Summary
The high-affinity cyclic adenosine monophosphate (cAMP) phosphodiesterase MoPdeH is important not only for cAMP signaling and pathogenicity, but also for cell wall integrity (CWI) maintenance in the rice blast fungus Magnaporthe oryzae. To explore the underlying mechanism, we identified MoImd4 as an inosine-5’-monophosphate dehydrogenase (IMPDH) homolog that interacts with MoPdeH. Targeted deletion of MoIMD4 resulted in reduced de novo purine biosynthesis and growth, as well as attenuated pathogenicity, which were suppressed by exogenous xanthosine monophosphate (XMP). Treatment with mycophenolic acid (MPA) that specifically inhibits MoImd4 activity resulted in reduced growth and virulence attenuation. Intriguingly, further analysis showed that MoImd4 promotes the MoPdeH phosphodiesterase activity thereby decreasing intracellular cAMP levels, and MoPdeH also promotes the IMP dehydrogenase activity of MoImd4. Our studies revealed the presence of a novel crosstalk between cAMP regulation and purine biosynthesis in M. oryzae and indicated that such a link is also important in pathogenesis of M. oryzae.
Keywords: Inosine-5’-monophosphate dehydrogenase, GTP biosynthesis, phosphodiesterase, pathogenicity, Magnaporthe oryzae
Introduction
In the rice blast fungus Magnaporthe oryzae, the cAMP signaling pathway plays important roles in vegetative growth, asexual/sexual development, cell wall integrity, appressorium formation and virulence (Lee & Dean, 1993, Ramanujam & Naqvi, 2010, Zhang et al., 2011a, Yang et al., 2017, Zhang et al., 2011b, Liu et al., 2016b, Yin et al., 2016). In M. oryzae, intracellular cAMP levels are governed by the dynamic balance between adenylyl cyclase MoMac1 that synthesizes cAMP and high-affinity phosphodiesterase MoPdeH and low-affinity MoPdeL that hydrolyze cAMP (Ramanujam & Naqvi, 2010, Zhang et al., 2011a, Yang et al., 2017, Woobong Choi 1997). We previously found that MoPdeH plays a role in hyphal autolysis, surface signal recognition, conidium morphology, cell wall integrity, and pathogenicity, while MoPdeL appears to affect conidial morphology (Zhang et al., 2011a). We also found that the HD and EAL domains of MoPdeH are critical for its phosphodiesterase activity (Yang et al., 2017). Interestingly, we found that the protein phosphatase MoYvh1 functions upstream of MoPdeH to regulate cell wall integrity (CWI) and pathogenicity (Liu et al., 2016b); however, the underlying mechanism of this regulation is unclear.
To explore how MoPdeH might affect CWI independent of cAMP signaling, we screened for proteins interacting with MoPdeH and identified MoImd4 as an inosine-5’-monophosphate dehydrogenase (IMPDH) homolog from M. oryzae. In eukaryotic cells, IMPDH is involved in de novo purine biosynthesis, a highly conserved biological process that provides ATP and GTP energy source for the cell (Elion, 1989). IMPDH catalyzes and hydrolyzed inosine monophosphate (IMP) as xanthosine monophosphate (XMP), a rate-limiting and first committed step in the de novo biosynthesis of GTP (Buey et al., 2015b). IMPDH is also related to provide the obligatory precursors for DNA and RNA biosynthesis and cell proliferation that may be linked to malignant cell transformation or tumor progression (Jackson et al., 1975, Shimura et al., 1983, Collart & Huberman, 1988, Buey et al., 2015b). In the budding yeast Saccharomyces cerevisiae, IMPDH is encoded by a family of four genes named ScIMD1 to ScIMD4 and loss of the ScIMD gene family resulted in cells auxotrophic for guanine. Despite encoding proteins with high amino acid sequence identity, their functions were distinct: ScIMD1 is a pseudogene, ΔScimd2 has intrinsic drug resistance, ΔScimd3 and ΔScimd4 could confer drug resistance lacking ScIMD2 (Hyle et al., 2003). In Cryptococcus neoformans, CnImd1 is important for growth, synthesis of the cryptococcal polysaccharide capsule and melanin, and virulence in mouse and nematode models (Morrow et al., 2012). In Ashbya gossypii, overexpression of the IMPDH gene increased metabolic flux through the guanine pathway that ultimately enhanced the riboflavin production (Buey et al., 2015a).
IMPDH contains two conserved tandem cystathionine β-synthase (CBS) subdomains (Bateman, 1997) that are also present in a variety of proteins, including voltage-gated chloride channels and the AMP-activated protein kinase (Ignoul & Eggermont, 2005, Baykov et al., 2011). The CBS domain mutation was related to many hereditary diseases of humans, such as homocystinuria, Wolff-Parkinson-White syndrome, and congenital myotonia (Scott et al., 2004, McGrew & Hedstrom, 2012). In Escherichia coli, CBS domains are essential for global regulation of purine utilization, ATP/GTP ratio, and functions on the enzymatic activity of IMPDH (Pimkin & Markham, 2008). However, not all IMPDH CBS domains are accountable for specific defects, such as in Pseudomonas aeruginosa and C. neoformans (Rao et al., 2013, Morrow et al., 2012). Hence, it seems that the physiological functions of CBS domains might vary considerably between different organisms.
Previous studies found that MoPdeH plays a multifaceted role in M. oryzae (Ramanujam & Naqvi, 2010, Zhang et al., 2011a, Yang et al., 2017, Woobong Choi 1997). Here we continued to investigate the mechanism linking MoPdeH to fungal pathogenesis. We identified a MoPdeH-interacting protein MoImd4 through a yeast two-hybrid screen and found that MoImd4 is involved in de novo purine metabolic pathway. MoImd4 and MoPdeH regulate their enzymatic activities mutually to promote the development and pathogenicity of M. oryzae. The finding of MoImd4 in association with MoPdeH provides a new link between cAMP signaling and purine biosynthesis pathway. It also reveals that MoImd4 has functions beyond guanine nucleotide biosynthesis.
Results
Identification of MoImd4 as a MoPdeH-interacting protein
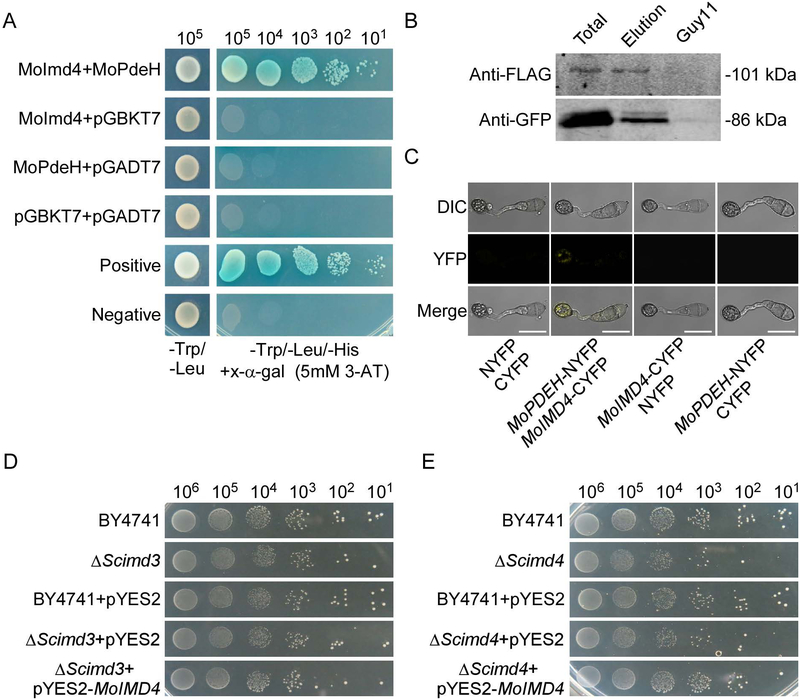
MoPdeH is a high-affinity phosphodiesterase that hydrolyzes intracellular cAMP required for vegetative growth, functional appressorium development and virulence of M. oryzae (Ramanujam & Naqvi, 2010, Zhang et al., 2011a, Yang et al., 2017). To further explore the molecular regulatory mechanism of MoPdeH, we screened a yeast two-hybrid cDNA library constructed with RNA pool from various stages including conidia and infectious hyphae (0, 2, 4, 8, 12 and 24 h). 19 putative MoPdeH-interacted proteins were identified. Among them, the fragments of MGG_03699 showed the highest frequency (12 times). Thus we chose MGG_03699 for further characterization. MGG_03699 shares high amino acid sequence homology with inosine monophosphate dehydrogenases (Imds) from various species (Fig. 1A, S2 and Table S1). To test if this Imd homolog encodes conserved functions, we expressed the protein with the yeast expression vector pYES2 in ΔScimd3 and ΔScimd4 strains and found that the M. oryzae protein could partially rescue the growth defect of ΔScimd4 (Fig. 1D, E). We named this single inosine-5’-monophosphate dehydrogenase orthologue of M. oryzae as MoImd4. We have also validated the interaction between MoImd4 and MoPdeH by Co-Immunoprecipitation (Co-IP) and bimolecular fluorescence complementation (BiFC) assays (Fig. 1B, C).
Fig. 1.
MoPdeH physically interacts with MoImd4. (A) Yeast two-hybrid assay for the interaction between MoImd4 and MoPdeH. MoIMD4 was inserted into vector pGADT7 and MoPDEH was inserted into vector pGBKT7. Both vectors were co-transferred into yeast AH109 cells and incubated on SD-Leu-Trp for 3 days prior to selection on SD-Leu-Trp-His medium with 1mM X-α-gal and 5mM 3-AT (3-amino-1,2,4-triazole) for 3 more days. (B) Co-immunoprecipitation (Co-IP) assay for the interaction between MoImd4 and MoPdeH. Plasmids of MoPDEH-Flag and MoIMD4-GFP were co-expressed in wild type Guy11 and proteins were detected using anti-Flag and anti-GFP antibodies. Lysed hyphal proteins were allowed to bind to Flag beads at 4˚C for 4 h and analyzed by immunoblot (IB) with appropriate antibodies. (C) Bimolecular fluorescence complementation (BiFC) assay for the interaction between MoImd4 and MoPdeH. Transformants expressing MoIMD4-CYFP and MoPDEH-NYFP were analyzed by DIC and epifluorescence microscopy following incubation on hydrophobic slides at 8 hpi. YFP, yellow fluorescent protein. Bar = 10 μm. (D and E) MoImd4 partially rescues the growth defect of ΔScimd4 but not ΔScimd3. Serial dilutions of BY4741, ΔScimd3, ΔScimd4, pYES2, and pYES2-MoIMD4 transformants were grown on SD-Leu-Met-Ura-His (galactose) plates at 30˚C for 6 days.
MoImd4 is involved in vegetative growth, conidiation, and virulence
To examine MoImd4 functions, we generated the ΔMoimd4 mutant by using the standard one-step gene replacement strategy and we also complemented the mutant with the wild type MoIMD4 gene (Fig. S1). Phenotypic analysis showed MoImd4 is required for mycelial growth, conidial formation, and pathogenicity. In comparison to Guy11, the ΔMoimd4 mutant showed significantly reduced growth on CM, MM, SDC and OM media plates and reduced biomass in liquid CM (Fig. S3A, B and Table 1). Conidiation was also significantly reduced to approximately 0.04-fold of the wild type and the complement strains following 10 day’s growth on SDC medium (Table 1).
Table 1.
The vegetative growth, biomass and conidiation analysis of the wild type, ΔMoimd4 mutant, and the complement strains.
| Strain | Colony diameter (cm) a | Biomass (g)b | Conidiation (x100/cm2)c |
|||
|---|---|---|---|---|---|---|
| CM | MM | OM | SDC | |||
| Guy11 | 4.9 ± 0.1 | 4.0 ± 0.1 | 4.4 ± 0.1 | 3.3 ± 0.1 | 0.0815 ± 0.0028 | 478.7 ± 36.2 |
| ΔMoimd4 | 2.8 ± 0.1* | 3.1 ± 0.2* | 3.8 ± 0.1* | 2.7 ± 0.1* | 0.0207 ± 0.0015* | 20.2 ± 5.0* |
| MoImd4 | 4.8 ± 0.1 | 3.9 ± 0.1 | 4.4 ± 0.1 | 3.3 ± 0.1 | 0.0803 ± 0.0012 | 490.4 ± 37.8 |
Colony diameter of the indicated strains on CM, MM, OM and SDC media after 7 days incubation at 28°C;
Dry weight of hyphal at day 2 after incubation in liquid complete medium by shaken at 160 rpm at 28°C.
