Abstract
More reliable and cheaper sequencing technologies have revealed the vast mutational landscapes characteristic of many phenotypes. The analysis of such genetic variants has led to successful identification of altered proteins underlying many Mendelian disorders. Nevertheless the simple one-variant one-phenotype model valid for many monogenic diseases does not capture the complexity of polygenic traits and disorders. Although experimental and computational approaches have improved detection of functionally deleterious variants and important interactions between gene products, the development of comprehensive models relating genotype and phenotypes remains a challenge in the field of genomic medicine.
In this context, a new view of the pathologic state as significant perturbation of the network of interactions between biomolecules is crucial for the identification of biochemical pathways associated with complex phenotypes. Seminal studies in systems biology combined the analysis of genetic variation with protein-protein interaction networks to demonstrate that even as biological systems evolve to be robust to genetic variation, their topologies create disease vulnerabilities. More recent analyses model the impact of genetic variants as changes to the ‘wiring’ of the interactome to better capture heterogeneity in genotype-phenotype relationships. These studies lay the foundation for using networks to predict variant effects at scale using machine-learning or algorithmic approaches. A wealth of databases and resources for the annotation of genotype-phenotype relationships have been developed to support developments in this area. This overview describes how study of the molecular interactome has generated insights linking the organization of biological systems to disease mechanism, and how this information can enable precision medicine.
Graphical Abstract
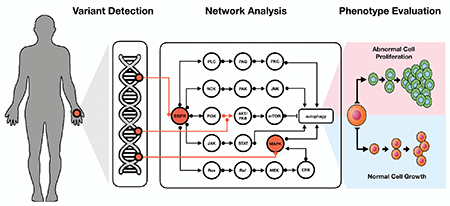
Conceptual representation of genome interpretation using biological networks.
Introduction
Recent advances in sequencing technologies have significantly reduced the costs of genome sequencing and genetic testing, allowing the detection of genetic variants at scale. In particular for humans, previous studies have aimed to identify genetic variants common to different populations 1 and nucleotide changes associated with phenotypes 2. Variant data collected in population studies have been used to describe the evolutionary history of humans 3 while, in medical settings, research has aimed to detect disease causing variants 4 and/or variants that increase susceptibility 5.
The analysis of genomic data and its relation to phenotypes is a fundamental step for enabling precision medicine 6. Although in the last decades many studies have uncovered genetic variants associated to diseases 7, these discoveries only partially explain the biological complexity of most human diseases 8. This observation is more evident in the case of polygenic disorders where the associated genetic variants are carried by only a fraction of the patients 9. Indeed many common and individually weak alleles have been detected for schizophrenia, bipolar disorder 10 and rheumatoid arthritis 11. The presence in the general population of large numbers of rare variants under strong selection suggests the hypothesis that these variants may contribute to a variety of diseases, potentially affecting many genes and pathways 12. Furthermore, it is well established that genes do not cause disease in isolation but rather encode elements that form a dynamic molecular network in which perturbations may result in different phenotypes 13, 14. As many disease mutations affect protein function or expression, this overview focuses on networks of proteins and their interactions. Indeed, knowledge of the protein-protein interaction network has proven relevant for understanding the mechanisms of many human disorders, including ataxia 15 (Kahle et al., 2011), autism 16, Huntington’s disease 17 and breast cancer 18. In addition, analysis of the interactome is important for the identification of cross-phenotype genetic associations 19, 20. This phenomenon, referred to as pleiotropy, was introduced more than 100 years ago by Ludwig Plate to describe cases where a mutation affecting the same gene results in clinically distinguishable phenotypes 21.
These observations sustain the need for more accurate tools for genome interpretation that consider the constellation of variants carried by an individual as possible perturbations of the underlying molecular interaction networks. In this review we summarize available resources, and describe how analysis of the interactome has led to an understanding of how the organization of biological systems leads to disease vulnerabilities. We discuss emerging strategies for predicting the impact of genetic variation using interactome networks and highlight future opportunities for network analysis of variants.
KEY CONCEPTS
In principle, genetic variants in the protein coding region of the genome can have a broad array of effects on protein activity, ranging from no consequence to severe alteration to the function and/or structure of a protein. The effect of a variant at the single protein level may not reflect the severity of the associated phenotype. Some loss of function mutations are well tolerated. Instead the severity of perturbation to the complex network of molecular interactions in the cell may more closely capture potential to generate a phenotype. Missense mutations that change only a single amino acid in the protein can significantly affect protein-protein, protein-DNA and enzyme-substrate interactions 22. Studying variants in the context of protein-protein interaction (PPI) networks and biochemical pathways can improve our understanding of the mechanisms underlying genotype-phenotype relationships.
Graphical modeling for computing on networks
Graphical models provide a mathematical framework for studying the architecture of biological systems. Biological systems can be represented as networks, wherein nodes usually represent biomolecules, and edges represent interactions among them. Graphical modeling then allows quantitative measures to be derived from the network topology for analyzing different aspects of biological systems. The network structure itself can be analyzed, or biological measurement data can be mapped onto network nodes and edges to facilitate integration or interpretation of those measurements in the context of the organization of the underlying system. In the context of the relating genotype to phenotype, genetic alterations are mapped onto their respective proteins to identify the PPIs and biochemical pathways that are potentially affected.
Network analysis measures
Various network measures have been developed to describe the characteristics of nodes within networks 23. In Figure 1 we summarize a few important network measures used to describe nodes, using the PPI network of the NTRK2 activation pathway as an example. Node degree describes the number of interaction partners, and can be used to designate proteins as hubs or peripheral nodes (Figure 1A). The clustering coefficient of a node describes how connected the immediate network neighborhood of a node is (Figure 1B). Various measures of centrality have been developed to capture the importance of a node to information flow in a network. For example, degree centrality captures how connected a node is to the rest of the network (Figure 1C), and betweenness centrality describes the number of shortest paths that traverse a node (Figure 1D). Nodes can also be assigned to modules within the network using community detection algorithms 24, 25. An example is represented in Figure 1 Panel D where NTRK2, BDNF, NTF4 constitute a module obtained using the Girvan-Newman algorithm 26 and NTRK2 is a bottleneck connecting two communities. The network measures described above can be used to identify nodes with key ‘roles’ within the network topology 27.
Figure 1. Network analysis measures.
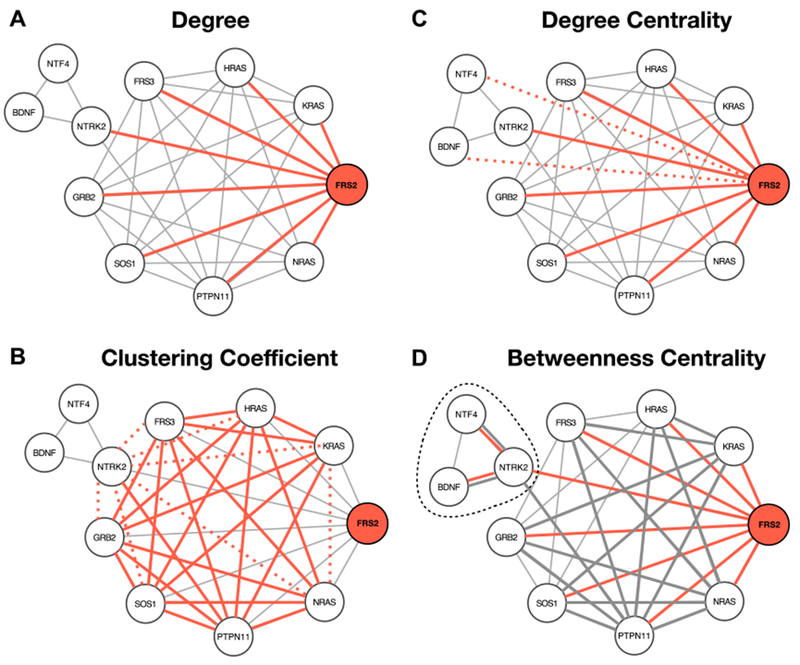
PPI network of the NTRK2 activation pathway through FRS2/FRS3. A) The degree (k) is the number of edges of a node. The degree of FRS2 is 8. The edges are highlighted in red. B) The clustering coefficient C of a node is calculated as the ratio between the connected triangles (delimited by the solid red and grey lines) and the total number of possible triangles k×(k-1). The dashed lines represent the unbound triangles. For FRS2, C is 0.75 (21/28). C) The degree centrality (CD) of a node is the number of edges divided by the total number of possible edges. The CD of FRS2 is 0.8 (8/10). Red dotted lines represent the missing edges. D) The betweenness centrality (B) is the sum over all the possible pairs of the fraction of shortest path passing through a node (red) divided by the total number of shortest paths. B of FRS2 is 9.167 (18×0.5+0.167). Grey edges are part of the shortest paths not passing through FRS2. In this example, edge length is determined by the layout algorithm and does not have a quantitative interpretation.
DATABASES AND RESOURCES
The development of new methods for analyzing and predicting the impact of genetic variants in the context of the protein-protein interaction network requires the collection of high-quality data from biological experiments and clinical reports. The primary sources of data needed for such analyses can be divided into three main categories: genetic variants, biological networks and disease association databases.
Genetic variant databases
There are a growing number of databases of genetic variants available on the internet. The most comprehensive database of small variants is dbSNP 28 which, in the current version (Build 151), includes more than 113 million validated genetic variants. In spite of the name, dbSNP does not contain only Single Nucleotide Polymorphisms (SNPs) but also rare and somatic variants. Considering these different types of genetic variation, single nucleotide variants account for ~90% of small variants. A significant amount of these data are the result of the 1000 Genomes project 1. The 1000 Genomes Consortium sequenced the whole genomes of more than 2,500 individuals from different populations allowing a more accurate characterization of the landscape of genetic variants in humans and better estimates of the average load of variants per individual 29.
Several data sources are more focused on collecting information about the phenotypic effects of genetic variants. For example, Clinvar hosts curated information about the health consequences of genetic variants 30. Clinvar includes ~440,000 variants with some supporting evidence of a relationship to human phenotypes (Clinvar release July 16, 2018). Focusing on the variants with clinical significance, Clinvar contains ~81,000 genetic variants classified either as “Pathogenic” or “Benign”. The Pathogenic subset consists of ~52,000 variants from ~3,500 genes which are associated with more than 4,400 Mendelian disorders. Clinvar also includes a small set of disease-associated variants in intronic and non-coding regions (~4,900), and pathogenic synonymous single nucleotide variants (~200). Another important database that contains information about the impact single amino acid variants (SAVs) and their relationship to human phenotype is SwissVar 31. SwissVar curators classify the impact of SAVs either as “Pathogenic” or “Polymorphism”, extracting relevant information from the literature. The current release of SwissVar database (release 18, July 2018) contains ~70,000 SAVs, 42% of which (~29,000) are “Pathogenic” in ~2,750 genes. These pathogenic SAVs are associated with more than 3,450 Mendelian diseases.
