Abstract
We have adapted the yeast 2-hybrid assay to simultaneously uncover dozens of transient and static protein interactions within a single screen utilizing high-throughput short-read DNA sequencing. The resulting sequence datasets can not only track what genes in a population that are enriched during selection for positive yeast 2-hybrid interactions, but also give detailed information about the relevant subdomains of proteins sufficient for interaction. Here we describe a full suite of stand-alone software programs that allow non-experts to perform all the bioinformatics and statistical steps to process and analyze DNA sequence fastq files from a batch yeast 2-hybrid assay. The processing steps covered by these software include: 1-mapping and counting sequence reads corresponding to each candidate protein encoded within a yeast 2-hybrid prey library; 2-a statistical analysis program that evaluates the enrichment profiles; and 3-tools to examine the translational frame and position within the coding region of each enriched plasmid that encodes the interacting proteins of interest.
Keywords: Protein Interaction, Next Generation Sequencing, DNA Sequence Analysis, Yeast 2-Hybrid
SHORT ABSTRACT:
Deep sequencing of yeast populations selected for positive yeast 2-hybrid interactions potentially yields a wealth of information about interacting partner proteins. Here, we describe the operation of specific bioinformatics tools and customized updated software to analyze sequence data from such screens.
INTRODUCTION:
One approach to discover protein interactions is the yeast 2-hybrid (Y2H) assay, which exploits engineered yeast cells that grow only when a protein of interest binds to a fragment of an interacting partner 1. Detection of multiple Y2H interactions can now be done with the help of massive parallel high-throughput sequencing. Several formats have been described 2–5 including one that we developed where populations are grown in batch under conditions that select for yeast containing plasmids that produce a positive Y2H interaction 6. The workflow we developed, termed DEEPN (Dynamic Enrichment for Evaluation of Protein Networks), identifies differential interactomes from the same prey libraries to identify proteins that interact with one protein (or domain) vs. another protein or a conformationally distinct mutant domain. One of the major steps in this workflow is proper processing and analysis of the DNA sequencing data. Some information can be gleaned by just counting the number of reads for each gene both before and after selection of Y2H interactions in a fashion analogous to an RNA-seq experiment. However, much more in-depth information can be extracted from these datasets including information on the subdomain of a given protein that is capable of producing a Y2H interaction. In addition, whereas the DEEPN approach is valuable, analyzing many sample replicates can be cumbersome and expensive. This problem is alleviated by using a statistical model that was developed specifically for DEEPN datasets where the number of replicates is limited 6. To make processing and analysis of DNA sequencing datasets reliable, complete, robust, and accessible for investigators without bioinformatics expertise, we developed a suite of software programs that cover all steps of analysis.
This suite of stand alone software programs that run on desktop computers includes MAPster, DEEPN, and Stat_Maker. Mapster is a graphic user interface that allows each fastq file queued for mapping to the genome using the HISAT2 program 7, producing a standard .sam file for use in downstream applications. DEEPN has several modules. It assigns and counts reads corresponding to particular gene similar to an RNA-seq type quantification using the module ‘Gene Count’. It also extracts the sequences corresponding to the junction between the Gal4 transcriptional domain and the prey sequence and collates the position of those junctions to allow their inspection by comparative tables and graphs (using the module ‘Junction_Make’) The module ‘Blast_Query’ allows easy inspection, quantitation, and comparison of the junction Gal4 junction sequences. Stat_Maker evaluates the reads per gene enrichment data statistically as a way of prioritizing likely Y2H hits. Here we describe how to use these software programs and to fully analyze the DNA sequence data from a DEEPN Y2H experiment. Versions of DEEPN are available to run on PC, Mac, and Linux systems. Other programs, such as the mapping program MAPster and the DEEPN statistics module Stat_Maker rely on subroutines that run under Unix and are available only on Mac and linux systems.
PROTOCOL:
1. Mapping fastq files
NOTE: DEEPN software as well as many bioinformatics programs use DNA sequence data wherein each sequence read has been mapped for its position in reference DNA. A variety of mapping programs can be used for this including the MAPster interface here that uses the HISTAT2 program to produce .sam files used in subsequent steps.
NOTE: It is imperative that the sequence data is mapped to the correct version of the genome. For Y2H libraries of mouse origin, use the UCSC mm10 genome; for those using human genes, use the UCSC hg38 reference genome, for Saccharomyces cerevisiae genes, use the UCSC SacCer3 reference genome.
-
1.1
Install MAPster.
Download MAPster software and install. The software can be found using a web browser at the following: https://github.com/emptyewer/MAPster/releases. HISAT2 runs on Unix-based systems such as an Apple Macintosh. Because of this, the MAPster program will only run on compatible systems such as Apple Macintosh and linux. System requirements for an Apple Mac are: OSX 10.10+, >4 Gb RAM, >500 Gb disk space, and internet access for downloading reference genomes. Users may need to consult with an institutional IT person if their enterprise has security protocols restricting administrator rights and permissions.
-
1.2
Entering required parameters.