Quantification of the conidial production of the indicated strains from SDC cultures in the dark for 7 d, followed by incubation under constant illumination for 3 d at room temperature.
±SD was calculated from three repeated experiments and asterisk indicates statistically significant differences (Duncan's new multiple range test,
means p<0.01).
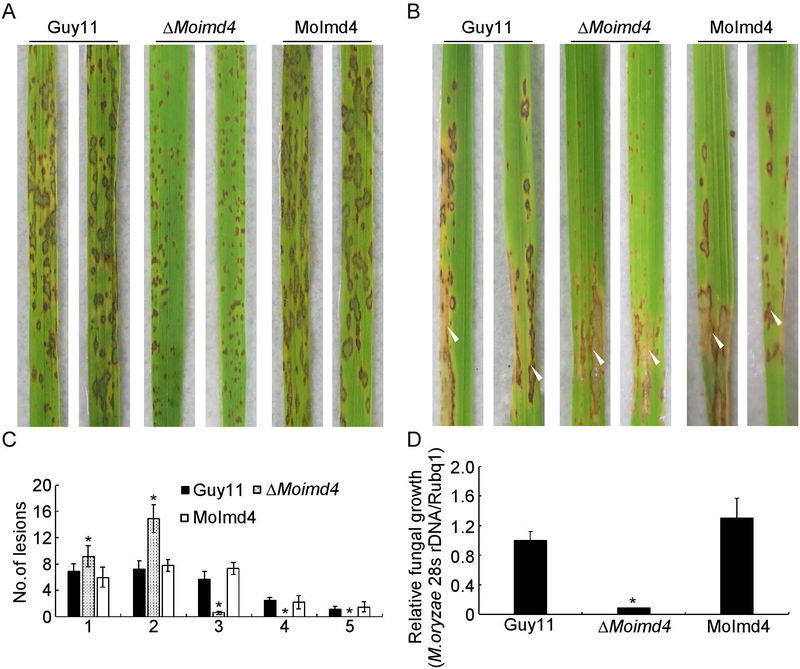
To test the role of MoImd4 in pathogenicity, conidial suspensions of the wild type Guy11, the ΔMoimd4 mutant and the complement strains were sprayed or injected into the susceptible rice seedlings CO-39. After 7 days in a chamber at 28˚C with 90% humidity, the ΔMoimd4 mutant produced smaller and needle-like lesions compared to the numerous typical lesions produced by wild type and the complement strains (Fig. 2A, B). Meanwhile, a ‘lesion type’ scoring assay according to Liu et al.(Liu et al., 2016b) showed that the ΔMoimd4 mutant formed type 1 and 2, and very few type 3, but no type 4 or 5 lesions (Fig. 2C). The results of fungal biomass assays in diseased leaves were in accordance with the spraying assays (Fig. 2D). In addition, appressorium formation on the inductive or non-inductive surface after 24 hours were examined that revealed no significant differences between the ΔMoimd4 mutant and the wild type strains (Table 2). Meanwhile, the turgor assay showed that the ΔMoimd4 mutant was more sensitive to 1-4 M glycerol than control strains (Table 2). Taken together, these results showed that MoImd4 is important for growth, conidiation, and pathogenicity of M. oryzae.
Fig. 2.
MoImd4 is required for pathogenicity. (A) Conidial suspensions (5 × 104 spores/ml) of Guy11, ΔMoimd4, and the complement strains were sprayed onto 11-day rice seedlings and photography were made 7 days after inoculation. (B) Conidial suspensions (15 × 104 spores/ml) were injected into 18-day old rice sheaths and photography were made 5 days post inoculation (dpi). (C) Lesions type statistical analysis. (0, no lesion; 1, pinhead-sized brown specks; 2, 1.5 mm brown spots; 3, 2–3 mm grey spots with brown margins; 4, many elliptical grey spots longer than 3 mm; 5, coalesced lesions infecting 50% or more of the leaf area). Lesions were photographed and were measured after 7 dpi. Experiments were repeated three times with similar results. Asterisk represents significant differences (Duncan’s new multiple range test, p<0.01). (D) Severity of lesions was analyzed by quantifying M. oryzae genomic 28S rDNA relative to rice genomic Rubq1 DNA. Experiments were repeated three times with similar results. Error bars represent the standard deviations and asterisk represents significant difference (Duncan’s new multiple range test, p<0.01).
Table 2.
Turgor assays and appressorium formation of the wild type, ΔMoimd4 mutant, and the complement strains.
| Strain | Appressorium exhibiting incipient cytorrhysis (%)a |
Appressorium formation (%)b |
||||
|---|---|---|---|---|---|---|
| 1 M | 2 M | 3 M | 4 M | Hydrophobic | Hydrophilic | |
| Guy11 | 30.7 ± 5.0 | 52.0 ± 5.7 | 67.0 ± 7.1 | 83.3 ± 4.2 | 99.1 ± 2.2 | 0 |
| ΔMoimd4 | 42.0 ± 3.2* | 65.0 ± 2.0* | 80.0 ± 2.1* | 92.0 ± 2.6* | 98.5 ± 2.8 | 0 |
| MoImd4 | 34.7 ± 4.2 | 56.0 ± 5.5 | 72.0 ± 1.5 | 79.4 ± 3.2 | 99.5 ± 3.2 | 0 |
Different concentrations of glycerol (1 to 4 M) to analysis incipient cytorrhysis. At least 100 appressoria were counted for each concentration.
Appressorium formation on hydrophilic or hydrophobic surfaces at 24 hours post incubation; ±SD was calculated from three repeated experiments and asterisk indicates statistically significant differences (Duncan's new multiple range test,
means p<0.01).
MoImd4 is important for xanthosine monophosphate biosynthesis
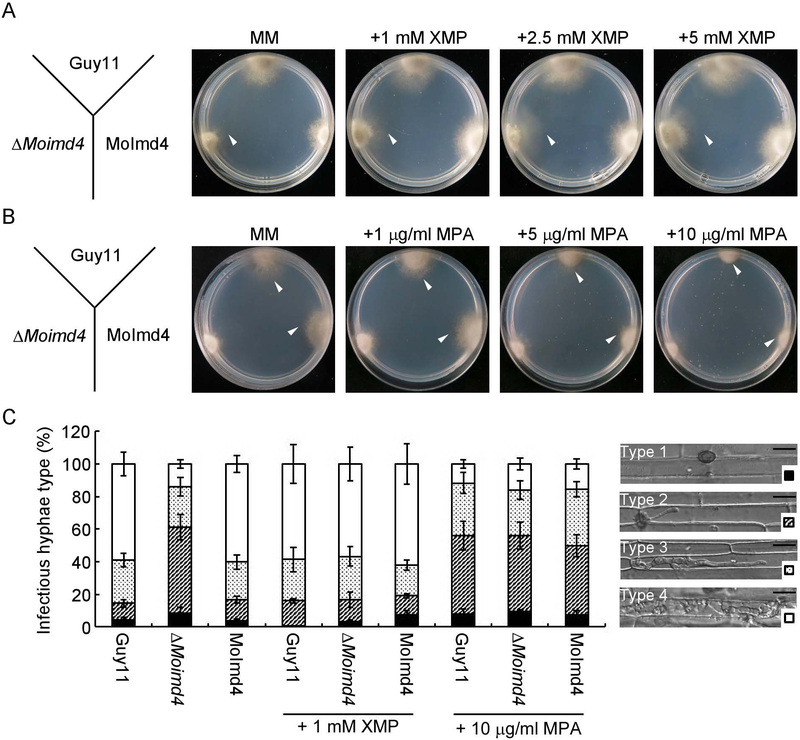
In S. cerevisiae, IMPDH catalyzes the hydrolysis of inosine monophosphate (IMP) to xanthosine monophosphate (XMP) (Hyle et al., 2003). To test if the ΔMoimd4 mutant failed to form XMP contributing to the defects in vegetative growth, conidial formation and virulence, we measured the intracellular level of XMP by HPLC and found that the ΔMoimd4 mutant has 0.15-fold of reduction compared with the wild type and complement strains (Fig. S4A). Consist with this result, the addition of 1, 2.5, and 5 mM XMP in the minimal medium (MM) all could restore the defects of the mutant, with the exception of conidial formation (Fig. 3A, S4B and Table 3). To further test virulence, we injected conidial suspension amended with 1 mM XMP in detached rice sheaths. We found that in Guy11 and the complement strain, nearly 80% invasion hyphal (IH) were type 4 and type 3, with 20% showing type 1 and type 2. Exogenous addition of 1 mM XMP could restore the expansion of IH of the ΔMoimd4 mutant to near the levels of the wild type and the complemented strains (80% of type 3 and type 4 IH). This is in contrast to that the ΔMoimd4 mutant normally produced 60% of type 2 IH and 40% of type 3 and type 4 IH (Fig. 3C). The results indicated that the hydrolytic activity of MoImd4 and XMP levels are critical for pathogenicity of M. oryzae.
Fig. 3.
XMP levels are critical for growth and virulence of M. oryzae. (A) Guy11, ΔMoimd4 and the complement strains were co-incubated on MM media treated with or without 1, 2.5, and 5 mM XMP at 28˚C in the dark for 7 days. White triangles represent the edges of mycelia. (B) Indicated strains were co-incubated on MM media with or without 1, 5, and 10 μg/ml MPA at 28˚C for 7 days in darkness. White triangles represent the edges of mycelia. (C) Four types of IH in rice sheath cells and four different shapes at bottom right corner represent the variant type of IH (Type 1, no penetration; Type 2, with a single invasive hypha; Type 3, with extensive hyphal growth in only one rice cell; Type 4, with extensive hyphal growth in neighboring rice cells). Statistical analysis the type of IH of the indicated strains treated with or without 1 mM XMP or 10 μg/ml MPA at 48 h, approximate 100 IH were counted and the experiments were repeated three times with similar results. The error bars indicate standard deviations of three replicates. Bar = 10 μm.
Table 3.
Phenotype analysis of the wild type, ΔMoimd4 mutant, the complement strains, point mutation mutants and three CBS domain deletion mutants.
| Strain | Colony diameter (CM)a | Colony diameter (MM)a | Colony diameter (MM + XMP)b | Conidiation (x100/cm2)c | Conidiation+XMP (x100/cm2)d |
|---|---|---|---|---|---|
| Guy11 | 4.8 ± 0.1A | 2.9 ± 0.1A | 3.0 ± 0.2A | 741.9 ± 50.9A | 746.3 ± 73.1A |
| ΔMoimd4 | 3.1 ± 0.1EF | 0.9 ± 0.1D | 2.8 ± 0.1A | 27.9 ± 2.5C | 25.1 ± 4.1C |
| T268AR269A | 3.0 ± 0.1FG | 1.3 ± 0.1C | 2.3 ± 0.3B | 29.3 ± 6.4C | 30.7 ± 3.8C |
| S345AC347A | 2.0 ± 0.1H | 1.4 ± 0.1B | 2.7 ± 0.2A | 46.3 ± 7.3C | 55.1 ± 5.0C |
| D380AG382A | 3.3 ± 0.2E | 0.8 ± 0.1D | 1.3 ± 0.1C | 41.9 ± 4.5C | 33.1 ± 8.2C |
| G403A | 3.2 ± 0.1EF | 0.8 ± 0.1D | 0.8 ± 0.1D | 46.8 ± 4.1C | 43.9 ± 7.6C |
| R458AY459A | 3.9 ± 0.1C | 0.4 ± 0.1E | 0.4 ± 0.1E | 33.0 ± 3.6C | 27.9 ± 6.1C |
| Y428A | 3.7 ± 0.1D | 0.3 ± 0.1E | 0.5 ± 0.1DE | 21.6 ± 3.8C | 21.8 ± 5.4C |
| ΔCBS1 | 4.5 ± 0.2B | 3.0 ± 0.2A | 3.0 ± 0.1A | 684.2 ± 40.1AB | 624.5 ± 50.2B |
| ΔCBS2 | 4.5 ± 0.1B | 2.9 ± 0.2A | 3.0 ± 0.2A | 624.3 ± 32.2B | 630.2 ± 35.1B |
| ΔCBS1CBS2 | 4.1 ± 0.1C | 0.4 ± 0.1E | 0.4 ± 0.1E | 33.7 ± 2.1C | 29.9 ± 6.5C |
| MoImd4 | 4.8 ± 0.2A | 2.9 ± 0.1A | 2.9 ± 0.2A | 726.3 ± 32.2A | 746.3 ± 15.0A |
Colony diameter of the indicated strains on CM or MM media after 7 days incubation at 28°C; ±SD was calculated from three repeated experiments and letters indicate statistically significant differences (Duncan's new multiple range test, letters mean p<0.01).