Since 2008, the published results of genome-wide association studies (GWAS) have been systematically reviewed to extract significant association between common variants (SNPs) and complex disorders. This data is hosted by the GWAS Catalogue 5 that contains significant SNP-Trait associations (p-value < 9e−6) for 50,900 unique genomic locations. About 63% of these loci are mapped to ~11,400 genes while the remaining variants are located in intergenic regions. Among complex diseases, cancer has been the focus of many sequencing studies 32 (Hudson et al., 2010). The analysis of genomic data from cancer patients resulted in the detection of a large number of somatic variants, found by comparing the genetic variants in tumor cells with those in the normal cells from the same individual. The somatic variants detected with this approach are collected in the COSMIC database33 (Forbes et al., 2017). Version 85 of the COSMIC database (May 2018) contains ~4.4 million variants from ~253,000 tumors samples across 45 primary sites. A small number of the variants reported in COSMIC (141) are additionally described as causing clinical resistance to pharmaceutical therapies. These 141 mutations affected 21 different genes, but were most prevalent in ABL1, EGFR and KIT.
Resources for biological network analysis
Biological networks can be built directly from expert knowledge, or in an unbiased fashion from large experimental screens34 (Rual et al., 2005) (see Sidebar 1). Perhaps the most intuitive biological network model is a protein-protein interaction (PPI) network, where nodes represent proteins and edges indicate physical interactions between proteins. Other common networks model intra-cellular signaling, mRNA co-expression, gene regulation or metabolic flux. A broad selection of pathways and networks are hosted via online databases such as KEGG 35, IntAct 36, iRefIndex 37, Reactome 38, Pathway Commons39, BioPlex 40, DIP41 and STRING 42. These databases can be classified according to the type of information collected. Although KEGG (Kyoto Encyclopedia of Genes and Genomes) collects many types of information, it serves as a reference database for biological pathways. The KEGG PATHWAY resource consists of graphical representations of cellular processes, such as metabolism, membrane transport, signal transduction and cell cycle. Recently, the MINT 43 and IntAct 44 databases merged their efforts to provide a curated repository of experimentally determined interactions. The current version of IntAct (August 2018) contains ~546,000 unique PPIs, 35% of which occur between ~23,500 human proteins. IntAct also contains a small set of proteins unlikely to be engaged in an interaction. This negative interaction set represents less than 0.2% of the total number of IntAct interactions. Other resources such as iRefIndex 37, Reactome 38, Pathway Commons 39 integrate pathways and/or protein-protein interaction data from several primary sources.
Sidebar 1: Interactome construction
PPIs can be detected in a variety of ways. The two most common technologies for high-throughput PPI screening are yeast two-hybrid (Y2H) and mass spectrometry (MS). These technologies have advantages and limitations that must be considered when analyzing the resulting interactomes. Y2H can effectively detect binary interactions, but cannot detect multi-protein complexes. MS can characterize the elements of multi-protein complexes, however it does not provide enough information to determine which proteins in the complex are in direct physical contact. Sometimes such complexes are depicted as ‘cliques’ in networks, such that all participating proteins in the complex are linked together by edges. Both technologies have associated false positive (finding an interaction when none exists) and false negative (failing to find an existing interaction) detection rates. Interactions can be also be obtained from low throughput experiments, for example co-crystal structures obtained via x-ray crystallography, cryo-electron microscopy or negative stain electron microscopy. Because many studies that probe protein interactions are not performed as high-throughput screens, the associated interactions are generally mined from the biomedical literature. Networks generated from literature mining are more susceptible to study bias than networks derived from high-throughput screens, however networks constructed from the literature tend to contain more interactions and those interactions may be less prone to random error 45.
Another widely used resource for protein-protein interactions is the STRING 42 database which integrates known and predicted interaction data across multiple organisms. Apart from experimentally verified PPIs, STRING collects data derived from different sources including gene co-expression analyses, automated text-mining and computational inference based on gene orthology. For human alone, the latest release of STRING (version 10.5, May 2017) contains more than 691,000 unique associations between ~18,700 genes.
New resources for studying biological networks, such as the Network Data Exchange (NDEx) 46 and the Cell Collective 47, are emerging to provide collaborative platforms for data sharing, analysis and model simulation. In particular NDEx implements a RESTful API which can be programmatically accessed by any application. In the most recent version of NDEx, the curators improved the quality and abundance of biological networks relevant to the cancer research community. Similarly, the Cell Collective platform enables users to build and analyze network models, and use them to run simulations via a web interface. This application can be used to simulate loss/gain of function and test possible scenarios in real time.
High-throughput experiments for detecting molecular interactions are important for mapping the interactome and are also useful for assessing the quality of available databases, validating the performance of methods that predicting PPIs and selecting sub-networks obtained from the same experimental technologies. A recent study based on affinity purification mass spectrometry detected more than 56,000 interactions among ~11,000 human proteins 40. The results of this experiment are accessible through the BioPlex website (http://bioplex.hms.harvard.edu/). Crosslink mass spectrometry is an alternative technique to profile PPIs 48, 49. A popular high-throughput approach for detecting interactions is by yeast two-hybrid (Y2H) experiments that detect binary protein-protein interactions 34, 50, 51. Many such interactions are available through the Human Reference Protein Interaction Mapping Project (HuRI; http://interactome.baderlab.org/).
The large number of available resources for studying biological networks poses a question about the implications of selecting a network for a particular study, including the reliability of particular networks for specific applications (see Sidebar 2). Focusing on the ability to recover disease gene sets, a recent study evaluated 21 human genome-wide interaction networks 52. This analysis showed that performance increased with network size. The STRING database had the best overall performance, however after correcting for size, the smaller network from the Database of Interacting Proteins (DIP) 41 had the highest per edge performance.
Sidebar 2: Selecting the right interactome
The availability of numerous biological networks constructed from different experimental techniques and literature mining poses a difficult question about the accuracy and reliability of networks for disease studies. Generally speaking the evaluation of the quality of the interaction networks is problem dependent. Focusing on the task of recovering disease-gene associations based on colocation and connectivity in the interactome, larger networks tend to achieve better overall performance (higher sensitivity) than smaller networks. Contrarily, if the analysis aims to minimize the number of false positive genes recovered, a network with a smaller and well-curated set of interactions on average scores with higher precision than larger networks 53. Interactions that have been detected by multiple technologies are often considered more reliable, thus some studies include interactions with multiple evidences 54–56. However this can throw away real interactions that could be informative for a particular study. As a rule of thumb, protein-protein interaction data from X-ray crystallography are more reliable than other types of data. An important limitation for evaluating the quality of PPI networks is the low number of known of non-interacting proteins (negative set). A fair assessment of the quality of a PPI network should include an analysis of the performance on a negative set.
Disease/Phenotype annotation and classification
Pivotal resources for studying the impact of genetic variants in the context biological networks are databases for the annotation and classification of diseases and phenotypes. A systematic classification of diseases and their genetic causes was carried out by McKusick, who developed the primary comprehensive curated repository for genotype/phenotype relationships. Available online since 1987, the Online Mendelian Inheritance in Man (OMIM) database 7 synthesizes and summarizes information extracted from the biomedical literature by careful curation. The OMIM database is freely available upon request for the academic community, and it its current version (August 2018) contains ~5,300 phenotypes with known molecular basis.
Cataloging and description of distinct phenotypes is a limiting step for their analysis and comparison. To overcome this limitation, different standardized vocabularies have been developed to ensure consistent, reusable and sustainable descriptions of human diseases. Initially the US National Library of Medicine (NLM) developed the Unified Medical Language System (UMLS) 57 which includes names, concepts and relationships from different biomedical vocabularies. Similarly, Medical Subject Headings (MeSH) 58 defines a hierarchically-organized terminology for indexing and cataloging biomedical information, and the Systematized Nomenclature of Medicine (SNOMED) provides a systematic, computer-processable collection of medical terms 59.
The medical terms developed by NLM curators are part of specific ontologies for the classification of disease and phenotype such as the Disease Ontology (DO) 60 and the Human Phenotype Ontology (HPO) 61. DO is a hierarchical disease-centric ontology collecting additional facts about disease. In the latest version, DO curators expanded the utilities for examination and comparison of genetic variants, phenotypes, proteins, drugs and epitopes. In contrast to DO, the HPO focuses on the analysis of phenotypic abnormalities. The HPO project is divided into three components: the phenotype vocabulary, disease-phenotype annotations and algorithms that operate on these. With respect to DO, HPO implements a better nomenclature for the description of rare diseases.
There are a number of additional resources for disease-gene associations such as DisGeNET 62, dSysMap 63 and the Comparative Toxicogenomics Database (CTD) 64. In particular, DisGeNET is one of the largest collections of genes and variants associated with human diseases, and integrates data from expert curated repositories, GWAS catalogues, animal models and the scientific literature. The current version of DisGeNET (v5.0) contains more than 560,000 gene-disease associations, between ~17,000 genes and more than 20,000 diseases and traits. In terms of variants, it contains more than 135,000 variant-disease associations, between ~83,000 variants and ~9,200 diseases and phenotypes. Focusing more on protein structure, dSysMap maps human disease-related mutations onto the structural interactome. In its latest version (April 2018), dSysMap contains ~29,000 mutations in ~2,700 proteins associated to ~3,600 phenotypes. Finally, the CTD is a database that aims to advance the understanding of the effect of environmental exposures on human health. The database is divided in six categories with eleven relationships among them. Apart from gene–disease associations and gene-gene interaction data, CTD collects associations between chemicals and diseases. The current version of the database (June 2018) is composed of ~37,500 curated gene–disease and ~211,500 curated chemical–disease associations. A summary of the resources described above is provided in Table 1.
Table 1.