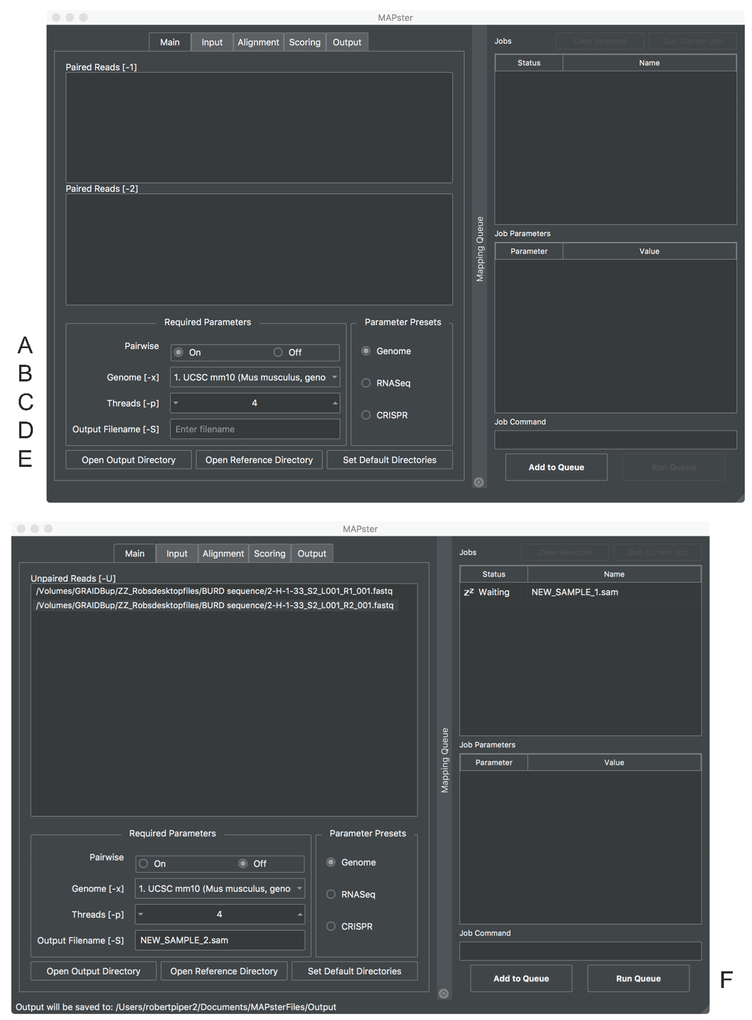
Required files and parameters are entered through the ‘Main’ tab (Figure 1). By selecting the appropriate “Pairwise” button, read files can be entered either as pairs or unpaired with FASTQ as the default file format.-
1.2.1For DEEPN analysis, turn the [Pairwise] option to ‘Off’ to run in single read format.
-
1.2.2Load files into MAPster simply by drag-and-drop into the appropriate window.
-
1.2.3Select a reference DNA/genome source that corresponds to the source of your Y2H prey library inserts. Indexed genomes from several model organisms are listed in the “Genome” box and can be automatically downloaded from the Johns Hopkins University Center for Computational Biology. Reference genomes will be stored locally for later use.
-
1.2.4Indicate the number of computer processes to be devoted to the mapping program under the ‘Threads’ box, since HISAT2 supports multi-threading. MAPster will search your computer and suggest the maximum number of processors available as a default.
-
1.2.5Specify an output file name. This file name will be used throughout the DEEPN process so a short yet descriptive name without space or special characters is recommended. Specify a folder to output the mapped files using the [Open Output Directory] button.
-
1.2.6Once the appropriate files and parameters have been selected, the mapping job must be added to the jobs queue using the [Add to Queue] button. The file names in the main window can be deleted and replaced with files corresponding to a new sample and they can be added to the queue after providing a corresponding output filename.
-
1.2.7Click the [Run Queue] button once all the jobs are entered into the job queue.NOTE: Once a mapping job has been placed in the queue, selecting that job causes the parameter settings to be displayed in the “Job Parameters” window and the command line statement with all arguments to be displayed in the “Job Command” window. The output options include directing whether to keep reads that fail to align and specifying the number of primary alignments allowed for each read. The default output file from MAPster is in SAM format (eg a ‘.sam’ file). It will contain all the sequence reads from the fastq files specified for that sample including those that were (mapped) and were not (unmapped) successfully mapped to the specified geome.
-
1.2.1
Figure 1: The MAPster interface.
Screen shot of the main window of MAPster. The boxes for entering required files and formats are shown. Turn “Pairwise’ (A) off to treat sequence files as single-end reads. The reference genome is selected with the ‘Genome’ menu bar (B). The number of processors used by HISAT2 is selected with the ‘Threads’ menu (C). The new sample name can be typed into ‘Output Filename’ text window. The directory for the output files can be designated in (E). Below is a window showing the queueing of single-end read files. After sample has been added to the queue, mapping can be initiated with the [Run Queue] button (F).