Colony diameter of the indicated strains on MM media which added 1mM XMP after 7 days incubation at 28°C; ±SD was calculated from three repeated experiments and letters indicate statistically significant differences (Duncan's new multiple range test, letters mean p<0.01).
Quantification of the conidial production of the indicated strains from SDC cultures in the dark for 7 d, followed by incubation under constant illumination for 3 d at room temperature; ±SD was calculated from three repeated experiments and letters indicate statistically significant differences (Duncan's new multiple range test, letters mean p<0.01).
Quantification of the conidial production of the indicated strains from SDC cultures which added 1mM XMP in conidial suspension in the dark for 7 d, followed by incubation under constant illumination for 3 days at room temperature; ±SD was calculated from three repeated experiments and letters indicate statistically significant differences (Duncan's new multiple range test, letters mean p<0.01).
Impaired XMP biosynthesis results in attenuated growth and virulence
In humans, mycophenolic acid (MPA) is an approved drug to target IMPDH as a method of immunosuppressive and antiviral chemotherapy (Chapuis et al., 2000, Umejiego et al., 2004, Gollapalli et al., 2010, Johnson et al., 2013, Morrow et al., 2012). Interestingly, 5 and 10 μg/ml of MPA induced the growth defect of the wild type and the complement strains in MM, similar to that of the ΔMoimd4 mutant (Fig. 3B, S4C). Further infection assay in rice sheaths showed that the infectious hyphae (IH) of Guy11 and the complement strain showed most of the type 2 kind when 10 μg/ml MPA was added to the conidial suspensions, similar to that by the ΔMoimd4 mutant (Fig. 3C). These results indicated that MPA specifically inhibits MoImd4 and attenuates growth and virulence of M. oryzae.
Inactivation of MPA binding sites attenuate MoImd4 activity
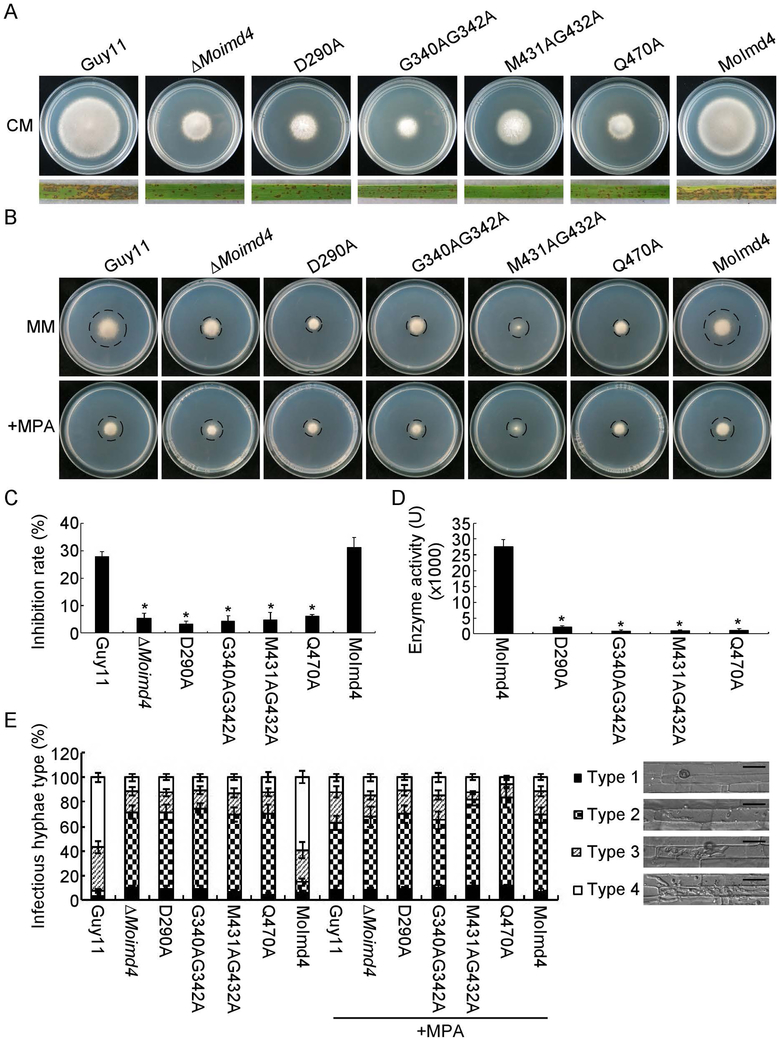
To further explore the function of MoImd4 in the XMP biosynthesis pathway, we modeled the three-dimensional structure of MoImd4 using the structure of CnImd1 (PDB entry 4af0.2.A) to which it shares 65.19% amino acid identity as the template (Arnold et al., 2006, Benkert et al., 2011, Biasini et al., 2014). MoImd4 forms a tetramer and has conserved potential MPA binding sites at D290, G340, G342, M431, G432 and Q470 (Fig. S5A, B). We then generated four point-mutation mutants: ΔMoimd4/MoIMD4D290A (D290A), ΔMoimd4/MoIMD4G340AG342A (G340AG342A), ΔMoimd4/MoIMD4M431AG432A (M431AG432A), ΔMoimd4/MoIMD4Q470A (Q470A) and tested their functions (Fig. S6). These mutants presented the similar phenotypic defects in vegetative growth on CM media and virulence with the ΔMoimd4 mutant (Fig. 4A). The vegetative growth and invasion assays in rice cell tests of these point mutation mutants treated with or without exogenous MPA (10 μg/ml) were consistent with that MPA cannot bind with these allele mutants of MoImd4 (Fig. 4B,C and E). Moreover, all these mutant proteins showed weakened Imd enzymatic activities of ~0.04-fold compared with that of wild-type (Fig. 4D).
Fig. 4.
Inactivation of MPA binding sites leads to attenuation of MoImd4 activities. (A) Vegetative growth of Guy11, ΔMoimd4, D290A, G340AG342A, M431AG432A, Q470A mutants, and the complement strains on CM media at 7 days in the darkness. Conidial suspensions (5 × 104 spores/ml) of the indicated strains were sprayed onto 11-day old rice seedlings. Photographs were taken 7 days after inoculation. (B) Vegetative growth of Guy11, ΔMoimd4, D290A, G340AG342A, M431AG432A, Q470A and the complement strains on MM media treated with or without 10 μg/ml MPA. (C) Inhibition rates of the indicated strains on MM media treated with or without 10 μg/ml MPA. Experiments were repeated three times with similar results. The error bars indicate standard deviation of three replicates. Asterisk indicates statistically significant differences (Duncan’s new multiple range test, p<0.01). (D) Detection of enzymatic activities of the indicated strains in vitro. Target proteins were expressed in E. coli BL21-CodonPlus (DE3) cells. We defined that the production of 1 mM XMP per milligram of protein per minute as one unit of enzyme activity (U). Experiments were repeated three times with similar results. The error bars indicate standard deviations of three replicates. Asterisks indicate statistically significant differences (Duncan’s new multiple range test, p<0.01). (E) Statistical analysis of IH of the indicated strains with or without 10 μg/ml MPA after 48 hpi, approximate 100 IH were counted and the experiments were repeated three times. The error bars indicate standard deviations of three replicates. Asterisks indicate statistically significant differences (Duncan’s new multiple range test, p<0.01). Four type grading standards as in Figure 3E. Bar = 10 μm.
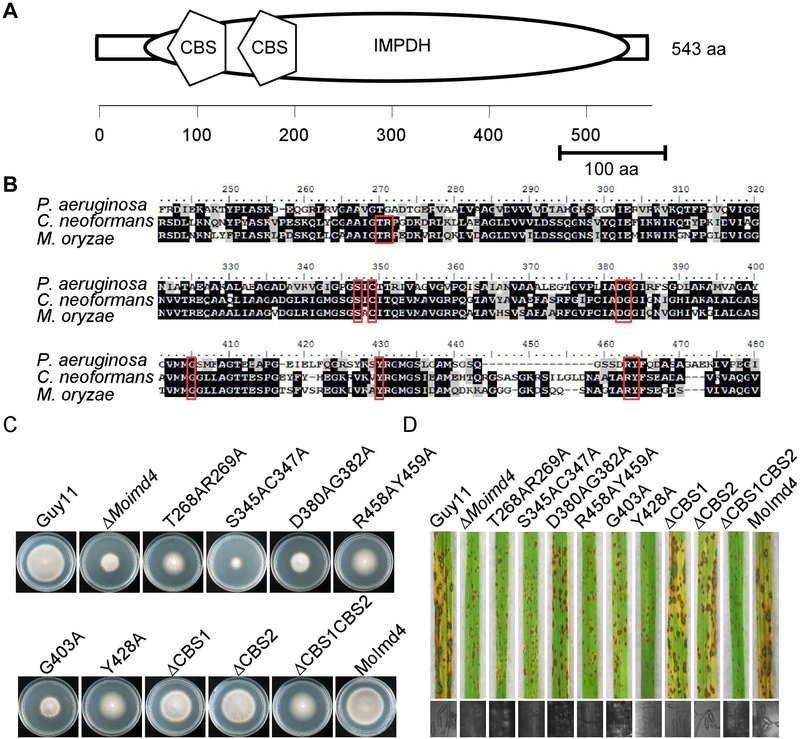
Functional characterization of different domains and reaction sites of MoImd4
MoImd4 has one conserved IMPDH domain (amino acids 47~533) and two tandem accessory cystathionine-ß-synthase (CBS) subdomains (CBS1, amino acids 136~187; CBS2, amino acids 199~247) (Fig. 5A). Previous studies showed that sites such as S317, C319, D358, G360, G381, Y405, R418 and Y419 of Pseudomonas aeruginosa are required for its catalytic function (Rao et al., 2013). Based on IMPDH of C. neoformans and P. aeruginosa, we predicted similar sites in MoImd4 (Fig. 5B) and generated six point-mutation mutants: ΔMoimd4/MoIMD4T268AR269A (T268AR269A), ΔMoimd4/MoIMD4S345AC347A (S345AC347A), ΔMoimd4/MoIMD4D380AG382A (D380AG382A), ΔMoimd4/MoIMD4R458AY459A (R458AY459A), ΔMoimd4/MoIMD4G403A (G403A), and ΔMoimd4/MoIMD4Y428A (Y428A). We have also generated three CBS-domain deletion mutants: ΔMoimd4/MoIMD4ΔCBS1 (ΔCBS1), ΔMoimd4/MoIMD4ΔCBS2 (ΔCBS2), and ΔMoimd4/MoIMD4ΔCBS1CBS2 (ΔCBS1CBS2) (Fig. S6). All of these mutant alleles, except ΔCBS1 and ΔCBS2, exhibited defects in vegetative growth, conidial formation, and virulence (Fig. 5C, D and Table 3). These results demonstrated that the tandem CBS domain and the reaction sites are important for MoImd4 function.
Fig. 5.
Functional characterization of CBS domains and reaction sites of MoImd4. (A) Schematic representation of MoImd4 IMPDH domain (oval) and tandem CBS subdomains (pentagon). Domains were predicted using the SMART program (http://smart.embl-heidelberg.de/). (B) Multiple alignments of P. aeruginosa, C. neoformans, and M. oryzae IMPDH proteins. Red boxes represent conservative functional sites involved in the interaction with the substrates. The amino acid identity of IMPDH was 37% between M. oryzae and P. aeruginosa, and 63% between M. oryzae and C. neoformans. (C) The wild type Guy11, ΔMoimd4, the complement strains, predicted sites mutation mutants, and CBS domain deletion mutants were incubated on CM media at 28˚C in the dark and photographed after 7 days incubation. (D) Pathogenicity test on rice seedlings of the indicated strains. Infected rice leaves were illuminated under the fluorescent light for 24 h to produce conidia. The lesions were observed under a light microscope.
