Abstract
Nuclear DNA replication requires the concerted action of hundreds of proteins to efficiently unwind and duplicate the entire genome while also retaining epigenetic regulatory information. Initiation of DNA replication is tightly regulated, rapidly firing thousands of origins once the conditions to promote rapid and faithful replication are in place, and defects in replication initiation lead to proliferation defects, genome instability, and a range of developmental abnormalities. Interestingly, DNA replication in metazoans initiates in actively transcribed DNA, meaning that replication initiation occurs in DNA that is co-occupied with tens of thousands of poised and active RNA polymerase complexes. Active transcription can induce genome instability, particularly during DNA replication, as RNA polymerases can induce torsional stress, formation of secondary structures, and act as a physical barrier to other enzymes involved in DNA metabolism. Here we discuss the challenges facing mammalian DNA replication, their impact on genome instability, and the development of cancer.
Keywords: DNA replication, Replication stress, Transcription, Origin licensing, R loop
Coordinating replication initiation to limit genome instability
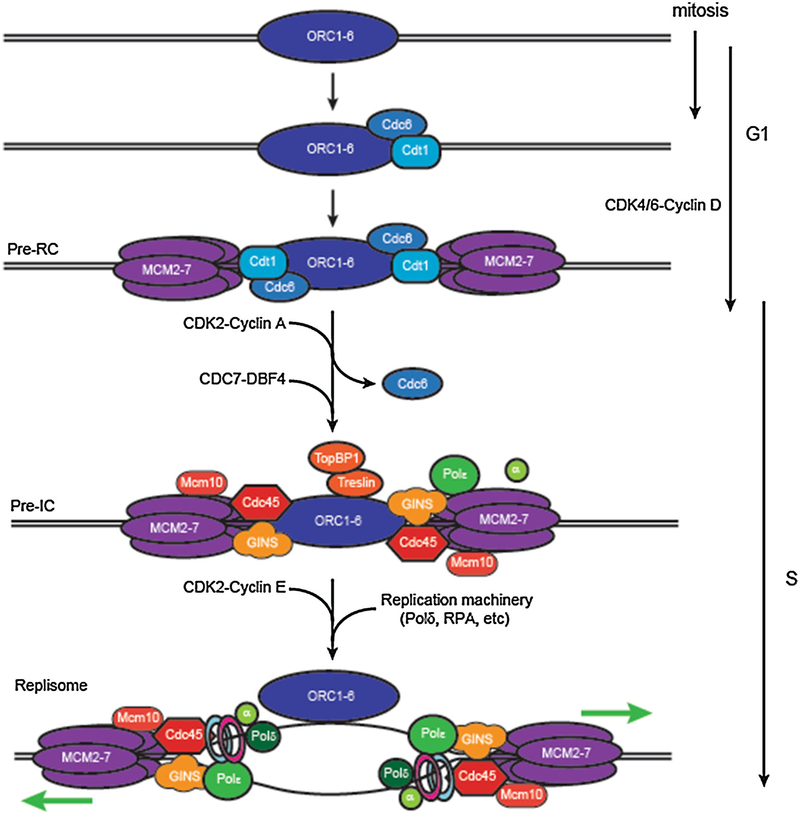
Regulating the initiation of DNA replication occurs at two different levels: (1) origin licensing where proteins necessary for replication initiation assemble on DNA to form pre-replicative (pre-RC) complexes, and (2) origin firing, where cyclin-dependent kinases drive initiation at a subset of these licensed origins. Origin licensing takes place in mitosis and G1 phases of the cells cycle, with the loading of the pre-replicative complex (pre-RC) proteins Orc1–6, Cdc6, Cdt1, followed by loading of the hexameric helicase Mcm2–7, Cdc45, and GINS [1, 2]. First, the highly conserved six-subunit origin recognition complex (ORC) binds to DNA (Fig. 1). ORC association with DNA provides a scaffold to recruit the rest of the pre-RC complex members, first recruiting Cdc6—an essential ATP-binding regulator of DNA replication. Cdc6 then recruits Cdt1, which licenses the loading of the replicative helicase composed of the minichromosome maintenance proteins 2–7 (MCM2–7), forming the pre-RC. Upon entry into the S phase, the assembled pre-RC recruits a number of new proteins required for origin firing, while Cdc6 and Cdt1 association is lost. The GINS complex—formed of four small subunits, Sld5, Psf1, Psf2, and Psf3—then associates with the pre-RC and helps recruit Mcm10. Cdc45 is loaded onto the pre-RC, and is thought to be a limiting factor to form the preinitiation (pre-IC) complex poised to begin DNA replication. GINS, Mcm10, and Cdc45 are involved in recruiting the DNA replication machinery—including the replication clamp PCNA, Polaα primase, the single-strand binding protein RPA, and the DNA polymerases Poleε and Poldδ—and forms a functional replisome [3, 4].
Fig. 1.
Assembling an origin of DNA replication. Following DNA segregation in mitosis, the heterohexameric ORC complex associates with DNA. Cdc6 and Cdt1 associate with DNA-bound ORC, which facilitates the loading of the MCM2–7 replicative helicase and forms the pre-replicative complex (pre-RC). In response to growth signals, the cell approaches the decision point to enter into S phase and commit to DNA replication. Cyclin A-CDK2 activity marks entry into S phase, phosphorylating Cdc6 leading to its relocalization to the cytoplasm. Cyclin E-CDK2 recruits Cdc45 to the assembled pre-RCs, which in turn leads to Mcm10 association, which is required for MCM2–7 activation. The GINS complex then binds Cdc45 and MCM2–7 to form a complete replicative helicase. The regulatory proteins Treslin and TopBP1 associate with pre-RCs, which are now capable of initiating replication, forming pre-initiation (pre-IC) complexes. Entry into S phase is also characterized by the activation of the Cdc7-Dbf4 kinase, which phosphorylates and activates many proteins associated with the pre-IC, and is required for origin firing. Supercoiling unwinds DNA at the origin in a Cdc45-dependent manner, and leads to the loading of downstream replisome components, including the clamp loader RFC (blue circle) and its target PCNA (pink circle), which associates directly with the DNA polymerases. Pola primase and the leading and lagging strand polymerases Pole and Pold all load onto the open DNA behind MCM2–7, forming a functional replisome
Entry into S phase and origin activation is governed by the cell cycle-regulated expression of proteins called cyclins, and the kinases they activate, cyclin-dependent kinases (CDKs). A variety of cyclins and CDKs promote entry into S phase and initiation of DNA replication. CDK4 and CDK6 in complex with cyclin D act to phosphorylate retinoblastoma protein in G1, pushing cells past a “restriction point,” committing the cell to enter S phase. CDK4,6-Cyclin D-mediated expression of the E2F transcription factors stimulate the expression of critical S phase proteins including ribonucleotide reductase (RNR), which is required for dNTP production, as well as Cyclin E and Cyclin A [5, 6]. CDK2-Cylin E drives the cell cycle towards the G1 to S transition point through further phosphorylation of Rb and E2F expression. Cyclin A expression starts to accumulate right before the G1 to S transition, and is critical for DNA synthesis. The Dbf4-dependent kinase (DDK) then phosphorylates multiple components of the assembled pre-RCs, inducing origin firing and initiating DNA replication. These sequential waves of cyclin expression and activation in G1 coordinate entry into S and replication origin firing with the expression of factors required for timely and faithful DNA synthesis.
Defining origins of replication
In bacteria and S. cerevisiae, the ORC proteins, DnaA and ORC1–6, respectively, bind short, AT-rich tracts of DNA in a sequence-specific manner. However, in fission yeast and higher eukaryotes, ORC proteins do not exhibit any sequence specificity to guide DNA binding in vitro or in vivo [2]. As a consequence, early studies in metazoans yielded very few well-mapped replication origins. Without inherent binding specificity, epigenetic marks and the binding of accessory proteins such as chromatin regulators likely promote ORC association and pre-RC assembly, thus defining the location of replication origins. Subsequent to ORC binding, the replication licensing factors Cdt1 and Cdc6 then assemble onto the pre-RC, and also bind DNA without any apparent sequence specificity in higher eukaryotes [2]. While the specificity and determinants of ORC association with DNA have been extensively studied, it is possible that Cdt1 or Cdc6 interact with epigenetic modifications such as histone methylation or additional DNA binding proteins to specify replication origin location and/or spacing in metazoans.