Selected databases and resources for variant interpretation in the context of biological interactions
| Database | Data | URL |
|---|---|---|
| Variant databases | ||
| 1000 Genomes | Whole genome and variants of >2500 individuals | http://www.internationalgenome.org |
| ClinVar | Human variants with clinical significance | https://www.ncbi.nlm.nih.gov/clinvar |
| COSMIC | Catalogue of somatic mutations in cancer | https://cancer.sanger.ac.uk/cosmic |
| dbSNP | Small variants from several organisms | https://www.ncbi.nlm.nih.gov/snp |
| GWAS Catalog | Disease-associated variants from published GWAS | https://www.ebi.ac.uk/gwas |
| SwissVar | Annotated single amino acid variants | https://swissvar.expasy.org |
| Network resources | ||
| BioPlex | Human PPIs from AP-MS | http://bioplex.hms.harvard.edu |
| HuRI | Human PPIs from Y2H | http://interactome.baderlab.org/ |
| IntAct | Manually curated PPIs from literature | https://www.ebi.ac.uk/intact |
| iRefIndex | Integration of PPIs from many databases | http://irefindex.org |
| KEGG | Reference database for biochemical pathways | https://www.genome.jp/kegg |
| NDEx | Platform for sharing and analyzing biological networks | http://www.ndexbio.org |
| Pathway Commons | Human PPIs and pathways from different sources | http://www.pathwaycommons.org |
| Reactome | Integration of PPIs and pathways from many databases | https://reactome.org |
| STRING | Experimental and predicted PPIs | https://string-db.org |
| Disease/Phenotype association and classification | ||
| CTD | Curated gene and chemical-phenotype associations | http://ctdbase.org |
| Disease Ontology | Hierarchical ontology for description of diseases. | http://disease-ontology.org |
| DisGeNet | Resource of variant and gene association to disease | http://www.disgenet.org |
| dSysMap | Maps of disease mutations on the structural interactome | https://dsysmap.irbbarcelona.org |
| HPO | Ontology for the description of phenotypic abnormalities | https://hpo.jax.org |
| OMIM | Database of genes implicated in Mendelian disorders | https://omim.org |
AP-MS: affinity purification-mass spectroscopy. PPI: Protein-Protein Interaction. Y2H: Yeast two-Hybrid
Computational methods for variant, network and disease annotation
Although not directly relevant to the current review, it is worth noting that a variety of computational tools have been developed to prioritize, annotate and extend the three categories of information required for network analysis of variants. Over the last decade many algorithms have been developed to predict the impact of single nucleotide variants (SNVs) 65, 66, protein-protein interactions 67, 68, and disease-gene associations 69, 70. In particular many machine learning methods are available to predict deleterious SNVs 65 and the effect of single amino acid substitutions on protein stability 66. The prediction of new protein-protein interactions can be performed using sequence and/or structure information 68. Some methods have also been trained to predict the interface residues that mediate the interactions between proteins 67. New associations between genes and diseases are frequent in the literature. Thus, methods for mining the literature to recover new disease-gene associations are essential 71 The majority of such tools use algorithms comparing regular text, specific ontologies and biological networks 69, 70. The computational methods described in the above-cited reviews represent important early attempts to bridge the gap between the vast numbers of catalogued genetic variants and their association with human phenotypes, and are frequently applied to inform mechanistic studies.
NETWORK TOPOLOGY AND DISEASE
Networks provide a versatile framework for modeling the architecture of biological systems. Biological network architectures arise through evolution which should select for characteristics that confer a fitness advantage to an organism. For example, protein interaction networks have evolved to be robust to random genetic variation 72–75. As a result, studying mutation rates together with location in PPI networks can provide information about evolutionary constraints on particular proteins. Proteins under stronger evolutionary constraint should be less tolerant to error, and mutations in those proteins should more likely be associated with extreme phenotypes. Thus topology should also be helpful for bridging the gap between genotype and phenotype. Indeed networks that recapitulate the organization of biological systems can be used to study the relevance of the constituent molecules and molecular interactions to fitness or disease 14, 76, 77.
Properties of biological interactome networks and how they relate to phenotype
Studies of PPI network topology have generated multiple insights linking protein location and connectivity within the network to particular phenotypes. The characteristics of PPI network topology that enable function and robustness of biological systems also create certain kinds of vulnerability. PPI networks tend to have a scale-free topology, such that the number of edges with degree k scales as a power-law distribution (p(k)~k−γ) 78 where the exponent (γ) typically ranges between 2 and 3. As a result, a minority of nodes are hubs with a very large number of interaction partners while most nodes participate in very few interactions. This is thought to render the system robust to random error, since genetic variants at random are more likely to affect a protein with few interactors, and thus cause only a minor perturbation the overall topology of the network. However, this leads to vulnerabilities, as mutations affecting a highly connected hub are likely to have a significant impact on the system 72.
In PPIs networks, on average, the shortest path length of edges separating a pair of randomly selected nodes grows proportionally to the logarithm of the number of nodes in the network (small-world network) 79. The shortest path length is thought to be important for efficient transfer of information and rapid response to perturbation. Redundant paths between nodes may confer robustness to genetic variation 79, 80. Although small-world properties would not be expected to generate a modular network topology per se 81, biological networks tend to be modular, with densely connected subnetworks that are linked into the global network architecture by a small number of connections 82. Indeed, there is selection against the formation of new interactions between nodes that are already highly connected. Instead, links between highly connected nodes and nodes with few interactions are favored 83. The observed balance between modularity and small-worldness in PPI networks may provide the optimal architecture for information flow 81, 84, 85. However this architecture also creates bottlenecks, nodes that bridge more clustered regions of the network, and as such may create additional vulnerabilities 86.
Integrating protein topology with other data layers, such as gene expression or protein structure can reveal more detail about how the elements of the network function together. Hubs can be further divided into party and date hubs, dependent on whether interaction partners are all co-expressed with the hub protein, or co-expressed at different times or locations 82. Incorporating protein structure into analysis of topology, Kim et al observed that hubs can be grouped according to whether interactors used different interfaces, allowing multiple simultaneous interactions, or used only a single interface, in which case interactions would be mutually exclusive 87. Interestingly, multi-interface hubs corresponded to party hubs, whereas hubs that interacted with multiple partners via a single interface were more likely to be date hubs. The differences in co-expression and interface usage by date and party hubs suggests that different evolutionary constraints may act on these subsystems. Indeed, in studies of the s. cerevisiae PPI, date hubs were found to evolve more rapidly than party hubs, and their removal had a more extreme effect on the average path length of the network 82, 87, 88. Similarly, multi-interface hubs were reported to evolve more slowly than single interface hubs 89. Bottlenecks are also reportedly significantly less coexpressed with other network nodes, suggesting they may play a more dynamic role in biological systems 86.
Network topology determines the potential of genes to drive phenotypes
Given the clear evidence that nodes with distinct characteristics in the network support different aspects of the function of biological systems and are under different evolutionary constraints, it makes sense to evaluate the implications for fitness-related phenotypes. Many studies have taken advantage of network measures to examine different classes of gene. Essential genes encode proteins that are required for organismal survival, such that loss of the gene is lethal. Essential genes are reported to have higher degree 90, higher clustering coefficients 91, 92 and higher betweenness centrality in the PPI network 86. Grouping high degree nodes according to their status as party and date hubs further revealed that party hubs are more often essential than date hubs 82. Cancer genes were also found to be enriched at hubs by several studies 93–96. In contrast, Mendelian disease genes were found to be less central in the network than essential and cancer genes 93, 94, particularly when essential Mendelian genes are excluded from the analysis 94. Interestingly, disease genes associated with dominant disorders had higher degree in the network than genes associated with recessive disorders (Feldman et al., 2008). In contrast, gene deletion at the periphery of the network was less frequently associated with an essential or disease phenotypes 93, 97. Figure 2 shows an example of analyzing network feature distributions for different classes of gene.
Figure 2. Exploring network topology as a determinant of gene-phenotype relationships.
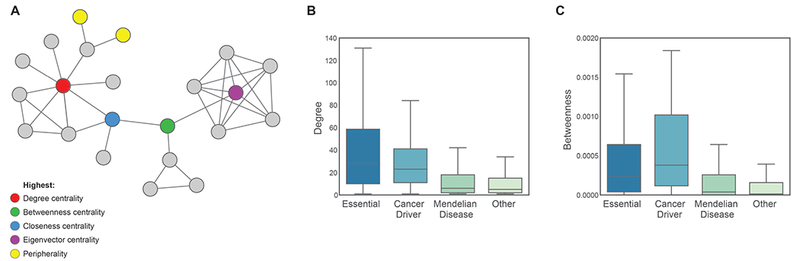
Topological location within the network has implications for biological function. A) Nodes can be described with respect to particular characteristics in the network, including high degree hubs (red), nodes at the periphery (yellow) and nodes with the highest centrality according to four popular measures of centrality. We calculated network measures including B) degree and C) betweenness centrality for four groups of genes: 1,371 essential genes 98, 125 cancer genes 99, 2,921 Mendelian disease genes 100, and 7,099 other genes based on the latest release of STRING 42 to illustrate the types of observation that have been revealed by systematic studies of genes with respect to location in the interactome.
The relationship between network location, gene essentiality and disease raises the possibility of inferring the importance of a gene using network measures. For example, Xu et al. used a k-nearest neighbors approach to implicate genes with similar network characteristics as likely Mendelian disease genes 101. Such approaches have also been generalized to predicting drug targets and toxicities 102, 103. Kotlyar et al. found that the centrality of genes regulated by a drug target was correlated with the toxicity of the drug 102.
The modular organization of the PPI network has been useful for implicating disease genes. According to the disease module hypothesis, genes related to a particular disease or symptom are likely to reside in the same region of the interactome 94, 104, 105. A variety of community detection algorithms are available to identify tightly clustered groups of nodes that are more likely to be functionally related. Approaches have included random walk-based algorithms 106 and non-negative matrix factorization 107. Other algorithms use modularity to implicate disease genes for various classes of genetic disease. For example, the HetRank method uses networks to rank candidate genes for monogenic diseases exhibiting locus heterogeneity 108. In the setting of complex multigenic risk for disorders such as obesity, heart disease or diabetes, disease modularity has been used to uncover shared biological mechanisms underlying diseases by mapping distant risk variants, such as are identified by genome wide association studies, to genes that are close together in the network. Under the assumption that risk genes for the same disorder are more likely to be functionally related, Tasan et al used a network of functionally associated genes to prioritize genes in disease-associated genomic regions 109.
Efforts to catalog mutations in thousands of tumor genomes have uncovered substantial genetic heterogeneity in cancer as well; despite their phenotypic similarities (cancer cells display certain hallmark behaviors 110), individual tumors rarely share the same mutations 111. Although there is very little overlap in the genes that are mutated between tumors, the genes affected by causal driver mutations tend to converge on a limited set of pathways 99, 112, 113. Since genes within a pathway also tend to cluster in biological networks, mutations can be mapped onto a network in order to identify sub-networks of genes that are enriched for alterations 114–116 or assess tumor similarity using the set of pathways or network regions mutated in common 117, 118. A more in depth discussion of network analysis for tumor genomes is provided by Ozturk et al 119.
A phenotype of great medical interest is synthetic lethality. In the setting of synthetic lethality, mutations impairing one gene render loss of function at another gene lethal to the cell 120. The most extensive studies of synthetic lethality have been performed by knocking out all genes individually and in pairwise combination is s. cerevisiae 121. Synthetic lethality occurs when cells are robust to knock out of each gene independently but sensitive to the loss of both. This raises the possibility of designing therapies that exploit pre-existing mutations in cells to selectively eliminate diseased cells, a strategy that has been successfully used to combat cancer 122–124. Analysis using the interactome network topology revealed that synthetic lethal genes pairs were frequently clustered and coded for functionally related proteins that shared interaction partners, implicating protein interactions as a major source of synthetic lethality 125. More recently, CRISPR-Cas9 was used to analyze the consequences of pairwise gene knockout in mammalian cells. Synthetic lethal pairs overlapped across three cell lines, but also showed significant differences, suggesting that lethality may vary considerably across cell types and conditions 120, 126.