MoImd4 is important in the purine metabolic pathway
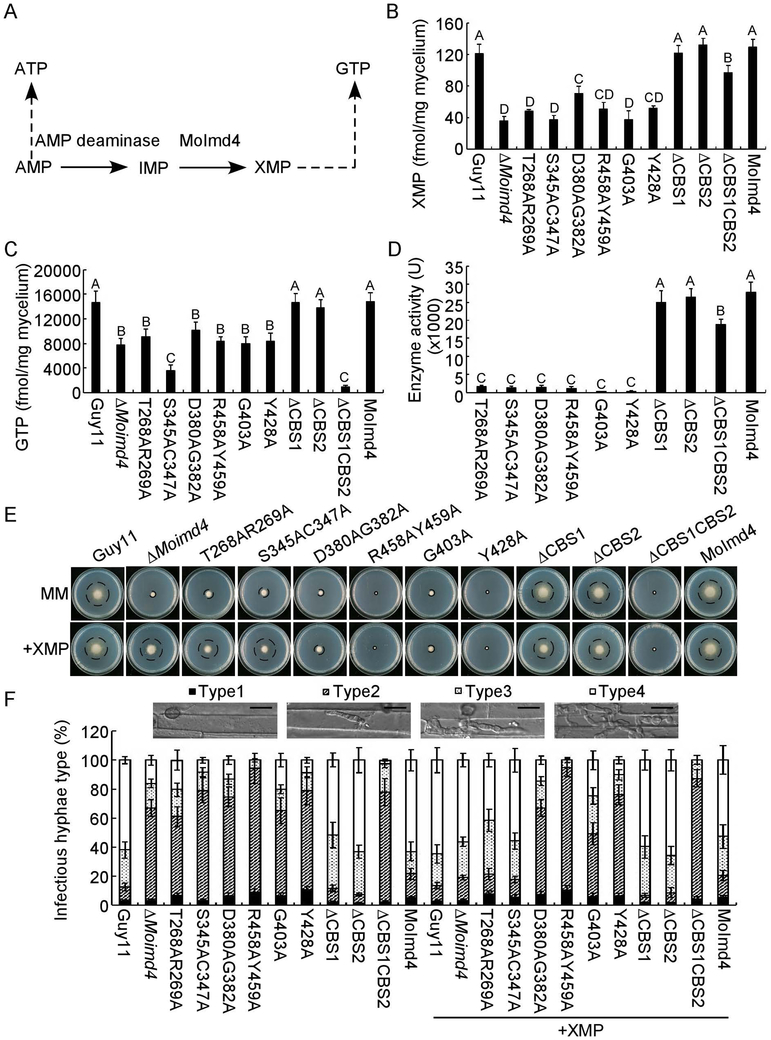
The purine metabolic pathway provides ATP and GTP essential for cellular processes and activities (Morrow et al., 2012). To evaluate the role of MoImd4 in the de novo purine biosynthesis pathway, we measured in vivo intracellular XMP and GTP levels in the ΔMoimd4 mutant, point mutation strains, CBS domain deletion mutants, and the wild type and complement strains (Fig. 6A). The XMP levels have reduced to 0.33-fold in the ΔMoimd4 mutant and all six point-mutants, and 0.83-fold in the tandem CBS deletion mutants when compared with the control strains. However, there was little difference between the solely CBS domain deletion mutants and wild type (Fig. 6B). We also found that GTP levels were remarkably reduced in most of the mutant strains in comparison to Guy11 and the complement strain, with the especially low levels of 0.2-fold and 0.06-fold found in S345AC347A and ΔCBS1CBS2 strains, respectively. However, the ΔCBS1 and ΔCBS2 mutants showed no attenuation (Fig. 6C). Moreover, we purified the point mutation proteins by His-tag and examined the enzymatic activities of these point mutation proteins and domain deletion proteins. The IMP dehydrogenase activity was nearly attenuated in these point-mutation mutants (Fig. 6D), despite that these mutations did not affect the three-dimensional structure (Fig. S7). The MoImd4 enzyme activity was reduced in ΔCBS1CBS2 but not in any of the CBS deletion mutants (Fig. 6D).
Fig. 6.
MoImd4 is required for the purine metabolic pathway in M. oryzae and exogenous XMP suppresses defects of the S345AC347A mutant in vegetative growth and virulence. (A) The de novo GTP/ATP biosynthesis pathway of M. oryzae. (B and C) Intracellular levels of XMP/GTP in mycelia of indicated strains by HPLC. Experiments were repeated three times with similar results. The error bars indicate standard deviations of three replicates. Letters indicate statistically significant differences (Duncan’s new multiple range test, p<0.01). (D) Detection of enzymatic activities of indicated strains in vitro. The target proteins were expressed in E. coli BL21-CodonPlus (DE3) cells. Experiments were repeated three times with similar results. The error bars indicate standard deviation of three replicates. Letters indicate statistically significant differences (Duncan’s new multiple range test, p<0.01). (E) Vegetative growth and statistical analysis of indicated strains on MM media treated with or without 1 mM XMP after 7 days incubation. Experiments were repeated three times with similar results. Error bars represent the standard deviations and asterisk denotes statistical significances (Duncan’s new multiple range test, p<0.01). (F) Statistical analysis type of IH of the indicated strains deal with or without 1 mM XMP after 48 hours post inoculation. Approximate 100 IH were counted and experiments were repeated three times. The error bars indicate standard deviations of three replicates. Please refer to Figure 3E for IH grading. Bar = 10 μm.
Exogenous XMP suppresses defects in vegetative growth and virulence of the S345AC347A mutant
To further understand which sites are critical for the function of MoImd4 in medicating XMP synthesis, XMP was added to MM medium. Only the S345AC347A point mutation mutant rescued the defects in vegetative growth and pathogenicity. However, the T268AR269A mutant was partially restored in vegetative growth (0.8-fold) and the formation of type 4 IH up to 0.4-fold (0.6-fold when compared with the ΔMoimd4 mutant and the S345AC347A mutant). None of the other mutants exhibited similar rescue of the defect (Fig. 6E, F and Table 3). It should be noted that, similar to the ΔMoimd4 mutant, none of these strains was rescued in conidial formation by exogenous XMP (Table 3). Together, these results showed that MoImd4 governs the production of XMP and S345 and C347 sites are the most critical for this activity.
MoImd4 interacts with MoPdeH to promote its phosphodiesterase activity
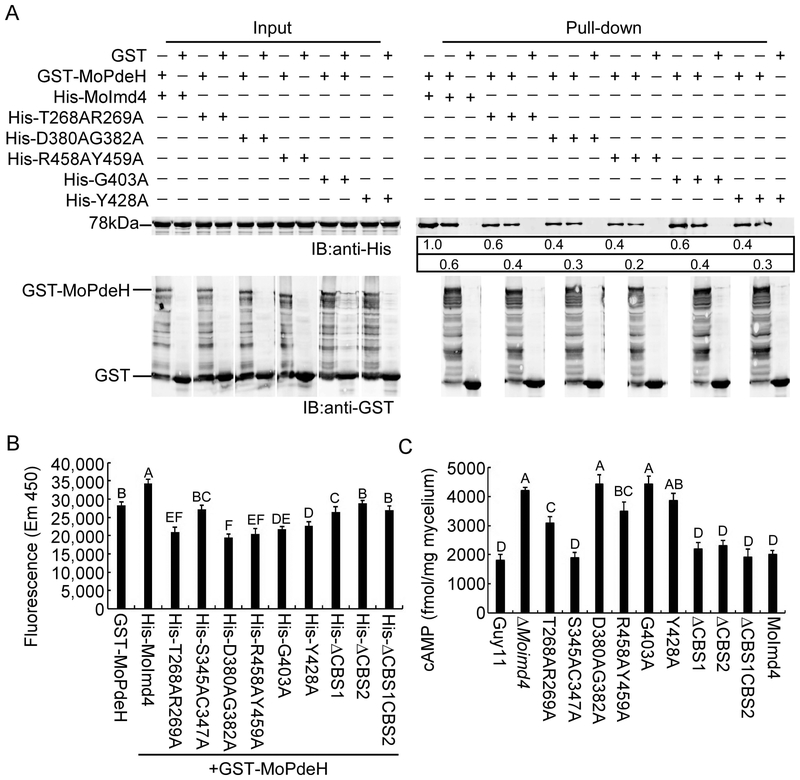
Owing to the interaction between MoPdeH and MoImd4, we hypothesized that MoImd4 affects the phosphodiesterase activity of MoPdeH. To test this, we first expressed GST-MoPdeH and His-MoImd4 fusion proteins and verified this interaction using GST-pull down assays (Fig. S8A). Then we measured the MoPdeH phosphodiesterase activity with and without the presence of MoImd4 using purified proteins in vitro and a fluorescence based assay method for free phosphate according to Yin et al. (Yin et al., 2018). Our results showed that the samples treated with MoPdeH had strong fluorescence, whereas samples treated with various concentrations of MoImd4 had more intense fluorescence (Fig. S8B), suggesting that MoImd4 could promote the phosphodiesterase activity of MoPdeH in vitro. Meanwhile, we also measured intracellular cAMP levels in Guy11, ΔMoimd4, and ΔMopdeH mutants and found that that cAMP levels in the ΔMoimd4 mutant was 2.0-fold higher than Guy11 but 0.5-fold lower when compared with that in the ΔMopdeH mutant (Fig. S8C), indicating that MoImd4 promotes the phosphodiesterase activity of MoPdeH.
T268, R269, D380, G382, R458, Y459, G403, Y428 of MoImd4 are important in promoting MoPdeH phosphodiesterase activities
As some of the point mutation mutants did not restore defects in vegetative growth or pathogenicity of the ΔMoimd4 mutant by exogenous XMP (Fig. 6E, F) and we have the evidence that MoImd4 promotes MoPdeH enzymatic activities, we hypothesized that some of these residues may have a role in interactions with MoPdeH. We used GST-Pull down assays again to find out that T268A, R269A, D380A, G382A, G403A, Y428A, R458A, Y459A of MoImd4 attenuated the interaction between MoImd4 and MoPdeH (Fig. 7A). We also measured the enzyme activities of MoPdeH in the presence of MoImd4 with various point mutations. MoImd4 promoted the enzyme activity of MoPdeH, while S345AC347A, ΔCBS2 and ΔCBS1CBS2 had no effects and T268AR269A, D380AG382A, G403A, Y428A, R458AY459A had only certain effects (Fig. 7B). We next detected the cAMP levels of Guy11, the ΔMoimd4 mutant, various point mutation mutants of MoIMD4, CBS domain deletion mutants, and the complement strains in vivo. Intriguingly, cAMP levels of the S345AC347A mutant and three CBS domain deletion mutants were similar to Guy11 and the complement strains. While the cAMP levels of T268AR269A and R458AY459A mutants were significantly different than the ΔMoimd4 mutant and were higher than Guy11 and the complement strains. Meanwhile, cAMP levels of D380AG382A, G403A and Y428A mutants were close to the ΔMoimd4 mutant and were about 2.0-fold higher than Guy11 (Fig. 7C). Taken together, these results provided direct evidence that T268, R269, D380, G382, G403, Y428, R458 and Y459 of MoImd4 are important in promoting the enzyme activity of MoPdeH.
Fig. 7.
T268, R269, D380, G382, R458, Y459, G403, Y428 of MoImd4 are important for promoting MoPdeH enzymatic activities. (A) GST pull-down assays for interactions between GST-MoPdeH and His-MoImd4, His-T268AR269A, His-D380AG382A, His-R458AY459A, His-G403A, and His-Y428A alleles of MoImd4. Equal MoPdeH-GST or GST protein were incubated with Glutathione Sepharose beads for 3 h at 4˚C prior to mixing with His infusion protein lysates for another 3 h at 4˚C. Equal His-tag infusion proteins from input were used as controls. Eluted proteins were detected by Western-blot analysis with anti-His and anti-GST antibodies. The number represents the intensity of eluted proteins detected by the anti-His antibody (Top-panel: elution protein; down-panel: elution protein after the same dilution). The intensity of the elutions from wild-type MoImd4 was set to 1. (B) Purified His-fusion expression proteins affecting the enzyme activities of MoPdeH. Equal amounts of His fusion proteins and GST-MoPdeH were mixed for measuring enzymatic activities, respectively. Fluorescence was read by a 10-min kinetic reaction at excitation 420 nm and emission 450 nm. Experiments were repeated three times with similar results. The error bars indicate standard deviations of three replicates. Letters indicate statistically significant differences (Duncan’s new multiple range test, p<0.01). (C) Intracellular cAMP level assay of indicated strains at the mycelial stage. Experiments were repeated three times with similar results. The error bars represent ±SD of three replicates. Letters indicate statistically significant differences (Duncan’s new multiple range test, p<0.01).
MoPdeH and MoImd4 show mutual regulations of their enzymatic activities.
