Abstract
Staphylococcus aureus is the leading cause of life-threatening infections, frequently originating from unknown or deep-seated foci. Source control and institution of appropriate antibiotics remain challenges, especially with infections due to methicillin-resistant S. aureus (MRSA). In this study, we developed a radiofluorinated analog of para-aminobenzoic acid (2-[18F]F-PABA) and demonstrate that it is an efficient alternative substrate for the S. aureus dihydropteroate synthase (DHPS). 2-[18F]F-PABA rapidly accumulated in vitro within laboratory and clinical (including MRSA) strains of S. aureus but not in mammalian cells. Biodistribution in murine and rat models demonstrated localization at infection sites and rapid renal elimination. In a rat model, 2-[18F]F-PABA positron emission tomography (PET) rapidly differentiated S. aureus infection from sterile inflammation and could also detect therapeutic failures associated with MRSA. These data suggest that 2-[18F]F-PABA has the potential for translation to humans as a rapid, noninvasive diagnostic tool to identify, localize, and monitor S. aureus infections.
Keywords: bacteria, PET, imaging, infection, folate, MRSA
Graphical Abstract
Staphylococcus aureus is the leading cause of serious deepseated infections such as osteomyelitis, disseminated musculoskeletal infections, abscesses, infective endocarditis, and bacteremia, as well as device-associated infections.1 Moreover, methicillin-resistant S. aureus (MRSA) continues to be one of the most prevalent antimicrobial-resistant pathogens, often requiring prolonged intravenous antibiotics.2 Current diagnostic methods depend on collecting tissue samples with suspected infection, but deep biopsies are complicated by surgical risk, time delays in diagnosis, and the imposition of additional costs. Traditional imaging techniques such as computed tomography (CT), magnetic resonance imaging, and ultrasonography provide anatomic information that lags behind physiological changes. Furthermore, these modalities lack specificity and cannot differentiate infection from sterile inflammatory processes.3 Therefore, noninvasive whole-body imaging with pathogen-specific agents could significantly improve patient outcomes by rapidly identifying a source of infection and monitoring the response to treatment, but no such technology is clinically available.
Positron emission tomography (PET) is becoming a routine clinical tool, particularly within oncology and neurology, and an emerging technology for infectious diseases with the advantages over traditional diagnostic methods of speed, sensitivity (nano- to picomolar),4 and whole-body analysis.5,6 Bacteria-specific PET imaging could help practicing infectious disease specialists and radiologists in differentiating active infection from other causes of inflammation. Previous efforts to develop pathogen-specific PET imaging methods have classically focused on radiolabeling existing antibiotics or antimicrobial peptides.7–10 While this approach builds upon known chemistry and target interaction, both antibiotics and antimicrobial peptides are pharmacologically active and may paradoxically destroy the bacteria and interfere with the signal. Also, these compounds lack the capacity for signal amplification that could be achieved with agents that accumulate in bacteria. Similarly, radiolabeled antibodies have also been evaluated as imaging agents in bacterial infection models.11–13 While promising, radiolabeled antibodies produce significant background noise and may require a longer time to clear from circulation. In contrast, small molecules may be designed to penetrate diseased tissue and rapidly clear from nontarget tissues. When combined with the short half-life isotopes (e.g., fluorine-18), such small molecules may substantially increase diagnostic speed and accuracy, as well as safety, by reducing radiation exposure to human subjects.
Recently, there have also been several efforts to develop bacteria-specific imaging agents based on metabolic substrates that are essential in prokaryotes.14–17 For example, 124I-FIAU, a nucleoside analog substrate for thymidine kinase, can be used to image S. aureus and other bacteria,18 but lack of specificity due to host metabolism by mitochondrial thymidine kinase limits its use.19 Maltose and maltohexaose analogs labeled with fluorine-18 have been evaluated for bacteria-specific imaging but have modest signal-to-noise ratios, likely due to host metabolism of the tracers,14,15 although a recently described second generation PET ligand (6-18F-fluoromaltotriose) has significantly better signal-to-noise ratios and is, therefore, more promising.17 Another recently described PET tracer, 18Ffluoropropyl-trimethoprim, which targets folate metabolism in bacteria, also suffers from poor signal-to-noise ratios and high background activity in nontarget tissues.20 We have also previously described 18F-fluorodeoxysorbitol (18F-FDS) to study Enterobacteriaceae (Gram-negative) infections in vivo,16 and 18F-FDS PET is currently being evaluated in human studies. However, 18F-FDS does not accumulate in Gram-positive bacteria, such as S. aureus. Previously, investigators have imaged S. aureus endocarditis using a labeled prothrombin analog that binds to the staphylococcal coagulase.21 However, this tracer is not specific for S. aureus. Instead, it is a prothrombin analog, which has the potential for nonspecific activation and false positive signaling by noninfectious processes and nontarget bacteria. Finally, optical imaging techniques have also been reported to detect S. aureus infections in small animals22,23 but have limited clinical translation due to the absorption of light by deep tissues.
We previously identified p-aminobenzoic acid (PABA) as a promising candidate for imaging a wide range of bacterial infections.24 PABA is the natural substrate for the bacterial enzyme dihydropteroate synthase (DHPS) which catalyzes the incorporation of PABA into folic acid.25 However, PABA is not significantly absorbed by mammalian cells, is apparently inert in humans, and is rapidly eliminated by renal clearance.26 It is commercially available as a nutritional supplement and topically in sunscreens. In this study, we developed a fast (90 min) automated radiosynthesis of 2-[18F]F-PABA in high radiochemical yield and demonstrate that 2-[18F]F-PABA is an efficient alternative substrate for the S. aureus dihydropteroate synthase (DHPS) and is rapidly accumulated in S. aureus cells (laboratory and clinical strains including MRSA) in vitro. We also performed whole-body 2-[18F]F-PABA PET to noninvasively differentiate infection from sterile inflammation as well as quantitatively monitor antibiotic efficacy in animal models of infection.
RESULTS AND DISCUSSION
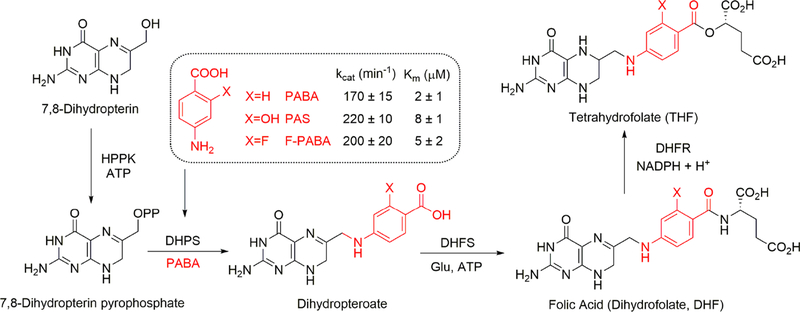
2-F-PABA Is an Alternative Substrate for DHPS.