2. Bioinformatic Processing using DEEPN software
NOTE: DEEPN software is currently compiled for use with prey libraries containing mouse cDNA sequences, human cDNA sequences, or S. cerevisiae genomic DNA sequences. DEEPN accepts the standard .sam file format and can accept a SAM (.sam) file containing both mapped and unmapped reads or separate files for each of the unmapped and mapped reads.
-
2.1
Download DEEPN software and install. The software can be found using a web browser at the following: https://github.com/emptyewer/DEEPN/releases. Select which version matches the computing platform and download. To install, open the downloaded install package.
NOTE: Versions of DEEPN are available for PC, Mac, and linux sysrems. Mac and PC systems should have >500 Gb hard disk space and >4 Gb RAM.
-
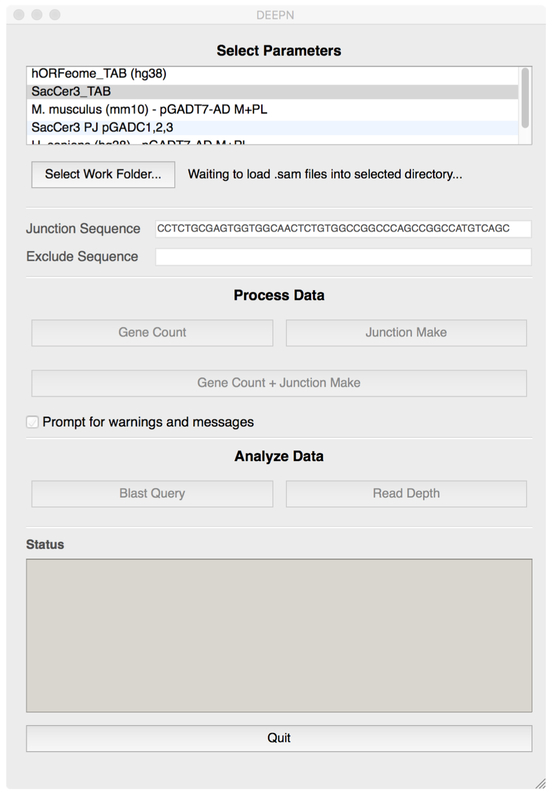
2.2
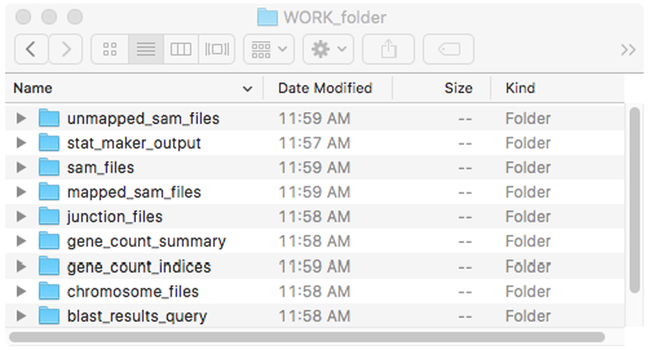
Open the DEEPN software. From the main window (Figure 2) select the corresponding prey library information from the top selection box. Select a folder where the processed files can go by clicking the [Work Folder] button and navigating to the folder/directory. One can create a new folder/directory if needed. Once a ‘work folder’ is selected, DEEPN will create three subfolders entitled unmapped_sam_files, mapped_sam_files, and sam_files. If using .sam files containing both mapped and unmapped reads such as those produced with default settings of the MAPster program, place them in the ‘sam_files’ folder. Otherwise place .sam files into the unmapped_sam_files and mapped_sam_files accordingly.
-
2.3
Initiate processing by clicking the [Gene Count+Junction Make] button. Processing will begin with the Gene Count module that will use mapping positions to count how many reads correspond to each gene. Junction Make will then extract junction sequences (the sequences fused directly downstream from the Gal4-activation domain) from the reads and identify them using the Blast algorithm. This will create a full set of folders pictured in Figure 3. Processing time depends on the size and number of sequence data files and processing speed of the computer used. Typical times range from 12–30 h for an experimental dataset of ~250 million reads. The Gene Count procedure and the Junction_Make procedure can be individually started by clicking the [Gene Count] button or the [Junction Make] button.
-
2.4Download and install Stat_Maker (https://github.com/emptyewer/DEEPN/releases). This is a statistical analysis package designed for DEEPN datasets that currently works only on Unix Mac systems.
-
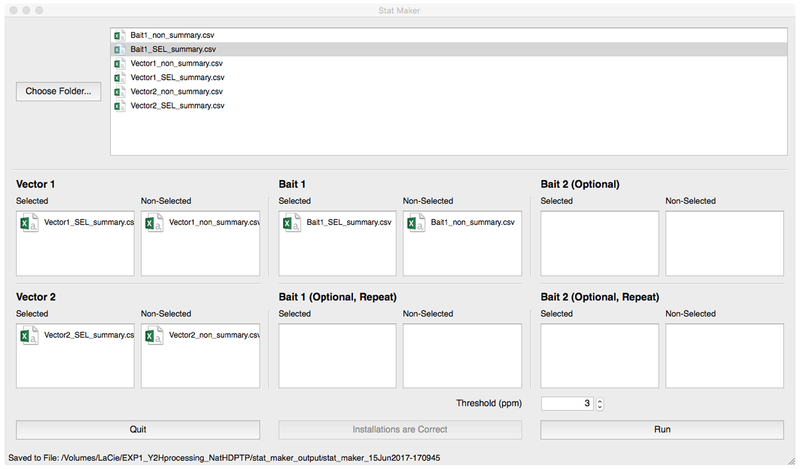
2.4.1Open Stat_Maker and click the button [Verify Installation](Figure 4). If running for the first time, Stat_Maker will automatically install R, JAGS, and Bioconductor by pulling these resources from the internet. Once R, JAGS, and Bioconductor are detected, Stat_Maker will become active and allow further user input.
