (eight with migraine and seven misdiagnosed with MS) cases. The fully automated technique replicated previous work that used manual adjudications7,11–16 by demonstrating that proportions of lesions with CVS differ significantly between MS and its mimics. Additionally, the automated biomarkers, ψi and were found to have strong diagnostic ability, with AUCs of 0.84 and 0.88 and optimal sensitivity/specificity of approximately 0.94/0.70. There is also great promise for this algorithm to perform consistently across study sites and MRI scanners, as in-house preprocessing and lesion segmentation methods can be easily substituted, and the remaining steps (obtaining vesselness scores, finding lesion centers, and calculating CVS probabilities) do not require parameter tuning.
Importantly, the automated biomarkers presented in this study did not perform as well as previously obtained proportions of CVS based on manual ratings of all lesions in patients’ scans. Specifically, the 40% and 50% cutoffs used in prior manually rated studies often achieved perfect discrimination between MS and non-MS cases7,18, which the automated biomarkers were not able to replicate. However, previous work in a subset of the current sample showed that manual ratings of all lesions did not fully distinguish migraine patients from patients with MS and no white-matter comorbidities15. This suggests that the non-MS cases present in the current sample might be more difficult to distinguish from MS using CVS alone than the non-MS cases present in the studies that did obtain perfect discrimination.
Additionally, although the sensitivity and specificity obtained by these biomarkers were lower than manually obtained CVS proportions, the biomarkers performed comparably to decision rules that use only a subset of lesions in a scan17. Thus, while automated adjudication of every lesion in a scan is not yet as accurate as manual adjudication of every lesion in a scan, the proposed automated method shows promise as an alternative to other clinically feasible methods for identifying inflammatory demyelination. Further study and refinement of this technique has the potential to yield biomarkers that are both feasible for use in the clinic and comparable in accuracy and reliability to CVS proportions obtained by manual adjudication.
There are a number of important limitations to the proposed algorithm. First, biomarker values were found to be lower than previously reported CVS proportions for MS patients, and higher than previously reported CVS proportions for non-MS patients. It is possible this effect is due to errors in lesion segmentation, which would pull the values of MS and non-MS participants toward each other due to the assessment of non-informative false-positive lesions. Because this method allows for in-house lesion segmentation algorithms to be applied, the impact of false- positive lesions could potentially be mitigated in practice. It is also possible that the effect is due to false-positives or false-negatives in automated CVS assessment. Future work will use manual lesion-level assessments to tease apart these potential sources.
Additionally, the exclusion of 9 of the 40 subjects due to noise in the T2*-EPI scan represents a potential weakness of this automated method. However, robustness analysis found that the performance of the method on the full sample was not drastically reduced compared to the high-quality subset. This finding suggests that, in clinical practice, a great deal of motion would not render a scan useless but instead may be an additional consideration for clinicians when interpreting the algorithm’s results.
Although the potential clinical implications of an automated tool for CVS adjudication call for further study and refinement of such techniques, the current study demonstrates the promising performance of a fully automated method for detecting CVS in white matter lesions. To our knowledge, this is the first automated technique for this challenging aspect of MS diagnosis, and represents an important step forward toward a specific MRI biomarker for MS lesions.
Acknowledgments
Funding
The project described is supported in part by the NIH grants RO1 NS085211, R01 NS060910, and R21 NS093349 from the National Institute of Neurological Disorders and Stroke (NINDS), R01 MH 112847 from the National Institute of Mental Health and R01 EB 017255 from the National Institute of Biomedical Imaging and Bioengineering. The research is also supported by the Intramural Research Program of NINDS (Z01 NS003119) and grants RG-1507–05243 and RG-1707–28586 from the National Multiple Sclerosis Society. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Abbreviations
- CVS
Central Vein Sign
- MIMoSA
Method for Inter-Modal Segmentation Analysis
- NAIMS
North American Imaging in Multiple Sclerosis
- ROC
Receiver Operating Characteristic
- AUC
Area Under the Curve
References
- 1.McDonald WI et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann. Neurol 50, 121–127 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Solomon AJ, Klein EP & Bourdette D ‘Undiagnosing’ multiple sclerosis: the challenge of misdiagnosis in MS. Neurology 78, 1986–1991 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solomon AJ et al. The contemporary spectrum of multiple sclerosis misdiagnosis: A multicenter study. Neurology 87, 1393–1399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aliaga ES & Barkhof F MRI mimics of multiple sclerosis. Handb. Clin. Neurol 122, 291–316 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Miller DH et al. Differential diagnosis of suspected multiple sclerosis: a consensus approach. Mult. Scler. Houndmills Basingstoke Engl 14, 1157–1174 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fog T On the vessel-plaque relationships in the brain in multiple sclerosis. Acta Neurol. Scand 40, 9–15 (1964). [PubMed] [Google Scholar]