PABA is the natural substrate for the bacterial DHPS which catalyzes the incorporation of PABA into tetrahydrofolate25 but is not accumulated by mammalian cells. PABA and its antibacterial structural analog, sulfonamide, enter bacterial cells by passive diffusion through nonspecific porin channels.27–29 In addition to acquiring PABA from the environment, bacteria are also capable of de novo PABA biosynthesis from glutamate and chorismate.25,30 Once inside, PABA is either incorporated into tetrahydrofolate by DHPS and retained inside the cells or pumped out by the AbgT transporters.29 We radiolabeled the electron-rich aromatic ring of PABA with fluorine-18 in the 2 position for several reasons. Our prior data demonstrate that, unlike fluorination at other positions, 2-F-PABA retained rapid, specific, and time-dependent bacterial accumulation.24 Moreover, 2-F-PABA uptake remains high, regardless of the growth phase, making it attractive for detecting metabolically quiescent bacteria often associated with implant-associated infections in the clinic.24 DHPS catalyzes the synthesis of 7,8dihydropteroate from PABA and 6-hydroxymethyl-7,8-dihydropterin pyrophosphate. We established an assay for the S. aureus DHPS reaction using 6-hydroxymethyl-7,8-dihydropterin as the initial substrate and involving the NADPHdependent reduction of 7,8-dihydropteroate by dihydrofolate reductase (DHFR). Using this assay, we demonstrated that 2F-PABA is an efficient alternative substrate for the S. aureus DHPS with similar values for kcat (200 ± 20 min−1) and KM (5 ± 2 μM) compared to PABA (kcat = 170 ± 15 min−1, KM = 2 ± 1 μM) and p-aminosalicylic acid (PAS) (kcat = 220 ± 10 min−1, KM = 8 ± 1 μM) (Figure 1). The ability of DHPS to accept 2F-PABA as an alternative substrate and the observation that the fluoro derivative of the DHPS product is a substrate for DHFR provides a mechanism for the accumulation of 2[18F]F-PABA in bacterial cells by the incorporation of this metabolite into tetrahydrofolate as shown previously for PAS in M. tuberculosis.31
Figure 1.
2-F-PABA is an alternative substrate for tetrahydrofolate (THF) biosynthesis. PABA and F-PABA are substrates for dihydropteroate synthase (DHPS) in the THF biosynthesis pathway (only the terminal portion of the pathway is shown). The incorporation of PABA and PABA analogs F-PABA and PAS into dihydropteroate was quantified using a coupled assay that omitted DHFS from the reaction mixture and instead involved the direct reduction of dihydropteroate by dihydrofolate reductase (DHFR).
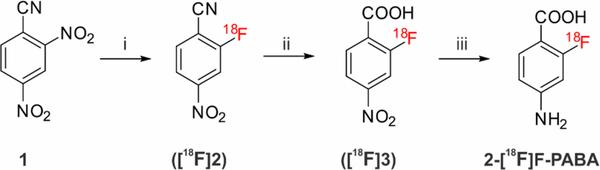
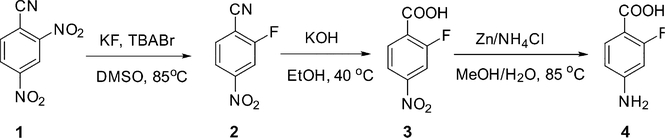
Radiosynthesis.
2-[18F]F-PABA was successfully synthesized as shown in Scheme 1. The radioactive product washaracterized by radio-HPLC (Figure S1) which determined the chemical and radiochemical impurities in the product and compared the retention time of the labeled material to a cold reference standard. The overall synthesis time was 90 min with a typical overall decay corrected radiochemical yield of 30− 40%. 2-[18F]F-PABA had a specific activity of 240.5 ± 77.7 GBq/μmole (6.5 ± 2.1 Ci/μmole, n = 4) and a radiochemical purity of 99.2 ± 0.7% (n = 4). We utilized a conventional nucleophilic aromatic substitution (SNAr) reaction for radiofluorination. The precursor, 2,4-dinitrobenzonitrile (1), is commercially available and is readily radiofluorinated to give intermediate [18F]2. Basic hydrolysis of the nitrile followed by reduction of the 4-nitro moiety proceeded rapidly to produce 2-[18F]F-PABA after purification by radio-HPLC. The entire radiosynthesis was automated using a GE Tracerlab FXN Pro Radiosynthesis Module which should facilitate the translation of 2-[18F]F-PABA to clinical applications.
Scheme 1.
Radiosynthesis of 2-[18F]F-PABAa
a Reagents and conditions: (i) [18F]potassium fluoride, Kryptofix222, potassium carbonate, dimethyl sulfoxide, r.t., 10 min; (ii) 2M potassium hydroxide in water, 105 °C, 10 min; (iii) zinc power, ammonium chloride, water, 105 °C, 5 min. Overall decay-corrected radiochemical yield: 30−40% (n = 3).
In Vitro Uptake of 2-[18F]F-PABA by S. aureus.
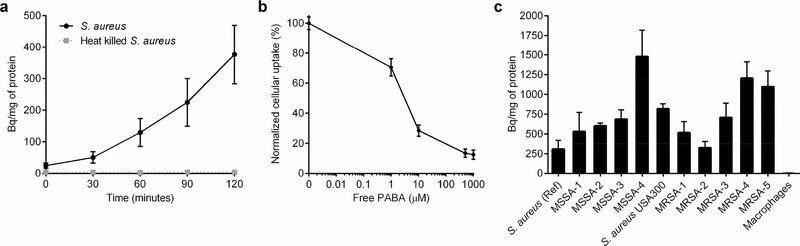
The accumulation of 2-[18F]F-PABA in bacterial cells was investigated in S. aureus. The tracer rapidly accumulated in viable cells but not in heat-killed bacteria (Figure 2a). After 2 h of incubation, the concentration of 2-[18F]F-PABA was significantly higher compared to heat-killed bacteria (P < 0.005). Co-incubation of 2-[18F]F-PABA with varying concentrations of PABA resulted in a dose-dependent inhibition of 2-[18F]F-PABA uptake (Figures 2b and S2), yielding a dose−response curve with an IC50 value of 1 ± 0.9 μM, which is similar to the KM value of 2-F-PABA (5 ± 2 μM) determined using the enzymatic assay. The concentrationdependent blocking of 2-[18F]F-PABA uptake in S. aureus by PABA supported our proposed mechanism for the specific accumulation of 2-[18F]F-PABA in bacteria. In order to further investigate the translational potential to clinically detect S. aureus infections, the accumulation of 2-[18F]F-PABA in various clinical strains was examined. As shown in Figure 2c, 2[18F]F-PABA accumulates in all the clinical strains tested including four methicillin-sensitive strains (MSSA) and six MRSA strains, one of which is USA300, which is the primary cause of community-acquired MRSA infections. The accumulation of 2-[18F]F-PABA in almost all the clinical isolated strains was similar or higher than that observed for the reference S. aureus ATCC 29213 (MSSA) strain. The variation in 2-[18F]F-PABA accumulation across the different strains of S. aureus may be due to different levels of DHPS and/or AbgT transporter expression or variation in the ability of the bacteria to undertake de novo PABA synthesis.
Figure 2.
Cell uptake of 2-[18F]F-PABA. (a) Time-dependent uptake of 2-[18F]F-PABA by viable (black) and heat-killed (dotted gray) S. aureus (ATCC 29213). (b) Concentration-dependent blocking of 2-[18F]F-PABA accumulation in viable S. aureus cells by PABA. (c) Accumulation of 2[18F]F-PABA in reference S. aureus (ATCC 29213) and various clinical MSSA and MRSA isolates, compared to mammalian cells (J774 murine macrophages).
N-Acetylation of 2-[18F]F-PABA.
PABA is known to be rapidly converted in the liver to N-acetyl-PABA by the arylamine N-acetyltransferase (NAT) as well as para-aminohippuric acid and N-acetyl-para-aminohippuric acid by theglycine N-acyltransferase.32,33 In humans, NAT type 1 is responsible for the N-acetylation of PABA and is enzymatically similar to the isoenzyme NAT type 2 in rodents.34 Similarly, 2[18F]F-PABA was also extensively N-acetylated (50.14%) in the rat model 2 min after intravenous injection (Figure S3). Although NAT enzymes have also been described in some bacterial species, N-acetylation of PABA does not occur in most of them.35
Whole-Body Biodistribution of 2-[18F]F-PABA in S. aureus-Infected Mice.