One major approach to determine where pre-RCs assemble in mammalian systems is to define ORC binding to DNA genome-wide by chromatin immunoprecipitation followed by deep sequencing (ChIP-Seq). These studies have been complicated by the replication-independent roles of ORC, in particular its role in the establishment and maintenance of DNA silencing [7]. ORC associates with a number of epigenetic marks associated with silencing including H3K9me3, H3K27me3, and H4K20me3, and interacts with proteins such as HMGA1a and HP1 which are involved in establishing and maintaining heterochromatin [8, 9]. Much of ORC binding occurs in transcriptionally silent heterochromatic regions, including centromeres and telomeres [2, 10]. ORC binding at telomeres is important for heterochromatin formation and telomere stability, yet whether some of these telomeric ORC sites can also function as pre-RCs is less clear [11, 12]. As origins, ORC bound at telomeres does not appear to initiate DNA replication, yet the telomere repeat binding factor Trf2 stimulates ORC binding and promotes replication initiation at the Epstein-Barr virus origin of replication, oriP [13].
Another approach for mapping replication initiation sites utilizes the biochemical purification of newly synthesized nascent DNA strands, and recent efforts couple this approach to genome-wide mapping technologies [14–17]. Unlike physical association of pre-RC components, isolation of newly replicated DNA specifically measures active origins of replication and can be used to define replication timing. In multiple genome-wide studies, researchers have determined that metazoan replication is organized into “zones” of early or late replication, ranging roughly from 0.4 to 1 Mb in size [18, 19]. Organization of replication into large domains suggests that replication timing correlates with chromatin architecture, which is similarly organized into large regions or loops sharing genetic and epigenetic features [20]. Indeed, 3-D genome organization as measured by genome-wide chromosome conformation capture (3C) techniques reveals a high correlation between replication zones and self-interacting nuclear domains [21]. Further, early replication zones occur in gene-rich, actively transcribed regions and are enriched for several epigenetic markers of euchromatin [16, 19, 22]. Therefore, accessible chromatin state promotes early replication, and early replication zones can differ between distinct cell types [20]. Intuitively, replication initiation within accessible chromatin is logical, as DNA binding requires less energy than heterochromatic regions. However, accessible chromatin is active chromatin, with the DNA template already bound by factors regulating and promoting transcription. Indeed, early replication zones correlate with many marks of active transcription, including sites of DNase I hypersensitivity (DHS), H3K4 trimethylation found at active promoters, and even Pol2 association. The positive correlation between active transcription marks and early DNA replication raises the question as to how transcriptional activity and replication initiation are coordinated. In response to DNA damage, transcription of nearby genes is temporarily inhibited to suppress conflicts between transcription and repair machinery [23]. Further, prolonged exposure to the replication stress agent hydroxyurea globally inhibits transcription ~10 %, yet a shorter exposure does not appear to have an effect [22, 24]. Thus, our current understanding of the effect of replication initiation on transcriptional activity is limited. Intriguingly, it has been demonstrated that double strand break repair suppresses ftranscription in cis of nearby loci in an ATM-dependent manner [25]. It will be interesting to determine if a similar process occurs during replication stress to promote efficient fork restart and suppress genome instability.
Though early replication and pre-RC assembly correlate with marks of active transcription, it seems highly unlikely that origins assemble at active promoter elements and transcribed exons. One possibility is that active RNA polymerases disrupt transient ORC binding within transcribed DNA, leaving intact only pre-RCs in adjacent chromatin. Another possibility is that pre-RC components are targeted to adjacent “inactive” DNA within the euchromatic domain by epigenetic, cis-, or trans-acting factors. Yet the initiation of DNA replication near active promoters coordinates the movement of replication and transcription in a co-directional manner, a compelling point raised by Knott and colleagues [26]. Furthermore, transcription factors can stimulate the activity of viral origins of replication from 2 to 50-fold [27]. It will be interesting to determine if general or tissue-specific transcription factors and coactivators also play a role in pre-RC assembly and replication initiation independent of their role in transcriptional regulation.
How dynamic is the process of pre-RC formation in G1? In mitosis, chromosomes are highly condensed and the DNA retains little to no higher chromatin organization, which is rapidly regained throughout G1 [28]. It is possible that pre-RC formation in mitosis and early G1 is highly dynamic, and transcriptional activity contributes to the removal of ORC from active DNA. In this model, pre-RC components loaded in telophase can load to active DNA, be displaced as transcription resumes, and reload prior to S phase. Time-course experiments examining ORC association and pre-RC formation can help determine if transcription shapes the origin licensing landscape. Future studies will determine whether pre-RCs are restricted from transcribed DNA by passive or active mechanisms, and identify the factors determining stable pre-RC assembly.
Origin licensing, activation, and replication stress
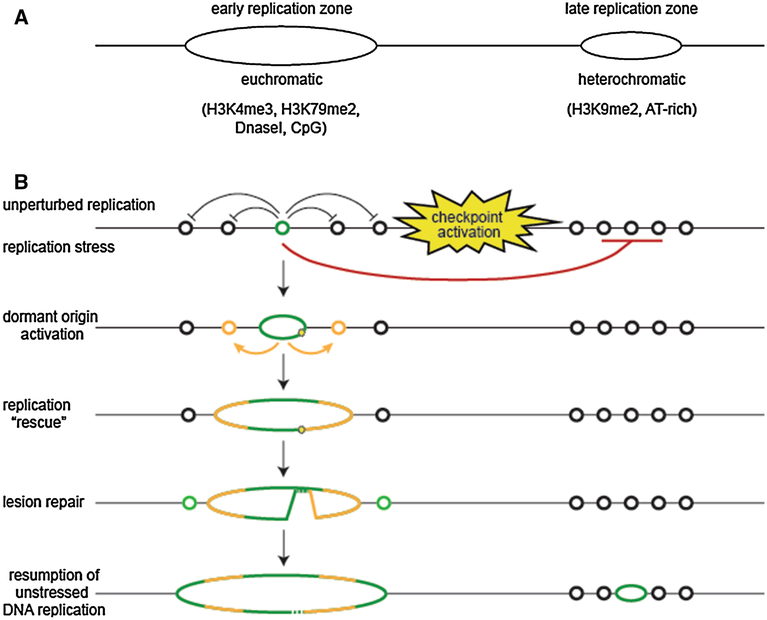
Origins fire in a stochastic manner each S phase and only a subset of origins are used in any given cell cycle [14, 29]. Multiple pre-RCs reside within each early and late replicating zone, which range from ~400 kb to 1 Mb in size [19]. Yet replication initiates from only one or a few widely spaced origin firing events within each zone, indicating that activated origins can suppress the firing of nearby pre-RCs ([30], Fig. 2). But what are all of these additional pre-RCs for? One primary role for excess origins is to provide a new initiation site adjacent to a replication fork experiencing stress. Replication stress is a phenomenon that arises when genetic or environmental conditions lead to the replicative polymerase to move slowly and/or stall, potentially leading to fork collapse and generating DNA damage and genome instability. It can arise spontaneously from defects in replication initiation and elongation components, similar to replication stress-inducing drugs such as aphidicolin that inhibit the activity of DNA polymerase. A variety of checkpoint proteins guard against genome instability arising at replication forks, including all three major DNA damage response kinases ATR, ATM, and DNA-PKcs [31, 32]. The fork protection complex composed of Timeless, Tipin and Claspin travels with the replisome, to sense and initiate repair of DNA damage present during replication and mutations in these proteins also lead to enhanced replication stress [31].
Fig. 2.
Organization of DNA replication in metazoans. a Metazoan replication begins with origin firing in early replication zones, which are co-incident with actively transcribed euchromatic DNA and marks associated with active transcription. Late replicating zones, which associate with heterochromatic regions, also contain replication origins that fire later in S phase to complete synthesis in a timely manner. b Regulation of replication initiation in response to stress. In the absence of replication stress, replication initiates (green circle) from only one or a few widely spaced origins in early replication zones. Many origins do not fire (black circles, left). Late-replicating regions also contain origins (black circles, right), which fire later in S phase to complete replication in heterochromatic zones. In response to replication stress early in S phase, additional origins near the stress site (yellow star) also fire (orange circles), while origin firing in late-replicating DNA is suppressed. Origin firing near damage sites can help “rescue” stalled or collapsed replication forks by reinitiating replication on the opposite side of the lesion, completing replication within the damaged region (left). Suppression of late-firing origins may also suppress genome instability by preventing the initiation of replication in new genomic locations during potentially unfavorable conditions. Origins in distant regions will fire after the stress signal abates, following successful repair or adaptation after prolonged checkpoint activation
Replicative stress arising early in S phase induces dormant origin firing in regions with active replication while origin activity in late replicating regions is suppressed [33, 34], and this process is dependent on the DNA damage checkpoint kinase ATR and activation of the downstream effectors CHK1, TopBP1 and the 9-1-1 checkpoint complex composed of Rad9, Hus1, and Rad1. ATR activation also stabilizes stalled replication forks by activating TopBP1 and the Timeless/Tipin/Claspin fork protection complex [35]. Thus, one important role for excess origin licensing is to re-initiate DNA replication near a stalled or collapsed fork to complete DNA synthesis surrounding the damage site (Fig. 2b). It is possible that not all of the potential pre-RCs, such as those located in the telomeric and centromeric heterochromatin, are able to initiate DNA replication. It would be interesting to determine if the origins in these highly repetitive regions are suppressed or fire more frequently following replication stress or local DNA damage.