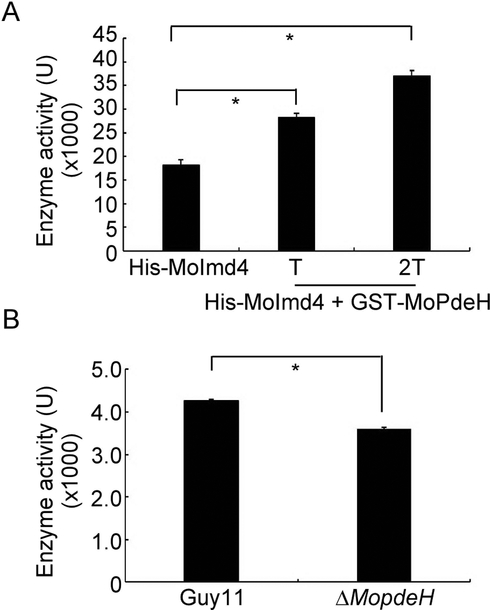
MoImd4 interacts with MoPdeH to promote its phosphodiesterase activity, however, the function of MoPdeH on the interaction with MoImd4 remained unclear. We purified the protein of GST-MoPdeH and His-MoImd4 in vitro and tested the enzymatic activity of MoImd4. The results showed that the MoImd4 activity without treatment was ~18 kU, but, it was continuously increasing when the purified exogenous MoPdeH protein was added (Fig. 8A). We also extracted total proteins from the wild type Guy11 and ΔMopdeH strains as the crude enzyme of MoImd4, and added them into the enzymatic reaction system. The result showed that the enzyme activity of MoImd4 in the Guy11 strain was significantly higher when compared with that in the ΔMopdeH mutant, indicating that MoPdeH also promotes the MoImd4 enzymatic activity.
Fig. 8.
MoPdeH promotes the enzymatic activity of MoImd4. (A) Enzymatic activity of purified His-MoImd4 proteins, with 1 T and 2 T of the purified GST-MoPdeH protein. The production of XMP was monitored by the absorbance at 290 nm. “T” represents the total quantity of the His-MoImd4 protein. Experiments were repeated three times with similar results. The error bars indicate standard deviation of three replicates. Asterisk indicates statistically significant differences (Duncan’s new multiple range test, p<0.01). (B) In vivo, we extracted the total protein from the wild type Guy11 and ΔMopdeH as the crude enzyme of MoImd4, then added them into the enzymatic reaction system for 5 min, the production of XMP was monitored by the absorbance at 290 nm. Experiments were repeated three times with similar results. The error bars indicate standard deviation of three replicates. Asterisk indicates statistically significant differences (Duncan’s new multiple range test, p<0.01).
Discussion
In eukaryotic cells, G-protein/cyclic AMP (cAMP)-dependent signaling pathway is involved in sensing of extracellular signals and integrating them into intrinsic pathways (Malbon, 2005). cAMP acts as a second messenger to transmit extracellular hormones and nutrients into the intracellular environment where it activates downstream targets (Daniel et al., 1998). In the rice blast fungus M. oryzae, high-affinity phosphodiesterase MoPdeH exhibits various regulatory functions in hyphal autolysis, spore morphology, cell wall integrity, and pathogenicity, as well as surface signal recognition (Ramanujam & Naqvi, 2010, Zhang et al., 2011a, Yang et al., 2017). To understand the underlying mechanisms, we searched for proteins that interact with MoPdeH and identified MoImd4. We characterized MoImd4 as an IMP dehydrogenase in the purine synthetic pathway and we found that MoImd4 functions in growth and pathogenicity of the fungus. We also revealed that MoImd4 interacted with MoPdeH to impact the development and pathogenicity of M. oryzae collectively.
MoIMD4 gene disruption leads to formation of atypical and restricted lesions in rice leaves, which is interesting, as IMPDH is not known to directly affect fungal virulence. We speculated that it could involve the following mechanisms: 1) Loss of MoImd4 severely impacts fungal growth and fitness levels; 2) Loss of MoImd4 may induce the host-derived defense that restricts infection. Generally speaking, the first layer of broad-spectrum defense against any pathogens is the conserved Pathogen-Associated Molecular Patterns (PAMPs) in the host that trigger PAMPs-triggered immunity (PTI). However, 3,3’-diaminobenzidine (DAB) staining of rice sheaths following infection showed no significant difference in reactive oxygen species (ROS) levels between the ΔMoimd4 mutant and wild type (Fig. S9). As MoImd4 is located in the cytoplasm than secreted into the host cell during infection and it regulates the conversion of IMP to XMP affecting the de novo purine metabolic pathway (Fig. S10), we considered that MoImd4 may function intracellularly to affect pathogenicity of M. oryzae. Given that the defect of growth and invasion in rice cells of the ΔMoimd4 mutant could be suppressed by addition of exogenous XMP, it is plausible that the uptake of XMP might be critical for M. oryzae. In accordance with this reasoning, virulence defect exhibited by the mutants with amino acid variants or purines could be supplemented by adding exogenous amino acids and purine relevance, including ILV2/6, LYS2/20, STR3/MET6/MET13, CPA2 and ADE1 (Rao et al., 2013, Chen et al., 2014, Wilson et al., 2012, Yan et al., 2013, Zhang et al., 2014, Liu et al., 2016a, Fernandez et al., 2013, Saint-Macary et al., 2015).
Tandem CBS subdomains of IMPDH exist extensively in eukaryotic organisms. In this study, we discovered that two CBS domains had overlap functions in development and tandem CBS deletion mutants caused defects in growth, conidial formation, and pathogenicity. The enzymatic activity of the MoImd4 ΔCBS1CBS2 allele and XMP contents were statistically reduced but the levels of GTP were also sharply reduced. This is consistent with the study of E. coli guaBΔCBS mutant (GuaB homologous MoImd4) (Pimkin & Markham, 2008). The three-dimensional protein structure of GuaB IMPDH was a tetramer with tandem CBS domains in each monomer (Pimkin & Markham, 2008). Homology modeling revealed that the tandem CBS domains were not present in the protein model owing to disorder in M. oryzae, similar to C. neoformans, and P. aeruginosa (Morrow et al., 2012, Rao et al., 2013). However, in A. gossypii, the regulatory CBS pair domains of IMPDH form octamers with GDP and GTP, resulting in decreased affinity between the catalytic domain and substrate IMP (Buey et al., 2015b, Buey et al., 2017). In consequence, tandem CBS domains take part in de novo purine metabolism and play an essential role in GTP level control. Nevertheless, it was debatable that the content of GTP and the location of CBS subdomains in protein structures were different in M. oryzae than from other organisms.
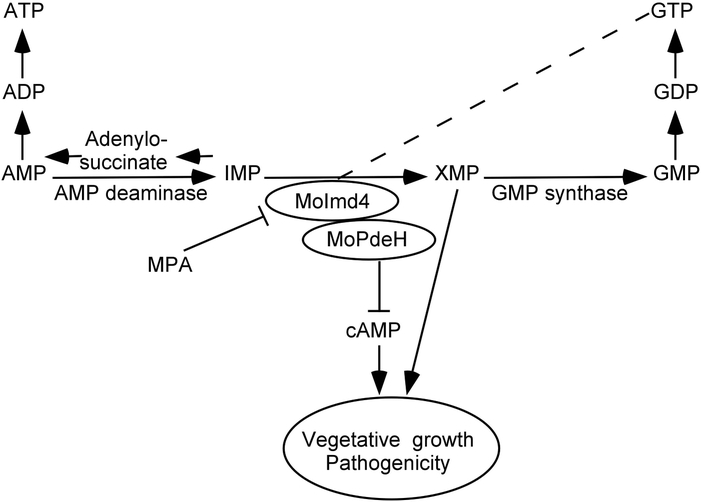
Our results revealed for the first time that MoPdeH interacts with MoImd4. In our study, sequence alignments of MoImd4 revealed several conserved amino acid sites whose mutations exhibited three different situations: 1) S345AC347A mutation affects the enzymatic activity of MoImd4, but not the interaction between MoImd4 and MoPdeH, and the phosphodiesterase activity of MoPdeH; yet the phenotypic defect is restored with exogenous XMP. This is similar to the recovery of the ΔMoimd4 mutant phenotype with XMP; 2) T268AR269A mutation affects the enzymatic activity of both MoImd4 and MoPdeH and the intensity of interaction between them, and exogenous XMP partially rescues the defects in growth and pathogenicity; 3) D380A, G382A, R458A, Y459A, G403A and Y428A mutations all affect enzymatic activities of MoImd4 and MoPdeH and the intensity of interaction between them, but exogenous XMP has no effects on their phenotypes. We thought that the reasons for this might derive from the ways of interaction between MoPdeH and MoImd4. We found that treated with MPA or inactivated the MPA-binding sites of MoImd4 showed no differences in the interaction between MoImd4 and MoPdeH (Fig. S11A, B). Further, inactivation MPA-binding sites didn’t affect the phosphodiesterase activity of MoPdeH which is similar to that of S345AC347A mutation (Fig. S11C). Based on these results, we speculated that when certain sites are changed, such as T268, R269, D380, G382, R458, Y459, G403, and Y428, they would interfere with the interaction between MoPdeH and MoImd4 attenuating the MoPdeH enzymatic activity, indicating that the interaction between MoImd4 and MoPdeH is important for the enzymatic activity of MoPdeH. Based on this reasoning, we proposed that MoImd4 mediates a crosstalk between the cAMP pathway and the de novo purine biosynthesis pathway that medicated the vegetative and virulence of M. oryzae collectively (Fig. 9).
Fig. 9.
A proposed model for crosstalk between the de novo purine metabolic pathway and the intracellular cAMP-signaling pathway in M. oryzae. Evidence supports a novel crosstalk pathway between de novo purine metabolism and cAMP signaling through MoPdeH and XMP regulating synergistically the growth and virulence of M. oryzae.
Why purine biosynthesis is linked to cAMP regulation? Previous work demonstrated that when the cytosolic pH value is maintained around neutral, adenylate cyclase is activated by increasing the affinity of the enzyme for ATP that induces cAMP accumulation (Purwin et al., 1986, Orij et al., 2011). In eukaryotic cells, ATP/GTP is involved in various cellular biological processes, including the signaling transduction, gene transcription, and cellular respiration (Pathak et al., 2013, Ganapathy-Kanniappan & Geschwind, 2013, Koopman et al., 2012). The levels of ATP/GTP are maintained by the purine nucleotide pool sizes, including transcriptional control and enzyme-level regulation of purine biosynthetic enzymes. Enzyme-level regulation works as the “first line of defense” to rapidly balance specific fluxes in the purine biosynthesis (Petersen, 1999; Yamaoka et al., 2001; Zalkin and Nygaard, 1996). Here, we found that MoImd4, which functions as the rate-limiting and first committed step in the de novo biosynthesis of GTP, controls not only the synthesis of ATP/GTP but also the balance between them in vivo. Thus, we speculated that the synthesis and hydrolysis of intracellular cAMP might need the energy that comes from the de novo purine biosynthesis system.
Mycophenolic acid (MPA) is a specific IMPDH inhibitor that perturbs the de novo purine metabolic pathway (Umejiego et al., 2004, Kohler et al., 2005, Johnson et al., 2013, Wei et al., 2016). Treatment with MPA attenuated the growth and virulence of M. oryzae and further assays confirmed that the inhibition constant was significantly reduced following mutations of MoImd4 in D290, G340, G342, M431, G432, and Q470 (Table 4). However, since IMPDH mediated de novo purine biosynthesis is highly conserved, questions emerge whether IMPDH gene(s) in rice would also be affected by MPA. Blast alignments found there was 42% amino acid sequence identity between MoImd4 and LOC_Os03g56800.1 encoding rice IMPDH (OsIMPDH). Homology modeling analysis showed that OsIMPDH could also form a tetramer (Fig. S12). However, differences, including kinetic profiles, were found between these two proteins (Table 4). Thus, while the possibility exists, MPA may exert more influences on MoImd4 than OsIMPDH. This is consistent with studies of Candida albicans and C. neoformans in which Km for both IMP and NAD was substantially different from human forms, despite sharing high amino acid sequence identities (Kohler et al., 2005, Morrow et al., 2012). Our additional studies showed that MPA didn’t affect appressorium formation or host invasion (Table S2).
Table 4.
Kinetic parameters of IMPDHs from M. oryzae and rice.
| parameter | Km(IMP)(μM) | Km(NAD)(μM) | Kii(NAD)(μM) |
|---|---|---|---|
| MoImd4 | 216.2 ± 56.4 | 857.8 ± 186.9 | 3.3 |
| D290A | 142.7 ± 15.3 | 304.9 ± 143.5 | 1.4 |
| G340AG342A | 91.8 ± 15.7 | 156.1 ± 76.2 | 1.7 |
| M431AG432A | 79.4 ± 12.2 | 293.8 ± 86.1 | 2.2 |
| Q470A | 151.9 ± 21.9 | 63.1 ± 7.8 | 1.1 |
| OsIMPDH | 76.5 ± 23.2 | 193.7 ± 98.2 | 0.4 |
Steady-state and inhibition constants for IMPDHs from M. oryzae and the rice.