-
2.4.2Click the [Choose Folder] button to navigate to the working folder that DEEPN processed. Stat_Maker will automatically find and list the files for statistical analysis in the window.
-
2.4.3Drag and drop the appropriate files from the file list window above into the file windows below for each vector and bait dataset and for each growth conditions: non-selected (His+ media) and selected (His-media). Importantly, Stat_Maker requires duplicate datasets for empty vector alone, two samples of non-selected populations and two samples of selected. This gives an estimate of variability within the experiment.
-
2.4.4Click the [Run] button. Depending on the speed of the computer, computation will take between 5–15 min.
-
2.4.1
-
2.5
Review results from the Stat_Maker output, which are placed in a new subfolder within the main work folder labeled ‘Stat_Maker results’. The results are found in a CSV (comma-separated values) file that can be opened in common spreadsheet programs such as Microsoft Excel. Stat_Maker will rank gene hits that are likely to be differentially enriched upon selection with the bait of interest over the empty pTEF-GBD (Figure 5). Also tabulated is the percentage of reads for each dataset where the gene insert is found upstream, downstream, or within the open reading frame and whether the gene is also found within the correct translational reading frame. Often, DEEPN will capture robust Y2H interactions of a bait with portions of a given cDNA that are out of the proper reading frame of the corresponding protein or are to a portion of the cDNA that is downstream of its corresponding open-reading frame. Scanning the combined output from Stat_Maker streamlines detection and elimination of these irrelevant hits.
-
2.6To review the data on each potential candidate, open the DEEPN software, select the corresponding prey library information and then the correct working folder using the [Work Folder].
-
2.6.1Click the [Blast Query] button. This loads a new window (Figure 6). In the top text box, type the gene name or GenBank NM number to select the candidate gene of interest. These gene names correspond to the names listed in the StatMaker output file. Type enter or return, which initiates retrieval of the gene of interest.
-
2.6.2Select which datasets will be used for the analysis using the ‘Select Dataset’ menus. Typically, these include the Vector only and Bait samples grown under non-selective conditions and the Bait sample grown under selection conditions. Initially, the datasets will take a few moments to load, however, subsequent query of the same datasets with different genes will go rapidly. Blast_Query will display the fusion points along the sequence of interest and how abundant each fusion point is. This can be displayed both in a table format using the [Results] tab or a graphic format using the [Plot] tab. These results can be exported to a .csv file by clicking the [Save .csv] button in the top right.
-
2.6.1
Figure 2: DEEPN interface.
Picture of the graphic user interface used to operate the DEEPN modules.
Figure 3: Completion of Processing.
Once DEEPN processes data, the following subfolders are created. These can be inspected, but downstream processes require that these subfolders remain within the main work folder and that they retain their contents and names.
Figure 4: Stat_Maker analysis.
Picture of the graphic user interface for Stat_Maker, which has been loaded with appropriate files to allow for processing. Top shows the initial view of Stat_Maker. Once the presence of underlying support data have been verified by clicking the [Verify Installation] button, and the proper work folder identified after clicking the [Choose Folder] button, the GUI will become active, allowing for loading files.
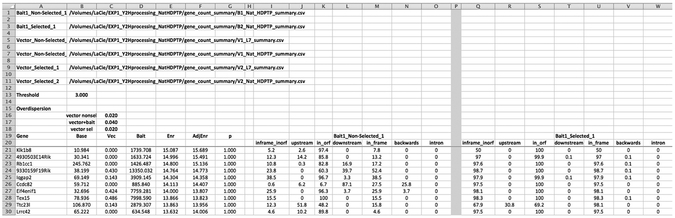
Figure 5: Excerpt from Stat_Maker output.
Portion of Stat_Maker output comparing the enrichment of prey candidates on a single bait protein to vector alone (empty pTEF-GBD). Also shown is the corresponding analysis of whether the plasmids corresponding to the prey candidate contain the proper open-reading frame. Each gene evaluated has several values: Base, Vec, Bait, and Enr. The ‘Base’ is the average proportion of reads (ppm) that were observed for the gene within the 2 datasets corresponding to the duplicate populations containing only vector alone and grown under non-selective conditions. ‘Vec’ refers to the average proportion of reads (ppm) that were observed for the gene within the 2 datasets corresponding to the duplicate populations containing only vector alone and grown under selective conditions (e.g. –His). ‘Bait’ refers to the proportion of reads (ppm) that were observed for the gene within the 2 datasets corresponding to the 2 populations containing the bait plasmid and grown under selective conditions (e.g. –His). ‘Enr’ (enrichement) is log2 ((Bs/Bn) / (Vs/Vn)) where Bs is the reads for bait under selection, Bn is reads for bait under non-selection, Vs is vector alone under selection, and Vn is vector alone under selection.