The biodistribution of 2-[18F]F-PABA in a mouse S. aureus thigh infection model was investigated by PET imaging and also by ex vivo tissue analysis (Figure S4). 2-[18F]F-PABA was found to distribute to all major organs and subsequently rapidly eliminated mainly through renal clearance. In addition, over the time course of the study, significantly higher levels of tracer were found in the infected thighs than in uninfected thighs.
2-[18F]F-PABA PET Can Detect and Localize S. aureus Infection in Vivo.
The ability of 2-[18F]F-PABA to detect and localize S. aureus infection in vivo was assessed using a rat triceps infection model. 2-[18F]F-PABA readily accumulated in the infected triceps (Figures S5 and S6), with higher tracer levels, compared to the uninfected triceps 10 min after tracer injection. The differential accumulation of tracer in the infected triceps compared to the uninfected triceps increased over the 60 min PET imaging time window, reaching a factor of 5.4 by the end of the study (0.35 ± 0.09%ID/cc vs 0.07 ± 0.03%ID/cc). 60 min after tracer administration, there was a significant difference between 2-[18F]F-PABA accumulation in the infected and uninfected triceps (P = 0.028) (Figure S5b). In addition, the activity in the infected triceps was greater than that found in the blood whereas the activity in the uninfected triceps was lower (Figure S5c). The infected triceps-to-blood ratio increased over the 60 min imaging time window while the uninfected triceps-to-blood ratio remained unchanged over this period. Importantly, the tracer was rapidly eliminated from blood, lungs, liver, and bone rapidly without specific accumulation (Figure S6). The activity in the infected triceps was 4.8-, 4.2-, and 3.4-fold higher than that of bone, lung, and liver, respectively, 60 min after tracer administration, while the activity in uninfected triceps was 0.9-, 0.7-, and 0.6-fold the amount in the bone, lung, and liver, respectively, 1 h after tracer injection (Figure S7). Ex vivo post-mortem analysis corroborated the PET imaging data (Figure S5d). Finally, infections with S. aureus are often fulminant and spread rapidly, and infection in the surrounding subcutaneous tissues was also noted (Figure S8).
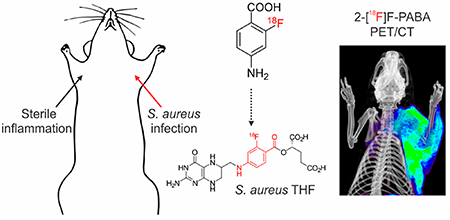
2-[18F]F-PABA PET Can Differentiate Infection from Sterile Inflammation.
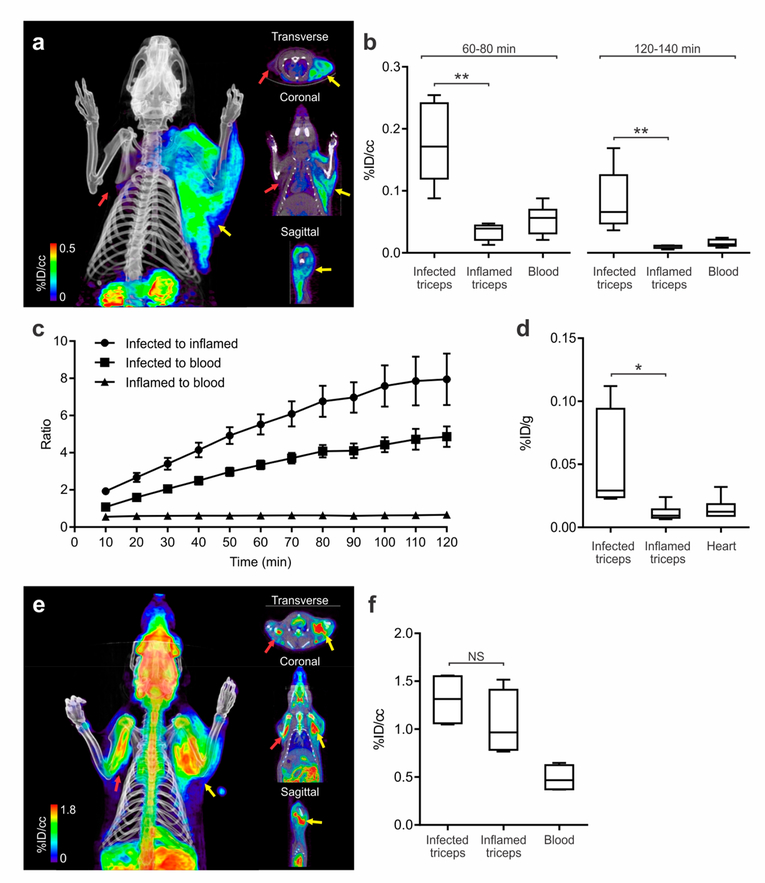
We next investigated whether 2[18F]F-PABA PET could differentiate between infection from a sterile inflammatory process. Bacterial infection was induced in the right triceps whereas heat-killed bacteria were used to generate an inflammatory response in the left triceps. Histological examination revealed extensive neutrophil infiltration in both the infected and inflamed triceps, while Gram staining demonstrated the presence of Gram-positive bacteria in the infected triceps (Figure S9). Subsequent PET/CT and ex vivo studies revealed significant differences in tracer accumulation between the infected and inflamed triceps (Figure 3). After 10 min, the accumulation of 2-[18F]FPABA was ~2-fold higher in the infected triceps compared to the inflamed triceps, and this ratio increased over time so that by 60 and 120 min the ratio was 5.52 and 7.95, respectively (Figures 3b,c and S10). Similar to the previous study, the activity in the infected triceps was higher than various other (uninfected) organs, with the activity in the infected triceps as a ratio to lung and liver increasing over the 120 min imaging time window (Figure S11). Ex vivo post-mortem analysis agreed with the tracer distribution from the PET studies (Figure 3d).
Figure 3.
2-[18F]F-PABA accumulation in infection compared to sterile inflammation. (a) 2-[18F]F-PABA PET/CT images of rats with S. aureus infection in the right triceps (yellow arrow) and sterile inflammation in the left triceps (red arrow). The images are a three-dimensional projection, transverse, coronal, and sagittal views 60−80 min post-tracer injection. (b) Comparison of 2-[18F]F-PABA accumulation represented as % injected dose per cc (%ID/cc) in infected triceps, inflamed triceps, and blood 60−80 and 120−140 min after tracer injection. **P < 0.001 from a two-tailed Mann−Whitney U Test (n = 7). (c) Time-dependent accumulation of 2-[18F]F-PABA in infected and uninfected triceps compared to the blood (left heart ventricle). Data represented as mean and standard error of the mean (n = 7). (d) Post-mortem ex vivo analysis of 2-[18F]F-PABA biodistribution in infected and uninfected triceps and heart represented as % injected dose per gram of tissue (%ID/g). The signal in infected triceps was 4.30 ± 0.82 higher compared to inflamed muscle. *P = 0.0043 from a two-tailed Mann−Whitney U Test (n = 6). (e) Threedimensional reconstruction, transverse, coronal, and sagittal views of the [18F]FDG PET/CT of a rat infected with S. aureus in the right triceps (yellow arrow) and sterile inflammation in the left triceps (red arrow), 60−80 min after injection of the tracer. (f) Comparison of [18F]F-FDG accumulation 60−80 min postinjection in rats infected with S. aureus and sterile inflammation, represented as %ID/cc. There was no difference between the signal of infected versus inflamed triceps (P = 0.2, from a two-tailed Mann−Whitney U Test, n = 4). NS, not significant.