NETWORK-INFORMED VARIANT INTERPRETATION
From the observed relationship between network topology and essential or disease genes, it follows that the potential of genetic variants to cause a phenotype is determined by the location of the altered protein within the network 14, 91, 127. Analysis of loss of function variants with and without pathogenic consequences confirms that interactome topology is a determinant of genotype to phenotype relationships. Garcia-Alonso et al reported that loss of function variants in healthy individuals were more frequently observed in genes located near the periphery of the interactome. In contrast, loss of function variants with pathological phenotypes were more central 93. Piñero et al found that network centrality was inversely correlated with tolerance to mutation and that this could be observed at different scales, global and local, within the interactome 128. Of note, the association between network centrality and tolerance to loss of function variants was found to hold for PPI and regulatory networks, but not metabolic networks 97. These studies suggest that the topological location of a variant within the network may be helpful in determining its pathogenicity.
Modeling variants as network perturbations
Most genetic variants are not loss of function events, but rather result in more subtle changes to protein sequences, and mutations within the same protein can have very different effects on its function 129. It has been shown experimentally that most nonsynonymous Mendelian disease mutations generate stable proteins, supporting that mutation effects on specific protein activities rather than absence of protein drives disease phenotypes 22. Mapping variants to nodes in the network cannot capture such subtle differences in variant effect, however the integration of information about protein structure and functional sites with protein interactions has made it possible to better discriminate variants in some cases by mapping them to network edges. Zhong et al introduced the concept of ‘edgetics’ to describe the potential of mutations to perturb distinct interactions in which a protein participates 13, 20, 130. Under this model, variants mapping to the core have the potential to eliminate all interactions by destabilizing the protein, while variants mapping to interaction interfaces have the potential to perturb specific subsets of interaction (Figure 3).
Figure 3. Conceptual framework of edgetics.
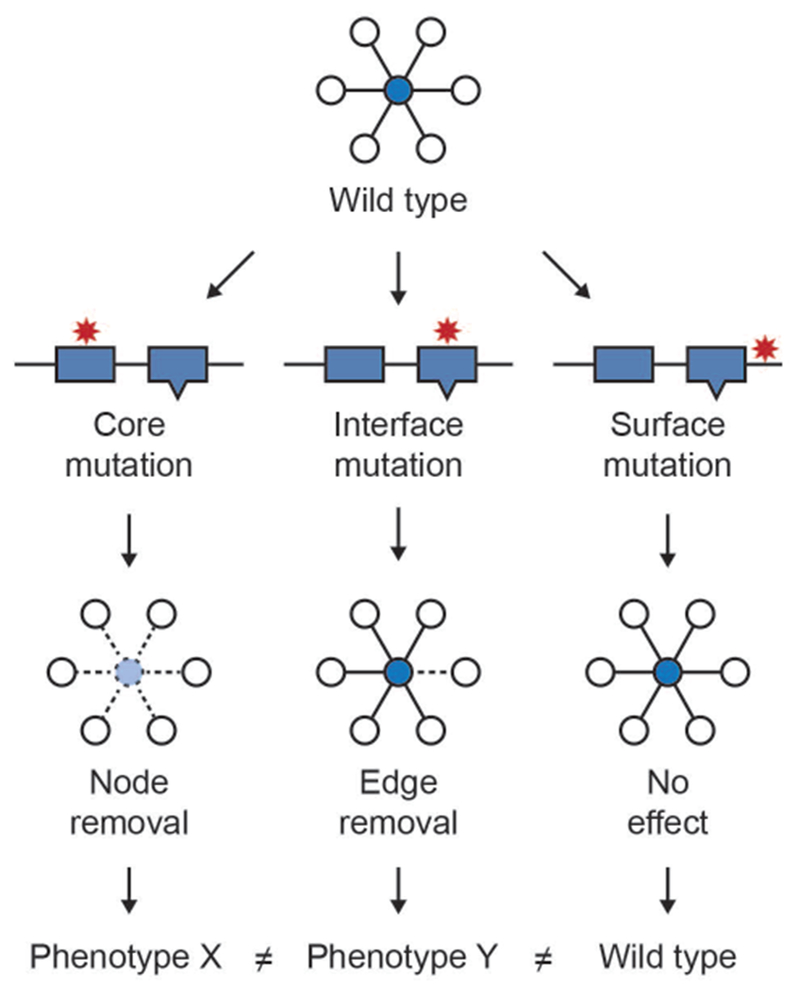
Location of variants within a protein has implications for their phenotypic consequences. Variants that map to the core of the protein are more likely to destabilize it, resulting in a loss of all interactions in which the protein participates. In contrast, mutations at protein interaction interfaces are more likely to perturb specific interactions. Variants mapping to the protein surfaces outside of binding interfaces are less likely to create a phenotype than core or interface variants.
Studies of edgetic effects requires information about the three-dimensional structure of protein complexes, so that amino acid residues can be labeled according to their location in the protein core, on the surface or at an interface between interacting proteins (Figure 4). The framework of edgetics thus allows variants to be studied not only in the context of their location in the network, but also according to their direct impact on network topology. Structurally resolved interactome networks, which integrate information about the domains or amino acid residues that physically interact, are increasingly available to explore the mechanisms by which mutations cause disease at scale 130–134. Multiple studies using structurally resolved networks revealed a statistical excess of known disease mutations at protein interaction interfaces135–137, with in-frame disease mutations enriched at interfaces relative to truncating mutations 137, confirming the utility of such networks for systematically investigating disease mechanism.
Figure 4. Mapping amino acid position to potential to interfere with protein interactions.
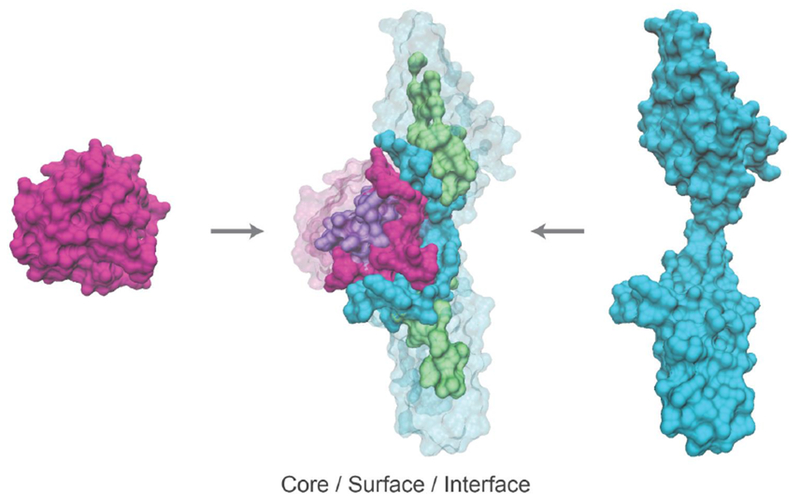
Protein structures of FGF2 and FGFR1 are shown on the left and right respectively, and as a complex in the center (Protein Data Bank structure ID: 1CVS). In the complex, residues are colored according to location in the protein core (purple and green), at the interface (pink and blue) or at the surface outside of the interface (transparent pink and blue) on the two proteins respectively.
Early edgetic analyses focused on Mendelian mutations and revealed several key advantages to the edgetic model. Zhong et al. proposed that edgetics had the potential to describe aspects of genetic disease that could not be captured by topological location of a protein alone (Figure 5). These aspects include: 1) allelic heterogeneity (or pleiotropy), in which a single gene is associated with multiple phenotypes, 2) locus heterogeneity, where a single disorder is caused by a mutation in one of several genes, 3) variable penetrance, wherein not all individuals with a variant have a disease, and 4) variable expressivity, wherein individuals with a disease are not affected equally 20. Indeed, classic examples of pleiotropy and locus heterogeneity could be explained by edgetics. Mutations in the WASP protein associated with Wiskott-Aldrich Syndrome versus X-linked Neutropenia were found to map to distinct interfaces, while mutations associated with hemolytic uremic syndrome in the C3 and CFH proteins were found to map to reciprocal interfaces on the two proteins 137. Guo et al. further investigated the relationship between edgetic effects and the inheritance mode of diseases caused by particular mutations 136, finding that recessive disease mutations affecting reciprocal interfaces were more likely to cause the same phenotype than similar mutations associated with dominant effects. They described cases where dominant truncating mutations removed some interfaces while preserving others, such as was found for TRIM27 mutations in ovarian cancer, supporting that truncating variants may also frequently have edgetic effects 136.
Figure 5. Modeling the edgetic effects of genetic variants supports exploration of disease mechanisms.
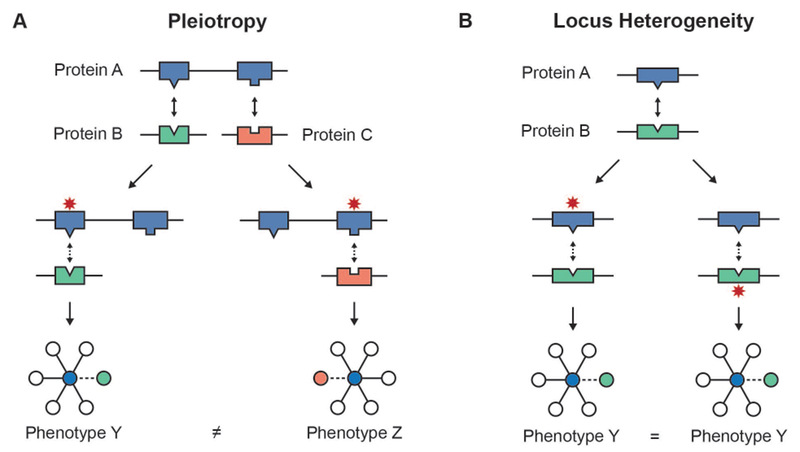
A) Pleiotropy can result when different variants in the same gene affect different interactions in which a protein participates. B) Variants at reciprocal interfaces of interacting proteins can contribute to locus heterogeneity.
Edgetics have also provided mechanistic insights for complex diseases including autism and cancer. For example, a study of 1733 de novo missense mutations from autism spectrum disorder exomes demonstrated a significant enrichment of missense mutations affecting protein interactions in probands relative to unaffected siblings 138. Experimental assessment using the CloneSeq pipeline to test the effect of mutations on binary protein interactions 139 found that 74/361 (20%) of tested interactions were altered in probands versus only 21/208 (10%) in unaffected siblings. Several studies have demonstrated that somatic mutations found in tumor genomes are overrepresented at protein interaction interfaces 140–143, suggesting that perturbations of protein interactions frequently contribute to tumor development. The Cancer Cell Map Initiative is now systematically experimentally mapping the impact of driver mutations on the interactome in cancer to further reveal the links between network rewiring and tumorigenesis 144.
Genetic variants perturbing network topology
While most edgetic variants would be expected to perturb existing interactions, it is also possible for variants to generate new interactions. For examples, the R273H amino acid substitution in TP53 was found to create a binding site for NRD1, and this novel interaction was found to promote cellular invasion in the setting of cancer 13, 145. Such novel interactions may be difficult to predict, and it remains unclear how frequently amino acid substitutions generate new interfaces. The simplest way for novel interactions to emerge is likely through modified specificity at existing binding sites, and such events may be most prevalent in proteins families with high functional similarity. For example, many mutations in cancer are thought to alter the specificity of kinases for their substrates 146, 147. Frequent cancer mutations were reported at acetylation and ubiquitination sites as well 148. Experimental evidence also suggested that many disease mutations affect the motif specificity of transcription factors, with many mutations reducing the specificity of DNA binding sites, thereby allowing more promiscuous binding 22.
