Abstract
Mass spectrometry (MS)-based integrated metaproteomic, metabolomic, and lipidomic (multi-omic) studies are transforming our ability to understand and characterize microbial communities in environmental and biological systems. These measurements are even enabling enhanced analyses of complex soil microbial communities, which are the most complex microbial systems known to date. Multi-omic analyses, however, do have sample preparation challenges, since separate extractions are typically needed for each omic study, thereby greatly amplifying the preparation time and amount of sample required. To address this limitation, a 3-in-1 method for the simultaneous extraction of metabolites, proteins, and lipids (MPLEx) from the same soil sample was created by adapting a solvent-based approach. This MPLEx protocol has proven to be both simple and robust for many sample types, even when utilized for limited quantities of complex soil samples. The MPLEx method also greatly enabled the rapid multi-omic measurements needed to gain a better understanding of the members of each microbial community, while evaluating the changes taking place upon biological and environmental perturbations.
Keywords: Chemistry, Issue 135, MPLEx, Metaproteomics, Lipidomics, Metabolomics, Multi-omics, Mass Spectrometry
Introduction
Evaluating soil microbial communities has important implications for understanding carbon cycling and climate change. Recent studies have however highlighted difficulties, such as the lack of sequenced genomes for microbiota in various soil types and the unknown function of many of the proteins detected. These challenges result due to soil being the most complex microbial community known to date1,2,3. Multi-omic analyses, which combine results from metagenomic, metatranscriptomic, metaproteomic, metabolomic, and lipidomic studies, have recently been implemented in numerous soil studies to gain a greater understanding into the microbes present, while obtaining comprehensive information about the molecular changes taking place due to environmental perturbations1,4,5. One challenge with multi-omic studies is that the mass spectrometry (MS)-based metaproteomic, metabolomic, and lipidomic measurements typically require a specific extraction process for each omic to be MS compatible6,7,8,9. These precise procedures make their implementation extremely difficult or impossible when only a limited quantity of sample is available. These challenges have prompted us to investigate a simultaneous metabolite, protein, and lipid extraction (MPLEx) method capable of using smaller sample volumes or masses, improving accuracy, and providing faster sample preparations for all three analyses10. To date, there are no alternate soil extraction procedures that can achieve all of these goals.
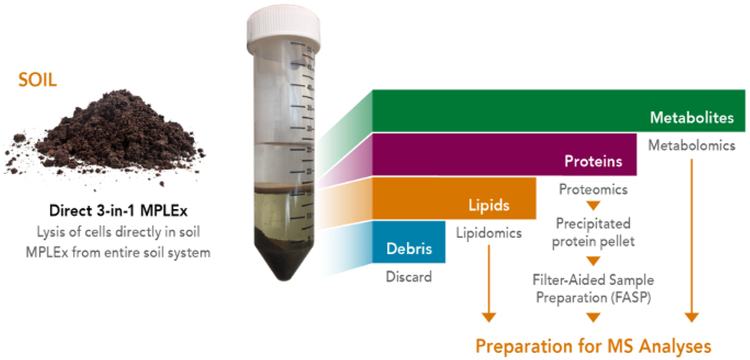
To enable global multi-omic analyses of a single soil sample, an organic solvent extraction protocol based upon chloroform, methanol, and water separations was utilized10. This method was originally developed for total lipid extractions9,11 and more recently was amended for the simultaneous extraction of metabolites, proteins, and lipids from a single sample12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30, enabling less sample quantity and experimental variability10. In the MPLEx protocol, chloroform is not miscible with water, which provides the basis for the triphasic chemical separation of sample constituents into distinct fractions. The top aqueous phase therefore contains the hydrophilic metabolites, followed by a protein disk, and then a lipid layer in the bottom chloroform phase (Figure 1). When MPLEx is applied to most soils, particulate debris accumulates at the very bottom of the sampling tubes and can be discarded after all layers are collected. Each soil type can be different, however, and in highly organic soil such as peat, the soil debris stays in the middle layer and does not fall to the bottom of the sampling tube. MPLEx provides several advantages when isolating multiple molecule types from the same sample such as 1) smaller sample quantities can be used for multi-omic analyses, 2) multi-omic extractions from the same sample decrease overall experimental variability, and 3) greater numbers of samples can be prepared much faster for higher throughput studies10. Together these benefits are vital for providing better measurement capabilities for evaluating soil samples and their complex microbial communities.