We next evaluated the effect of unlabeled F-PABA on the accumulation of 2-[18F]F-PABA in infected tissue by repeating the imaging study after adding 2 mg of 19F-PABA to the solution of the radiotracer. Two hours after the injection of 2[18F]F-PABA containing 19F-PABA, the ratio of 2-[18F]FPABA in infected triceps compared to inflamed muscle increased to 9.38 ± 2.43 (Figure S12). As described above, 2-[18F]F-PABA is rapidly converted to metabolites such as the N-acetyl derivative by liver NAT enzymes. The increase in signal from the infected tissues is thus consistent with the competition of 19F-PABA for saturable metabolic processes such as N-acetylation, thereby increasing the concentration of intact 2-[18F]F-PABA at the infection site and the subsequent accumulation of radiotracer in the bacteria.
Finally, we also used [18F]fluorodeoxyglucose ([18F]FDG), a glucose analog and a highly sensitive imaging technique increasingly being used for imaging infections, in our rat infection model. The [18F]FDG PET signal at the infection site was not significantly different compared to the site of sterile inflammation (P = 0.2, n = 4) (Figure 3e,f), consistent with the knowledge that [18F]FDG PET is taken up by a wide-range of metabolically active host cells, including those involved in the inflammatory response with high glycolytic activity,36–39 and cannot differentiate inflammatory versus infectious processes.
Monitoring Therapeutic Efficacy of Antibacterial Agents.
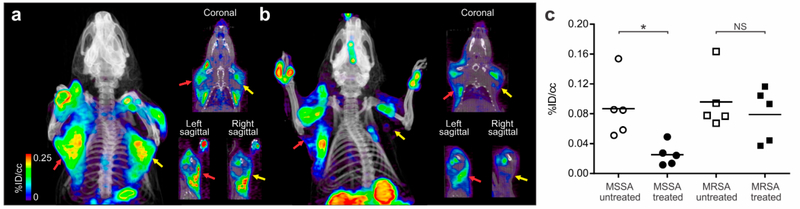
We next investigated the ability of 2-[18F]F-PABA PET to monitor the therapeutic efficacy of antibacterial agents in situ and correlate with the bacterial burden. Briefly, rats were infected with MSSA in the right triceps and MRSA in the left triceps and received either no treatment (untreated group) or four doses of 200 mg/kg oxacillin treatment (treatment group) for 20 h following infection. Oxacillin is highly effective against MSSA but not against MRSA. As shown in Figure 4, significant levels of 2-[18F]F-PABA accumulated in both the right (0.087 ± 0.041%ID/cc) and left triceps (0.096 ± 0.039%ID/cc) in the untreated group. Conversely, antibiotic treatment led to a decrease in the 2-[18F]F-PABA signal in the right triceps (0.025 ± 0.015%ID/cc) whereas tracer accumulation in the left triceps, with the resistant strain (MRSA), did not change significantly (0.079 ± 0.036%ID/cc) (Figure 4b,c). The 2[18F]F-PABA PET signal from the right triceps (MSSA infection) of the treated animals was significantly lower than the right triceps (MSSA infection) of the untreated animals (P = 0.008), whereas there was no difference in the PET signal from the treated and untreated left triceps (MRSA infection) (P = 0.841) (Figure 4c). The change in PET activity correlated with the bacterial burden determined by ex vivo microbiological examination: in the right triceps (MSSA infection), the bacterial burden was significantly higher in the untreated versus treated animals (8.4 ± 0.1 and 6.4 ± 0.4 log10 CFU/g, respectively), whereas the bacterial burden in the left triceps (MRSA infection) was similar in the untreated and treated animals (9.0 ± 0.2 and 8.2 ± 0.3 log10 CFU/g, respectively). While data on the bacterial burden in clinical infections is sparse, one report indicates that clinically relevant infections due to S. aureus have high bacterial burdens averaging 8.3 log10 CFU/mL, suggesting that 2-[18F]F-PABA has sufficient diagnostic sensitivity to be clinically relevant.40
Figure 4.
2-[18F]F-PABA PET correlates with the therapeutic efficacy of antibacterial agents. To evaluate if the signal of 2-[18F]F-PABA correlated with bacterial burden, rats were infected with MSSA in the right triceps (yellow arrows) and MRSA in the left triceps (red arrows). (a) Representative three-dimensional projection, coronal and sagittal views of the 2-[18F]F-PABA PET/CT 60 min postinjection of tracer in the untreated group of rats infected with both MSSA and MRSA. The PET signal in both infection sites was similar. (b) 2-[18F]F-PABA PET/CT of a representative oxacillin-treated (200 mg/kg, four doses subcutaneously) rat infected with MSSA and MRSA as described above. The images are a three-dimensional projection, coronal and sagittal views of the PET 60−80 min post-tracer injection, where the signal in the MSSA infection site was lower compared to the MRSA infection (c) 2-[18F]F-PABA PET was able to differentiate between treated and untreated groups in the MSSA infection site (*P = 0.008) but not between treated and untreated MRSA infections (P = 0.841). P values calculated using a two-tailed Mann− Whitney U Test (n = 4). NS, not significant.
Compartment Pharmacokinetic (PK) Model Demonstrates Specific Accumulation of 2-[18F]F-PABA at the Site of Infection.
The dynamic 2-[18F]F-PABA PET data in S. aureus-infected rats was analyzed using a compartment PK model which assumes that the tracer is irreversibly trapped in bacteria at the site of infection (Figure S13). After determining the values of the distribution rate constants by fitting the data from blood and infected tissues (right triceps) to the PK model, simulations of the changes in specific and nonspecific activities within both sites were performed.41,42 After 2 h, ~60% of the total activity observed at the site of infection results from specific accumulation in the infection site which we hypothesize is due to the incorporation of 2-[18F]F-PABA into the bacterial folate biosynthesis pathway.
CONCLUSIONS
We have developed a novel, noninvasive diagnostic tool for detecting, localizing, and monitoring S. aureus infections which are a leading cause of serious deep-seated as well as deviceassociated infections. 2-[18F]F-PABA is clinically relevant as it can be easily synthesized in 90 min and is rapidly accumulated by clinical isolates, including drug-resistant S. aureus (MRSA) with the potential for detecting metabolically quiescent bacteria often associated with clinical infections. Therefore, 2-[18F]F-PABA has a high potential for a safe translation to humans to provide a rapid, noninvasive diagnostic tool to locate infections and guide antimicrobial selection. While we have focused on S. aureus in this study, 2-[18F]F-PABA is taken up by many different pathogenic bacteria24 and therefore could prove to be useful for a wide-range of bacterial infections.
METHODS
Study Design.
The objective of this study was to synthesize and test 2-[18F]F-PABA as a noninvasive PET imaging diagnostic tool that identifies, localizes, and monitors S. aureus infections in vivo. All protocols were approved by the Stony Brook and Johns Hopkins Biosafety, Radiation Safety, and Animal Care and Use Committees.
Synthesis of 2-F-PABA.
2-F-PABA was synthesized from 2, 4-dinitrobenzonitrile (1) as shown in Scheme 2.
Scheme 2.
Synthesis of 2-F-PABA from 2,4-Dinitrobenzonitrile
2-Fluoro-4-nitrobenzonitrile (2).