Nonsynonymous variants and small insertions and deletions have been the focus of most edgetic studies, however there are other types of alteration that have the potential to ‘rewire’ the interactome. Alternative splicing generates different proteins from the same gene, and dependent on which exons are included, these protein isoforms can include different binding interfaces. Yang et al used experimental approaches to map the interactome of 1,423 protein isoforms 149, and subsequently Ghadie et al. used in silico analysis of interface containing domain usage by various splice isoforms to construct an isoform-specific interactome 150. Both studies found that patterns of protein interaction with different isoforms was associated with divergent disease phenotypes, creating additional opportunity for pleiotropy 149, 150. In cancer, aberrant splicing was enriched at domain families that mediate protein interactions which also frequently harbored other types of mutation 151.
Not all genes create equal opportunity for ‘rewiring’ biological networks. In cancer, mutations often affect genes capable of causing large changes in the network. For example, cancer mutations frequently affect genes involved in chromatin remodeling, resulting in widespread changes to gene expression 113. Fusion proteins are another common event in tumor genomes that can result in network rewiring. A recent pan-cancer analysis found that fusions often involved genes with high degree in the network and that did not interact prior to the fusion event 152. Some recurrent cancer fusions involve genes that regulate the activity of multiple targets such as transcription factors, (e.g. RUNX1, ERG) or kinases (e.g. ABL1, NTRK1). Mutations with large impacts on network architecture are likely to be uncommon in inherited disease, as their consequences may be too severe to support early development in multicellular organisms. The hypothesis is consistent with reports that Mendelian disease genes are less central in the PPI network and participate in fewer protein interactions than cancer genes 93.
Studying variant accumulation at network edges instead of nodes
Studying proteins in the context of their location in the network focuses analysis on node-level properties. In contrast, in the framework of edgetics, variants are viewed based on their ability to perturb network edges. From this perspective it becomes possible to analyze the accumulation of variation affecting network edges rather than network nodes in order to better understand disease mechanism. Many cases of mutations affecting interacting genes and leading to similar phenotypes such as Branchiootic syndrome, Charcot‐Marie‐Tooth disease and Polycystic kidney disease have been reported153. Several groups have developed scoring strategies to prioritize network edges that are enriched for disease mutation. This requires first mapping variants to interface residues or interacting domains of proteins. Some methods evaluate the ratio of observed to expected variants in interface regions controlling or the size of the region relative to the size of the protein 142 and or the size and the amino acid composition 143. An alternative approach uses the nonsynonymous to synonymous (dN/dS) ratio at interfaces, a signature of selective pressure that has been used to identify cancer genes 154, to evaluate whether interfaces are unexpectedly biased toward functional variants 140. Mechismo evaluates the consequences of amino acid substitutions at interfaces in terms of their impact on the pair potentials relative to what is expected 131, enabling an assessment of the likely effect of the variant on the interface. Other analyses used methods such as FoldX 155 to estimate the impact of the amino acid substitution on the stability of the protein complex.
Using networks to predict the phenotypic consequences of variants
Since networks influence the potential of variants to have a phenotypic effect, and variants result in phenotypic effects by perturbing biological networks in different ways, network measures should be informative for computational tasks relating to variant prioritization and interpretation. Some methods have begun to incorporate network information into classification tasks. Khurana et al. combined network and evolutionary properties to build a classifier capable of predicting gene tolerance to loss of function mutations, enabling automated prioritization of loss of function events according to their potential to have phenotypic consequences 97. Location of a variant at a protein interface was found to be one of the most informative features for discriminating driver missense mutations from neutral passenger mutations in analysis of tumor genomes 156. Gao et al. used features derived from gene regulatory networks to predict the functional consequences of non-coding variants 157 under the assumption that the causal effects of gene-expression altering variants must be transmitted through this network. There may be various ways to derive features from interactome networks for the purpose of predicting variant effects. Yu et al derived ‘ontotypes’, a signature of nodes reached by a gene in an s. cerevisiae-specific hierarchical interactome network, to predict the impact of specific gene knockouts on colony growth 158.
Several approaches have used the structure of the network more directly in predicting variant effects. To capture the potential for variants in the same gene to have different effects on biological processes, Engin et al mapped cancer mutations affecting different interfaces on the same protein onto different perturbed network architectures. Network propagation was applied to identify downstream subnetworks specifically affected by each mutation 159 (Figure 6). These subnetworks could then be analyzed for functional enrichment to suggest mechanisms by which mutations perturbing distinct activities of a protein could result in different phenotypes. In a different study, Poole et al evaluated the statistical association of clusters of somatic mutations within selected proteins to pathway level changes in gene expression in tumors 160, raising the possibility of aggregating mutations with common edgetic effects across samples and simultaneously analyzing them using networks.
Figure 6. Propagating variant effects on networks.
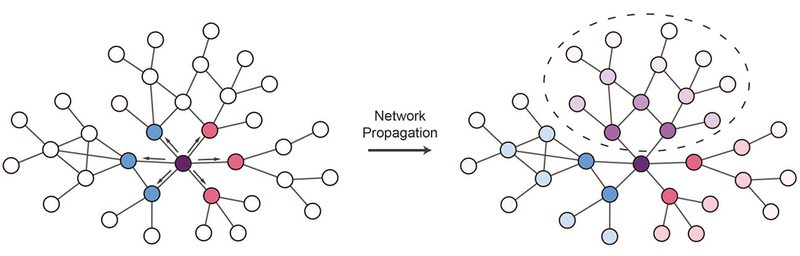
Variants can be used as signal sources for network propagation in order to identify network neighborhoods affected by variants. Edgetic effects can be used to constrain network propagation according to the effects of variants on specific protein interactions. On the left side of this schematic, two variants to the purple node affect interactions with different subsets of partners (indicated by blue and pink nodes respectively). Network propagation can be used to implicate network regions likely to be affected by each variant, and these can be contrasted to identify regions perturbed by both variants that could explain shared phenotypes (right network, circled purple shaded nodes), or regions affected specifically by each variant (right network, blue and pink shaded regions) which could help explain pleiotropic effects.
Recently, Ma et al. developed DeepCell wherein the hierarchical interactome was used to constrain the architecture of a deep convolutional neural network that was trained to predict growth effects from genotype in s. cerevisiae 161. DeepCell was found to effectively simulate growth phenotypes observed in the laboratory directly from genotype. Furthermore, the weights learned by the neural network could be used to develop testable hypotheses about the mechanisms by which variants generated growth phenotypes, with some variants displaying Boolean-type effects on specific subsystems. Thus DeepCell mines the multi-scale, hierarchical organization of the interactome to extract novel information linking genotype to phenotype.
In summary, multiple strategies have been developed to annotate variants according their consequences for biological networks to support their interpretation in the context of various phenotypes. These works lay the foundation for the next generation of automated variant interpretation tools that will integrate information about the architecture of the biological system and the potential of variants to perturb it. Strategies for quantifying variant effects on the network, or using network information to predict variant activity have thus far focused largely on single variants, however we note that multiple variants can simultaneously be mapped to network effects. This raises the possibility of using networks to predict the joint effects of combinations of variants, such as occur in complex polygenic diseases.
CHALLENGES AND FUTURE DIRECTIONS
While early works integrating networks with variant information to understand the mechanisms driving genetic disease show great promise, many challenges remain to be addressed. At times, analyses of network topologies have generated contradictory findings 162–166, suggesting that careful consideration must be given to the limitations of the available data and the implications of choices made in constructing and analyzing network models when drawing biological conclusions.
Data availability and quality is an important consideration for network analyses. PPI networks remain incomplete and may contain many false positive connections. In addition, networks assembled from various published experiments may exhibit literature bias; proteins associated with certain phenotypes may be more studied and as a result, may appear more connected in the network, giving the illusion that higher degree nodes are associated with phenotypes of interest 45, 91, 101. Choice of network may thus influence the biological conclusions drawn from network analysis. Indeed, Huang et al showed that network performance at recovering known disease genes varied according to the disease, and no single network performed best for all diseases 52. To partially address this issue, several methods for selecting high-quality PPI datasets and score the reliability of an interaction have been developed 53.
Models frequently focus on specific aspects of biology while ignoring others. For example, many molecular interactome networks ignore regulatory interactions by which distant proteins in the network can influence each other, and often do not account for post-translational modifications. Hybrid networks that incorporate both physical and regulatory interactions can be constructed167 but statistical analysis may be complicated by the inclusion of different types of relationship. More sophisticated applications of natural language processing may support direct inference of more complex network models directly from the literature168. Many of the approaches discussed in this overview treat proteins as static network nodes and do not attempt to incorporate protein levels. Integrating interactome networks with more quantitative modeling techniques that rely on differential equations169 or agent based models170 could present a pathway for including quantitative and dynamic information about protein levels, however these methods are frequently computationally intensive and may not be practical for large networks.
Structure is not available for the majority of human proteins, limiting the investigation of variants affecting protein interactions. The extent to which networks themselves are complete also remains poorly understood. Many conclusions have been drawn based on the architecture of the human interactome, however some estimates suggest that at most 20% of interactions have been experimentally measured 105. In addition, gene expression patterns differ widely across cell types, suggesting that for accurate inference, network architectures need to be cell-type specific. Indeed, disease network modules tend to include genes that are coexpressed in specific tissues 171, and several groups have now constructed tissue-specific networks to study disease variation 172, 173. Many of the analyses described here have yet to be revisited in a tissue- or cell-type specific setting. Finally, network representations are usually static, whereas the biological networks that they represent are dynamic and conditional. Protein interactions often require particular localization or post-translational modification. Novel technologies such as APEX, a proximity labeling technique recently developed to enable spatially resolved analysis of protein interaction networks 174, may provide a solution to further resolve cell-type specific interactions and subcellular location thereof. Distinguishing between constitutive and transient interactions, and cell-state specific interactions may be important for further understanding the potential of variants to generate relevant phenotypes 175. Thus, the Stable Isotope Labeling by Amino acids in Cell culture (SILAC) method, a mass spectrometry technique able to detect differences in protein abundance among samples using non-radioactive isotopic labeling, has been used to measure stability of protein-protein interaction 176.
New technologies are emerging that can accelerate the pace of interaction profiling and that will create more complete networks and new opportunities for analysis. Next generation sequencing-based interaction screening technologies such as BFG-Y2H 85 and CrY2H-seq 177 allow higher throughput screening for binary interaction. Combining such technologies with techniques like deep mutational scanning 178 could allow more systematic profiling of the edgetic effects of variants. Technologies such as Perturb-seq also allow high-throughput profiling of the effects of variants on single cell transcriptional profiles, creating opportunities to quantify the broader effects of variants at the resolution of specific cell types 179. Interactome networks have the potential to play a key role in the interpretation of such assays.