In summary, our results revealed that MoImd4 plays a critical role in growth, development, and virulence of M. oryzae by mediating the de novo purine biosynthesis pathway. MoImd4 promotes the enzymatic activity of MoPdeH to influence intracellular cAMP levels. Our results provided new insight into how cAMP signaling and purine metabolism collectively regulate the growth and pathogenicity of M. oryzae.
Experimental Procedures
Strains and cultures
Guy11 was used as the wild type in this work. All strains were cultured on CM and MM media at 28°C in the darkness. Mycelia were harvested from strains grown in liquid CM for 48 h for DNA and RNA extraction. Protoplasts were prepared and transformed as described previously (Sweigard et al., 1992). Transformants were selected on TB3 medium (3 g of yeast extract, 3 g of casamino acids, 200 g of sucrose, and 7.5 g of agar in 1 L of distilled water) with 300 μg/ml hygromycin B (Roche) or 200 μg/ml zeocin (Invitrogen). For conidial formation, mycelial blocks were incubated -on SDC media at 28°C for 7 days in the dark followed by 3 day’s continuous illumination under the fluorescent light.
Targeted gene deletion assay
MoIMD4 deletion mutant was generated using the standard one-step gene replacement strategy. The gene deletion vector was constructed by PCR amplification using two 1.0 kb of sequences flanking the targeted gene and primer pairs (Table S3); Resulting PCR products were digested with restriction endonucleases and ligated with the hygromycin resistance cassette (HPH) released from pCX62. Candidate mutants were first screened by PCR and later confirmed by Southern blot analysis.
Generation of the complement, the point mutation and the CBS deficient GFP or His fusion constructs
In order to generate GFP fusion constructs, target genes containing the native promoter of MoIMD4 were amplified by PCR (Table S3) and inserted into pYF11 plasmid (bleomycin resistance) using the yeast gap repair approach (Bruno et al., 2004). The plasmids were verified by sequencing prior to being introduced into the protoplasts of the ΔMoimd4 mutant. For D290, G340, G342, M431, G432, Q470, T268, R269, S345, C347, D380, G382, G403, Y428, R458, Y459 point mutation vector construction, we introduced Alanine to specific inactivate the sites and imminent adjacent residues to generate double point mutations, for example, G340AG342A, M431AG432A, T268AR269A, S345AC347A, and R458AY459A. CBS deletion mutants were generated using the similar approach as above described. The zeocin-resistant transformants were confirmed by the presence of GFP signals and by Western blot analysis. To generate His-fusion constructs, cDNA was amplified by PCR using Phanta® Super-Fidelity DNA Polymerase (Vazyme Biotech Co., Ltd) and cloned into pET-32a vector for expression in E. coli (BL21-CodonPlus).
Vegetative growth assays
For vegetative growth, mycelium blocks were cut from the edge of 5-day-old cultures and placed onto media in the darkness at 28˚C for 7 days (Zhang et al., 2010) prior to evaluation. To test the effect of exogenous XMP, strains were cultured on MM supplemented with XMP at the final concentrations of 1, 2.5, and 5 mM. For inhibitor assays, strains were inoculated on MM treated with 1, 5, and 10 μg/ml MPA in the darkness at 28˚C for 7 days.
Conidial and appressorial formation, turgor pressure, pathogenicity, and rice sheath penetration assays
Conidia were induced by culturing on SDC media in darkness at 28°C for 7 days followed by a constant illumination for 3 days at room temperature. Conidia were collected and 1×105 spores/ml conidia were used for appressorium formation. More than 200 appressoria were counted for each strain after 24 hpi and the experiments were repeated at least three times. For appressorium turgor assays, 1-4 M glycerol solution were used to measure the collapse of appressoria as described (Liu et al., 2016b).
For spraying assays, conidia were collected and adjusted to 5×104 spores/ml in a 0.2% (w/v) gelatin solution. Eleven-day rice seedlings (Oryza sativa cv CO-39) were sprayed and incubated in a chamber at 28˚C with 90% humidity in the dark for the first 24 h, followed by a 12/12 h light/dark cycle for 7 days. The rice sheath penetration assays were carried out as described previously (Liu et al., 2016b). For XMP or MPA, they were added to the conidia suspension and hyphal expansions were assessed after 48 hpi.
Pull-down assays
Glutathione-S-transferase (GST), GST-MoPdeH, His-MoImd4, His-T268AR269A, His-D380AG382A, His-R458AY459A, His-G403A, His-Y428A were expressed in E. coli BL21-CodonPlus (DE3) cells (Stratagene, Cedar Creek, Texas, USA) and proteins were induced as described previously (Yang et al., 2017, Yin et al., 2016). Supernatants of lysed cells of purified GST-MoPdeH- or GST proteins were incubated with 30 μl of Glutathione Sepharose™ 4B (GE Healthcare Lot 10148479) incubated at 4°C for 3 h. Then, centrifugation (500 rcf, 2 min). Lysed His-infusion proteins were re-incubated with Glutathione Sepharose™ beads at 4°C for another 3 h. Finally, the beads were washed with washing buffer [20 mM Tris (pH 7.5), 0.25 mM NaCl, 2 mM EDTA, 2 mM EGTA] for five times prior to elution with elution buffer [20 mM Tris (pH 7.5), 0.25 mM NaCl, 2 mM EDTA, 2 mM EGTA, 1 mM reduced glutathione, PH 8.0]. Eluted proteins were analyzed by Western blotting with anti-His and anti-GST antibodies.
Measurements of IMP dehydrogenase in vitro activities, Km constant and Kii constant
Proteins of His, His-MoImd4, His-T268AR269A, His-S345AC347A, His-D380AG382A, His-R458AY459A, His-G403A, His-Y428A, His-D290A, His-G340AG342A, His-M431AG432A, His-Q470A, His-ΔCBS1, His-ΔCBS2 and His-ΔCBS1CBS2, GST-MoPdeH were expressed and purified with Ni-NTA Agarose (QLAGEN Lot No.151047819) and Glutathione Sepharose™ 4B (GE Healthcare Lot 10148479). Enzyme activities were estimated in 100 mM Tris-HCl, pH 8.1, 10 mM KCl, 0.1 mM DTT in the presence of 250 μM NAD (Solarbio Lot.No.427D0213) and 500 μM IMP (absin abs42019840) and purified protein of His, His-MoImd4 with or without equal protein of GST-MoPdeH for 5 min. The production of XMP was monitored by the absorbance at 290 nm (Pimkin & Markham, 2008) in a 96 well cell culture plate (CELLTER CS016-0096). A standard curve of absorbance vs concentration was established using commercial xanthosine-5’-monophosphate disodium (Alading X113495). The amount of purified His-fusion protein was kept the same. Protein concentrations were estimated using standard Bradford protein assay Kit (Beyotime P0006). For Km value estimation, IMP and NAD concentrations were adjusted over a range from 100 μM to 8,000 μM, with 250 μM NAD and 500 μM IMP, respectively (Morrow et al., 2012). When one substrate remains the same and the other changes over different concentrations, the reaction velocity equals to the linear slope of XMP increase by a 10-min kinetic reaction (Green et al., 2012). Nonlinear fitting of the data to the Michaelis-Menton equation (Equation 1) and the uncompetitive substrate inhibition equation (Equation 2) were used Origin 8.0:
| (1) |
| (2) |
Where V is the initial velocity; Vmax is the maximum velocity; Km is the Michaelis constant; [S] is substrate concentration; [I] is inhibitor concentration; Kii is inhibitor constant.
Intracellular XMP, GTP and cAMP measurements
Indicated strains were cultured on CM media and a certain amount of mycelium was grown in liquid CM for two days. Then, the mycelium was pressed dry and quickly grinded into powder with liquid nitrogen before lyophilizing for 24 h. Each mycelium sample was treated following previously established procedures (Liu et al., 2016b). Samples were quantified by HPLC (High Performance Liquid Chromatography) analysis as described previously (Liu et al., 2016b). The peak flowing out of the standard of XMP (Alading X113495) was at ~6.4 min, the standard of GTP (Santa Cruz sc-203062) was at ~2.6 min, and the standard of cAMP (Meilune P/N MB3159) was at ~13.8 min. Calculation of XMP, GTP and cAMP were done according the method previously reported (Liu et al., 2016b).
Effects of MoImd4 and point mutant proteins on in vitro MoPdeH phosphodiesterase activitiy
GST-MoPdeH, His-MoImd4, His-T268AR269A, His-D380AG382A, His-R458AY459A, His-G403A, His-Y428A, His-D290A, His-G340AG342A, His-M431AG432A, His-Q470A and His were induced and purified as described before (Yang et al., 2017). The phosphodiesterase activity was detected by fluorescence intensity. The MDCC fluorophore coupled with phosphate and the fluorescence of Phosphate Sensor increases approximately 6- to 8-fold which can be measured in real time (Brune et al., 1994). The reaction system was: 5 cM cAMP (Meilune P/N MB3159), purified GST-MoPdeH with or without equal purified His-fusion proteins, separately, 10 mU/mL alkaline phosphatase (Calbiochem P/N 524545) were incubated in PDE enzymatic reaction buffer for 60 min at room temperature (PDE enzymatic reaction buffer: 50 mM Tris pH 7.6, 100 mM NaCl, 10 mM MgCl2, 0.01% Triton® X-100, and 0.5 mM DTT), then added 0.5 μM Phosphate Sensor (Invitrogen P/N PV4406) in Phosphate Sensor detection buffer to each well (Phosphate Sensor detection buffer: 20 mM Tris pH 7.6 and 0.05% Triton® X-100). The plate was mixed and read by a 10-min kinetic reaction at excitation 420 nm and emission 450 nm. Positive and negative controls were included as previously described (Yang et al., 2017). The experiments were repeated three times and each experiment had three replicates.
Homology modeling assay
Amino acid sequence of MoImd4 and OsIMPDH were submitted to the online platform SWISS-MODEL (https://www.swissmodel.expasy.org/interactive/xBVTUG/templates/) to predict protein structures and the structures were analyzed by PyMOL.
Supplementary Material
Fig. S1 Southern blot analysis of the MoIMD4 deletion mutant. (A) The strategy of MoIMD4 gene replacement. Fragment of the MoIMD4 coding region were replaced with hygromycin phosphotransferase (HPH) fragment. (B) Southern blot analysis of the MoIMD4 knockout mutant with specific probe (probe 1) and HPH probe (probe 2).
Fig. S2 Phylogenetic analysis of MoImd4 and other Imd proteins by CLUSTAL_W and MEGA 6 programs. Species names and accession numbers are as followings: M. oryzae (Magnaporthe oryzae XP_003716182.1), M. robertsii (Metarhizium robertsii XP_007816906.1), C. purpurea (Claviceps purpurea CCE29804.1), T. reesei (Trichoderma reesei XP_006966877.1), C. orbiculare (Colletotrichum orbiculare ENH80323.1), G. tritici (Gaeumannomyces tritici XP_009224448.1), S. cerevisiae (Saccharomyces cerevisiae NP_013536.3), C. neoformans (Cryptococcus neoformans XP_012046718.1), H. sapiens (Homo sapiens NP_000875.2), P. aeruginosa (Pseudomonas aeruginosa WP_070336446.1), B. anthracis (Bacillus anthracis WP_047401536.1), S. pyogenes (Streptococcus pyogenes WP_002991454), T. foetus (Tritrichomonas foetus OHT09030.1).
Fig. S3 MoImd4 is involved in vegetative growth. (A) The wild type Guy11, ΔMoimd4 and the complement strains were cultured on CM, MM, SDC, and OM media at 28˚C in the darkness for 7 days. (B) Mycelial pellets in liquid CM medium. Mycelia of Guy11, ΔMoimd4 and the complement strains were inoculated in liquid CM medium with shaking condition (160 rpm) for 48 h at 28°C. Experiments were performed three times with similar results.
Fig. S4 MoImd4 is important for xanthosine monophosphate (XMP) biosynthesis. (A) Intracellular levels of XMP in Guy11, ΔMoimd4, and the complement strains estimated by HPLC. Experiments were repeated three times with the similar results. Error bars represent the standard deviations and asterisk denotes statistical significances (Duncan’s new multiple range test, p<0.01). (B) Statistical analysis of colony radius on MM media with or without XMP. Experiments were repeated three times with similar results. Error bars represent the standard deviations and asterisk denotes statistical significances (Duncan’s new multiple range test, p<0.01). (C) Statistical analysis of colony radius on MM medium with or without MPA. Error bars represent the standard deviations and asterisk denotes statistical significances (Duncan’s new multiple range test, p<0.01).