2,4-Dinitrobenzonitrile (1) (500 mg, 2.59 mmol), KF (450.75 mg, 7.77 mmol), and tetra-n-butylammonium bromide (TBABr, 167.02 mg, 0.52 mmol) were added to a 100 mL round-bottom flask (RBF) and dried under vacuum for 1 h. After filling the RBF with N2, 50 mL of dry DMSO (4 Å molecular sieves) was added, and the reaction mixture was heated to 85 °C and then refluxed for 30 min under N2. After the reaction was shown to be complete by TLC (20% ethyl acetate in hexane), the mixture was cooled to RT and iced water was added; the reaction mixture was then extracted with ethyl acetate. The organic extracts were combined, dried with anhydrous MgSO4, and evaporated in vacuo to yield the crude product which was then purified by Combiflash using a silica gel column and petroleum ether and EtOAc (20% ethyl acetate in hexane) as the mobile phase to yield compound 2. EI-MS calculated for molecular ion C7H3FN2O2 ([M]+•): m/z = 166.02, found m/z = 166.05, found m/z = 120.03 for [M − NO2]+• (C7H3FN). 1H NMR (400 MHz, CDCl3): δ 8.19 (ddd, J = 8.5, 2.1, 0.9 Hz, 1H), 8.13 (dd, J = 8.4, 2.1 Hz, 1H), 7.91 (dd, J = 8.5, 6.2 Hz, 1H). 19F NMR (400 MHz, CDCl3): δ −101.0492 to −101.09 (m).
2-Fluoro-4-nitrobenzoic Acid (3).
2-Fluoro-4-nitrobenzonitrile (2) (200 mg, 1.20 mmol) was added to 10 mL of 2 M KOH, followed by the addition of 2 mL of EtOH. The reaction mixture was stirred for 4 h at 40 °C, and after the reaction was shown to be complete by TLC (5% methanol in dichloromethane), the reaction mixture was concentrated in vacuo. Subsequently, the pH of the reaction mixture was adjusted to around 1 using 6 M HCl and then extracted with EtOAc. The organic layer was dried with anhydrous MgSO4, filtered, and evaporated in vacuo to yield the crude product which was then purified by Combiflash using a silica gel column and 5% MeOH in CH2Cl2 containing 0.1% CH3COOH as the eluent to yield compound 3. ESI-MS calculated for molecular ion C7H3FNO4 ([M − H]−): m/z = 184.01, found m/z = 184.01. 1H NMR (400 MHz, CDCl3 and CD3OD): δ 8.16−8.12 (m,1H), 8.06 (dd, J = 8.6, 1.7 Hz, 1H), 8.00 (dd, J = 9.9, 1.9 Hz, 1H). 19F NMR (400 MHz, CDCl3 and CD3OD): δ 104.90−104.95 (m). (Commercially available standard 2-fluoro-4nitrobenzoic acid (3). 1H NMR (400 MHz, CDCl3 and CD3OD): δ 8.14 (dd, J = 8.6, 7.0 Hz, 1H), 8.06 (ddd, J = 8.6, 2.1, 0.8 Hz, 1H), 8.00 (dd, J = 9.9, 2.1 Hz, 1H). 19F NMR (400 MHz, CDCl3 and CD3OD): δ 104.78−104.82 (m)).
2-Fluoro-para-aminobenzoic Acid (2-F-PABA, 4).
2-Fluoro-4-nitrobenzoic acid (3) (100 mg, 0.54 mmol), Zn powder (353.20 mg, 5.40 mmol), and NH4Cl (433.53 mg, 8.10 mmol) were added to a 25 mL RBF containing 4.5 mL of methanol and 1.0 mL of water. The reaction mixture was heated to 80 °C and then refluxed for 20 min. After the reaction was shown to be complete by TLC (5% methanol in dichloromethane), the reaction mixture was concentrated in vacuo and the pH was adjusted to 4 using a saturated solution of NaHCO3 in water. The solution was then extracted with EtOAc, and the organic layers were combined, dried with anhydrous MgSO4, and filtered. The organic extract was evaporated in vacuo to yield the crude product which was then purified by Combiflash (Teledyne Isco) using a silica gel column and 5% MeOH in CH2Cl2 containing 0.1% CH3COOH as the eluent to yield compound 4. ESI-MS calculated for molecular ion C7H6FNO2 ([M − H]−): m/z = 154, found m/z = 154 in negative mode. Calculated for molecular ion C7H8FNO2 ([M + H]+): m/z = 156, found m/z = 156 in positive mode ESI-MS. 1H NMR (400 MHz, CD3OD): δ 7.66 (dd, J = 8.6, 8.6 Hz, 1H), 6.42 (dd, J = 2.2, 8.6 Hz), 6.32 (dd, J = 2.2, 13.8 Hz). 19F NMR (400 MHz, CD3OD): δ −110.5213 to −110.5825 (m). 13C NMR (400 MHz, CD3OD): δ 168.32 (d, J = 3.7 Hz, 7′-C), 166.05 (d, J = 254.7 Hz, 2′-C), 156.95 (d, J = 12.5 Hz, 4′-C), 134.98 (d, J = 3.1 Hz, 6′-C), 110.63 (d, J = 1.6 Hz, 5′-C), 06.51 (d, J = 9.5 Hz, 1′-C), 101.51 (d, J = 25.6 Hz, 3′-C).
Kinetic Parameters for DHPS.
Expression and Purification of 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase (HPPK).
A pET15b vector carrying the gene for E. coli HPPK in frame with an N-terminal His-tag (GenScript USA Inc.) was transformed into competent E. coli BL21(DE3) cells and plated on Lysogeny Broth (LB) agar containing 100 μg/ mL ampicillin. A single colony was then used to inoculate 10 mL of LB media containing 100 μg/mL ampicillin, which was then incubated overnight at 37 °C with 250 rpm agitation. This culture was then used to inoculate 2 L of LB media containing ampicillin in 4 L flasks, which were grown at 37 °C for ~2.5 h until the OD600 reached ~0.8. Subsequently, protein expression was induced by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and the culture was shaken overnight at 20 °C. The cells were harvested by centrifugation at 5000 rpm for 10 min and stored at −20 °C until purification. Cell pellets were then resuspended in 20 mM TRIS buffer pH 8 containing 300 mM NaCl and lysed by sonication. The lysate was clarified by ultracentrifugation at 40 000 rpm for 1 h and applied to Ni-NTA resin equilibrated with resuspension buffer. The bound protein was washed with increasing concentrations of imidazole (10 mM, 20 mM) in resuspension buffer and eluted using 250 mM imidazole in resuspension buffer. The eluent was collected in 1 mL fractions, pooled, and dialyzed against 50 mM TRIS buffer pH 8, containing 150 mM NaCl and stored at −80 °C with 10% glycerol. The protein was shown to be >95% pure by SDS-PAGE.
Expression and Purification of Dihydropteroate Synthase (DHPS).
A pET16b plasmid carrying the gene for S. aureus DHPS was generously provided by Dr. Richard E. Lee (St. Jude Children’s Research Hospital). The plasmid was transformed into E. coli BL21(DE3) cells and expressed and purified as described above for HPPK.
Determination of Kinetic Parameters for DHPS.
The steady-state kinetic parameters for DHPS were determined on a Cary 100 Bio (Varian) spectrometer at 25 °C. Reactions were performed in 50 mM TRIS buffer pH 8.0 containing 5 mM 2-mercaptoethanol, 5 mM MgCl2, 10 μM 6-hydroxymethyl-7,8-dihydropterin hydrochloride (Schircks Laboratories), 1 mM ATP, 9 μM DHFR, 100 μM NADPH, 5 μM HPPK, and 20 nM DHPS. The reaction was initiated by the addition of the substrate (PABA, F-PABA, or PAS), and the progress of the reaction was then followed by monitoring the oxidation of NADPH to NADP+ at 340 nm (ε = 6220 M−1 cm−1). Initial velocities were measured at varying concentrations of the substrates (0.5, 1, 2, 5, 10, 20, 50, and 80 μM of PABA, F-PABA, or PAS), and kinetic parameters (kcat and KM) were determined by fitting the data to the Michaelis−Menten equation.
Radiosynthesis of 2-[18F]F-PABA.