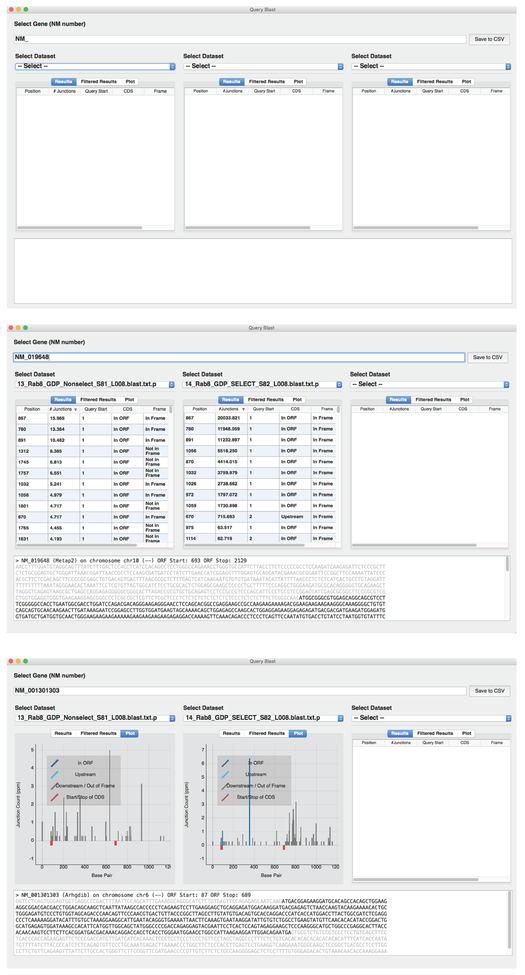
Figure 6: Display of Blast_Query.
Output of Blast_Query from 3 different views. Top is the initial view of Stat_Maker before the datasets of candidate are selected. The middle panel is an example view of the data table displaying information on a given candidate for two different datasets. Bottom shows a graphic view of the tabular data, plotting the number of particular junction points along the gene/cDNA of interest.
3. Verification of candidates identified by DEEPN
NOTE: The purpose of DEEPN and Stat_Maker is to identify candidate genes that give a positive Y2H interaction. Verifying such Y2H interactions can be done using a traditional binary Y2H format using the bait plasmid of interest paired with the empty Gal4-activation domain ‘prey’ plasmid as well as paired with the prey plasmid carrying the gene/cDNA fragment of interest. It is not feasible to isolate the actual plasmid of interest within the mixture of DNA isolated from the yeast population subjected to Y2H selection. However, one can computationally reconstruct what the gene/cDNA fragment is that produces the Y2H interaction, design primers for the 5’ and 3’ ends of that fragment, and amplify that fragment from the DNA isolated from the yeast population. This section describes how to find the 5’ and 3’ end of the candidate prey fragment.
-
3.1.
Open the DEEPN software and choose the parameters [Select Parameter] and the work folder [Select Work Folder] corresponding to the project. Launch the Blast_Query module by clicking the [Blast Query] button.
-
3.2.
Type the name of the gene of interest or its GenBank ‘NM’ number in the top text box. Select from the pull-down menu the dataset that corresponds to the selected yeast population for the bait of interest to retrieve the table of junction positions under the ‘Results’ tab. By default, Blast_Query will order the different positions according their abundance in the dataset, quantified by the ppm of the total number of junctions found within the database. Find the most abundant position that is ‘In ORF’ and ‘In Frame’. The value for position corresponds to the nucleotide position of the gene with the NCBI Reference Sequence (‘NM’ number) found in the top text box. This sequence can be retrieved from GenBank (https://www.ncbi.nlm.nih.gov/nuccore/) or copied from the lower text box in the Blast_Query window.
NOTE: An example can be found in Figure 6, middle panel. In the center dataset, the ‘Results’ show as the most abundant junction: ‘Position’: 867; ‘#Junctions’: 20033.821; ‘Query Start’, 1; CDS: In ORF; and ‘Frame’: In Frame. Nucleotide 867 of the GenBank NCBI reference sequence NM_019648 is the start of the prey fragment.
-
3.3.