Many drivers regulating entry into S phase are protooncogenes, and induce DNA damage from replication stress when overexpressed [36]. Overexpression of oncogenic RAS suppresses expression of the RNR component RRM2, leading to entry into S phase without sufficiently high levels of dNTP pools and results in replication fork stalling and genome instability that can be suppressed by the addition by exogenous nucleosides [37]. Replication defects and the resulting DNA damage foci induced by cyclin E overexpression can be alleviated by inhibition of transcription by cordycepin or the CDC7/CDK9 inhibitor PHA-767491, respectively [38]. These results directly link the replicative stress associated with oncogene activation to conflicts between DNA replication and transcription. Indeed, highly transcribed early replicating zones in murine B cells harbor replication stress-sensitive fragile sites that are commonly rearranged in B cell lymphoma and these sites are hyper-sensitive to overexpression of the oncogene c-Myc [22].
Chromatin accessibility promotes replication initiation and genome stability
Similar to transcriptional activation, origin licensing also involves the concerted action of multiple chromatin modifying enzymes. While ORC can bind euchromatic and heterochromatic DNA, a more “accessible” chromatin state is necessary for the efficient loading of the MCM helicase. Recruitment of the histone acetyltransferase HBO1 to the pre-RC is required for chromatin decondensation and subsequent MCM2–7 helicase loading by acetylation of histone H4 [39, 40]. The chromatin remodeler Snf2 h also promotes MCM association and pre-RC licensing in a Cdt1-dependent manner [41]. Chromatin modifiers have also been shown to play an important role in origin activation. The histone deacetylase Rpd3 is a transcriptional regulator, acting as both a general and specific inhibitor of transcription as part of two functionally distinct complexes. While the “large” Rpd3 complex targets specific promoters to silence transcription, a “small” Rpd3 containing complex also inhibits spurious initiation at cryptic start sites following transcriptional elongation. Yeast lacking Rpd3 show increased DNA replication at early time points at normally late-firing origins, suggesting a role for H3 and H4 acetylation in regulating global replication timing [42].
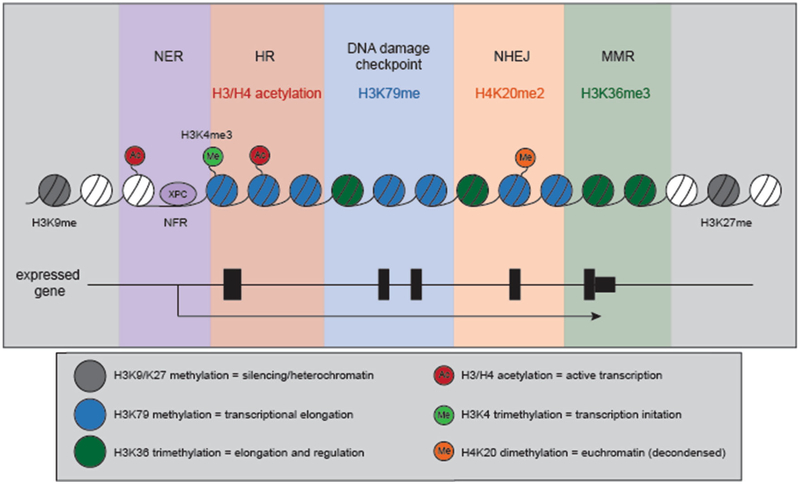
Epigenetic marks, primarily histone methylation, also contribute to efficient DNA replication progression and genome stability. H3 lysine 4 trimethylation (H3K4me3) is increased upon replication stress by treatment with HU, and elevated H3K4me3 also associates with later replication origins, normally suppressed during replication stress [34, 43]. The histone methyltransferase MLL1 induces H3K4 trimethylation, a mark associated with active gene promoters. MLL1 protein levels fluctuate with the cell cycle, undergoing proteasome-mediated degradation in S and M phases, and are stabilized in response to replication stress by ATR [43, 44]. In response to HU, Mll1 depleted cells no longer exhibit increased H3K4me3, and can no longer suppress DNA replication [43]. Thus, Mll1 protein stabilization by ATR is a key component of the intra-S phase checkpoint to replication stress, preventing the firing of late origins and maintaining genomic stability. Studies from the Aladjem lab have observed that H3K79 dime-thylation is also found adjacent to replication initiation sites (Fig. 3). Furthermore, depletion of DOT1L—the sole methyltransferase responsible for H3K79 methylation—triggers limited overreplication and activation of the DNA damage checkpoint [45]. Thus, there is a complex interplay between multiple epigenetic marks and chromatin remodeling events normally involved in transcriptional regulation that dictates origin location as well as influence firing and replication timing, and perturbations in these processes lead to replication stress-induced genome instability.
Fig. 3.
Histone modifications associated with both transcription and DNA replication/repair processes. A variety of epigenetic modifications link transcription with DNA replication and repair processes. A subset of proteins and histone modifications appear to play dual roles, and may allow for crosstalk in S phase. The promoters of active genes predominantly contain a nucleosome free region (NFR) 5’ of the transcription start site (TSS). Interestingly, the DNA repair proteins involved in transcription-coupled repair XPC, XPG and XPF have been found to play a role in regulating transcriptional activity at a subset of genes, and occupy promoters at the NFR, like many transcription factors (purple panel). Acetylation of histones H3 and H4 promotes transcriptional activity by enhancing chromatin accessibility and the adoption of a euchromatic state. H3/H4 acetylation also increases with homologous recombination (HR). Similar to its role in transcription, H3/H4 acetylation makes the DNA surrounding the DSB “accessible” for repair. HR requires the invasion of a homologous template (either the sister chromatid or homologous chromosome) to prime new DNA synthesis and repair the damaged region. Thus, “open” chromatin surrounding the DNA break and the repair template would increase repair efficiency (red panel). H3 methylation on lysine 79 (H3K79me1/2) is important for telomeric silencing and heterochromatin maintenance, but also promotes transcriptional elongation. It also correlates with replication initiation regions, and depletion of the sole H3K79 methyltransferase DOT1L can induce over-replication and genome instability (blue panel). Lysine 20 dimethylation of H4 (H4K20me2) is a common mark of euchromatin, and is bound by the Tudor domain of the DNA repair regulator 53BP1. 53BP1 binding promotes NHEJ by limiting DNA resection, suppressing RPA association and the recruitment of HR proteins (orange panel). H3 lysine 36 trimethylation (H3K36me3) increases along a gene body, peaking in the 3’end of a transcribed region. Recent work has uncovered a role for H3K36me3 in regulating DNA mismatch repair (MMR), recruiting MutSa (Msh2-Msh6) through direct interaction with the Msh6 subunit even in the absence of DNA mismatches ([109], green panel). In the diagram of a gene body in the lower panel, black boxes mark exons while the thin black line represents non-coding DNA
The interplay between replication initiation, transcriptional activity, and chromatin state
Why do metazoans initiate replication in transcriptionally active zones, rather than retain the discrete origins of replication based on sequence specificity as in lower eukaryotes? Unlike lower eukaryotes, metazoans undergo extensive development in embryogenesis from a single cell to differentiate into hundreds of distinct tissue types in the adult. Each cell type exhibits a unique transcriptional “signature” and therefore the most used genes can differ widely between cell types. Indeed, studies comparing replication patterns throughout development and between tissue types show a strong correlation to transcriptional activity and open chromatin state [46, 47]. The replication timing of the immunoglobulin heavy chain locus (IgH) changes throughout B cell development. At early B cell stages, prior to programmed VDJ recombination events to produce a functional antibody, the entire IgH region replicates early. However, in late stages of B cell development after successful VDJ rearrangement, the downstream variable genes that do not form part of the antibody transition to a late replication time, concurrent with gene inactivation [48]. Thus, it is likely that the rapid completion of DNA replication within heavily transcribed genomic regions is evolutionarily advantageous. In this manner, replication timing, while potentially more variable in middle and late-replicating regions, remains relatively constant within genomic regions where high rates of transcription are essential for cellular identity and function.
Replication timing and differentiation; chicken or the egg?