2-[18F]F-PABA was synthesized using a GE TracerLab FXN pro radio-synthesis box. [18F]Fluoride was produced at an offsite cyclotron and arrived at our facility around 4 h EOB. [18F]Fluoride (200− 1000 mCi) was first trapped on an Acell Plus QMA cartridge (Waters), and the cartridge was flushed with nitrogen for 1 min. [18F]Fluoride was then released with 1 mL of a 96% CH3CN solution containing 14.4 mg of kryptofix222 and 3.0 mg of K2CO3 and collected in a 10 mL glassy carbon reaction vessel. The solvent was removed under vacuum, and a stream of nitrogen for 7 min at 70 °C followed by 1 min at 100 °C. The reaction vessel was cooled to 35 °C; 2 mg of 2,4dinitrobenzonitrile was added in 1.0 mL of dry DMSO, and the solution stirred for 5 min. The reaction mixture was diluted with 8 mL of water, and the desired 2-[18F]fluoro-4nitrobenzonitrile was trapped on tandem oasis HLB plus and Sep-Pak light C18 cartridges (Waters). The cartridges were flushed with N2 for 2 min before being backflushed with 3 mL of acetonitrile into the same reaction vessel. The solvent was removed at 60°C for 5 min under vacuum and a stream of N2. The reaction vessel was cooled to 35 °C before 1.0 mL of 2 M KOH was then added, and the reaction mixture was heated at 105 °C for 10 min. The reaction mixture was then cooled, diluted with 5 mL of water, and acidified with 2 mL of 2.0 M CH3COOH. The product 2-[18F]fluoro-4-nitrobenzoic acid was trapped on the same oasis HLB and C18 Sep-Pak cartridges which had been rewashed with 8 mL of water. The cartridges were flushed with N2 for 2 min before being back flushed with 1.5 mL of CH3CN into a 3 mL quartz glass reactor containing 10 mg of metallic Zn powder. The solvent was removed at 60 °C for 5 min under vacuum and a stream of N2. The reaction vessel was cooled to 35 °C before 1.0 mL of a 60 mg/mL solution of NH4Cl and 0.1 mL of 2.0 M CH3COOH were added. The reaction vessel was then heated to 105 °C for 5 min. Following this final reaction, the solution was cooled to 35 °C and diluted with 4.5 mL of water before being passed through a 0.22 μm vented filter (Millex GS). The filtered reaction mixture was then purified by RP HPLC (Phenomenex Luna C18(2), 100 Å, 250 × 10 mm) and an eluent of 0.5% CH3COOH and 5% ethanol at 5 mL/min. The desired product, 2-[18F]fluoro-p-aminobenzoic acid, eluted at around 16 min.
The purified product, 2-[18F]fluoro-p-aminobenzoic acid, was neutralized with 0.2 M sodium bicarbonate before being used for further studies. QC was performed on the product as well a second solution which had additional unlabeled 2-fluoro-p-aminobenzoic acid. These solutions were analyzed with HPLC (Phenomenex Luna, C18(2), 100 Å, 250 × 4 mm) with an eluent of 0.1% TFA and 8% acetonitrile at 1 mL/min. The eluate was monitored for UV absorption at 280 nm as well as for radioactivity to determine the chemical identity, chemical purity, radiochemical purity, and specific activity.
In Vitro Uptake Assays.
Bacterial reference strains from the American Type Culture Collection (ATCC) and random, consecutive clinical isolates (Johns Hopkins Hospital) were utilized for the in vitro uptake assays. All bacteria were aerobically grown to OD600 1.0 in LB media at 37 °C. Murine macrophages J774.1 (ATCC) were cultured in RPMIGlutaMAX (Thermo Fisher Scientific) with 10% heatinactivated fetal bovine serum at 37 °C with 5% CO2. To test the uptake of 2-[18F]F-PABA, bacterial cultures were incubated with 10 kBq/mL of the radiotracer at 37°C with rapid agitation. Heat-killed (90°C for 30 min) bacteria were similarly incubated with 2-[18F]F-PABA. Bacteria were pelleted by centrifugation and washed three times with phosphate buffered saline (PBS). Total radioactivity was measured using an automated gamma counter (1282 Compugamma CS Universal gamma counter, LKB Wallac). A minimum of six replicates was used for each assay and time point and presented as a percentage or normalized to total protein (Bradford assay, Sigma-Aldrich).
Animal Experiments.
Rats Triceps Infection Model.
An overnight culture of S. aureus (Newman) or MRSA (ATCC BAA1762) was initiated by inoculating 10 mL of fresh Mueller Hinton cation-adjusted (CAMH) broth with 5 μL of bacteria from a glycerol stock that had been stored at −80 °C. The culture was agitated on an orbital shaker at 37 °C overnight, and 100 μL of the overnight culture was then transferred to 10 mL of fresh CAMH media. Bacterial cells were harvested by centrifugation (11 000 rpm/7500g, 3 min) and washed with sterile brain heart infusion (BHI) broth. Final inoculums were generated by diluting bacteria to the desired concentrations in BHI broth. Female Sprague−Dawley (SD) rats (10 weeks old, Charles River) were injected with 50 μL of the S. aureus (Newman) culture in the right triceps. Similarly, for the studies following oxacillin treatment, 50 μL of the MRSA culture was injected into the left triceps. In each case, the bacteria were injected to give an initial bacterial load of 8 log10 CFU. The triceps infections were allowed to incubate for 20 to 24 h before PET/CT imaging. After imaging, the rats were euthanized; tissues were harvested, and the associated radioactivity was quantified using a gamma counter (Wizard 2480, PerkinElmer). After gamma counting, tissue samples were homogenized in PBS at 4 °C, serially diluted, and plated in duplicate onto solid MH medium. After overnight incubation at 37 °C, CFU were quantified by enumeration. For histological examination, tissue samples were also obtained from some of the infected animals, fixed overnight in 4% formaldehyde before being embedded in paraffin, sectioned at 4 μm, and stained with hematoxylin and eosin (H&E) or Gram staining.
Rats Triceps Inflammation Model.
S. aureus inoculum was prepared as described above except that an additional culture was heat inactivated to generate a sample of heat-killed bacteria. One mL of the heat-killed bacteria was subsequently isolated by centrifugation and suspended in 1 mL of sterile BHI broth. Female SD rats (10 weeks old, Charles River) were injected with 50 μL of the viable S. aureus (Newman) culture in the right triceps and 50 μL of the heat-killed BHI suspension in the left triceps. After incubation for 20 to 24 h, the biodistribution of 2-[18F]F-PABA was determined by PET/CT imaging. The rats were then euthanized; tissues were harvested, and the associated radioactivity was quantified using a gamma counter (Wizard 2480, PerkinElmer). The tissues were processed for CFU enumeration and histological examination as described above.
Mouse Thigh Infection Model.
S. aureus (Newman) inoculums were prepared as described above. Female CBA/j mice (10 weeks old, Jackson Laboratory) were injected with 50 μL of the bacterial culture in the right thighs to give an initial bacterial load of 7 log10 CFU. After 8 to 12 h, the biodistribution of 2-[18F]F-PABA was determined by PET/ CT imaging. The mice were then euthanized; tissues were harvested, and the associated radioactivity was quantified using a gamma counter (Wizard 2480, PerkinElmer).
Metabolite Analysis.
Uninfected rats were injected IV with 25.9−33.3 MBq of 2-[18F]F-PABA. After 2 or 20 min, the animals were sacrificed and the blood was collected through cardiac puncture. Unlabeled 19F-PABA and N-acetyl-2-F-PABA were added to the blood samples which were subsequently centrifuged to obtain plasma. To remove proteins, plasma was mixed with acetonitrile and both the supernatant and precipitate were collected; the radioactivity was counted. The supernatant was filtered and analyzed using a radio-HPLC system (mobile phase: 12% acetonitrile/water). The radioactive HPLC peaks that corresponded to intact 2-[18F]F-PABA and N-acetyl-2-[18F]F-PABA were collected and quantified by gamma counting (Wizard 2480, PerkinElmer).