Figure 1. A schematic showing the MPLEx process, in which metabolites, proteins and lipids are simultaneously extracted from the same soil sample for MS analyses.
Protocol
NOTE: Very wet soils can be lyophilized prior to extraction without detriment to the effectiveness of the extraction. Wet soil can also be used but should be considered when adding reagents at specific ratios.
NOTE: It is recommended to use 20 g of dry soil weight per extraction, which must be split between two 50 mL tubes (maximum of 10 g soil per 50 mL tube). Extractions can be scaled up or down dependent upon available sample.
NOTE: Dry soil samples can be sieved through a 3 mm screen in order to homogenize and remove small roots and rocks. Do not sieve wet soil samples, as the sample will get stuck in the screen.
1. Soil Cell Lysis and Extraction of Metabolites, Proteins, and Lipids (Timing ~1 d)
-
Per 20 g soil sample, weigh 10 g into two separate 50 mL tubes that are methanol/chloroform compatible. Be extremely careful with the tubes chosen, as chloroform will leach most plastics, contaminating the samples. Use glass whenever possible or plastic tubes made from polypropylene or polytetrafluoroethylene (PTFE).
NOTE: Keep the soil on ice and as cool as possible during the initial weighing and homogenization steps.
Add 10 mL of chloroform-washed stainless steel and garnet beads to each tube. On ice, add 4 mL of cold ultrapure water (see Table of Materials for purification system) to each tube (one sample split between two tubes) and transfer the samples in an ice bucket into a fume hood.
-
Using a 25 mL glass serological pipette, quickly add 20 mL of ice-cold 2:1 chloroform:methanol (v/v).
Caution: Chloroform:methanol has acute potential health effects: skin irritation, possible chemical burns, and irritation to the respiratory system. It may affect the kidneys, liver, and heart. Wear suitable protective glasses, clothing, and gloves, and always work in a fume hood.
Tighten the lids and vortex into solution. 2:1 choloroform:methanol helps break down the cell wall of prokaryotes and also inactivates enzymatic activities.
Attach the tubes to 50 mL tube vortex attachments and horizontally vortex for 10 min at 4 °C inside a fridge if possible. Then, place the samples inside a −80 °C freezer for ~15 min in order to cool them down completely.
Using a probe sonicator inside a fume hood, sonicate each sample with a 6 mm (1/4”) probe at amplitude 60% for 30 s each on ice. Caution: It is recommended to use a sound abating enclosure and ear protection while sonicating. High voltage is present in the power supply and high frequency cable. Avoid touching the bottom or sides of the sample vessel with an active probe; it may crack or melt the plastic.
Place the samples in the −80 °C freezer for ~15 min, as sonicating can generate a lot of heat.
-
Repeat steps 1.5 – 1.7.
NOTE: Make sure the samples stay cold throughout the lysis procedure.
Centrifuge the samples at 4,000 × g for 5 min, 4 °C. At this point, the sample will be separated into the upper metabolite layer, the protein interlayer, and the lower lipid layer (and remaining debris pellet). See Figure 1.
-
Place the samples on ice in an ice bucket and, inside a fume hood and using a 10 mL glass serological pipette, remove the upper metabolite layers from the two 50 mL tubes into one large glass vial.
NOTE: Avoid aspirating any of the protein layer into the pipette; leave a small layer of metabolite if necessary.
Slightly tilt the 50 mL tube to release the protein interlayer so that it is free floating upon the lower lipid layer. Using a clean stainless steel flat-head lab spatula, carefully scoop both of the protein interlayers and place them together into one new 50 mL tube.
-
Using a 25 mL glass serological pipette, remove the lower lipid layers into one large glass vial.
NOTE: Try to avoid aspiring soil particles; however, some soil debris and particles aspired along with the lipids will be removed at a later time.