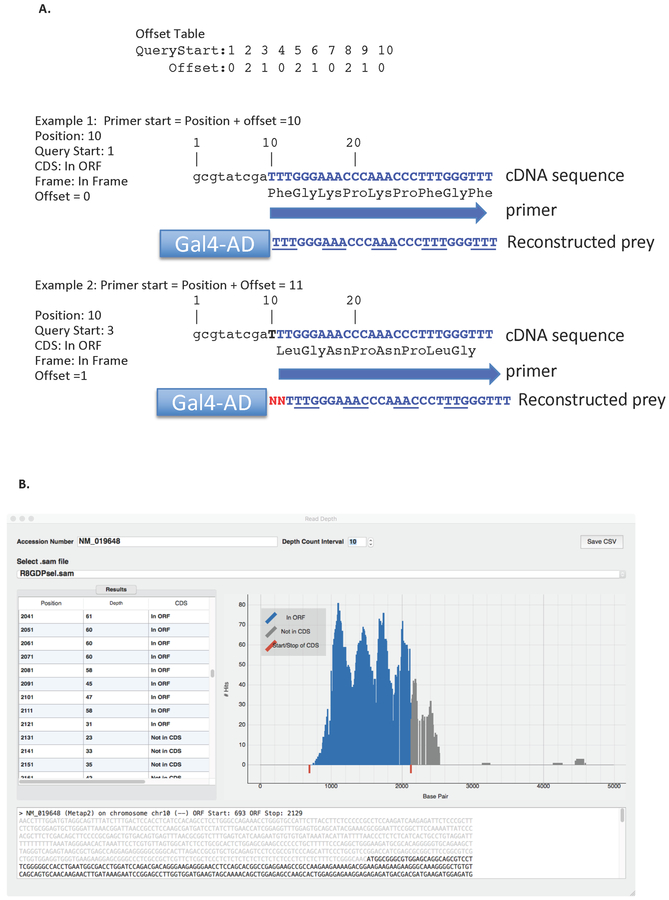
If the Query Start is 1, design the 5’ end of the primer to include the nucleotide corresponding to the position number and extend 25 nucleotides downstream from that position (Figure 7). If Query Start is more than 1, it indicates that there are extra nucleotides between the Gal4 activation domain and the prey sequence of interest and that the primer should start further downstream according to the Query Start value.
-
3.4.
From the DEEPN window click the [Read Depth] button under ‘Analyze Data’. Once the Read Depth window is open, type the NCBI reference sequence (NM) number or gene name into the top text box. Use the pull-down menu to select the relevant dataset that contains the enriched gene of interest. Use the Table on the left and the graphics display on the right to determine how many reads were found in the data that correspond to the gene of interest (Figure 7B).
-
3.5.
Design a 3’ end primer that will capture the sequence of the gene fragment computed by Read Depth. If the abundance of reads goes beyond the ORF and stop codon, design the primer so that it includes the stop codon and the region just upstream of the stop codon. If the sequences for the gene do not extend to past the stop codon, use the Results Table to find the most distant 3’ region that can be detected and use this position as the furthest 3’ position to place the primer.
NOTE: The Read Depth program scans in intervals to find sequences that match the specified gene/cDNA of interest. This helps predict where the 5’ and 3’ end of the most abundant prey fragment is for that gene in the sample. Fluctuations in the read depth along the length of the sequence are normal, as can be seen in Figure 7. If the read depth is clearly past the stop codon, it indicates that the prey fragment extends beyond the stop codon and thus the 3’ primer can simply correspond the region around the stop codon.
-
3.6.
Perform a 50 µL PCR reactions per gene. Each reaction contains 25 pmoles of each forward and reverse primer matching the prey-library plasmid (see materials). Reactions also contain 25 µL High-Fidelity 2× PCR Master Mix, 5 µg of DNA sample, and water up to 50 µL. Reactions are amplified for 25 cycles with extension times of 3 min at 72 °C, annealing temperature of 55 °C for 30 s, and denaturing at 98 °C for 10 s. Cycling is preceded by a 30 s denaturation at 98 °C and is followed by a 5 min incubation at 72 °C.
Figure 7: Finding the 5’ and 3’ primers to amplify.
(A) shows a hypothetical sequence and how to design the 5’ oligo to capture the correct frame and fusion point between the Gal4-activation domain and the prey sequence of interest. In Example 1, the position of fusion point is at the 10th nucleotide with a Q start of 1. Using the above offset Table, 0 nucleotides are to be added to find the 5’ start position of the primer. The reconstructed prey plasmid fusion point shows that the Gal4 activation domain is fused directly to the prey at nucleotide 10. In Example 2, the Query Start is 3, which requires an offset of 1 nucleotide in order to capture the correct starting point and frame of the prey insert. The schematic of the reconstructed prey shows that there are 2 nucleotides between the Gal4 activation domain and the known position of the prey insert that must be accounted for. (B) Shows the Read Depth window. The textbox at the top is used to enter the NCBI reference sequence number and the pull-down menu under ‘Select .sam file’ is used to select the data for the sample containing the enriched interacting gene if interest. Read Depth shows how many sequences (Y axis) were found in the data that correspond to the nucleotide positions of the sequence of interest (X-axis).