In embryonic development, cell differentiation, and somatic cell reprogramming, alterations in both the transcriptional expression patterns and replication timing profiles occur at the same time [46, 47]. Defects in replication initiation and progression have profound effects on development organismal viability; knockouts of cyclin E in mice result in embryonic lethality from defective placental development. Cellular proliferation also contributes to somatic cell reprogramming and induced pluripotent stem (iPS) cell fate, as more rapidly dividing cells are more easily induced to the iPS state. Further, mutations enhancing cycling also lead to more efficient reprogramming, and even stimulation to proliferate via growth factors and cytokines enhances the reprogramming capacity of normally inefficient hematopoietic stem cells (HSCs) [49]. These exciting results correlate high cellular proliferation with a potential for pluripotency, and suggest that DNA replication contributes to the establishment and/or maintenance of epigenetic alterations associated with pluripotency.
Establishing sufficient origins for early cell proliferation contributes to growth control and embryonic development, as hypomorphic mutations in replication initiation from mutations in pre-RC components lead to Meier-Gorlin syndrome, a form of primordial dwarfism and developmental abnormalities [47]. Replication elongation and maintaining fork integrity throughout S phase is also important for human development and disease. The helicase RECQL4 plays an essential role in replication and genome stability, and mutations in humans have been linked to Rothmund-Thompson syndrome, characterized by developmental abnormalities, premature aging, and skeletal defects [47]. Thus, defective or dysregulated replication initiation and elongation lead to distinct tissue-specific defects, and this may be a consequence of the intimate interplay between replication initiation and transcriptional activity.
Regulation of replication timing: a crossroads for transcription and DNA repair
The evidence tying transcriptional activity to replication timing is largely correlative; however chromatin state has been shown to directly influence replication timing, as histone acetylation promotes both an open chromatin state as well as early origin firing. Indeed, histone acetyltransferase (HAT) recruitment to an origin promotes firing from yeast to humans [40, 50, 51]. Further, recruitment of a histone deacetylase to an origin suppresses firing and delays replication [52]. Together, these results suggest that altering chromatin state influences origin firing and replication timing. However, recent studies in S. cerevisiae have shown a more direct role for transcriptional regulators in replication timing. The transcription factors Fkh1 and Fkh2, which regulate transcriptional elongation and silencing, promote early origin firing by recruiting a key initiation factor Cdc45 to pre-RCs [53]. Importantly, the role of Fkh1/2 in replication timing is independent of transcriptional activation, and provides the first direct evidence for transcription factors regulating replication timing.
First identified as a telomere binding protein in yeast, the DNA repair protein Rif1 also regulates DNA replication timing in both yeast and mammals [54, 55]. Mice deficient in Rif1 are born at a severely reduced frequency, and the few mice born exhibit lifespan and fertility defects. Further, Rif1−/− cells are hypersensitive to replication stress, suggesting that Rif1 plays an important role in DNA replication [56]. Indeed, Rif1 depletion in mouse and human cells leads to a global alteration in replication timing, similar to results in the fission yeast Schizosaccharomyces pombe (S. pombe) [54, 55, 57]. Interestingly, Rif1 was shown to bind near late-replicating origins in S. pombe, potentially suppressing their activity while early replicating Rif1-deficient origins are activated [54]. Rif1 has also been shown to govern the choice between homologous recombination (HR) and non-homologous end-joining (NHEJ) in double-strand break (DSB) repair [58–61]. Together with 53BP1, Rif1 suppresses DNA end resection at DSBs during G1, inhibiting Brca1 recruitment and promoting NHEJ-mediated repair. It remains unclear whether the role of RIF1 during replication and DNA repair are linked, though it is tantalizing to hypothesize that Rif1 association at DNA damage sites may delay replication initiation to promote repair prior to replication, suppressing genome instability.
DNA topology and replication stress
RNA polymerase translocates along the DNA concurrent with or in opposition to the replication fork, dependent upon the orientation of the coding DNA template. Multiple polymerase complexes may co-occupy a single gene body, and RNA polymerases can also stably bind promoter regions in a “poised” state, all of which may present problems for DNA replication progression. The highly dynamic and processive nature of RNA polymerase can induce multiple stresses on the transcribed DNA template that can be acutely problematic during DNA replication. The ribosomal DNA (rDNA) array in S. cerevisiae is one of the most well-characterized examples of replication stress generated by transcriptional activity. The rDNA provides all of the translational machinery for the cell, is composed of ~150 highly expressed short coding regions packed together in the subnuclear compartment, the nucleolus. A physical barrier resides between each repeat unit—the replication fork block (RFB)—to prevent head-on collisions between RNA and DNA polymerase [35]. The protein Fob1 binds a specific DNA sequence at the 3’ end of the 35S gene, altering the chromatin state and blocking replication progression from the 3’ direction. Deletion of Fob1 or the RFB leads to extensive genome instability in the rDNA, resulting in deletions, amplifications, and episomal rDNA circles [62, 63]. Further, elegant studies in bacteria have shown that inverting transcriptional units to induce head-on conflicts with replication forks leads to proliferation defects and genome instability [64]. These defects can be suppressed by slower growth conditions like minimal media, suggesting that increasing the cell cycle time can alleviate such pressure. Thus, coordinating origin firing, replication timing, and transcriptional activity appears to be a conserved mechanism to maintain proliferative advantage and suppress genome instability.
Transcriptional activity can also induce topological strain on duplex DNA induced by unwinding to access the template strand, particularly in genomic regions containing multiple actively transcribed units. DNA supercoiling and unwinding can accumulate in regions of high transcription, at sites of transcription termination or start sites, respectively. Further, DNA replication stress correlates with convergent or divergent transcriptional activity, where either transcription termination sites or promoter elements reside very close to one another (>5 kb; [22]). These observations argue that both supercoiling and under-wound DNA both contribute to genome instability in the context of DNA replication. Additional factors can exacerbate DNA stress at genomic locations generating supercoiling or under-wound DNA. Supercoiling is normally relieved by the action of topoisomerase enzymes that nick either one or both strands of the double helix to unwind or “relax” the DNA in an ATP-dependent manner. Topoisomerase poisons such as camptothecin (CPT) are commonly used chemotherapy agents. CPT intercalates at the DNA cleavage site and “traps” covalently bound Topo1 to nicked DNA, leading to genome instability in replicating cells [65]. Divergent transcription can also give rise to secondary DNA structures conferring genome instability. Here, closely located transcription initiation sites can lead to the appearance of long stretches of unwound DNA with ssDNA regions. Regions prone to forming secondary structures, such as hairpins, stem-loops or G-quadruplex regions form more readily in under-wound DNA. These hyper-stable structures rely on specialized enzymes such as the helicase Pif1 for unwinding [66], and can block DNA replication progression due to the enhanced energy required for unwinding. Further, secondary structures also leave the unpaired strand as free ssDNA, which is much more vulnerable to damage from reactive oxygen species (ROS) or nucleophilic attack. Importantly, mapped human common fragile sites are highly enriched for these secondary structures, suggesting that they present potent blocks to DNA replication progression [67].
Transcriptional activity, R loop formation, and replication fork collapse
The negative supercoiling behind elongating polymerase can also promote the formation of RNA:DNA hybrids, or R loops, by opening the duplex DNA and allowing for reannealing of the RNA to the template strand [68]. In particular, transcription within DNA exhibiting GC-skew where the template strand is rich in guanine is extremely prone to R loop formation [69]. R loops are an extremely common phenomenon and perform important functions in regulating transcription and maintaining genomic architecture. R loops form at the 3’ end of genes in the untranslated region (UTR) and promote transcriptional termination and the release of mRNA molecules from template DNA [70]. Additionally, R loops form at CpG islands found in gene promoters, protecting from gene silencing mediated by DNMT3B1-dependent de novo methylation [69]. R loops exhibit stability greater than duplex DNA, and can impede both transcription elongation and replication if not efficiently processed.
A number of enzymes promote R loop dissolution, and suppress transcriptionally associated genome instability. Members of the THO/TREX complex, which couples transcription with mRNA export, were some of the first identified mutations linking RNA metabolism to genome instability [71]. Replication fork progression is impaired in THO complex mutants from increased R loops, and the resulting genome instability can be suppressed by overexpression of RnaseH1 [72, 73]. The helicase senataxin (SETX) unwinds R loops in both yeast and mammalian cells [74–76]. SETX acts in R loop unwinding to promote transcriptional termination, but also can suppress replication stress generated by R loops. Additionally, both the monomeric Rnase H1 and heterotrimeric Rnase H2 enzymes recognize R loops, cleaving the RNA component to leave intact duplex DNA. In mouse, deletion of the RnaseH2C subunit of Rnase H2 results in early embryonic lethality and genomic instability, stemming from increased ribonucleotide load in duplex DNA that triggers ssDNA breaks and the DNA damage response, similar to observations made in yeast [77, 78]. RNA splicing factors also contribute to R loop dissolution, as ASF/SF1 depletion leads to increased R loop accumulation and genome instability [79]. Further, Rnase H1 overexpression suppresses the mutagenesis and can delay the cell death following ASF/SF1 depletion, further supporting a role for alternative splicing in R loop dissolution. Another member of the TREX complex—DSS1—interacts with and stabilizes the HR protein BRCA2 [80, 81]. R loop accumulation has been observed in cells deficient for DSS1 or BRCA2, leading to defects in DNA replication and genome instability [82]. Importantly, BRCA2-deficient cells accumulate R loops in both replicating and non-replicating cells, indicating that BRCA2 plays an important role in R loop dissolution independent from the induction of replication stress.