Place breathable membranes over the top of the metabolite and lipid glass vials and dry in a vacuum concentrator until dryness (lipids ~4 – 5 h, metabolites overnight).
To the protein sample tube and the remaining debris pellet tubes, add 20 mL of ice-cold methanol to each and vortex. This is done to the debris pellets to rinse off chloroform before attempting to solubilize any possible remaining protein out of the debris before tossing it.
Centrifuge the debris pellets and protein interlayer at 4,000 × g for 5 min, 4 °C, then decant the methanol into a hazardous waste container inside a fume hood.
-
Freeze the protein and debris pellets in liquid nitrogen and dry in a lyophilizer (with a collector temperature capable of −105 °C due to methanol having a very low freezing point of −98 °C) for ~2 hours.
NOTE: Do not over-dry the pellets, as they will be more difficult to solubilize.
-
Add 10 mL of protein solubilization buffer (4% SDS, 100mM DTT (DL-dithiothreitol) in 50mM tris buffer, pH 8.0; see Supplementary Reagent Set-up) to the protein interlayer tube and 20 mL to each of the debris pellets.
Caution: SDS causes acute toxicity and is flammable. It is a skin, eye, and airway irritant. Wear gloves and safety glasses.
In the fume hood, probe sonicate the samples at 20% amplitude for 30 s to bring them into solution. Vortex for 2 min.
Place the protein interlayer sample into a lab tube rotator for 30 min at 300 rpm, 50 °C, to solubilize the protein.
Horizontally vortex the debris samples for an additional 10 min to lyse any remaining intact cells, then rotate with the protein interlayer samples for the remaining 20 min.
Centrifuge all of the samples at 4,500 × g for 10 min, room temperature (RT), and collect the supernatant from each tube per sample into two 50 mL tubes.
Add 10 mL of solubilization buffer to the protein interlayer pellet, then sonicate and vortex back into solution and centrifuge as before.
-
Combine the supernatants into the two 50 mL tubes equally and centrifuge at 8,000 × g for 10 min, 4 °C, in a fixed angle bucket rotor.
NOTE: A final centrifugation is necessary to remove excessive contaminating humic substances.
-
Decant the supernatants into two 50 mL tubes so that there is 30 mL in each. Then, using a 10 mL glass serological pipette, add 7.5 mL of TCA (trichloroacetic acid) to each tube. Vortex into solution. This makes 20% TCA in each 30 mL sample (adjust accordingly),
Caution: TCA is caustic, toxic and may cause skin burns. Wear gloves and safety glasses.
-
Place the samples in a −20 °C freezer for 2 h to overnight.
NOTE: Proteins will typically precipitate within 1 h but can be left overnight (up to 18 h).
NOTE: Do not let the TCA extraction go longer than 18 h due to possible acid hydrolysis of the protein. If the sample is frozen, thaw it on ice; do not let the sample warm up past thaw.
To pellet the precipitated protein, centrifuge the sample at 4,500 × g for 10 min, 4 °C and decant the supernatant into waste.
Add 10 mL of 100% ice-cold acetone to each protein pellet, vortex and combine like pellets into one tube.
-
Centrifuge the tube containing the combined pellet (using a balance), and then decant the supernatant into waste.
Caution: Acetone may cause respiratory tract and skin and eye irritation, and is a flammable liquid and vapor. Wear safety glasses gloves and a lab coat, work in a fume hood.
Wash the pellet twice using 1.5mL of acetone and finally transferring to a 2 mL tube for the final spin at 10,000 × g for 5 min.
Decant the supernatant into waste and allow the pellet to dry inverted on a paper wipe in a fume hood for ~20 min or under a nitrogen stream until the pellet slightly begins to crack.
-
Add 100 – 200 μL of the protein solubilization buffer, depending on the size of the pellet.
NOTE: Keep the volume of solubilization buffer added to the sample as low as possible. Subsequent Filter Aided Sample Preparation (FASP) can only use up to 50 μL of solubilized sample per column; any more must be split into multiple FASP columns.
Sonicate and vortex the pellet into solution. Note that the sample may be viscous due to humic substances precipitating along with the protein, which will be removed with a subsequent centrifugation.