REPRESENTATIVE RESULTS:
Mapping fastq data: the first step
In practically all NGS applications including DEEPN the initial output is a file of short sequence reads that must be mapped by alignment to genomic, transcriptomic, or other reference DNA 8. Recently, the HISAT2 alignment program was developed that uses state-of-the-art indexing algorithms to dramatically increase the mapping speed 7,9. HISAT2 runs efficiently on a desktop computer and can map a typically sized read file in minutes. This allowed us to wrap HISAT2 into a graphic user interface called MAPster that can map fastq files locally, allowing users to avoid relying on remote high-performance computer clusters that typically operate with command-line language (Figure 1). Important features of MAPster include the presence of preset parameters for RNA-seq and whole genome mapping experiments, the ability to queue multiple jobs, and access to a full set of easily adjustable HISAT2 parameters for expert users and for customized applications. In order to illustrate MAPster’s functionality, a publically available eHAP cell RNA-seq data file was mapped to the Ensemble GRChg38 genome plus transcript reference DNA. The eHAP A11 replicate 1 FASTQ file was downloaded from the NCBI Sequence Read Archive and contained 38.3 million reads. MAPster was run on an Apple iMac with 3.5 GHz Intel Core i7 processor using default RNA-seq parameters for the unpaired read file. The mapping was completed in less than five minutes. The overall alignment rate was 96.6%. Similar results are found with typical DEEPN datasets of 15–25 million reads/sample, although the overall alignment rate is lower due to the presence of vector sequence from the Y2H prey plasmid.
Finding candidate hits with the help of Stat_Maker.
The StatMaker program produces an excel-viewable file that summarizes most of the pertinent information needed to identify candidate interacting proteins. Because Stat_Maker makes use of unix-based subroutines, it will run on a Mac (OS10.10+) but not PC. First, it summarizes the reads in ppm for each gene for both vector control and bait populations and also produces a probability ranking whether the enrichment of a particular gene when selected for Y2H interaction with the bait of interest is truly greater than the enrichment of that gene when selected for interaction with the vector-only control (Figure 5). Second, StatMaker performs the BlastQuery module computations on every gene evaluated and tabulates the percentage of junction reads that are in the correct translational frame and the coding sequence which would be required for a bonafide biologically relevant interactor. This combined output makes it possible to quickly sort and filter candidates to identify those that can be inspected closer by BlastQuery. With this output, one can first sort for those candidates with the highest probabily of being enriched during selection for Y2H interaction on the bait protein of interest and not when selected for interaction on the vector plasmid alone. In practice, we find that P > 0.95 works well. Then candidates can be ranked for those that have the most junction reads that are both in the coding region and in the proper reading frame using a simple sorting function. Here, candidates with >85% of junctions that are in the correct translational frame and are found either within the open reading frame/protein coding region (in ORF) or that begin just upstream of the start codon (upstream). This latter filter eliminates 60–80% of candidates that have an acceptable P value, producing a list that is much more biologically relevant and manageable for further inspection.
The DEEPN software.
The core DEEPN software bundles several computational modules together to integrate all the bioinformatics steps using SAM files. Gene_Count provides the number of reads per gene, performing a calculation similar to an RNA-seq quantitation. Other programs that perform this type of calculation could be used as well, however, the file format would need to be altered to be compatible with other DEEPN modules and the Stat_Maker program. Alternatively, the Gene_Count module could be used to quantify RNAseq experiments, however, other packages intergrated with specific statistics programs have been developed 10. The process of matching a particular mapped read with its corresponding gene of interest has been improved since the initial DEEPN software by using a data tree structure for gene assignment. The effect of this was to greatly accelerate the speed of processing such that a typical dataset containing 10 million mapped reads takes 5–10 minutes on desktop computer with minimal system requirements. Other analyses, in particular the analysis of junction reads that span the Gal4-activation domain and the interacting candidate of interest, are self-contained. They are packaged with the BLAST alogorithm that runs locally and have parsing procedures to correctly collate all the junction reads and their positions for all the given genes. One of drawbacks of the DEEPN software is that it makes use of special formatted databases that define which exons in the reference genomes are used to define cDNAs or coding regions, and formatted databases that specify the sequence and translational start and stops of each cDNA/genes used. We found that it was difficult to retrieve all the database information DEEPN requires in a reliable format that lacked some of the spurious mistakes we encountered with the indexing of particular genes. Thus, we assembled new databases that we quality controlled and embedded them into the DEEPN software for consistent internal reference. Currently, mouse, human, and Saccaromyces cerevisiae Y2H prey libraries are supported by the included databases provided that the DNA fastq files are mapped against the mm10, hg38, or SacCer3 reference databases available from UCSC. Y2H libraries from different organisms can be processed by DEEPN provided that similar databases are built and placed into the DEEPN software. Overall, however, the self-contained packaging of all the DEEPN modules, databases, and other programs make these bioinformatic analyses accessible to investigators at all levels of expertise.