But how do R loops generate DNA damage? Genetic studies performed in yeast demonstrated that proteins governing the intra-S phase checkpoint are required for THO mutant viability in response to replication stress [83]. More recently, evidence from E. coli shows that R loop mediated genome instability requires active replication [84]. Further, human cells depleted for the splicing factor SRSF1 (ASF/SF1) also induce a DNA damage response specifically in replicating cells. Indeed, a genome-wide screen for factors that increase phosphorylation of histone variant H2AX—an early mark of the DNA damage response—isolated multiple components of RNA processing, including proteins involved in splicing and spliceosome assembly [85]. Together, these results suggest that stabilized R loops induce DNA damage during replication, and R loop-mediated S phase genome instability is conserved from bacteria to humans. More recently, R loops have also been shown to induce genome instability during meiosis. Yeast lacking THO/TREX components accumulate unrepaired DSBs, exhibit both an activated DNA damage response and a decrease in successful meiotic cell divisions [86]. Further, C. elegans harboring a mutation in the THO homolog Thoc-2 are infertile, and exhibit defective meiotic DNA replication. Other enzymes involved in R-loop dissolution also lead to meiotic defects. Male mice deficient for SETX are infertile, and female fertility is markedly decreased, leading to decreased litter sizes [75]. Spermatocytes in SETX-deficient males arrest prior to meiosis I with unrepaired double-strand breaks (DSBs) and the absence of chiasmata that normally form after successful double-strand break repair. Together, these results highlight an important role for R loop dissolution in meiosis as well as mitosis.
While secondary DNA structures and R loop formation are two extensively studied inducers of replication stress and DNA damage, additional transcriptionally associated phenomena may also contribute to genome instability. Tightly bound proteins at unwound actively transcribed sites such as paused Pol2 or transcription factors may also impede replication progression, and further studies will dissect whether additional components associated with active transcription and repair also contribute to replicative stress and genome instability.
Dual-purpose proteins: transcriptional regulators involved in DNA repair
Multiple components of the PolII preinitiation complex (PIC) general transcription factor TFIIH are enzymes involved in nucleotide excision repair (NER). Mutations in these genes can give rise to a variety of diseases and cancer predisposition syndromes including xeroderma pigmentosum (XP) characterized by photosensitivity, Cockayne syndrome (CS), which also exhibits premature aging and impaired nervous system development. In transcription-coupled NER (TC-NER), DNA damage impeding transcription elongation is sensed directly by TFIIH and CSB, leading to PolII pausing and the recruitment of repair factors [87]. Genotoxic agents impeding replication fork progression such as camptothecin also inhibit RNA polymerase II elongation [88]. Interestingly, the NER helicase UvrD can “push” PolII backwards in bacteria, uncovering damaged DNA for repair [89].
Recently, it has also been shown that NER components that are not a part of TFIIH are also found at sites of transcription, independent of DNA damage. The DNA damage sensor XPC is recruited to promoters upon gene activation, enhancing both H3K4 methylation and H3K9/K14 acetylation and optimal RNA transcription [90]. Further analysis revealed that the structure-specific nucleases XFP and XPG are found at promoters and termination sites, interacting with the chromatin architecture protein CTCF to promote DNA looping [91]. Both XPC-mediated histone methylation and XPF-XPG-induced looping promote transcriptional activity. Intriguingly, XPG nuclease activity is required for looping and CG demethylation observed at both the promoter and terminator sequences of the RARα gene upon transcriptional activation. It would be interesting to determine whether R loops correlate with NER-occupied promoters, and if these two pathways interact in transcriptional activation. It is possible that NER proteins may act on R loops present at promoter sequences that influence DNA methyltransferase activity and regulate transcriptional activity [69].
Early replicating fragile sites and transcriptional activity
Recently, we have identified a novel class of fragile sites associated with early replication (ERFS) that exhibit high rates of genome instability in response to replication stress. Interestingly, ERFS correlate with actively transcribed genes as measured by RNA-Seq, as well as chromatin signatures found at active promoters, including Dnase I hypersensitivity and H3K4 trimethylation [22]. Indeed, decreased transcriptional activity at discrete ERFS diminishes the genome instability, suggesting that firing origins in active chromatin can act as a double-edged sword under conditions of replicative stress. Promoter regions are more highly correlated with ERFS than exons, 3’ untranslated regions or termination sites. Further, ERFS are enriched for GC content and CpG islands, which both correlate with gene-rich DNA and promoter sequences. Such a bias in where replication stress-induced DNA breaks occur may explain how translocations driving constitutive oncogene expression are generated. It will be interesting to determine which aspects of transcription contribute to or suppress genome instability in S phase.
ERFS do not correlate solely with high transcription, suggesting additional factors contributing to replication-induced genome instability. ERFS map to similar genomic loci in WT and HR-deficient XRCC2 knockout B cells, though DNA aberrations occur at a higher frequency in the latter. Spontaneous DNA breaks also occur at ERFS in XRCC2−/− cells, suggesting that ERFS experience replication stress even in the absence of exogenous agents, and spontaneous ERFS damage is repaired by the HR machinery. Fragile sites adjacent to early firing origins have also been identified in S. cerevisiae in mutant strains deficient for the S phase checkpoint kinases Mec1 or Rad53 [92]. Like ERFS, these sites also correlate with an increased incidence of repetitive elements, and one of the strongest sites identified—adjacent to ARS 310—contains two Ty retrotransposons in a head-to-head orientation [93]. ERFS in mammalian cells are also highly enriched for repetitive DNA elements including highly transcribed short RNAs, suggesting that they can present difficulties for replication fork progression, possibly due to the formation of secondary structures that are difficult to unwind, such as DNA hairpins.
ERFS, genome instability and B cell lymphoma
In B lymphocytes, R loop formation may play a particularly important role in genome instability. Mature B lymphocytes express activation-induced cytidine deaminase (AID) in response to cytokine stimulation, initiating class switch recombination (CSR) a programmed DNA rearrangement at the immunoglobulin heavy chain locus (IgH). AID deaminates cytidine residues in exposed single-strand DNA regions, resulting in abasic sites that are excised form the DNA backbone and converted into ssDNA nicks by the apyrimidinic/apurinic (AP) endonucleases or the mismatch repair machinery [94, 95]. In CSR, AID induces DNA double strand breaks at two or more IgH loci, and the resulting double-strand breaks are repaired by NHEJ, deleting the intervening DNA. Strikingly, both active transcription and the formation of R loops are important for targeting AID to the repetitive, GC-rich switch regions upstream of the heavy chain coding exons. AID directly interacts with PolII, mediated by the transcription elongation factor Spt5 [96, 97]. Thus, the GC-rich nature of switch region DNA performs a two-fold task: (1) forming R loops to expose ssDNA loops and (2) provide a C-rich substrate for AID activity. Further, these results suggest that AID has the potential to interact with any actively transcribed genomic regions promoting R loop formation, and can lead to DNA damage outside the IgH locus. Yet R loops are not strictly necessary for AID activity and functional CSR. Unlike humans and mice, the Xenopus laevis switch region is AT-rich and not prone to R loop formation, yet can functionally substitute for a mouse switch region and stimulate CSR in the mouse in vivo [98]. Thus, while R loop formation increases the time the switch region DNA exists as ssDNA loops, it is not the sole mechanism responsible for AID targeting. Rather, R loops likely increase AID occupancy time by further stabilizing ssDNA loops initially created by active transcription then subsequently bound by RPA [94].
Recently, a number of groups have used genome-wide mapping techniques to identify potential “off-target” AID damage sites. These studies have identified a number of genomic loci susceptible to AID-mediated DNA damage and translocation events to IgH, a hallmark of B cell lymphoma [99–102]. Outside of IgH, AID localizes to other highly transcribed genomic regions, correlating genome-wide with ssDNA and paused PolII signals, indicating the machinery used for CSR also leads to DNA damage that can result in potentially oncogenic DNA translocations [101, 102]. Indeed, some ERFS are also AID “off-site” targets, and these shared “hits” are likely due to their mutual association with high transcription. It will be interesting to determine if replication and deamination act synergistically at these sites, generating high levels of genome instability in activated B cells.