Shake the sample in an incubator/shaker for 30 min at 300 rpm, 40 °C, to solubilize the protein into solution and proceed to protein digestion.
Snap freeze the sample in liquid nitrogen and store in a −80 °C freezer until ready for protein digestion.
2. Lipid Preparation (Timing ~20 min)
Once the lipid samples are dry, add 200 μL of 2:1 chloroform:methanol to the vial, vortex into solution and transfer into a 1.5 mL polypropylene tube, add an additional 200 μL to the glass vial, vortex and pipette the remaining lipids and transfer to the tube.
Centrifuge the debris out of the sample at 12,000 × g for 5 min at 4 °C.
Transfer the supernatant into a glass lipid vial and store at −20 °C until LC-MS/MS analysis (additional details for LC-MS/MS in Supplementary Methods).
If the samples cannot be analyzed immediately after preparation, the lipids need to be stored in solvent at −20 °C to prevent oxidation and degradation.
3. Metabolite Preparation and Derivatization (Timing ~5 h)
On the day following extraction, remove the metabolite samples from the Speed Vac and store dry at −20 °C until ready for derivatization and analysis on the GC. If not running the metabolites on a GC, then prepare them using the appropriate protocol for the instrument.
Immediately before derivatizing the metabolites for analysis on the GC, transfer them from the large glass vials by adding 200 μL of methanol, vortexing and adding to a 1.5 mL polypropylene tube. Repeat once.
Centrifuge the tube at 12,000 × g for 10 min, 4 °C. Transfer the supernatant into smaller glass vials, add a breathable membrane to the top, and completely dry in a Speed Vac.
-
Derivatize the metabolites by adding 20 μL of methoxyamine solution to the sample vial and vortex for 30 s on a vortexer at medium speed.
Caution: Methoxyamine hydrochloride causes severe burns and serious damage to eyes, may cause sensitization by skin contact. Wear safety glasses, gloves and lab coat, and work in a fume hood.
Use a bath sonicator to ensure the sample is completely dissolved.
Incubate the sample in an incubator with a condensation prevention lid maintained at 37 °C for 1 h 30 min with 1,000 rpm shaking.
Invert the vial one time to mix the samples with condensed drops at the cap surface. Spin the sample down at 1,000 × g for 1 min, RT.
-
Perform silylation by adding 80 μL (using a syringe) of N-Methyl-N-(trimethylsilyl)trifluoroacetamide with 1% trimethylchlorosilane. Vortex for 10s.
Caution: MSTFA + 1% TMCS can cause skin corrosion, serious eye damage, and specific target organ toxicity, and is a flammable liquid and vapor. Wear safety glasses, gloves and lab coat, and work in a fume hood.
Incubate the sample in an incubator with a condensation prevention lid maintained at 37 °C for 30 min with 1,000 rpm shaking.
Invert the vial one time to mix the samples with condensed drops at the cap surface.
Spin the sample down for 5 min at 2,000 × g, RT.
Transfer the reacted solution into vials appropriate for the GC-MS analysis.
4. Protein Digestion (Timing ~1 d)
Centrifuge the sample at 15,000 × g for 5 min, RT, to pellet any debris.
- Perform Filter-Aided-Sample-Preparation (FASP) for the digestion using FASP kits (see Table of Materials) following modified manufacturer’s instructions31,32:
- Add 400 μL of the 8 M urea buffer into a FASP column (500 μL 30K MWCO spin filter).
-
Once the sample has finished centrifuging, pipette off the supernatant (discard the pellet) and add up to 50 μL of the sample to the 400 μL of 8 M urea in the column.NOTE: Add only up to 50 μL of sample per column. If there is more than 50 μL of sample, use multiple columns and combine the peptides after digestion. It’s best to leave this volume as low as possible (30 μL is ideal) in order to ensure the FASP column will remove all of the SDS which can dramatically interfere with mass spec analysis.
- Place the column into the included 1.5 mL tube and centrifuge the sample at 14,000 × g for 30 min, RT.
- Remove the flow-through into waste and add 400 μL of 8 M urea to the column and centrifuge again.