- 7.Tallantyre EC et al. Ultra-high-field imaging distinguishes MS lesions from asymptomatic white matter lesions. Neurology 76, 534–539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reichenbach JR, Venkatesan R, Schillinger DJ, Kido DK & Haacke EM Small vessels in the human brain: MR venography with deoxyhemoglobin as an intrinsic contrast agent. Radiology 204, 272–277 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Sati P, George IC, Shea CD, Gaitán MI & Reich DS FLAIR*: A Combined MR Contrast Technique for Visualizing White Matter Lesions and Parenchymal Veins. Radiology 265, 926–932 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campion T et al. FLAIR* to visualize veins in white matter lesions: A new tool for the diagnosis of multiple sclerosis? Eur. Radiol 27, 4257–4263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilsdonk ID et al. Improved differentiation between MS and vascular brain lesions using FLAIR* at 7 Tesla. Eur. Radiol 24, 841–849 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Kister I, Herbert J, Zhou Y & Ge Y Ultrahigh-Field MR (7 T) Imaging of Brain Lesions in Neuromyelitis Optica. Mult. Scler. Int 2013, 398259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mistry N et al. Imaging central veins in brain lesions with 3-T T2*-weighted magnetic resonance imaging differentiates multiple sclerosis from microangiopathic brain lesions. Mult. Scler. Houndmills Basingstoke Engl 22, 1289–1296 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Sinnecker T et al. Distinct lesion morphology at 7-T MRI differentiates neuromyelitis optica from multiple sclerosis. Neurology 79, 708–714 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Solomon AJ et al. ‘Central vessel sign’ on 3T FLAIR* MRI for the differentiation of multiple sclerosis from migraine. Ann. Clin. Transl. Neurol 3, 82–87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wuerfel J et al. Lesion morphology at 7 Tesla MRI differentiates Susac syndrome from multiple sclerosis. Mult. Scler. Houndmills Basingstoke Engl 18, 1592–1599 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Solomon AJ et al. Diagnostic performance of central vein sign for multiple sclerosis with a simplified three-lesion algorithm. Mult. Scler. Houndmills Basingstoke Engl 1352458517726383 (2017). doi: 10.1177/1352458517726383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maggi P et al. Central vein sign differentiates Multiple Sclerosis from central nervous system inflammatory vasculopathies: Central Vein Sign. Ann. Neurol (2018). doi: 10.1002/ana.25146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sati P et al. The central vein sign and its clinical evaluation for the diagnosis of multiple sclerosis: a consensus statement from the North American Imaging in Multiple Sclerosis Cooperative. Nat. Rev. Neurol 12, 714–722 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Frangi AF, Niessen WJ, Vincken KL & Viergever MA Multiscale vessel enhancement filtering. in Medical Image Computing and Computer-Assisted Intervention — MICCAI’98 (eds. Wells WM, Colchester A & Delp S) 1496, 130–137 (Springer Berlin; Heidelberg, 1998). [Google Scholar]
- 21.Valcarcel AM et al. MIMoSA: An Automated Method for Intermodal Segmentation Analysis of Multiple Sclerosis Brain Lesions: Method For Inter-Modal Segmentation Analysis. J. Neuroimaging (2018). doi: 10.1111/jon.12506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2015). [Google Scholar]
- 23.Sweeney EM et al. OASIS is Automated Statistical Inference for Segmentation, with applications to multiple sclerosis lesion segmentation in MRI. NeuroImage Clin 2, 402–413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dworkin JD et al. An Automated Statistical Technique for Counting Distinct Multiple Sclerosis Lesions. Am. J. Neuroradiol (2018). doi: 10.3174/ajnr.A5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oguz I et al. Dice Overlap Measures for Objects of Unknown Number: Application to Lesion Segmentation. in Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries (eds. Crimi A, Bakas S, Kuijf H, Menze B & Reyes M) 10670, 3–14 (Springer International Publishing, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Brady M & Smith S Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 20, 45–57 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Perona P & Malik J Scale-space and edge detection using anisotropic diffusion. IEEE Trans. Pattern Anal. Mach. Intell 12, 629–639 (1990). [Google Scholar]
- 28.Sati P et al. Rapid, high-resolution, whole-brain, susceptibility-based MRI of multiple sclerosis. Mult. Scler. J 20, 1464–1470 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tustison NJ et al. N4ITK: Improved N3 Bias Correction. IEEE Trans. Med. Imaging 29, 1310–1320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carass A et al. A joint registration and segmentation approach to skull stripping. in Biomedical Imaging: From Nano to Macro, 2007. ISBI 2007. 4th IEEE International Symposium on 656–659 (IEEE, 2007). [Google Scholar]
- 31.DeLong ER, DeLong DM & Clarke-Pearson DL Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44, 837–845 (1988). [PubMed] [Google Scholar]
- 32.Robin X et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]