Competition with 19F-PABA.
After infecting the rats as described above, 2-[18F]F-PABA (29.6−37 MBq) spiked with 2 mg of 19F-PABA was injected via the tail vein. Two hours postinjection, the animals were sacrificed and the organs harvested to quantify the radioactivity using a gamma counter (Wizard 2480, PerkinElmer) while the bacterial load was determined by plating and CFU enumeration.
Oxacillin Treatment.
Oxacillin treatment was initiated 2 to 4 h after bacterial infection. The rats were treated with 4 doses of 200 mg/kg oxacillin (Sigma-Aldrich) through subcutaneous injection over 20 h before imaging. PET/CT imaging was performed 2 to 4 h after the last oxacillin treatment.
Imaging.
Mice.
Mice were anesthetized by isoflurane inhalation and placed on the scanner bed. An 8 min fullbody CT scan using a SIEMENS Inveon Docked PET/SPECT/CT was performed. Then, 2-[18F]F-PABA (3.7 to 14.8 MBq in 100 to 300 μL of saline) was injected via a tail vein catheter under anesthesia. A 120 min PET scan was initiated as the injection began, followed by a full-body CT scan. The CT data were reconstructed in voxel numbers that were automatically calculated by the software. After the imaging, the PET data were binned into designated time frames, followed by reconstruction using the OSEM 3D method. During PET reconstruction, attenuation correction was performed using the reconstructed CT data. The images were subsequently analyzed by Amide version 1.0.4 (http://www.amide.sourceforge.net).
Rats.
2-[18F]F-PABA (22.2 to 44.4 MBq in 100 to 800 μL of saline) was injected into the rats via a tail vein catheter under anesthesia. PET/CT imaging was performed using a SIEMENS Inveon Docked PET/SPECT/CT. For PET imaging, a dynamic scan was performed 10 min after tracer administration for 60 or 120 min, followed by a full-body CT scan. The image data were reconstructed and analyzed as described above.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant GM-102864 (P.J.T.), an NIH Director’s Transformative Research Award R01EB020539 (S.K.J.), and an NIH New Innovator Award DP2OD006492 (S.K.J.). J.N.I. was supported by an NIH Chemistry-Biology Interface training grant (T32-GM092714).
Footnotes
Notes
The authors declare the following competing financial interest(s): E.A.W., A.A.O., and S.K.J. are inventors for an international patent PCT/US13/059897: Bacteria-specific labeled substrates as imaging biomarkers to diagnose, locate and monitor infections filed by Johns Hopkins University.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.8b00182.
Additional information regarding the HPLC analysis of 2-[18F]F-PABA and in vitro and animal PET imaging experiments (PDF)
REFERENCES
- (1).Tong SY, Davis JS, Eichenberger E, Holland TL, and Fowler VG Jr. (2015) Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28 (3), 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray BE,J Rybak M, Talan DA, and Chambers HF (2011) Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary. Clin. Infect. Dis. 52 (3), 285. [DOI] [PubMed] [Google Scholar]
- (3).Chen DL, Scherer PM, and Palestro CJ (2017) In Imaging Infections: From Bench to Bedside (Jain SK,, Ed.), Springer International Publishing, Cham; DOI: 10.1007/978-3-319-54592-9_29_2. [DOI] [Google Scholar]
- (4).James ML, and Gambhir SS (2012) A molecular imaging primer: modalities, imaging agents, and applications. Physiol. Rev. 92 (2), 897. [DOI] [PubMed] [Google Scholar]
- (5).Jain SK (2017) The Promise of Molecular Imaging in the Study and Treatment of Infectious Diseases. Mol. Imaging Biol. 19 (3), 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Signore A, and Glaudemans AW (2011) The molecular imaging approach to image infections and inflammation by nuclear medicine techniques. Ann. Nucl. Med. 25 (10), 681. [DOI] [PubMed] [Google Scholar]
- (7).Gemmel F, Dumarey N, and Welling M (2009) Future diagnostic agents. Semin. Nucl. Med. 39 (1), 11. [DOI] [PubMed] [Google Scholar]
- (8).van Oosten M, Hahn M, Crane LM, Pleijhuis RG, Francis KP, van Dijl JM, and van Dam GM (2015) Targeted imaging of bacterial infections: advances, hurdles and hopes. FEMS Microbiol Lett. 39 (6), 892. [DOI] [PubMed] [Google Scholar]
- (9).Ordonez AA, Bambarger LE, Murthy N, Wilson DM, and Jain SK Bacterial Imaging (2017) In Imaging Infections: From Bench to Bedside (Jain SK, Ed.), Springer International Publishing, Cham; DOI: 10.1007/978-3-319-54592-9_6. [DOI] [Google Scholar]
- (10).Ordonez AA, and Jain SK (2018) Pathogen-specific bacterial imaging in nuclear medicine. Semin. Nucl. Med. 48 (2), 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Pinkston KL, Singh KV, Gao P, Wilganowski N, Robinson H, Ghosh S, Azhdarinia A, Sevick-Muraca EM, Murray BE, and Harvey BR (2014) Targeting Pili in Enterococcal Pathogenesis. Infect. Immun. 82 (4), 1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Wiehr S, Warnke P, Rolle AM, Schutz M, Oberhettinger P, Kohlhofer U, Quintanilla-Martinez L, Maurer A, Thornton C, Boschetti F, Reischl G, Autenrieth IB, Pichler BJ, and Autenrieth SE (2016) New pathogen-specific immunoPET/MR tracer for molecular imaging of a systemic bacterial infection. Oncotarget 7 (10), 10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Pastrana FR, Thompson JM, Heuker M, Hoekstra H, Dillen CA, Ortines RV, Ashbaugh AG, Pickett JE, Linssen MD, Bernthal NM, Francis KP, Buist G, van Oosten M, van Dam GM, Thorek DLJ, Miller LS, and van Dijl JM (2018) Noninvasive optical and nuclear imaging of Staphylococcus-specific infection with a human monoclonal antibody-based probe. Virulence 9, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Gowrishankar G, Namavari M, Jouannot EB, Hoehne A, Reeves R, Hardy J, and Gambhir SS (2014) Investigation of 6[18F]-fluoromaltose as a novel PET tracer for imaging bacterial infection. PLoS One 9 (9), e107951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ning X, Seo W, Lee S, Takemiya K, Rafi M, Feng X, Weiss D, Wang X, Williams L, Camp VM, Eugene M, Taylor WR, Goodman M, and Murthy N (2014) PET imaging of bacterial infections with fluorine-18-labeled maltohexaose. Angew. Chem., Int. Ed. 53 (51), 14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Weinstein EA, Ordonez AA, DeMarco VP, Murawski AM, Pokkali S, MacDonald EM, Klunk M, Mease RC, Pomper MG, and Jain SK (2014) Imaging Enterobacteriaceae infection in vivo with 18F-fluorodeoxysorbitol positron emission tomography. Sci. Transl. Med. 6 (259), 259ra146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Gowrishankar G, Hardy J, Wardak M, Namavari M, Reeves RE, Neofytou E, Srinivasan A, Wu JC, Contag CH, and Gambhir SS (2017) Specific Imaging of Bacterial Infection Using 6’’-18F-Fluoromaltotriose: A Second-Generation PET Tracer Targeting the Maltodextrin Transporter in Bacteria. J. Nucl. Med. 58 (10), 1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Bettegowda C, Foss CA, Cheong I, Wang Y, Diaz L, Agrawal N, Fox J, Dick J, Dang LH, Zhou S, Kinzler KW, Vogelstein B, and Pomper MG (2005) Imaging bacterial infections with radiolabeled 1-(2′-deoxy-2′-fluoro-beta-D-arabinofuranosyl)-5iodouracil. Proc. Natl. Acad. Sci. U. S. A. 102 (4), 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhang XM, Zhang HH, McLeroth P, Berkowitz RD, Mont MA, Stabin MG, Siegel BA, Alavi A, Barnett TM, Gelb J, Petit C, Spaltro J, Cho SY, Pomper MG, Conklin JJ, Bettegowda C, and Saha S (2016) [124I]FIAU: Human dosimetry and infection imaging in patients with suspected prosthetic joint infection. Nucl. Med. Biol. 43 (5), 273. [DOI] [PubMed] [Google Scholar]