-
Repeat previous step one more time for a total of 3 urea rinses.NOTE: Make sure the sample goes close to dryness on the column, if there is any more than ~ 30 μL remaining on the filter after each spin, continue centrifuging.
- Add 400 μL of 50 mM NH4HCO3 and centrifuge at 14,000 × g for 20 min, discard the waste, and repeat once.
- Transfer the columns to a clean 1.5 mL centrifuge tube.
- Add 75 μL of trypsin digestion buffer to the columns and incubate at 37 °C for 3 h in an incubator with a condensation prevention lid at 750 rpm.
- Add 40 μL of 50 mM NH4HCO3 to the columns.
- Centrifuge the sample at 14,000 × g for 15 min to collect the peptides into the tube while keeping the high molecular weight contaminants on top of the column.
- Add 40 μL of 50mM NH4HCO3 to the column and centrifuge again.
Discard the column, dry down the peptides in the tube in a Speed Vac to ~30 μL, and BCA assay (Bicinchoninic acid protein assay kit; see Table of Materials) the peptides.
Optionally, perform a clean-up step if SDS contamination is suspected (visible bubbles)33. A solid phase extraction must be done afterwards if choosing this option. Detailed information about this clean-up method is available in the Supplementary Methods.
Dilute the sample for MS analysis or optionally proceed to HPLC fractionation (next two steps).
To fractionate, dilute samples to a volume of 400 μL with 10 mM ammonium formate buffer (pH 10.0).
- Resolve on an C18 column (see Table of Materials) by separating at 0.5 mL/min using an HPLC system with mobile phases (A) 10 mM ammonium formate, pH 10.0 and (B) 10 mM ammonium formate, pH 10.0/acetonitrile (10:90).
- Adjust the gradient from at 100% A to 95% A over the first 10 min, 95% A to 65% A over min 10 to 70, 65% A to 30% A over min 70 to 85, maintain at 30% A over min 85 to 95.
- Re-equilibrate with 100% A over min 95 to 105 and hold at 100% A until min 120.
- Collect fractions every 1.25 min (96 fractions over the entire gradient) and finally combine every row for a total of 12 samples or every other row for 24 samples (each with n = 8 or n = 4 fractions pooled).
Dry all fractions under vacuum and add 15 μL of ultrapure water to each for storage at −20 °C until LC-MS/MS analysis.
Representative Results
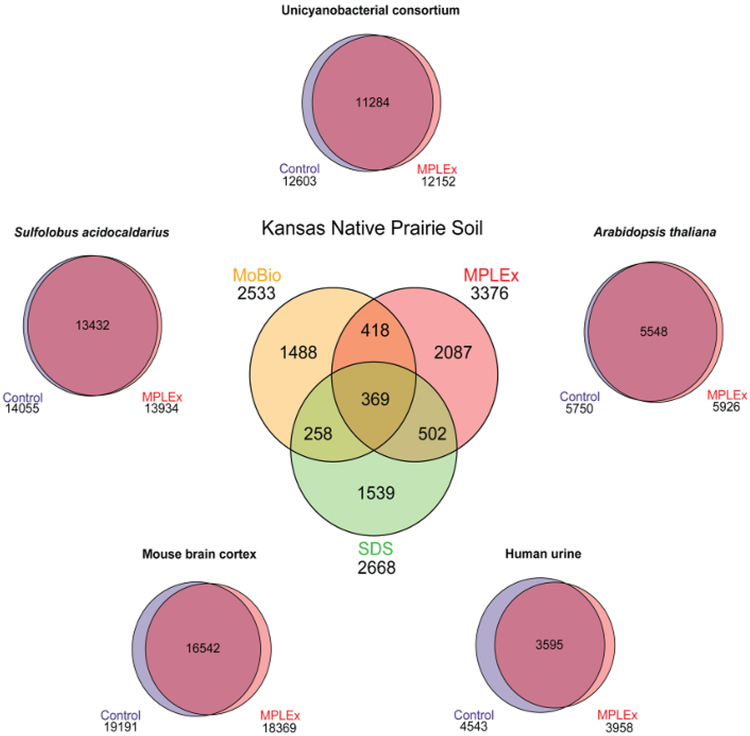
When the MPLEx protocol was used to extract molecules from Kansas native prairie soil (a Mollisol soil), the triplicate analyses provided results for 3376 peptides, 105 lipids, and 102 polar metabolites (all unique identifications). While the MPLEx protocol has been well-established for general extraction of lipids and metabolites12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,34, its comparison to common soil protein extraction methods for microbial analyses, such as soil protein extraction kits (see Table of Materials) and SDS (sodium dodecyl sulfate) extractions35, is further evaluated here. To assess these techniques, Kansas native prairie soil proteins were extracted with each approach and analyzed directly with reversed-phase LC-MS/MS using a UPLC system coupled with a hybrid quadrupole/Orbitrap mass spectrometer. Detailed information about the method parameters are available in the Supplementary Methods. The resulting experimental peptide MS/MS spectra from each extraction approach were compared with predicted peptide sequences from the representative Kansas native prairie metagenome2 using a stringent MSGF+ spectral probability cutoff of 1×10−10 36,37 and a mass error cutoff of less than 5 ppm. It