However AID is not the sole source of transcriptionally associated DNA damage in B lymphocytes. In B cells lacking AID, extensive DNA aberrations are still observed at multiple ERFS in response to replication stress. Of note, the proto-oncogene Bcl2 is rearranged in many B cell cancers and frequently involved in activating translocations to IgH, yet the cause of these rearrangements was unknown. Interestingly, the genomic region harboring Bcl2 is acutely prone to replication stress, resulting in genomic aberrations and rearrangement events even in the absence of AID [103]. Further, constitutive expression resulting from translocations to IgH is thought to be the primary cause of follicular lymphoma [104]. Bcl2 is a potent oncogene, whose overexpression blocks apoptotic cell death of lymphocytes. Interestingly, Bcl2 has recently shown to induce replication stress through inhibition of RNR [105]. Thus, it is tempting to speculate that replication stress first induces DNA rearrangements leading to constitutive expression, which in turn generates additional genome instability. Furthermore, we observed a high overlap of ERFS with copy number variations mapped in human diffuse large B cell lymphoma (DLBCL) samples, but not with DNA deletions and/or amplifications mapped in T lineage acute lymphoblastic leukemia (T-ALL). Together, these observations suggest that ERFS represent sites of both “spontaneous” damage and of frequent rearrangement in lymphoma. These results suggest that the replication and transcriptional patterns specific to each cell type influence where DNA rearrangements occur, and may explain why cancers arising from different cell types share few if any genetic aberrations.
Concluding remarks/discussion
The processes of DNA replication and transcription are heavily intertwined in metazoans, particularly upon origin firing and entry into S phase, where highly transcribed euchromatic regions are the first genomic regions duplicated. Regulation of origin licensing, entry into S phase, and replication initiation are essential to genome stability, as dysregulation of these processes result in developmental defects, enhanced DNA damage response, and a predisposition to cancer. Strict regulation of these processes is of particular importance in highly proliferating cells such as B lymphocytes. Stimulated B cells undergo an enormous burst of transcriptional activity and massive proliferative expansion, and replicative stress induces potentially oncogenic genome rearrangements and translocation events [22]. Indeed, genes regulating entry into S phase are commonly dysregulated in a variety of cancers, including hematological malignancies [106–108]. Conventional cancer treatments including radiation therapy and chemo-therapeutic drugs preferentially eliminate tumor cells by exploiting the DNA damage sensitivity of rapidly proliferating cells. Furthermore, specific cell types each have a distinct transcriptional profile that heavily influences both chromatin architecture and replication timing. These differences can influence where DNA damage occurs, rendering some regions more susceptible to breakage, as well as how lesions are repaired. Future studies will underscore the role replicative stress plays in the initiation and evolution of cancer, and how transcriptional activity contributes to replication-mediated genome instability in a cell type-specific manner. Such studies will delineate how cancer cells escape and adapt to existing cancer treatments, and will lead to the development of more effective therapies.
Acknowledgments
The authors would like to thank Sam John and Serena Tan for critical reading of the manuscript, and apologize for not being able to cite more primary literature due to space considerations. Work in the laboratory of A. N. is supported by the Intramural Research Program of the National Institutes of Health, the National Cancer Institute, and the Center for Cancer Research.
References
- 1.Diffley JF (2004) Regulation of early events in chromosome replication. Curr Biol 14(18):R778–R786. doi:10.1016/j.cub.2004.09.019 [DOI] [PubMed] [Google Scholar]
- 2.Bell SP (2002) The origin recognition complex: from simple origins to complex functions. Genes Dev 16(6):659–672. doi:10.1101/gad.969602 [DOI] [PubMed] [Google Scholar]
- 3.Siddiqui K, On KF, Diffley JF (2013) Regulating DNA replication in Eukarya. Cold Spring Harb Perspect Biol 5(9). doi:10.1101/cshperspect.a012930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costa A, Hood IV, Berger JM (2013) Mechanisms for initiating cellular DNA replication. Annu Rev Biochem 82:25–54. doi:10.1146/annurev-biochem-052610-094414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chabes AL, Bjorklund S, Thelander L (2004) S Phase-specific transcription of the mouse ribonucleotide reductase R2 gene requires both a proximal repressive E2F-binding site and an upstream promoter activating region. J Biol Chem 279(11): 10796–10807. doi:10.1074/jbc.M312482200 [DOI] [PubMed] [Google Scholar]
- 6.Blow JJ, Dutta A (2005) Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Bio 6(6):476–486. doi:10.1038/Nrm1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraborty A, Shen Z, Prasanth SG (2011) “ORCanization” on heterochromatin: linking DNA replication initiation to chromatin organization. Epigenetics 6(6):665–670 [DOI] [PubMed] [Google Scholar]
- 8.Prasanth SG, Shen Z, Prasanth KV, Stillman B (2010) Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc Natl Acad Sci USA 107(34):15093–15098. doi:10.1073/pnas.1009945107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomae AW, Pich D, Brocher J, Spindler MP, Berens C, Hock R, Hammerschmidt W, Schepers A (2008) Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proc Natl Acad Sci USA 105(5): 1692–1697. doi:10.1073/pnas.0707260105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scholefield G, Veening JW, Murray H (2011) DnaA and ORC: more than DNA replication initiators. Trends Cell Biol 21(3):188–194. doi:10.1016/j.tcb.2010.10.006 [DOI] [PubMed] [Google Scholar]
- 11.Tatsumi Y, Ezura K, Yoshida K, Yugawa T, Narisawa-Saito M, Kiyono T, Ohta S, Obuse C, Fujita M (2008) Involvement of human ORC and TRF2 in pre-replication complex assembly at telomeres. Genes Cells 13(10):1045–1059. doi:10.1111/j.1365-2443.2008.01224.x [DOI] [PubMed] [Google Scholar]
- 12.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM (2009) TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell 35(4):403–413. doi:10.1016/j.molcel.2009.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atanasiu C, Deng Z, Wiedmer A, Norseen J, Lieberman PM (2006) ORC binding to TRF2 stimulates OriP replication. EMBO Rep 7(7):716–721. doi:10.1038/sj.embor.7400730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert DM (2010) Evaluating genome-scale approaches to eukaryotic DNA replication. Nat Rev Genet 11(10):673–684. doi:10.1038/nrg2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mesner LD, Valsakumar V, Karnani N, Dutta A, Hamlin JL, Bekiranov S (2011) Bubble-chip analysis of human origin distributions demonstrates on a genomic scale significant clustering into zones and significant association with transcription. Genome Res 21(3):377–389. doi:10.1101/gr.111328.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA (2010) Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci USA 107(1):139–144. doi:10.1073/pnas.0912402107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farkash-Amar S, Simon I (2010) Genome-wide analysis of the replication program in mammals. Chromosome Res 18(1):115–125. doi:10.1007/s10577-009-9091-5 [DOI] [PubMed] [Google Scholar]
- 18.Pope BD, Hiratani I, Gilbert DM (2010) Domain-wide regulation of DNA replication timing during mammalian development. Chromosome Res 18(1):127–136. doi:10.1007/s10577-009-9100-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farkash-Amar S, Lipson D, Polten A, Goren A, Helmstetter C, Yakhini Z, Simon I (2008) Global organization of replication time zones of the mouse genome. Genome Res 18(10): 1562–1570. doi:10.1101/gr.079566.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pope BD, Gilbert DM (2013) The replication domain model: regulating replicon firing in the context of large-scale chromo-some architecture. J Mol Biol 425(23):4690–4695. doi:10.1016/j.jmb.2013.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moindrot B, Audit B, Klous P, Baker A, Thermes C, de Laat W, Bouvet P, Mongelard F, Arneodo A (2012) 3D chromatin conformation correlates with replication timing and is conserved in resting cells. Nucleic Acids Res 40(19):9470–9481. doi:10.1093/nar/gks736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barlow JH, Faryabi RB, Callen E, Wong N, Malhowski A, Chen HT, Gutierrez-Cruz G, Sun HW, McKinnon P, Wright G, Casellas R, Robbiani DF, Staudt L, Fernandez-Capetillo O, Nussenzweig A (2013) Identification of early replicating fragile sites that contribute to genome instability. Cell 152(3):620–632. doi:10.1016/j.cell.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svejstrup JQ (2010) The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem Sci 35(6):333–338. doi:10.1016/j.tibs.2010.02.001 [DOI] [PubMed] [Google Scholar]
- 24.Cui P, Lin Q, Xin C, Han L, An L, Wang Y, Hu Z, Ding F, Zhang L, Hu S, Hang H, Yu J (2010) Hydroxyurea-induced global transcriptional suppression in mouse ES cells. Carcino-genesis 31(9):1661–1668. doi:10.1093/carcin/bgq106 [DOI] [PubMed] [Google Scholar]