DISCUSSION:
The software suite described here allows one to completely process and analyze high throughput DNA sequencing data from a DEEPN experiment. The first program used is MAPster, which takes the DNA sequence reads in standard fastq files and maps their position onto a reference DNA for downstream processing by a whole host of informatics programs including the DEEPN software. The utility of the MAPster interface and its ability to queue multiple jobs, combine input files, coveniently name output files, coupled with the speed of the underlying HISAT2 program 7 it controls provides an easy-to-use tool mapping for a variety of applications beyond DEEPN. MAPster can access several parameters of the HISAT2 program that are suited for other types of data analysis besides DEEPN. Some of these features include preset parameters for RNA-seq and whole genome mapping experiments and access to a full set of easily adjustable HISAT2 parameters for expert users and for customized applications. For instance, the RNA-seq button adds formatting that would facilitate transcript assembly. The CRISPR button blocks alignment to the reverse complement strand as would be appropriate for a reference DNA file derived from guide RNA sequences. The optional parameters are found under four tabs labeled, “Input”, “Alignment”, “Scoring”, and “Output.” The input options include the ability to change input file formats and to specify basic read trimming options. The alignment and scoring tabs include the options to select only one strand on the reference DNA and to set the gap and mismatch penalties for the alignment scores. The ability to conveniently queue multiple mapping jobs each with distinct parameter setting should make MAPster of interest to both expert and non-expert users pursuing complex NGS applications.
The DEEPN and Stat_Maker software programs are dedicated to the specific bioinformatics analysis of data from batch Y2H screens. This is accessible to a broad range of investigators and constitutes a contiguous bioinformatic software package run through a graphic user interface. This package has been further optimized and integrated from its original description 6 so that it runs faster and analysis of candidate hits is streamlined. All the bioinformatics steps can be run on a desktop computer. The main DEEPN software takes these map positions to calculate how many reads correspond to each gene thereby forming the basis for how a given gene is enriched upon selection. This software also finds the ‘junction’ sequences that correspond to the insert of interest as it is fused to the transcriptional activation domain of the prey plasmid and tabulates these results so that one can visualize all the different portions of a particular ORF or cDNA that is sufficient for interaction. In addition, this also provides information to verify the reading frame of each insert. The third arm of the bioinformatic software is Stat_Maker, which uses output files processed by DEEPN to calculate the statistical relevance of gene enrichments resulting from interactions with a given bait protein vs. the Gal4-DNA-binding domain vector alone (empty pTEF-GBD). A recent improvement is that Stat_Maker not only provides a statistical ranking of each candidate, but also tabulates the corresponding information extracted from the corresponding junction sequences, making them available in a single file making it much easier for investigators to survey and review the results.
ACKNOWLEDGMENTS:
This work was supported by National Institutes of Health: NIH R21 EB021870–01A1 and by NSF Research Project Grant: 1517110.
Footnotes
DISCLOSURES:
The authors have nothing to disclose
Contributor Information
Venkatramanan Krishnaman, Molecular Physiology and Biophysics, University of Iowa, Iowa City, IA USA 52242, venky.agas@gmail.com.
Tabitha A. Peterson, Molecular Physiology and Biophysics, University of Iowa, Iowa City, IA USA 52242, tabitha-peterson@uiowa.edu
Robert C. Piper, Molecular Physiology and Biophysics, University of Iowa, Iowa City, IA USA 52242, Robert-Piper@uiowa.edu
Mark A. Stamnes, Molecular Physiology and Biophysics, University of Iowa, Iowa City, IA USA 52242, Mark-Stamnes@uiowa.edu
REFERENCES
- 1.Fields S & Song O A novel genetic system to detect protein-protein interactions. Nature. 340 (6230), 245–246, doi: 10.1038/340245a0, (1989). [DOI] [PubMed] [Google Scholar]
- 2.Rajagopala SV Mapping the Protein-Protein Interactome Networks Using Yeast Two-Hybrid Screens. Adv Exp Med Biol. 883 187–214, doi: 10.1007/978-3-319-23603-2_11, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weimann M et al. A Y2H-seq approach defines the human protein methyltransferase interactome. Nat Methods. 10 (4), 339–342, doi: 10.1038/nmeth.2397, (2013). [DOI] [PubMed] [Google Scholar]
- 4.Yachie N et al. Pooled-matrix protein interaction screens using Barcode Fusion Genetics. Mol Syst Biol. 12 (4), 863, doi: 10.15252/msb.20156660, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trigg SA et al. CrY2H-seq: a massively multiplexed assay for deep-coverage interactome mapping. Nat Methods. doi: 10.1038/nmeth.4343, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pashkova N et al. DEEPN as an Approach for Batch Processing of Yeast 2-Hybrid Interactions. Cell Rep. 17 (1), 303–315, doi: 10.1016/j.celrep.2016.08.095, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D, Langmead B & Salzberg SL HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 12 (4), 357–360, doi: 10.1038/nmeth.3317, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinert K, Langmead B, Weese D & Evers DJ Alignment of Next-Generation Sequencing Reads. Annu Rev Genomics Hum Genet. 16 133–151, doi: 10.1146/annurev-genom-090413-025358, (2015). [DOI] [PubMed] [Google Scholar]
- 9.Pertea M, Kim D, Pertea GM, Leek JT & Salzberg SL Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 11 (9), 1650–1667, doi: 10.1038/nprot.2016.095, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conesa A et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 17 13, doi: 10.1186/s13059-016-0881-8, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]