should be noted that the analytes extracted from soil using the MPLEx protocol are suitable for MS-based studies and other analytical techniques. When initially comparing three replicates of the known methods of MoBio and SDS, only 12% of the peptides overlapped between the two techniques showing the complexity of the microbial community and different extraction effects (Figure 2). Upon comparison of MPLEx with MoBio and SDS extracts, ~38% of the peptides observed using the MPLEx method were also detected when using the SDS and/or MoBio extractions (Figure 2). Considering the number of species in the Kansas soil bacterial community and its metagenome2, the overlap of peptide identifications is reasonable and due to its complexity, the combination of the approaches extracts more peptides than each alone35. Also, since these samples were unfractionated and analyzed by only 1-dimensional LC-MS/MS, the extremely high sample complexity was possibly causing some bias in detection due to different peptide concentrations extracted by each approach. The MPLEx protocol has also been applied to other soil types such as permafrost and wetlands, and in all cases the preliminary proteomic results are of similar quality to that of the Kansas native prairie soil data presented here.
Figure 2. Venn diagrams illustrating the peptide overlap for a diverse set of samples extracted using MPLEx and a standard urea-based protein extraction method normally performed.
Data from MPLEx extraction of the archaeon S.acidocaldarius, the unicyanobacterial consortium, human urine, mouse brain cortex, and A.thaliana plant leaves adapted from Nakayasu, E. S. et al. 201610. Please click here to view a larger version of this figure.
The broad applicability of the MPLEx approach, has been previously evaluated using a diverse set of samples, including the archaeon Sulfolobus acidocaldarius, a unicyanobacterial consortium38, mouse brain cortex tissue, human urine, and leaves from Arabidopsis thaliana (Figure 2)10.
For all samples except A.thaliana, urea extractions are the most common way to extract proteins, so they were used as the control approach for evaluation. The A. thaliana MPLEx results were compared to an extraction performed with trichloroacetic acid (TCA) because plant leaves are rich in phenolic compounds that need to be removed prior to MS analyses39. In all cases, the number of proteins observed from the MPLEx method was similar to that for the controls10, indicating its utility for many different sample types. Also, there were no trends associated with the specific protein classes extracted in all the sample types (including soil). Therefore, the broad applicability of MPLEx to numerous biological and environmental systems makes it a promising approach.
Discussion
It is important to note that not all laboratories will have the same available equipment so certain methods, for example the lysis step, can be adapted. Here we use vortexing and sonicating, however the use of a large 50 mL bead beater would work. If a lyophilizer with a collector temperature capable of −105 °C is not available, then samples can be dried under a nitrogen stream. Also soil types vary greatly and can include sand, silt, clay, peat, and loam (etc.), and they can also vary based on pH, salinity, and organic matter richness. Each of these variances can have an impact on the protocol, so it is important to be flexible and to make adjustments when needed. It is critical, however, that the ratios and percentages remain the same.