- (20).Sellmyer MA, Lee I, Hou C, Weng CC, Li S, Lieberman BP, Zeng C, Mankoff DA, and Mach RH (2017) Bacterial infection imaging with [18F]fluoropropyl-trimethoprim. Proc. Natl. Acad. Sci. U. S. A. 114 (31), 8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Panizzi P, Nahrendorf M, Figueiredo JL, Panizzi J, Marinelli B, Iwamoto Y, Keliher E, Maddur AA, Waterman P, Kroh HK, Leuschner F, Aikawa E, Swirski FK, Pittet MJ, Hackeng TM, Fuentes-Prior P, Schneewind O, Bock PE, and Weissleder R (2011) In vivo detection of Staphylococcus aureus endocarditis by targeting pathogen-specific prothrombin activation. Nat. Med. 17 (9), 1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).van Oosten M, Schafer T, Gazendam JA, Ohlsen K, Tsompanidou E, de Goffau MC, Harmsen HJ, Crane LM, Lim E, Francis KP, Cheung L, Olive M, Ntziachristos V, van Dijl JM, and van Dam GM (2013) Real-time in vivo imaging of invasive- and biomaterial-associated bacterial infections using fluorescently labelled vancomycin. Nat. Commun. 4, 2584. [DOI] [PubMed] [Google Scholar]
- (23).Hernandez FJ, Huang L, Olson ME, Powers KM, Hernandez LI, Meyerholz DK, Thedens DR, Behlke MA, Horswill AR, and McNamara JO II (2014) Noninvasive imaging of Staphylococcus aureus infections with a nuclease-activated probe. Nat. Med. 20 (3), 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ordonez AA, Weinstein EA, Bambarger LE, Saini V, Chang YS, DeMarco VP, Klunk MH, Urbanowski ME, Moulton KL, Murawski AM, Pokkali S, Kalinda AS, and Jain SK (2017) A Systematic Approach for Developing Bacteria-Specific Imaging Tracers. J. Nucl. Med. 58 (1), 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wegkamp A, van Oorschot W, de Vos WM, and Smid EJ (2007) Characterization of the role of para-aminobenzoic acid biosynthesis in folate production by Lactococcus lactis. Appl. Environ. Microbiol. 73 (8), 2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Johansson G, Bingham S, and Vahter M (1999) A method to compensate for incomplete 24-h urine collections in nutritional epidemiology studies. Public Health Nutr 2 (4), 587. [DOI] [PubMed] [Google Scholar]
- (27).Zarfl C, Matthies M, and Klasmeier J (2008) A mechanistical model for the uptake of sulfonamides by bacteria. Chemosphere 70 (5), 753. [DOI] [PubMed] [Google Scholar]
- (28).Hancock R (1997) The bacterial outer membrane as a drug barrier. Trends Microbiol. 5 (1), 37. [DOI] [PubMed] [Google Scholar]
- (29).Delmar JA, and Yu EW (2016) The AbgT family: A novel class of antimetabolite transporters. Protein Sci. 25 (2), 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Green JM, Merkel WK, and Nichols BP (1992) Characterization and sequence of Escherichia coli pabC, the gene encoding aminodeoxychorismate lyase, a pyridoxal phosphatecontaining enzyme. J. Bacteriol. 174 (16), 5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Chakraborty S, Gruber T, Barry CE, Boshoff HI, and Rhee KY (2013) Para-aminosalicylic acid acts as an alternative substrate of folate metabolism in Mycobacterium tuberculosis. Science 339 (6115), 88−91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Laurieri N, Dairou J, Egleton JE, Stanley LA, Russell AJ, Dupret J-M, Sim E, and Rodrigues-Lima F (2014) From arylamine N-acetyltransferase to folate-dependent acetyl CoA hydrolase: impact of folic acid on the activity of (HUMAN) NAT1 and its homologue (MOUSE) NAT2. PLoS One 9 (5), e96370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Furuya KN, Durie PR, Roberts EA, Soldin SJ, Verjee Z, Yung-Jato L, Giesbrecht E, and Ellis L (1995) Glycine conjugation of para-aminobenzoic acid (PABA): a quantitative test of liver function. Clin. Biochem. 28 (5), 531. [DOI] [PubMed] [Google Scholar]
- (34).Imai T, Tanaka K, Yonemitsu T, Yakushiji Y, and Ohura K (2017) Elucidation of the intestinal absorption of para-aminobenzoic acid, a marker for dietary intake. J. Pharm. Sci. 106 (9), 2881. [DOI] [PubMed] [Google Scholar]
- (35).Delomenie C, Fouix S, Longuemaux S, Brahimi N, Bizet C, Picard B, Denamur E, and Dupret JM (2001) Identification and functional characterization of arylamine N-acetyltransferases in eubacteria: evidence for highly selective acetylation of 5-aminosalicylic acid. J. Bacteriol. 183 (11), 3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Stumpe KDM, Dazzi H, Schaffner A, and von Schulthess GK (2000) Infection imaging using whole-body FDG-PET. Eur. J. Nucl. Med. Mol. Imaging 27 (7), 822. [DOI] [PubMed] [Google Scholar]
- (37).del Rosal T, Goycochea WA, Mendez-Echevarria A, Garcia-Fernandez de Villalta M, Baquero-Artigao F, Coronado M, Marin MD, and Albajara L (2013) 18F-FDG PET/CT in the diagnosis of occult bacterial infections in children. Eur. J. Pediatr. 172 (8), 1111. [DOI] [PubMed] [Google Scholar]
- (38).Simons KS, Pickkers P, Bleeker-Rovers CP, Oyen WJ, and van der Hoeven JG (2010) F-18-Fluorodeoxyglucose positron emission tomography combined with CT in critically ill patients with suspected infection. Intensive Care Med. 36 (3), 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Heysell S, Thomas T, Sifri C, Rehm P, and Houpt E (2013) 18-Fluorodeoxyglucose positron emission tomography for tuberculosis diagnosis and management: a case series. BMC Pulm. Med. 13 (1), 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Konig C, Simmen HP, and Blaser J (1998) Bacterial concentrations in pus and infected peritoneal fluid–implications for bactericidal activity of antibiotics. J. Antimicrob. Chemother. 42 (2), 227. [DOI] [PubMed] [Google Scholar]
- (41).Morris ED, Endres CJ, Schmidt KC, Christian BT, Muzic RF, and Fisher RE (2004) Kinetic modeling in positron emission tomography In Emission Tomography: The Fundamentals of PET and SPECT, Academic Press, San Diego; DOI: 10.1016/B978012744482-6.50026-0. [DOI] [Google Scholar]
- (42).Bertoldo A, Peltoniemi P, Oikonen V, Knuuti J, Nuutila P, and Cobelli C (2001) Kinetic modeling of [18F] FDG in skeletal muscle by PET: a four-compartment five-rate-constant model. Am. J. Physiol Endocrinol Metab 281 (3), E524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.