Conclusion
A rapidly growing body of research has demonstrated the utility of network analysis for understanding disease mechanism. Many early studies relied on interactome networks derived from model organisms, such as s. cerevisiae, but the less complete human interactome has been instrumental for studying the relationships between genetic variation, genes and disease. Many insights have been gained from the study of genes with clear phenotypes, including essential genes, Mendelian disease genes and cancer genes. Interactome networks have also been successfully used to identify drug targets and study mechanisms underlying toxicities. However, a significant proportion of human genetic disease remains poorly understood, particularly for complex multigenic disorders.
As new high-throughput experimental assays emerge, databases of genetic variation and network models will become increasingly available and more complete. Large consortia are generating invaluable multi-layer datasets that can create new opportunities for integrated analysis for variant interpretation. For example, the eGTEx project will add epigenetic measurements to complement the library of tissue-specific expression data in GTEx 180, enabling integration of epigenetic factors and tissue specific gene regulation into models for interpreting disease variants 181, 182. These new methodologies and datasets create opportunities for the development of computational models that support network-informed inference to predict the phenotypic consequences of genetic variation and reveal the mechanisms by which variants contribute to complex human diseases.
Recent technological advances are enabling the development of emerging research fields such as the Molecular Pathological Epidemiology (MPE) incorporating interpersonal heterogeneity of a disease process into epidemiology 183. In this framework the integration of data capturing the complex combination of genetic heterogeneity (endogenous factors) and the environment (exogenous factors) is providing novel insights underlying etiologic mechanisms of different cancer types184 and define new therapeutic strategies 185.
Although these early efforts show great promise, new technological and algorithmic advances will be required to realize the full potential of networks to inform variant interpretation and precision medicine.
Acknowledgments
This work was supported by NIH grants DP5-OD017937 and a CIFAR fellowship to H.C. EC is supported by an FFABR grant from the Ministry of Education, Universities and Research (MIUR). EC acknowledges the CINECA award under the ISCRA initiative, for the availability of high performance computing resources and support. HC and EC acknowledge the support from the University of Bologna through the funding of the UNIBO-UCSD International Cooperation Project.
Contributor Information
Emidio Capriotti, Department of Pharmacy and Biotechnology (FaBiT), University of Bologna, via F. Selmi 3, Bologna, 40126, Italy emidio.capriotti@unibo.it..
Kivilcim Ozturk, Bioinformatics and Systems Biology Program, University of California, San Diego, La Jolla, CA 92093, USA kozturk@eng.ucsd.edu..
Hannah Carter, Department of Medicine and Institute for Genomic Medicine, University of California, San Diego, La Jolla, CA 92130, USA hkcarter@ucsd.edu..
References
- 1.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA, Consortium GP. A map of human genome variation from population-scale sequencing. Nature 2010, 467:1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush WS, Oetjens MT, Crawford DC. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat Rev Genet 2016, 17:129–145. [DOI] [PubMed] [Google Scholar]
- 3.Schraiber JG, Akey JM. Methods and models for unravelling human evolutionary history. Nat Rev Genet 2015, 16:727–740. [DOI] [PubMed] [Google Scholar]
- 4.Ku CS, Naidoo N, Pawitan Y. Revisiting Mendelian disorders through exome sequencing. Hum Genet 2011, 129:351–370. [DOI] [PubMed] [Google Scholar]
- 5.MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales J, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 2017, 45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernald GH, Capriotti E, Daneshjou R, Karczewski KJ, Altman RB. Bioinformatics challenges for personalized medicine. Bioinformatics 2011, 27:1741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 2015, 43:D789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature 2009, 461:747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bomba L, Walter K, Soranzo N. The impact of rare and low-frequency genetic variants in common disease. Genome Biol 2017, 18:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P, Consortium IS. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009, 460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stahl EA, Wegmann D, Trynka G, Gutierrez-Achury J, Do R, Voight BF, Kraft P, Chen R, Kallberg HJ, Kurreeman FA, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat Genet 2012, 44:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 2010, 11:446–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahni N, Yi S, Zhong Q, Jailkhani N, Charloteaux B, Cusick ME, Vidal M. Edgotype: a fundamental link between genotype and phenotype. Curr Opin Genet Dev 2013, 23:649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell 2011, 144:986–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kahle JJ, Gulbahce N, Shaw CA, Lim J, Hill DE, Barabasi AL, Zoghbi HY. Comparison of an expanded ataxia interactome with patient medical records reveals a relationship between macular degeneration and ataxia. Hum Mol Genet 2011, 20:510–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakai Y, Shaw CA, Dawson BC, Dugas DV, Al-Mohtaseb Z, Hill DE, Zoghbi HY. Protein interactome reveals converging molecular pathways among autism disorders. Sci Transl Med 2011, 3:86ra49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalathur RKR, Pedro Pinto J, Sahoo B, Chaurasia G, Futschik ME. HDNetDB: A Molecular Interaction Database for Network-Oriented Investigations into Huntington’s Disease. Sci Rep 2017, 7:5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, Bull S, Pawson T, Morris Q, Wrana JL. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol 2009, 27:199–204. [DOI] [PubMed] [Google Scholar]
- 19.Hackinger S, Zeggini E. Statistical methods to detect pleiotropy in human complex traits. Open Biol 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong Q, Simonis N, Li QR, Charloteaux B, Heuze F, Klitgord N, Tam S, Yu H, Venkatesan K, Mou D, et al. Edgetic perturbation models of human inherited disorders. Mol Syst Biol 2009, 5:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stearns FW. One hundred years of pleiotropy: a retrospective. Genetics 2010, 186:767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahni N, Yi S, Taipale M, Fuxman Bass JI, Coulombe-Huntington J, Yang F, Peng J, Weile J, Karras GI, Wang Y, et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell 2015, 161:647–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman MEJ. Networks: an introduction. 2010.
- 24.Leung IX, Hui P, Lio P, Crowcroft J. Towards real-time community detection in large networks. Phys Rev E Stat Nonlin Soft Matter Phys 2009, 79:066107. [DOI] [PubMed] [Google Scholar]
- 25.Newman ME. Modularity and community structure in networks. Proc Natl Acad Sci U S A 2006, 103:8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girvan M, Newman ME. Community structure in social and biological networks. Proc Natl Acad Sci U S A 2002, 99:7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guimera R, Amaral LA. Cartography of complex networks: modules and universal roles. J Stat Mech 2005, 2005:nihpa35573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001, 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA, Consortium GP. An integrated map of genetic variation from 1,092 human genomes. Nature 2012, 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 2016, 44:D862–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mottaz A, David FP, Veuthey AL, Yip YL. Easy retrieval of single amino-acid polymorphisms and phenotype information using SwissVar. Bioinformatics 2010, 26:851–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.International Cancer Genome C Hudson TJ Anderson W Artez A Barker AD Bell C Bernabe RR Bhan MK Calvo FE Erola I, et al. International network of cancer genome projects. Nature 2010, 464:993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 2017, 45:D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature 2005, 437:1173–1178. [DOI] [PubMed] [Google Scholar]
- 35.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017, 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, Campbell NH, Chavali G, Chen C, del-Toro N, et al. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res 2014, 42:D358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics 2008, 9:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res 2018, 46:D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res 2011, 39:D685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545:505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res 2004, 32:D449–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017, 45:D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res 2012, 40:D857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res 2012, 40:D841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, Aronow BJ, Jegga AG. Disease candidate gene identification and prioritization using protein interaction networks. BMC Bioinformatics 2009, 10:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pratt D, Chen J, Welker D, Rivas R, Pillich R, Rynkov V, Ono K, Miello C, Hicks L, Szalma S, et al. NDEx, the Network Data Exchange. Cell Syst 2015, 1:302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Helikar T, Kowal B, McClenathan S, Bruckner M, Rowley T, Madrahimov A, Wicks B, Shrestha M, Limbu K, Rogers JA. The Cell Collective: toward an open and collaborative approach to systems biology. BMC Syst Biol 2012, 6:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaake RM, Wang X, Burke A, Yu C, Kandur W, Yang Y, Novtisky EJ, Second T, Duan J, Kao A, et al. A new in vivo cross-linking mass spectrometry platform to define protein-protein interactions in living cells. Mol Cell Proteomics 2014, 13:3533–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zybailov BL, Glazko GV, Jaiswal M, Raney KD. Large Scale Chemical Cross-linking Mass Spectrometry Perspectives. J Proteomics Bioinform 2013, 6:001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cafarelli TM, Desbuleux A, Wang Y, Choi SG, De Ridder D, Vidal M. Mapping, modeling, and characterization of protein-protein interactions on a proteomic scale. Curr Opin Struct Biol 2017, 44:201–210. [DOI] [PubMed] [Google Scholar]
- 51.Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, et al. A proteome-scale map of the human interactome network. Cell 2014, 159:1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang JK, Carlin DE, Yu MK, Zhang W, Kreisberg JF, Tamayo P, Ideker T. Systematic Evaluation of Molecular Networks for Discovery of Disease Genes. Cell Syst 2018, 6:484–495 e485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng X, Wang J, Peng W, Wu FX, Pan Y. Protein-protein interactions: detection, reliability assessment and applications. Brief Bioinform 2017, 18:798–819. [DOI] [PubMed] [Google Scholar]
- 54.Chou KC, Cai YD. Predicting protein-protein interactions from sequences in a hybridization space. J Proteome Res 2006, 5:316–322. [DOI] [PubMed] [Google Scholar]
- 55.Sharan R, Suthram S, Kelley RM, Kuhn T, McCuine S, Uetz P, Sittler T, Karp RM, Ideker T. Conserved patterns of protein interaction in multiple species. Proc Natl Acad Sci U S A 2005, 102:1974–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trivodaliev K, Bogojeska A, Kocarev L. Exploring function prediction in protein interaction networks via clustering methods. PLoS One 2014, 9:e99755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bodenreider O The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004, 32:D267–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rogers FB. Medical subject headings. Bull Med Libr Assoc 1963, 51:114–116. [PMC free article] [PubMed] [Google Scholar]