The MPLEx protocol shows great promise for application in numerous biological and environmental systems and the ability to enable multi-omic analyses of soil microbial communities. Previous comparisons of MPLEx to other extraction protocols for diverse systems illustrated a high degree of peptide overlap10. However, one observed limitation was that MPLEx was not applicable to blood plasma proteins that need to be depleted. In these cases, the precipitated proteins were not well recognized by the antibodies in the immunodepletion columns needed for broad coverage of the plasma proteome. Therefore, more standard extraction approaches should be used under these circumstances. One additional limitation noted for the MPLex extraction approach is that it does not discern between intracellular and extracellular protein, however this is true for all other soil protein extraction protocols as well.
In this manuscript, MPLEx provided results for 3376 peptides, 105 lipids, and 102 polar metabolites in Kansas native prairie soil using the extraction and analysis methods detailed in the protocol section. Better overlap was observed between the peptides extracted in the control and MPLEx approaches of individual systems compared to the soil microbial community (Figure 2). This is not a limitation, but one consequence of working with the extremely complicated soil microbial communities. These results are further noted as the well-known soil extraction techniques of SDS and MoBio did not overlap well either due to the great diversity and difficulties extracting soil proteins equally. Interestingly, the MPLEx protocol allowed the identification of more total peptides than either SDS or MoBio and also detected additional components not seen in either analysis.
These observations make MPLEx a very promising technique for working with small samples sizes, reducing overall multi-omic experimental variability, and reducing sample preparation time. The advantages possible with MPLEx look to enable the multi-omic capabilities and analyses required for large-scale microbial community studies.
Supplementary Material
Acknowledgements
The authors would like to thank Nathan Johnson for his assistance in preparing the figures. This research was supported by the Pan-omics Program that is funded by the U.S. Department of Energy’s Office of Biological and Environmental Research (Genomic Science Program), the Microbiomes in Transition (MinT) Laboratory Directed Research Development Initiative at the Pacific Northwest National Laboratory, as well as the National Institutes of Health National Institute of Environmental Health Sciences (R01 ES022190) and NIH (P42 ES027704). KEBJ would like to thank R21 HD084788 for financial support to develop and validate novel multi-omic extraction techniques. This work was performed in the W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a DOE national scientific user facility at the Pacific Northwest National Laboratory (PNNL). PNNL is a multi-program national laboratory operated by Battelle for the DOE under Contract DE-AC06–76RL01830.
Footnotes
Video Link
The video component of this article can be found at https://www.jove.com/video/57343/
Disclosures
The authors declare that they have no competing financial interests.
References
- 1.Hultman J et al. Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature. 521, 208–212 (2015). [DOI] [PubMed] [Google Scholar]
- 2.White RA 3rd et al. Moleculo long-read sequencing facilitates assembly and genomic binning from complex soil metagenomes. mSystems. 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White RA, Callister SJ, Moore RJ, Baker ES, & Jansson JK The past, present and future of microbiome analyses. Nat Protoc. 11, 4–8 (2016). [Google Scholar]
- 4.Ritchie MD, Holzinger ER, Li R, Pendergrass SA, & Kim D Methods of integrating data to uncover genotype-phenotype interactions. Nat Rev Genet. 16, 85–97 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Jansson JK, & Baker ES A multi-omic future for microbiome studies. Nat Microbiol. 1 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Domon B, & Aebersold R Options and considerations when selecting a quantitative proteomics strategy. Nat Biotechnol. 28, 710–721 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Marx V Targeted proteomics. Nat Methods. 10, 19–22 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Roberts LD, Souza AL, Gerszten RE, & Clish CB Targeted metabolomics. Curr Protoc Mol Biol. Chapter 30, Unit 30 32 31–24 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folch J, Lees M, & Sloane Stanley GH A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 226, 497–509 (1957). [PubMed] [Google Scholar]
- 10.Nakayasu ES et al. MPLEx: a Robust and Universal Protocol for Single-Sample Integrative Proteomic, Metabolomic, and Lipidomic Analyses. mSystems. 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bligh EG, & Dyer WJ A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