Fig. S5 The structure of MoImd4 in binding with MPA and the binding sites of MPA. (A) Predicted the structure of MoImd4 was a tetramer binding with MPA. Green represents the three-dimensional structure of MoImd4; Red represents inhibitor MPA. (B) Enlarged drawing of MPA was added in one of the ligand on the structure of MoImd4. Around the MPA, different-color curves are predicted the binding sites of MPA, includes D290, G340, G342, M431, G432 and Q470.
Fig. S6 Western blot analyzes the GFP expression of the point mutation and CBS deficient mutants. Total proteins of the indicated strains were extracted from 2-day old mycelia in liquid CM media. The anti-GFP was used to analyze by Western blotting.
Fig. S7 Homology modeling of key sites and point mutants of MoImd4. The target protein sequence of MoImd4 was predicted by SWISS-MODEL and point mutants were analyzed by PyMOL. Red curves represent key sites, gray curves were after point mutants, red stars stand for the sites of red curves and gray curves.
Fig. S8 MoImd4 promotes MoPdeH phosphodiesterase activities. (A) In vitro GST pull-down assays for MoImd4 and MoPdeH. The expression of GST-MoPdeH and His-MoImd4 was induced with 0.1 mM IPTG. GST-MoPdeH or GST were incubated with Glutathione Sepharose beads for 3 h at 4°C, precipitated, and co-incubated with lysates containing His-MoImd4 for additional 3 h at 4°C. Eluted proteins were detected by Western-blot analysis with anti-His and anti-GST antibodies. (B) Phosphodiesterase activities of purified GST-MoPdeH proteins, with 0.1 T, 0.2 T, and 1 T of the purified His-MoImd4 protein. Fluorescence was detected by a 10-min kinetic reaction at excitation 420 nm and emission 450 nm. “T” represents total quantity of the GST-MoPdeH protein. Experiments were repeated three times with similar results. The error bars indicate standard deviation of three replicates. Asterisk indicates statistically significant differences (Duncan’s new multiple range test, p<0.01). (C) In vivo cAMP levels measurements of Guy11, ΔMoimd4, MoImd4, ΔMopdeH, and MoPdeH at the stage of mycelia. Experiments were repeated three times with similar results. The error bars represent ±SD of three replicates. Asterisk indicates statistically significant differences (Duncan’s new multiple range test, p<0.01).
Fig. S9 MoImd4 didn’t participate in ROS scavenging. (A) Conidial suspensions of Guy11, ΔMoimd4 mutant, and the complement strains were injected into separated rice sheaths. After 24 hpi, DAB was used to dye the sheaths for 8 h. (B) Percentages of cells with IH were dyed by DAB. Means were calculated from three independent replicates. There were little significant differences between the strains. Bar = 10 μm.
Fig. S10 Locations of MoImd4-GFP in the ΔMoimd4 mutant at the stages of sporulation and infection.
Fig. S11 Interaction between MoPdeH and MoImd4 treated with MPA. (A) Lysed cells of purified GST-MoPdeH or GST proteins were incubated with Glutathione SepharoseTM 4B at 4°C for 3 h and centrifugation. Then, lysed cells of purified MoImd4-His with 1, 5, and 10 μg/ml MPA were re-incubated with GST beads at 4°C for another 3 h and then eluted. Equal His-tag infusion proteins from input were used as controls. Eluted proteins were detected by Western-blot analysis with anti-His and anti-GST antibodies. The number represents the intensity of eluted proteins detected by the anti-His antibody. The intensity of the elutions from wild-type MoImd4 was set to 1. (B) Lysed cells of purified GST-MoPdeH or GST proteins were incubated with Glutathione SepharoseTM 4B at 4°C for 3 h and centrifugation. Then, lysed cells of inactivation MPA-binding sites proteins (D290A, G340AG342A, M431AG432A and Q470A) were re-incubated with GST beads at 4°C for another 3 h and then eluted. Equal His-tag infusion proteins from input were used as controls. Eluted proteins were detected by Western-blot analysis with anti-His and anti-GST antibodies. The number represents the intensity of eluted proteins detected by the anti-His antibody. The intensity of the elutions from wild-type MoImd4 was set to 1. Protein of S345AC347A was as a control that mutation these sites didn’t affect the interaction between MoPdeH and MoImd4. (C) Purified His-fusion expression proteins affecting the enzyme activities of MoPdeH. Equal amounts of His fusion proteins and GST-MoPdeH were mixed for measuring enzymatic activities, respectively. Fluorescence was read by a 10-min kinetic reaction at excitation 420 nm and emission 450 nm. Experiments were repeated three times with similar results. The error bars indicate standard deviations of three replicates. Asterisk indicates statistically significant differences (Duncan’s new multiple range test, p<0.01).
Fig. S12 The predicted three-dimensional structure of OsIMPDH.
Table S1 Potential interacting protein list of MoPdeH identified by yeast two-hybrid screen
Table S2 Appressorium formation on the hydrophobic surface of the wild type strain in the presence of MPA.
Table S3 Primers used in this study.
Acknowledgments
This research was supported by the key program of Natural Science Foundation of China (Grant No: 31530063, ZZ), the Fundamental Research Funds for the Central Universities (Grant No: KYT201805), Natural Science Foundation of China (Grant No: 31470248, XZ), and Innovation Team Program for Jiangsu Universities (2017). The Wang lab research was supported by US NIH grant AI121451. We thank Baodian Guo, Yeqiang Xia, Jie Huang and Shuaishuai Wang for their technical expertise.
References
- Arnold K, Bordoli L, Kopp J and Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics, 22, 195–201. [DOI] [PubMed] [Google Scholar]
- Bateman A (1997) The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci, 22, 12–13. [DOI] [PubMed] [Google Scholar]
- Baykov AA, Tuominen HK and Lahti R (2011) The CBS domain: a protein module with an emerging prominent role in regulation. ACS Chem Biol, 6, 1156–1163. [DOI] [PubMed] [Google Scholar]
- Benkert P, Biasini M and Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics, 27, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L and Schwede T (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res, 42, W252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune M, Hunter JL, Corrie JE and Webb MR (1994) Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry, 33, 8262–8271. [DOI] [PubMed] [Google Scholar]
- Bruno KS, Tenjo F, Li L, Hamer JE and Xu JR (2004) Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot Cell, 3, 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buey RM, Fernandez-Justel D and Marcos-Alcalde I (2017) A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases. Sci Rep, 7, 2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buey RM, Ledesma-Amaro R, Balsera M, de Pereda JM and Revuelta JL (2015a) Increased riboflavin production by manipulation of inosine 5’-monophosphate dehydrogenase in Ashbya gossypii. App Microbiol Biotechnol, 99, 9577–9589. [DOI] [PubMed] [Google Scholar]
- Buey RM, Ledesma-Amaro R, Velazquez-Campoy A, Balsera M, Chagoyen M, de Pereda JM, and Revuelta JL (2015b) Guanine nucleotide binding to the Bateman domain mediates the allosteric inhibition of eukaryotic IMP dehydrogenases. Nat Commun, 6, 8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuis AG, Paolo Rizzardi G, D’Agostino C, Attinger A, Knabenhans C, Fleury S, Ach-Orbea H and Pantaleo G (2000) Effects of mycophenolic acid on human immunodeficiency virus infection in vitro and in vivo. Nat Med, 6, 762–768. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhai S, Zhang H, Zuo R, Wang J, Guo M, Zheng X, Wang P and Zhang Z (2014) Shared and distinct functions of two Gti1/Pac2 family proteins in growth, morphogenesis and pathogenicity of Magnaporthe oryzae. Environ Microbiol, 16, 788–801. [DOI] [PubMed] [Google Scholar]
- Collart FR and Huberman E (1988) Cloning and sequence analysis of the human and Chinese hamster inosine-5’-monophosphate dehydrogenase cDNAs. J Biol Chem, 263, 15769–15772. [PubMed] [Google Scholar]
- Daniel PB, Walker WH and Habener JF (1998) Cyclic AMP signaling and gene regulation. Annu Rev Nutr, 18, 353–383. [DOI] [PubMed] [Google Scholar]
- Elion GB (1989) Nobel Lecture. The purine path to chemotherapy. Biosci Rep, 9, 509–529. [DOI] [PubMed] [Google Scholar]
- Fernandez J, Yang KT, Cornwell KM, Wright JD and Wilson RA (2013) Growth in rice cells requires de novo purine biosynthesis by the blast fungus Magnaporthe oryzae. Sci Rep, 3, 2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathy-Kanniappan S and Geschwind JF (2013) Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer, 12, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollapalli DR, Macpherson IS, Liechti G, Gorla SK, Goldberg JB and Hedstrom L (2010) Structural determinants of inhibitor selectivity in prokaryotic IMP dehydrogenases. Chem Biol, 17, 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green LS, Chun LE, Patton AK, Sun X, Rosenthal GJ and Richards JP (2012) Mechanism of inhibition for N6022, a first-in-class drug targeting S-nitrosoglutathione reductase. Biochemistry, 51, 2157–2168. [DOI] [PubMed] [Google Scholar]
- Hyle JW, Shaw RJ and Reines D (2003) Functional distinctions between IMP dehydrogenase genes in providing mycophenolate resistance and guanine prototrophy to yeast. J Biol Chem, 278, 28470–28478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignoul S and Eggermont J (2005) CBS domains: structure, function, and pathology in human proteins. Am J Physiol Cell Physiol, 289, C1369–1378. [DOI] [PubMed] [Google Scholar]
- Jackson RC, Weber G and Morris HP (1975) IMP dehydrogenase, an enzyme linked with proliferation and malignancy. Nature, 256, 331–333. [DOI] [PubMed] [Google Scholar]
- Johnson CR, Gorla SK, Kavitha M, Zhang M, Liu X, Striepen B, Mead JR, Cuny GD and Hedstrom L (2013) Phthalazinone inhibitors of inosine-5’-monophosphate dehydrogenase from Cryptosporidium parvum. Bioorg Med Chem Lett, 23, 1004–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler GA, Gong X, Bentink S, Theiss S, Pagani GM, Agabian N, and Hedstrom L (2005) The functional basis of mycophenolic acid resistance in Candida albicans IMP dehydrogenase. J Biol Chem, 280, 11295–11302. [DOI] [PubMed] [Google Scholar]
- Koopman WJ, Willems PH and Smeitink JA (2012) Monogenic mitochondrial disorders. New Engl J Med, 366, 1132–1141. [DOI] [PubMed] [Google Scholar]
- Lee YH and Dean RA (1993) cAMP regulates infection structure formation in the plant pathogenic fungus Magnaporthe grisea. Plant Cell, 5, 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Cai Y, Zhang X, Zhang H, Zheng X and Zhang Z (2016a) Carbamoyl phosphate synthetase subunit MoCpa2 affects development and pathogenicity by modulating arginine biosynthesis in Magnaporthe oryzae. Front Microbiol, 7, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Qian B, Gao C, Huang S, Cai Y, Zhang H, Zheng X, Wang P and Zhang Z (2016b) The putative protein phosphatase MoYvh1 functions upstream of MoPdeH to regulate the development and pathogenicity in Magnaporthe oryzae. Mol Plant Microbe Interact, 29, 496–507. [DOI] [PubMed] [Google Scholar]
- Malbon CC (2005) G proteins in development. Nat Rev Mol Cell Biol, 6, 689–701. [DOI] [PubMed] [Google Scholar]
- McGrew DA and Hedstrom L (2012) Towards a pathological mechanism for IMPDH1-linked retinitis pigmentosa. Adv Exp Med Biol, 723, 539–545. [DOI] [PubMed] [Google Scholar]
- Morrow CA, Valkov E, Stamp A, Chow EW, Lee IR, Wronski A, Williams SJ, Hill JM, Djordjevic JT, Kappler U, Kobe B and Fraser JA (2012) De novo GTP biosynthesis is critical for virulence of the fungal pathogen Cryptococcus neoformans. PLoS Pathog, 8, e1002957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orij R, Brul S and Smits GJ (2011) Intracellular pH is a tightly controlled signal in yeast. Biochim Biophys Acta, 1810, 933–944. [DOI] [PubMed] [Google Scholar]