- 59.Ruch P, Gobeill J, Lovis C, Geissbuhler A. Automatic medical encoding with SNOMED categories. BMC Med Inform Decis Mak 2008, 8 Suppl 1:S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kibbe WA, Arze C, Felix V, Mitraka E, Bolton E, Fu G, Mungall CJ, Binder JX, Malone J, Vasant D, et al. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res 2015, 43:D1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Köhler S, Vasilevsky NA, Engelstad M, Foster E, McMurry J, Aymé S, Baynam G, Bello SM, Boerkoel CF, Boycott KM, et al. The Human Phenotype Ontology in 2017. Nucleic Acids Res 2017, 45:D865–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, García-García J, Sanz F, Furlong LI. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 2017, 45:D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mosca R, Tenorio-Laranga J, Olivella R, Alcalde V, Ceol A, Soler-Lopez M, Aloy P. dSysMap: exploring the edgetic role of disease mutations. Nat Methods 2015, 12:167–168. [DOI] [PubMed] [Google Scholar]
- 64.Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, Wiegers J, Wiegers TC, Mattingly CJ. The Comparative Toxicogenomics Database: update 2017. Nucleic Acids Res 2017, 45:D972–D978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Capriotti E, Nehrt NL, Kann MG, Bromberg Y. Bioinformatics for personal genome interpretation. Brief Bioinform 2012, 13:495–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Compiani M, Capriotti E. Computational and theoretical methods for protein folding. Biochemistry 2013, 52:8601–8624. [DOI] [PubMed] [Google Scholar]
- 67.Esmaielbeiki R, Krawczyk K, Knapp B, Nebel JC, Deane CM. Progress and challenges in predicting protein interfaces. Brief Bioinform 2016, 17:117–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gonzalez MW, Kann MG. Chapter 4: Protein interactions and disease. PLoS Comput Biol 2012, 8:e1002819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chapter Bromberg Y. 15: disease gene prioritization. PLoS Comput Biol 2013, 9:e1002902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tranchevent LC, Capdevila FB, Nitsch D, De Moor B, De Causmaecker P, Moreau Y. A guide to web tools to prioritize candidate genes. Brief Bioinform 2011, 12:22–32. [DOI] [PubMed] [Google Scholar]
- 71.Gonzalez GH, Tahsin T, Goodale BC, Greene AC, Greene CS. Recent Advances and Emerging Applications in Text and Data Mining for Biomedical Discovery. Brief Bioinform 2016, 17:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature 2000, 406:378–382. [DOI] [PubMed] [Google Scholar]
- 73.Félix MA, Barkoulas M. Pervasive robustness in biological systems. Nat Rev Genet 2015, 16:483–496. [DOI] [PubMed] [Google Scholar]
- 74.Kaneko K Phenotypic plasticity and robustness: evolutionary stability theory, gene expression dynamics model, and laboratory experiments. Adv Exp Med Biol 2012, 751:249–278. [DOI] [PubMed] [Google Scholar]
- 75.Payne JL, Wagner A. Mechanisms of mutational robustness in transcriptional regulation. Front Genet 2015, 6:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barabási AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet 2004, 5:101–113. [DOI] [PubMed] [Google Scholar]
- 77.Ideker T, Krogan NJ. Differential network biology. Mol Syst Biol 2012, 8:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barabasi AL, Albert R. Emergence of scaling in random networks. Science 1999, 286:509–512. [DOI] [PubMed] [Google Scholar]
- 79.Albert R Scale-free networks in cell biology. J Cell Sci 2005, 118:4947–4957. [DOI] [PubMed] [Google Scholar]
- 80.Papin JA, Palsson BO. Topological analysis of mass-balanced signaling networks: a framework to obtain network properties including crosstalk. J Theor Biol 2004, 227:283–297. [DOI] [PubMed] [Google Scholar]
- 81.Gallos LK, Makse HA, Sigman M. A small world of weak ties provides optimal global integration of self-similar modules in functional brain networks. Proc Natl Acad Sci U S A 2012, 109:2825–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP, et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 2004, 430:88–93. [DOI] [PubMed] [Google Scholar]
- 83.Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science 2002, 296:910–913. [DOI] [PubMed] [Google Scholar]
- 84.Jarman N, Steur E, Trengove C, Tyukin IY, van Leeuwen C. Self-organisation of small-world networks by adaptive rewiring in response to graph diffusion. Sci Rep 2017, 7:13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yachie N, Petsalaki E, Mellor JC, Weile J, Jacob Y, Verby M, Ozturk SB, Li S, Cote AG, Mosca R, et al. Pooled-matrix protein interaction screens using Barcode Fusion Genetics. Mol Syst Biol 2016, 12:863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol 2007, 3:e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Science 2006, 314:1938–1941. [DOI] [PubMed] [Google Scholar]
- 88.Bertin N, Simonis N, Dupuy D, Cusick ME, Han JD, Fraser HB, Roth FP, Vidal M. Confirmation of organized modularity in the yeast interactome. PLoS Biol 2007, 5:e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Biswas K, Acharya D, Podder S, Ghosh TC. Evolutionary rate heterogeneity between multi- and single-interface hubs across human housekeeping and tissue-specific protein interaction network: Insights from proteins’ and its partners’ properties. Genomics 2017. [DOI] [PubMed] [Google Scholar]
- 90.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature 2001, 411:41–42. [DOI] [PubMed] [Google Scholar]
- 91.Feldman I, Rzhetsky A, Vitkup D. Network properties of genes harboring inherited disease mutations. Proc Natl Acad Sci U S A 2008, 105:4323–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Said MR, Begley TJ, Oppenheim AV, Lauffenburger DA, Samson LD. Global network analysis of phenotypic effects: protein networks and toxicity modulation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2004, 101:18006–18011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Garcia-Alonso L, Jiménez-Almazán J, Carbonell-Caballero J, Vela-Boza A, Santoyo-López J, Antiñolo G, Dopazo J. The role of the interactome in the maintenance of deleterious variability in human populations. Mol Syst Biol 2014, 10:752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL. The human disease network. Proc Natl Acad Sci U S A 2007, 104:8685–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jonsson PF, Bates PA. Global topological features of cancer proteins in the human interactome. Bioinformatics 2006, 22:2291–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sun J, Zhao Z. A comparative study of cancer proteins in the human protein-protein interaction network. BMC Genomics 2010, 11 Suppl 3:S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Khurana E, Fu Y, Chen J, Gerstein M. Interpretation of genomic variants using a unified biological network approach. PLoS Comput Biol 2013, 9:e1002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 2015, 163:1515–1526. [DOI] [PubMed] [Google Scholar]
- 99.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science 2013, 339:1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, Phillips AD, Cooper DN. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet 2017, 136:665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu J, Li Y. Discovering disease-genes by topological features in human protein-protein interaction network. Bioinformatics 2006, 22:2800–2805. [DOI] [PubMed] [Google Scholar]
- 102.Kotlyar M, Fortney K, Jurisica I. Network-based characterization of drug-regulated genes, drug targets, and toxicity. Methods 2012, 57:499–507. [DOI] [PubMed] [Google Scholar]
- 103.Sun J, Zhu K, Zheng W, Xu H. A comparative study of disease genes and drug targets in the human protein interactome. BMC Bioinformatics 2015, 16 Suppl 5:S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bauer-Mehren A, Bundschus M, Rautschka M, Mayer MA, Sanz F, Furlong LI. Gene-disease network analysis reveals functional modules in mendelian, complex and environmental diseases. PLoS One 2011, 6:e20284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Menche J, Sharma A, Kitsak M, Ghiassian SD, Vidal M, Loscalzo J, Barabási AL. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science 2015, 347:1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rosvall M, Bergstrom CT. Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci U S A 2008, 105:1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang S, Wang RS, Zhang XS. Uncovering fuzzy community structure in complex networks. Phys Rev E Stat Nonlin Soft Matter Phys 2007, 76:046103. [DOI] [PubMed] [Google Scholar]
- 108.Dand N, Schulz R, Weale ME, Southgate L, Oakey RJ, Simpson MA, Schlitt T. Network-Informed Gene Ranking Tackles Genetic Heterogeneity in Exome-Sequencing Studies of Monogenic Disease. Hum Mutat 2015, 36:1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Taşan M, Musso G, Hao T, Vidal M, MacRae CA, Roth FP. Selecting causal genes from genome-wide association studies via functionally coherent subnetworks. Nat Methods 2015, 12:154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011, 144:646–674. [DOI] [PubMed] [Google Scholar]
- 111.Tian R, Basu MK, Capriotti E. Computational methods and resources for the interpretation of genomic variants in cancer. BMC Genomics 2015, 16 Suppl 8:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ali MA, Sjöblom T. Molecular pathways in tumor progression: from discovery to functional understanding. Mol Biosyst 2009, 5:902–908. [DOI] [PubMed] [Google Scholar]
- 113.Garraway LA, Lander ES. Lessons from the cancer genome. Cell 2013, 153:17–37. [DOI] [PubMed] [Google Scholar]
- 114.Leiserson MD, Vandin F, Wu HT, Dobson JR, Eldridge JV, Thomas JL, Papoutsaki A, Kim Y, Niu B, McLellan M, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet 2015, 47:106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vandin F, Upfal E, Raphael BJ. Algorithms for detecting significantly mutated pathways in cancer. J Comput Biol 2011, 18:507–522. [DOI] [PubMed] [Google Scholar]
- 116.Zhang J, Zhang S, Wang Y, Zhang XS. Identification of mutated core cancer modules by integrating somatic mutation, copy number variation, and gene expression data. BMC Syst Biol 2013, 7 Suppl 2:S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hofree M, Shen JP, Carter H, Gross A, Ideker T. Network-based stratification of tumor mutations. Nat Methods 2013, 10:1108–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhong X, Yang H, Zhao S, Shyr Y, Li B. Network-based stratification analysis of 13 major cancer types using mutations in panels of cancer genes. BMC Genomics 2015, 16 Suppl 7:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ozturk K, Dow M, Carlin DE, Bejar R, Carter H. The Emerging Potential for Network Analysis to Inform Precision Cancer Medicine. J Mol Biol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shen JP, Ideker T. Synthetic Lethal Networks for Precision Oncology: Promises and Pitfalls. J Mol Biol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science 2010, 327:425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434:913–917. [DOI] [PubMed] [Google Scholar]
- 123.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434:917–921. [DOI] [PubMed] [Google Scholar]
- 124.Lord CJ, Tutt AN, Ashworth A. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annu Rev Med 2015, 66:455–470. [DOI] [PubMed] [Google Scholar]
- 125.Talavera D, Robertson DL, Lovell SC. The role of protein interactions in mediating essentiality and synthetic lethality. PLoS One 2013, 8:e62866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, Licon K, Klepper K, Pekin D, Beckett AN, et al. Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nat Methods 2017, 14:573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Carter H, Hofree M, Ideker T. Genotype to phenotype via network analysis. Curr Opin Genet Dev 2013, 23:611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Piñero J, Berenstein A, Gonzalez-Perez A, Chernomoretz A, Furlong LI. Uncovering disease mechanisms through network biology in the era of Next Generation Sequencing. Sci Rep 2016, 6:24570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Taipale M Disruption of protein function by pathogenic mutations: common and uncommon mechanisms. Biochem Cell Biol 2018. [DOI] [PubMed] [Google Scholar]