- 12.Pomraning KR et al. Multi-omics analysis reveals regulators of the response to nitrogen limitation in Yarrowia lipolytica. BMC Genomics. 17, 138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tisoncik-Go J et al. Integrated Omics Analysis of Pathogenic Host Responses during Pandemic H1N1 Influenza Virus Infection: The Crucial Role of Lipid Metabolism. Cell Host Microbe. 19, 254–266 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kyle JE et al. Uncovering biologically significant lipid isomers with liquid chromatography, ion mobility spectrometry and mass spectrometry. Analyst. 141, 1649–1659 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovelace ES et al. Silymarin Suppresses Cellular inflammation by inducing reparative stress signaling. J Nat Prod. 78, 1990–2000 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YM et al. Diel metabolomics analysis of a hot spring chlorophototrophic microbial mat leads to new hypotheses of community member metabolisms. Front Microbiol. 6, 209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pomraning KR et al. Comprehensive Metabolomic, Lipidomic and microscopic profiling of Yarrowia lipolytica during lipid accumulation identifies targets for increased lipogenesis. PLoS One. 10, e0123188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang EL et al. The fungus gardens of leaf-cutter ants undergo a distinct physiological transition during biomass degradation. Environ Microbiol Rep. 6, 389–395 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Deatherage Kaiser BL et al. A Multi-Omic View of Host-Pathogen-Commensal Interplay in Salmonella-Mediated Intestinal Infection. PLoS One. 8, e67155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim YM et al. Salmonella modulates metabolism during growth under conditions that induce expression of virulence genes. Mol Biosyst. 9, 1522–1534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ansong C et al. A multi-omic systems approach to elucidating Yersinia virulence mechanisms. Mol Biosyst. 9, 44–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bordbar A et al. Model-driven multi-omic data analysis elucidates metabolic immunomodulators of macrophage activation. Mol Syst Biol. 8, 558 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu ZP et al. Metabolomic response of human skin tissue to low dose ionizing radiation. Mol Biosyst. 8, 1979–1986 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Perera R et al. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 8, e1002584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao X et al. A reversed-phase capillary ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) method for comprehensive top-down/bottom-up lipid profiling. Anal Bioanal Chem. 402, 2923–2933 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorensen CM et al. Perturbations in the lipid profile of individuals with newly diagnosed type 1 diabetes mellitus: Lipidomics analysis of a Diabetes Antibody Standardization Program sample subset. Clin Biochem. 43, 948–956 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diamond DL et al. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 6, e1000719 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alquier T et al. Deletion of GPR40 impairs glucose-induced insulin secretion in vivo in mice without affecting intracellular fuel metabolism in islets. Diabetes. 58, 2607–2615 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding J et al. Application of the accurate mass and time tag approach in studies of the human blood lipidome. J Chromatogr B Analyt Technol Biomed Life Sci. 871, 243–252 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasmussen AL et al. Systems virology identifies a mitochondrial fatty acid oxidation enzyme, dodecenoyl coenzyme A delta isomerase, required for hepatitis C virus replication and likely pathogenesis. J Virol. 85, 11646–11654 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manza LL, Stamer SL, Ham AJ, Codreanu SG, & Liebler DC Sample preparation and digestion for proteomic analyses using spin filters. Proteomics. 5, 1742–1745 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Wisniewski JR, Zougman A, Nagaraj N, & Mann M Universal sample preparation method for proteome analysis. Nat Methods. 6, 359–362 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Zhou JY et al. Simple sodium dodecyl sulfate-assisted sample preparation method for LC-MS-based proteomics applications. Anal Chem. 84, 2862–2867 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson JC et al. Decreased abundance of type III secretion system-inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae. Proc Natl Acad Sci U S A. 111, 6846–6851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chourey K et al. Direct cellular lysis/protein extraction protocol for soil metaproteomics. J Proteome Res. 9, 6615–6622 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Kim S, Gupta N, & Pevzner PA Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J Proteome Res. 7, 3354–3363 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S, & Pevzner PA MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun. 5, 5277 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cole JK et al. Phototrophic biofilm assembly in microbial-mat-derived unicyanobacterial consortia: Model systems for the study of autotroph-heterotroph interactions. Front Microbiol. 5, 109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isaacson T et al. Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat Protoc. 1, 769–774 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.