- 25.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA (2010) ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141(6):970–981. doi:10.1016/j.cell.2010.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knott SR, Viggiani CJ, Aparicio OM (2009) To promote and protect: coordinating DNA replication and transcription for genome stability. Epigenetics 4(6):362–365 [DOI] [PubMed] [Google Scholar]
- 27.Guo ZS, DePamphilis ML (1992) Specific transcription factors stimulate simian virus 40 and polyomavirus origins of DNA replication. Mol Cell Biol 12(6):2514–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J (2013) Organization of the mitotic chromosome. Science 342(6161):948–953. doi:10.1126/science.1236083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ (2006) Excess Mcm2–7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol 173(5):673–683. doi:10.1083/jcb.200602108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbert DM (2001) Making sense of eukaryotic DNA replication origins. Science 294(5540):96–100. doi:10.1126/science.1061724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marechal A, Zou L (2013) DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 5(9). doi:10.1101/cshperspect.a012716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S, Yajima H, Huynh H, Zheng J, Callen E, Chen HT, Wong N, Bunting S, Lin YF, Li M, Lee KJ, Story M, Gapud E, Sleckman BP, Nussenzweig A, Zhang CC, Chen DJ, Chen BP (2011) Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J Cell Biol 193(2):295–305. doi:10.1083/jcb.201009074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santocanale C, Sharma K, Diffley JF (1999) Activation of dormant origins of DNA replication in budding yeast. Genes Dev 13(18):2360–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karnani N, Dutta A (2011) The effect of the intra-S-phase checkpoint on origins of replication in human cells. Genes Dev 25(6):621–633. doi:10.1101/gad.2029711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leman AR, Noguchi E (2013) The replication fork: understanding the eukaryotic replication machinery and the challenges to genome duplication. Genes 4(1):1–32. doi:10.3390/genes4010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halazonetis TD, Gorgoulis VG, Bartek J (2008) An oncogene-induced DNA damage model for cancer development. Science 319(5868):1352–1355. doi:10.1126/science.1140735 [DOI] [PubMed] [Google Scholar]
- 37.Aird KM, Zhang G, Li H, Tu Z, Bitler BG, Garipov A, Wu H, Wei Z, Wagner SN, Herlyn M, Zhang R (2013) Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep 3(4): 1252–1265. doi:10.1016/j.celrep.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones RM, Mortusewicz O, Afzal I, Lorvellec M, Garcia P, Helleday T, Petermann E (2012) Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. doi:10.1038/onc.2012.387 [DOI] [PubMed] [Google Scholar]
- 39.Iizuka M, Matsui T, Takisawa H, Smith MM (2006) Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol 26(3):1098–1108. doi:10.1128/MCB.26.3.1098-1108.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miotto B, Struhl K (2008) HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev 22(19):2633–2638. doi:10.1101/gad.1674108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugimoto N, Yugawa T, Iizuka M, Kiyono T, Fujita M (2011) Chromatin remodeler sucrose nonfermenting 2 homolog (SNF2H) is recruited onto DNA replication origins through interaction with Cdc10 protein-dependent transcript 1 (Cdt1) and promotes pre-replication complex formation. J Biol Chem 286(45):39200–39210. doi:10.1074/jbc.M111.256123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knott SR, Viggiani CJ, Tavare S, Aparicio OM (2009) Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes Dev 23(9):1077–1090. doi:10.1101/gad.1784309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu H, Takeda S, Kumar R, Westergard TD, Brown EJ, Pandita TK, Cheng EH, Hsieh JJ (2010) Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature 467(7313):343–346. doi:10.1038/nature09350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, Cheng EH, Hsieh JJ (2007) Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes Dev 21(19):2385–2398. doi:10.1101/gad.1574507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu H, Maunakea AK, Martin MM, Huang L, Zhang Y, Ryan M, Kim R, Lin CM, Zhao K, Aladjem MI (2013) Methylation of histone H3 on lysine 79 associates with a group of replication origins and helps limit DNA replication once per cell cycle. PLoS Genet 9(6):e1003542. doi:10.1371/journal.pgen.1003542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, Rathjen PD, Gilbert DM (2010) Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res 20(2):155–169. doi:10.1101/gr.099796.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nordman J, Orr-Weaver TL (2012) Regulation of DNA replication during development. Development 139(3):455–464. doi:10.1242/dev.061838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guan Z, Hughes CM, Kosiyatrakul S, Norio P, Sen R, Fiering S, Allis CD, Bouhassira EE, Schildkraut CL (2009) Decreased replication origin activity in temporal transition regions. J Cell Biol 187(5):623–635. doi:10.1083/jcb.200905144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo S, Zi X, Schulz VP, Cheng J, Zhong M, Koochaki SH, Megyola CM, Pan X, Heydari K, Weissman SM, Gallagher PG, Krause DS, Fan R, Lu J (2014) Nonstochastic reprogramming from a privileged somatic cell state. Cell 156(4):649–662. doi:10.1016/j.cell.2014.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M (2002) Histone acetylation regulates the time of replication origin firing. Mol Cell 10(5):1223–1233 [DOI] [PubMed] [Google Scholar]
- 51.Goren A, Tabib A, Hecht M, Cedar H (2008) DNA replication timing of the human beta-globin domain is controlled by histone modification at the origin. Genes Dev 22(10):1319–1324. doi:10.1101/gad.468308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aggarwal BD, Calvi BR (2004) Chromatin regulates origin activity in drosophila follicle cells. Nature 430(6997):372–376. doi:10.1038/nature02694 [DOI] [PubMed] [Google Scholar]
- 53.Knott SR, Peace JM, Ostrow AZ, Gan Y, Rex AE, Viggiani CJ, Tavare S, Aparicio OM (2012) Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell 148(1–2):99–111. doi:10.1016/j.cell.2011.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayano M, Kanoh Y, Matsumoto S, Renard-Guillet C, Shirahige K, Masai H (2012) Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev 26(2):137–150. doi:10.1101/gad.178491.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cornacchia D, Dileep V, Quivy JP, Foti R, Tili F, Santarella-Mellwig R, Antony C, Almouzni G, Gilbert DM, Buonomo SB (2012) Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J 31(18):3678–3690. doi:10.1038/emboj.2012.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buonomo SB, Wu Y, Ferguson D, de Lange T (2009) Mammalian Rif1 contributes to replication stress survival and homology-directed repair. J Cell Biol 187(3):385–398. doi:10.1083/jcb.200902039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamazaki S, Hayano M, Masai H (2013) Replication timing regulation of eukaryotic replicons: Rif1 as a global regulator of replication timing. Trends Genet 29(8):449–460. doi:10.1016/j.tig.2013.05.001 [DOI] [PubMed] [Google Scholar]
- 58.Di Virgilio M, Callen E, Yamane A, Zhang W, Jankovic M, Gitlin AD, Feldhahn N, Resch W, Oliveira TY, Chait BT, Nussenzweig A, Casellas R, Robbiani DF, Nussenzweig MC (2013) Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 339(6120):711–715. doi:10.1126/science.1230624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T (2013) 53BP1 regulates DSB repair using Rif1 to control 5’ end resection. Science 339(6120):700–704. doi:10.1126/science.1231573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, Cook MA, Rosebrock AP, Munro M, Canny MD, Xu D, Durocher D (2013) A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell 49(5):872–883. doi:10.1016/j.molcel.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 61.Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ (2013) RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell 49(5):858–871. doi:10.1016/j.molcel.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L (1999) Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell 3(4):447–455 [DOI] [PubMed] [Google Scholar]
- 63.Johzuka K, Horiuchi T (2002) Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells 7(2):99–113 [DOI] [PubMed] [Google Scholar]
- 64.Srivatsan A, Tehranchi A, MacAlpine DM, Wang JD (2010) Co-orientation of replication and transcription preserves genome integrity. PLoS Genet 6(1):e1000810. doi:10.1371/journal.pgen.1000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pommier Y (2013) Drugging topoisomerases: lessons and challenges. ACS Chem Biol 8(1):82–95. doi:10.1021/cb300648v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA (2013) Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 497(7450):458–462. doi:10.1038/nature12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dillon LW, Pierce LC, Ng MC, Wang YH (2013) Role of DNA secondary structures in fragile site breakage along human chromosome 10. Hum Mol Genet 22(7):1443–1456. doi:10.1093/hmg/dds561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aguilera A, Garcia-Muse T (2012) R loops: from transcription byproducts to threats to genome stability. Mol Cell 46(2): 115–124. doi:10.1016/j.molcel.2012.04.009 [DOI] [PubMed] [Google Scholar]