- 130.Das J, Fragoza R, Lee HR, Cordero NA, Guo Y, Meyer MJ, Vo TV, Wang X, Yu H. Exploring mechanisms of human disease through structurally resolved protein interactome networks. Mol Biosyst 2014, 10:9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Betts MJ, Lu Q, Jiang Y, Drusko A, Wichmann O, Utz M, Valtierra-Gutiérrez IA, Schlesner M, Jaeger N, Jones DT, et al. Mechismo: predicting the mechanistic impact of mutations and modifications on molecular interactions. Nucleic Acids Res 2015, 43:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Meyer MJ, Das J, Wang X, Yu H. INstruct: a database of high-quality 3D structurally resolved protein interactome networks. Bioinformatics 2013, 29:1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mosca R, Céol A, Aloy P. Interactome3D: adding structural details to protein networks. Nat Methods 2013, 10:47–53. [DOI] [PubMed] [Google Scholar]
- 134.Vázquez M, Valencia A, Pons T. Structure-PPi: a module for the annotation of cancer-related single-nucleotide variants at protein-protein interfaces. Bioinformatics 2015, 31:2397–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.David A, Razali R, Wass MN, Sternberg MJ. Protein-protein interaction sites are hot spots for disease-associated nonsynonymous SNPs. Hum Mutat 2012, 33:359–363. [DOI] [PubMed] [Google Scholar]
- 136.Guo Y, Wei X, Das J, Grimson A, Lipkin SM, Clark AG, Yu H. Dissecting disease inheritance modes in a three-dimensional protein network challenges the “guilt-by-association” principle. Am J Hum Genet 2013, 93:78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang X, Wei X, Thijssen B, Das J, Lipkin SM, Yu H. Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nat Biotechnol 2012, 30:159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen S, Fragoza R, Klei L, Liu Y, Wang J, Roeder K, Devlin B, Yu H. An interactome perturbation framework prioritizes damaging missense mutations for developmental disorders. Nat Genet 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wei X, Das J, Fragoza R, Liang J, Bastos de Oliveira FM, Lee HR, Wang X, Mort M, Stenson PD, Cooper DN, et al. A massively parallel pipeline to clone DNA variants and examine molecular phenotypes of human disease mutations. PLoS Genet 2014, 10:e1004819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Engin HB, Kreisberg JF, Carter H. Structure-Based Analysis Reveals Cancer Missense Mutations Target Protein Interaction Interfaces. PLoS One 2016, 11:e0152929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kamburov A, Lawrence MS, Polak P, Leshchiner I, Lage K, Golub TR, Lander ES, Getz G. Comprehensive assessment of cancer missense mutation clustering in protein structures. Proc Natl Acad Sci U S A 2015, 112:E5486–5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Porta-Pardo E, Garcia-Alonso L, Hrabe T, Dopazo J, Godzik A. A Pan-Cancer Catalogue of Cancer Driver Protein Interaction Interfaces. PLoS Comput Biol 2015, 11:e1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Raimondi F, Singh G, Betts MJ, Apic G, Vukotic R, Andreone P, Stein L, Russell RB. Insights into cancer severity from biomolecular interaction mechanisms. Sci Rep 2016, 6:34490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Krogan NJ, Lippman S, Agard DA, Ashworth A, Ideker T. The cancer cell map initiative: defining the hallmark networks of cancer. Mol Cell 2015, 58:690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Coffill CR, Muller PA, Oh HK, Neo SP, Hogue KA, Cheok CF, Vousden KH, Lane DP, Blackstock WP, Gunaratne J. Mutant p53 interactome identifies nardilysin as a p53R273H-specific binding partner that promotes invasion. EMBO Rep 2012, 13:638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Creixell P, Schoof EM, Simpson CD, Longden J, Miller CJ, Lou HJ, Perryman L, Cox TR, Zivanovic N, Palmeri A, et al. Kinome-wide decoding of network-attacking mutations rewiring cancer signaling. Cell 2015, 163:202–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Reimand J, Wagih O, Bader GD. The mutational landscape of phosphorylation signaling in cancer. Sci Rep 2013, 3:2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Narayan S, Bader GD, Reimand J. Frequent mutations in acetylation and ubiquitination sites suggest novel driver mechanisms of cancer. Genome Med 2016, 8:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yang X, Coulombe-Huntington J, Kang S, Sheynkman GM, Hao T, Richardson A, Sun S, Yang F, Shen YA, Murray RR, et al. Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell 2016, 164:805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ghadie MA, Lambourne L, Vidal M, Xia Y. Domain-based prediction of the human isoform interactome provides insights into the functional impact of alternative splicing. PLoS Comput Biol 2017, 13:e1005717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Climente-González H, Porta-Pardo E, Godzik A, Eyras E. The Functional Impact of Alternative Splicing in Cancer. Cell Rep 2017, 20:2215–2226. [DOI] [PubMed] [Google Scholar]
- 152.Latysheva NS, Oates ME, Maddox L, Flock T, Gough J, Buljan M, Weatheritt RJ, Babu MM. Molecular Principles of Gene Fusion Mediated Rewiring of Protein Interaction Networks in Cancer. Mol Cell 2016, 63:579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Oti M, Snel B, Huynen MA, Brunner HG. Predicting disease genes using protein-protein interactions. J Med Genet 2006, 43:691–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature 2007, 446:153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res 2005, 33:W382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tokheim C, Karchin R. Enhanced context reveals the scope of somatic missense mutations driving human cancers. bioRxiv 2018:313296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gao L, Uzun Y, Gao P, He B, Ma X, Wang J, Han S, Tan K. Identifying noncoding risk variants using disease-relevant gene regulatory networks. Nat Commun 2018, 9:702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yu MK, Kramer M, Dutkowski J, Srivas R, Licon K, Kreisberg J, Ng CT, Krogan N, Sharan R, Ideker T. Translation of Genotype to Phenotype by a Hierarchy of Cell Subsystems. Cell Syst 2016, 2:77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Engin HB, Hofree M, Carter H. Identifying mutation specific cancer pathways using a structurally resolved protein interaction network. Pac Symp Biocomput 2015:84–95. [PMC free article] [PubMed] [Google Scholar]
- 160.Poole W, Leinonen K, Shmulevich I, Knijnenburg TA, Bernard B. Multiscale mutation clustering algorithm identifies pan-cancer mutational clusters associated with pathway-level changes in gene expression. PLoS Comput Biol 2017, 13:e1005347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ma J, Yu MK, Fong S, Ono K, Sage E, Demchak B, Sharan R, Ideker T. Using deep learning to model the hierarchical structure and function of a cell. Nat Methods 2018, 15:290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Coulomb S, Bauer M, Bernard D, Marsolier-Kergoat MC. Gene essentiality and the topology of protein interaction networks. Proc Biol Sci 2005, 272:1721–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gandhi TK, Zhong J, Mathivanan S, Karthick L, Chandrika KN, Mohan SS, Sharma S, Pinkert S, Nagaraju S, Periaswamy B, et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet 2006, 38:285–293. [DOI] [PubMed] [Google Scholar]
- 164.Ivanic J, Yu X, Wallqvist A, Reifman J. Influence of protein abundance on high-throughput protein-protein interaction detection. PLoS One 2009, 4:e5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jordan IK, Wolf YI, Koonin EV. No simple dependence between protein evolution rate and the number of protein-protein interactions: only the most prolific interactors tend to evolve slowly. BMC Evol Biol 2003, 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. High-quality binary protein interaction map of the yeast interactome network. Science 2008, 322:104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rioualen C, Da Costa Q, Chetrit B, Charafe-Jauffret E, Ginestier C, Bidaut G. HTS-Net: An integrated regulome-interactome approach for establishing network regulation models in high-throughput screenings. PLoS One 2017, 12:e0185400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gyori BM, Bachman JA, Subramanian K, Muhlich JL, Galescu L, Sorger PK. From word models to executable models of signaling networks using automated assembly. Mol Syst Biol 2017, 13:954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Schnoerr D, Sanguinetti G, Grima R. Approximation and inference methods for stochastic biochemical kinetics—a tutorial review. Journal of Physics A: Mathematical and Theoretical 2017, 50:093001. [Google Scholar]
- 170.Abar S, Theodoropoulos GK, Lemarinier P, O’Hare GMP. Agent Based Modelling and Simulation tools: A review of the state-of-art software. Computer Science Review 2017, 24:13–33. [Google Scholar]
- 171.Kitsak M, Sharma A, Menche J, Guney E, Ghiassian SD, Loscalzo J, Barabási AL. Tissue Specificity of Human Disease Module. Sci Rep 2016, 6:35241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, Zhang R, Hartmann BM, Zaslavsky E, Sealfon SC, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet 2015, 47:569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pierson E, Koller D, Battle A, Mostafavi S, Ardlie KG, Getz G, Wright FA, Kellis M, Volpi S, Dermitzakis ET, et al. Sharing and Specificity of Co-expression Networks across 35 Human Tissues. PLoS Comput Biol 2015, 11:e1004220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lobingier BT, Huttenhain R, Eichel K, Miller KB, Ting AY, von Zastrow M, Krogan NJ. An Approach to Spatiotemporally Resolve Protein Interaction Networks in Living Cells. Cell 2017, 169:350-360 e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Perkins JR, Diboun I, Dessailly BH, Lees JG, Orengo C. Transient protein-protein interactions: structural, functional, and network properties. Structure 2010, 18:1233–1243. [DOI] [PubMed] [Google Scholar]
- 176.Budayeva HG, Cristea IM. A mass spectrometry view of stable and transient protein interactions. Adv Exp Med Biol 2014, 806:263–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Trigg SA, Garza RM, MacWilliams A, Nery JR, Bartlett A, Castanon R, Goubil A, Feeney J, O’Malley R, Huang SC, et al. CrY2H-seq: a massively multiplexed assay for deep-coverage interactome mapping. Nat Methods 2017, 14:819–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Fowler DM, Fields S. Deep mutational scanning: a new style of protein science . Nat Methods 2014, 11:801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 2016, 167:1853-1866.e1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013, 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.eGTEx Project. Enhancing GTEx by bridging the gaps between genotype, gene expression, and disease. Nat Genet 2017, 49:1664–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Li X, Kim Y, Tsang EK, Davis JR, Damani FN, Chiang C, Hess GT, Zappala Z, Strober BJ, Scott AJ, et al. The impact of rare variation on gene expression across tissues. Nature 2017, 550:239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hamada T, Keum N, Nishihara R, Ogino S. Molecular pathological epidemiology: new developing frontiers of big data science to study etiologies and pathogenesis. J Gastroenterol 2017, 52:265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ogino S, Nowak JA, Hamada T, Milner DA Jr, Nishihara R Insights into Pathogenic Interactions Among Environment, Host, and Tumor at the Crossroads of Molecular Pathology and Epidemiology. Annu Rev Pathol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ogino S, Nowak JA, Hamada T, Phipps AI, Peters U, Milner DA Jr, Giovannucci EL, Nishihara R, Giannakis M, Garrett WS, et al. Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut 2018, 67:1168–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further Reading
- Bartocci E, Lió P. (2016). Computational Modeling, Formal Analysis, and Tools for Systems Biology. PLoS Comput Biol. 12(1):e1004591. doi: 10.1371/journal.pcbi.1004591. PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DY, Kim YA, Przytycka TM. (2012), Chapter 5: Network biology approach to complex diseases. PLoS Comput Biol. 8(12):e1002820. doi: 10.1371/journal.pcbi.1002820. PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller D, & Friedman N (2009). Probabilistic graphical models: principles and techniques. MIT press. [Google Scholar]
- Mezlini AM, Goldenberg A. (2017). Incorporating networks in a probabilistic graphical model to find drivers for complex human diseases. PLoS Comput Biol. 13(10):e1005580. doi: 10.1371/journal.pcbi.1005580.PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]