- Pathak D, Berthet A and Nakamura K (2013) Energy failure: does it contribute to neurodegeneration? Ann Neurol, 74, 506–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimkin M and Markham GD (2008) The CBS subdomain of inosine 5’-monophosphate dehydrogenase regulates purine nucleotide turnover. Mol Microbiol, 68, 342–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwin C, Nicolay K, Scheffers WA and Holzer H (1986) Mechanism of control of adenylate cyclase activity in yeast by fermentable sugars and carbonyl cyanide m-chlorophenylhydrazone. J Biol Chem, 261, 8744–8749. [PubMed] [Google Scholar]
- Ramanujam R and Naqvi NI (2010) PdeH, a high-affinity cAMP phosphodiesterase, is a key regulator of asexual and pathogenic differentiation in Magnaporthe oryzae. PLoS Pathog, 6, e1000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VA, Shepherd SM, Owen R and Hunter WN (2013) Structure of Pseudomonas aeruginosa inosine 5’-monophosphate dehydrogenase. Acta Crystallogr Sect F Struct Biol Cryst Commun, 69, 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Macary ME, Barbisan C, Gagey MJ, Frelin O, Beffa R, Lebrun MH, and Droux M (2015) Methionine biosynthesis is essential for infection in the rice blast fungus Magnaporthe oryzae. PLoS One, 10, e0111108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG and Hardie DG (2004) CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest, 113, 274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura K, Okada M, Shiraki H and Nakagawa H (1983) IMP dehydrogenase. I. Studies on regulatory properties of crude tissue extracts based on an improved assay method. J Biol Chem, 94, 1595–1603. [PubMed] [Google Scholar]
- Sweigard JA, Chumley FG and Valent B (1992) Disruption of a Magnaporthe grisea cutinase gene. Mol Gen Genet, 232, 183–190. [PubMed] [Google Scholar]
- Umejiego NN, Li C, Riera T, Hedstrom L and Striepen B (2004) Cryptosporidium parvum IMP dehydrogenase: identification of functional, structural, and dynamic properties that can be exploited for drug design. J Biol Chem, 279, 40320–40327. [DOI] [PubMed] [Google Scholar]
- Wei Y, Kuzmic P, Yu R, Modi G and Hedstrom L (2016) Inhibition of inosine-5’-monophosphate dehydrogenase from Bacillus anthracis: mechanism revealed by pre-steady-state kinetics. Biochemistry, 55, 5279–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RA, Fernandez J, Quispe CF, Gradnigo J, Seng A, Moriyama E, and Wright JD (2012) Towards defining nutrient conditions encountered by the rice blast fungus during host infection. PLoS One, 7, e47392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Woobong, R. A. D. (1997) The adenylate cyclase gene MACI of Magnaporthe grisea. Plant Cell, 9, 1973–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Que Y, Wang H, Wang C, Li Y, Yue X, Ma Z, Talbot NJ and Wang Z (2013) The MET13 methylenetetrahydrofolate reductase gene is essential for infection-related morphogenesis in the rice blast fungus Magnaporthe oryzae. PLoS One, 8, e76914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LN, Yin Z, Zhang X, Feng W, Xiao Y, Zhang H, Zheng X and Zhang Z (2017) New findings on phosphodiesterases, MoPdeH and MoPdeL in Magnaporthe oryzae revealed by structure analysis. Mol Plant Pathol. 19(5):1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Tang W, Wang J, Liu X, Yang L, Gao C, Zhang J, Zhang H, Zheng X, Wang P and Zhang Z (2016) Phosphodiesterase MoPdeH targets MoMck1 of the conserved mitogen-activated protein (MAP) kinase signalling pathway to regulate cell wall integrity in rice blast fungus Magnaporthe oryzae. Mol Plant Pathol, 17, 654–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Zhang X, Wang J, Yang L, Feng W, Chen C, Zhang H, Zheng X, Wang P and Zhang Z (2018) MoMip11, a MoRgs7-interacting protein, functions as a scaffolding protein to regulate cAMP signaling and pathogenicity in the rice blast fungus Magnaporthe oryzae. Environ. Microbiol doi: 10.1111/1462-2920.14102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu K, Zhang X, Song W, Zhao Q, Dong Y, Zheng X and Zhang Z (2010) A two-component histidine kinase, MoSLN1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Curr Genet, 56, 517–528. [DOI] [PubMed] [Google Scholar]
- Zhang H, Liu K, Zhang X, Tang W, Wang J, Guo M, Zhao Q, Zheng X, Wang P and Zhang Z (2011a) Two phosphodiesterase genes, PDEL and PDEH, regulate development and pathogenicity by modulating intracellular cyclic AMP levels in Magnaporthe oryzae. PLoS One, 6, e17241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ma H, Xie X, Ji J, Dong Y, Du Y, Tang W, Zheng X, Wang P and Zhang Z (2014) Comparative proteomic analyses reveal that the regulators of G-protein signaling proteins regulate amino acid metabolism of the rice blast fungus Magnaporthe oryzae. Proteomics, 14, 2508–2522. [DOI] [PubMed] [Google Scholar]
- Zhang H, Tang W, Liu K, Huang Q, Zhang X, Yan X, Chen Y, Wang J, Qi Z, Wang Z, Zheng X, Wang P and Zhang Z (2011b) Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae. PLoS Pathog, 7, e1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Southern blot analysis of the MoIMD4 deletion mutant. (A) The strategy of MoIMD4 gene replacement. Fragment of the MoIMD4 coding region were replaced with hygromycin phosphotransferase (HPH) fragment. (B) Southern blot analysis of the MoIMD4 knockout mutant with specific probe (probe 1) and HPH probe (probe 2).
Fig. S2 Phylogenetic analysis of MoImd4 and other Imd proteins by CLUSTAL_W and MEGA 6 programs. Species names and accession numbers are as followings: M. oryzae (Magnaporthe oryzae XP_003716182.1), M. robertsii (Metarhizium robertsii XP_007816906.1), C. purpurea (Claviceps purpurea CCE29804.1), T. reesei (Trichoderma reesei XP_006966877.1), C. orbiculare (Colletotrichum orbiculare ENH80323.1), G. tritici (Gaeumannomyces tritici XP_009224448.1), S. cerevisiae (Saccharomyces cerevisiae NP_013536.3), C. neoformans (Cryptococcus neoformans XP_012046718.1), H. sapiens (Homo sapiens NP_000875.2), P. aeruginosa (Pseudomonas aeruginosa WP_070336446.1), B. anthracis (Bacillus anthracis WP_047401536.1), S. pyogenes (Streptococcus pyogenes WP_002991454), T. foetus (Tritrichomonas foetus OHT09030.1).
Fig. S3 MoImd4 is involved in vegetative growth. (A) The wild type Guy11, ΔMoimd4 and the complement strains were cultured on CM, MM, SDC, and OM media at 28˚C in the darkness for 7 days. (B) Mycelial pellets in liquid CM medium. Mycelia of Guy11, ΔMoimd4 and the complement strains were inoculated in liquid CM medium with shaking condition (160 rpm) for 48 h at 28°C. Experiments were performed three times with similar results.
Fig. S4 MoImd4 is important for xanthosine monophosphate (XMP) biosynthesis. (A) Intracellular levels of XMP in Guy11, ΔMoimd4, and the complement strains estimated by HPLC. Experiments were repeated three times with the similar results. Error bars represent the standard deviations and asterisk denotes statistical significances (Duncan’s new multiple range test, p<0.01). (B) Statistical analysis of colony radius on MM media with or without XMP. Experiments were repeated three times with similar results. Error bars represent the standard deviations and asterisk denotes statistical significances (Duncan’s new multiple range test, p<0.01). (C) Statistical analysis of colony radius on MM medium with or without MPA. Error bars represent the standard deviations and asterisk denotes statistical significances (Duncan’s new multiple range test, p<0.01).
Fig. S5 The structure of MoImd4 in binding with MPA and the binding sites of MPA. (A) Predicted the structure of MoImd4 was a tetramer binding with MPA. Green represents the three-dimensional structure of MoImd4; Red represents inhibitor MPA. (B) Enlarged drawing of MPA was added in one of the ligand on the structure of MoImd4. Around the MPA, different-color curves are predicted the binding sites of MPA, includes D290, G340, G342, M431, G432 and Q470.
Fig. S6 Western blot analyzes the GFP expression of the point mutation and CBS deficient mutants. Total proteins of the indicated strains were extracted from 2-day old mycelia in liquid CM media. The anti-GFP was used to analyze by Western blotting.
Fig. S7 Homology modeling of key sites and point mutants of MoImd4. The target protein sequence of MoImd4 was predicted by SWISS-MODEL and point mutants were analyzed by PyMOL. Red curves represent key sites, gray curves were after point mutants, red stars stand for the sites of red curves and gray curves.
Fig. S8 MoImd4 promotes MoPdeH phosphodiesterase activities. (A) In vitro GST pull-down assays for MoImd4 and MoPdeH. The expression of GST-MoPdeH and His-MoImd4 was induced with 0.1 mM IPTG. GST-MoPdeH or GST were incubated with Glutathione Sepharose beads for 3 h at 4°C, precipitated, and co-incubated with lysates containing His-MoImd4 for additional 3 h at 4°C. Eluted proteins were detected by Western-blot analysis with anti-His and anti-GST antibodies. (B) Phosphodiesterase activities of purified GST-MoPdeH proteins, with 0.1 T, 0.2 T, and 1 T of the purified His-MoImd4 protein. Fluorescence was detected by a 10-min kinetic reaction at excitation 420 nm and emission 450 nm. “T” represents total quantity of the GST-MoPdeH protein. Experiments were repeated three times with similar results. The error bars indicate standard deviation of three replicates. Asterisk indicates statistically significant differences (Duncan’s new multiple range test, p<0.01). (C) In vivo cAMP levels measurements of Guy11, ΔMoimd4, MoImd4, ΔMopdeH, and MoPdeH at the stage of mycelia. Experiments were repeated three times with similar results. The error bars represent ±SD of three replicates. Asterisk indicates statistically significant differences (Duncan’s new multiple range test, p<0.01).
Fig. S9 MoImd4 didn’t participate in ROS scavenging. (A) Conidial suspensions of Guy11, ΔMoimd4 mutant, and the complement strains were injected into separated rice sheaths. After 24 hpi, DAB was used to dye the sheaths for 8 h. (B) Percentages of cells with IH were dyed by DAB. Means were calculated from three independent replicates. There were little significant differences between the strains. Bar = 10 μm.
Fig. S10 Locations of MoImd4-GFP in the ΔMoimd4 mutant at the stages of sporulation and infection.
Fig. S11 Interaction between MoPdeH and MoImd4 treated with MPA. (A) Lysed cells of purified GST-MoPdeH or GST proteins were incubated with Glutathione SepharoseTM 4B at 4°C for 3 h and centrifugation. Then, lysed cells of purified MoImd4-His with 1, 5, and 10 μg/ml MPA were re-incubated with GST beads at 4°C for another 3 h and then eluted. Equal His-tag infusion proteins from input were used as controls. Eluted proteins were detected by Western-blot analysis with anti-His and anti-GST antibodies. The number represents the intensity of eluted proteins detected by the anti-His antibody. The intensity of the elutions from wild-type MoImd4 was set to 1. (B) Lysed cells of purified GST-MoPdeH or GST proteins were incubated with Glutathione SepharoseTM 4B at 4°C for 3 h and centrifugation. Then, lysed cells of inactivation MPA-binding sites proteins (D290A, G340AG342A, M431AG432A and Q470A) were re-incubated with GST beads at 4°C for another 3 h and then eluted. Equal His-tag infusion proteins from input were used as controls. Eluted proteins were detected by Western-blot analysis with anti-His and anti-GST antibodies. The number represents the intensity of eluted proteins detected by the anti-His antibody. The intensity of the elutions from wild-type MoImd4 was set to 1. Protein of S345AC347A was as a control that mutation these sites didn’t affect the interaction between MoPdeH and MoImd4. (C) Purified His-fusion expression proteins affecting the enzyme activities of MoPdeH. Equal amounts of His fusion proteins and GST-MoPdeH were mixed for measuring enzymatic activities, respectively. Fluorescence was read by a 10-min kinetic reaction at excitation 420 nm and emission 450 nm. Experiments were repeated three times with similar results. The error bars indicate standard deviations of three replicates. Asterisk indicates statistically significant differences (Duncan’s new multiple range test, p<0.01).
Fig. S12 The predicted three-dimensional structure of OsIMPDH.
Table S1 Potential interacting protein list of MoPdeH identified by yeast two-hybrid screen
Table S2 Appressorium formation on the hydrophobic surface of the wild type strain in the presence of MPA.
Table S3 Primers used in this study.