- 69.Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F (2012) R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45(6):814–825. doi:10.1016/j.molcel.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Skourti-Stathaki K, Proudfoot NJ, Gromak N (2011) Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 42(6):794–805. doi:10.1016/j.molcel.2011.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A (2000) A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J 19(21): 5824–5834. doi:10.1093/emboj/19.21.5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dominguez-Sanchez MS, Barroso S, Gomez-Gonzalez B, Luna R, Aguilera A (2011) Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet 7(12):e1002386. doi:10.1371/journal.pgen.1002386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gomez-Gonzalez B, Garcia-Rubio M, Bermejo R, Gaillard H, Shirahige K, Marin A, Foiani M, Aguilera A (2011) Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J 30(15):3106–3119. doi:10.1038/emboj.2011.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ (2011) Yeast Sen1 Helicase protects the genome from transcription-associated instability. Mol Cell 41(1):21–32. doi:10.1016/j.molcel.2010.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Becherel OJ, Yeo AJ, Stellati A, Heng EY, Luff J, Suraweera AM, Woods R, Fleming J, Carrie D, McKinney K, Xu X, Deng C, Lavin MF (2013) Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet 9(4):e1003435. doi:10.1371/journal.pgen.1003435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alzu A, Bermejo R, Begnis M, Lucca C, Piccini D, Carotenuto W, Saponaro M, Brambati A, Cocito A, Foiani M, Liberi G (2012) Senataxin associates with replication forks to protect fork integrity across RNA-Polymerase-II-transcribed genes. Cell 151(4):835–846. doi:10.1016/j.cell.2012.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Williams JS, Smith DJ, Marjavaara L, Lujan SA, Chabes A, Kunkel TA (2013) Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol Cell 49(5):1010–1015. doi:10.1016/j.molcel.2012.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA (2010) Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol 6(10):774–781. doi:10.1038/nchembio.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li X, Manley JL (2005) Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122(3): 365–378. doi:10.1016/j.cell.2005.06.008 [DOI] [PubMed] [Google Scholar]
- 80.Marston NJ, Richards WJ, Hughes D, Bertwistle D, Marshall CJ, Ashworth A (1999) Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol 19(7):4633–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li J, Zou C, Bai Y, Wazer DE, Band V, Gao Q (2006) DSS1 is required for the stability of BRCA2. Oncogene 25(8): 1186–1194. doi:10.1038/sj.onc.1209153 [DOI] [PubMed] [Google Scholar]
- 82.Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A (2014) BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. doi:10.1038/nature13374 [DOI] [PubMed] [Google Scholar]
- 83.Gomez-Gonzalez B, Felipe-Abrio I, Aguilera A (2009) The S-phase checkpoint is required to respond to R-loops accumulated in THO mutants. Mol Cell Biol 29(19):5203–5213. doi:10.1128/MCB.00402-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X (2011) R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev 25(19):2041–2056. doi:10.1101/gad.17010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, Meyer T, Cimprich KA (2009) A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35(2):228–239. doi:10.1016/j.molcel.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Castellano-Pozo M, Garcia-Muse T, Aguilera A (2012) R-loops cause replication impairment and genome instability during meiosis. EMBO Rep 13(10):923–929. doi:10.1038/embor.2012.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vermeulen W, Fousteri M (2013) Mammalian transcription-coupled excision repair. Cold Spring Harb Perspect Biol 5(8):a012625. doi:10.1101/cshperspect.a012625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ljungman M, Hanawalt PC (1996) The anti-cancer drug camptothecin inhibits elongation but stimulates initiation of RNA polymerase II transcription. Carcinogenesis 17(1):31–35 [DOI] [PubMed] [Google Scholar]
- 89.Epshtein V, Kamarthapu V, McGary K, Svetlov V, Ueberheide B, Proshkin S, Mironov A, Nudler E (2014) UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature 505(7483):372–377. doi:10.1038/nature12928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Le May N, Mota-Fernandes D, Velez-Cruz R, Iltis I, Biard D, Egly JM (2010) NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol Cell 38(1):54–66. doi:10.1016/j.molcel.2010.03.004 [DOI] [PubMed] [Google Scholar]
- 91.Le May N, Fradin D, Iltis I, Bougneres P, Egly JM (2012) XPG and XPF endonucleases trigger chromatin looping and DNA demethylation for accurate expression of activated genes. Mol Cell 47(4):622–632. doi:10.1016/j.molcel.2012.05.050 [DOI] [PubMed] [Google Scholar]
- 92.Raveendranathan M, Chattopadhyay S, Bolon YT, Haworth J, Clarke DJ, Bielinsky AK (2006) Genome-wide replication profiles of S-phase checkpoint mutants reveal fragile sites in yeast. EMBO J 25(15):3627–3639. doi:10.1038/sj.emboj.7601251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD (2005) Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120(5):587–598. doi:10.1016/j.cell.2004.12.039 [DOI] [PubMed] [Google Scholar]
- 94.Dudley DD, Chaudhuri J, Bassing CH, Alt FW (2005) Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv Immunol 86:43–112. doi:10.1016/S0065-2776(04)86002-4 [DOI] [PubMed] [Google Scholar]
- 95.Stavnezer J, Guikema JE, Schrader CE (2008) Mechanism and regulation of class switch recombination. Annu Rev Immunol 26:261–292. doi:10.1146/annurev.immunol.26.021607.090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B (2013) Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 152(3):417–429. doi:10.1016/j.cell.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, Resch W, Yamane A, Reina San-Martin B, Barreto V, Nieland TJ, Root DE, Casellas R, Nussenzweig MC (2010) Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell 143(1):122–133. doi:10.1016/j.cell.2010.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zarrin AA, Alt FW, Chaudhuri J, Stokes N, Kaushal D, Du Pasquier L, Tian M (2004) An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat Immunol 5(12):1275–1281. doi:10.1038/ni1137 [DOI] [PubMed] [Google Scholar]
- 99.Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R (2011) Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol 12(1):62–69. doi:10.1038/ni.1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, McBride KM, Klein IA, Stone G, Eisenreich TR, Ried T, Nussenzweig A, Nussenzweig MC (2009) AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell 36(4):631–641. doi:10.1016/j.molcel.2009.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, Casellas R, Nussenzweig MC (2011) Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 147(1):95–106. doi:10.1016/j.cell.2011.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, Neuberg D, Monti S, Giallourakis CC, Gostissa M, Alt FW (2011) Genome-wide translocation sequencing reveals mechanisms of chromo-some breaks and rearrangements in B cells. Cell 147(1): 107–119. doi:10.1016/j.cell.2011.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lindsley RC, LaCasce AS (2012) Biology of double-hit B-cell lymphomas. Curr Opin Hematol 19(4):299–304. doi:10.1097/MOH.0b013e328353bbbd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kridel R, Sehn LH, Gascoyne RD (2012) Pathogenesis of follicular lymphoma. J Clin Investig 122(10):3424–3431. doi:10.1172/JCI63186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xie M, Yen Y, Owonikoko TK, Ramalingam SS, Khuri FR, Curran WJ, Doetsch PW, Deng X (2014) Bcl2 induces DNA replication stress by inhibiting ribonucleotide reductase. Cancer Res 74(1):212–223. doi:10.1158/0008-5472.CAN-13-1536-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Malumbres M, Barbacid M (2001) To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer 1(3):222–231. doi:10.1038/35106065 [DOI] [PubMed] [Google Scholar]
- 107.Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, Wright G, Shaffer AL, Hodson DJ, Buras E, Liu X, Powell J, Yang Y, Xu W, Zhao H, Kohlhammer H, Rosenwald A, Kluin P, Muller-Hermelink HK, Ott G, Gascoyne RD, Connors JM, Rimsza LM, Campo E, Jaffe ES, Delabie J, Smeland EB, Ogwang MD, Reynolds SJ, Fisher RI, Braziel RM, Tubbs RR, Cook JR, Weisenburger DD, Chan WC, Pittaluga S, Wilson W, Waldmann TA, Rowe M, Mbulaiteye SM, Rickinson AB, Staudt LM (2012) Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 490(7418):116–120. doi:10.1038/nature11378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Choi YJ, Li X, Hydbring P, Sanda T, Stefano J, Christie AL, Signoretti S, Look AT, Kung AL, von Boehmer H, Sicinski P (2012) The requirement for cyclin D function in tumor maintenance. Cancer Cell 22(4):438–451. doi:10.1016/j.ccr.2012.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li F, Mao G, Tong D, Huang J, Gu L, Yang W, Li GM (2013) The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell 153(3): 590–600. doi:10.1016/j.cell.2013.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]