Abstract
DNA methylation is an epigenetic modification that is implicated in transcriptional silencing. It is becoming increasingly clear that both correct levels and proper interpretation of DNA methylation are important for normal development and function of many organisms, including humans. In this review we focus on recent advances in understanding of how proteins that bind to methylated DNA recognize their binding sites and translate the DNA methylation signal into functional states of chromatin. Although the function of methyl-CpG binding proteins in transcriptional repression has been attributed to their cooperation with corepressor complexes, additional roles for these proteins in chromatin compaction and spatial organization of nuclear domains have also been proposed. Finally, we provide a brief overview of how methyl-CpG proteins contribute to human disease processes such as Rett Syndrome and cancer.
Keywords: DNA methylation, methyl-CpG binding proteins, MBD, chromatin
Introduction
Post-synthetic modification of DNA by methylation is found in most living organisms from bacteria to mammals. In prokaryotes, methylation occurs both on adenine and cytosine nucleotides and is involved in regulation of DNA replication timing, DNA repair and defence against invasion by foreign DNA [1]. The genomes of eukaryotes are modified exclusively at cytosine and in vertebrates only in the context of CpG dinucleotides [2]. Fungi, such as Neurospora crassa, and plants also contain methylated cytosine in non-CpG context [3,4]. Not all eukaryotes have methylated DNA. Species such as yeast and many invertebrates, including the nematode C . elegans and the fly D. melanogaster, contain either no or barely detectable amounts of methylated cytosine in their genomes [2,5].
DNA methylation is introduced into DNA by enzymes of the DNA cytosine methyltransferases family. In vertebrates, these are represented by DNMT1, DNMT3A and DNMT3B [6]. DNMT1, the maintenance DNA methyltransferase, works most efficiently on hemi-methylated DNA and functions to restore symmetrically methylated cytosine on daughter DNA strands generated during replication [7,8]. DNMT3 enzymes, also referred to as de novo methyltransferases, can work on fully unmodified DNA and are essential for establishment of DNA methylation patterns during embryogenesis [9].
Most genomes with high levels of DNA methylation are depleted of CpGs due to the frequent deamination of methyl-cytosine into thymidine. This generates mCpG:TpG mismatches which, if unrepaired, are further stabilized by DNA replication [2,10]. The remaining CpGs are unevenly distributed throughout the genome. Most gene promoters (~70% in mammals) are imbedded in an unmethylated stretches of DNA with high CpG density, also known as CpG islands [2,11,12]. How the CpG islands are maintained in an unmethylated state and what protects them from the action of DNA methyltransferases is currently unclear. Nevertheless, these sequences are not intrinsically unmethylatable since some of them acquire DNA methylation in differentiated cells and can be found aberrantly methylated in cancers [13,14]. The lack of methylation at CpG islands and heavily methylated coding and intergenic regions generate patterns of DNA methylation that are heritably maintained in somatic cell lineages [2].
Biological functions of DNA methylation have been intensively studied over several decades. It is now well established that DNA methylation generally associates with silent chromatin which is inhibitory to transcriptional initiation [11,15]. Important processes such as monoallelic expression of imprinted genes in plants and placental animals, X-chromosome inactivation in mammals and suppression of transposable elements in complex genomes include DNA methylation as part of more complex regulatory functions [16,17]. Given that most of the gene promoters are methylation-free, the question of whether DNA methylation is essential for regulation of gene expression on a global scale has been a subject of debate [2,16,18]. Nevertheless, mice with disrupted alleles of Dnmt1 or double null for Dnmt3a and Dnmt3b die early in embryogenesis and show inappropriate expression of large number of genes [9,19,20]. Xenopus laevis embryos depleted of DNMT1 initiate zygotic transcription 2-3 cell cycles earlier than it normally occurs at the midblastula transition and display aberrant expression of developmentally decisive genes [21,22]. Therefore, at least in vertebrates, DNA methylation plays a conserved role in maintaining stable patterns of gene expression. Additional evidence from plants and the primitive chordate C. intestinalis shows that methylated cytosines are enriched within the coding regions of genes and may regulate transcriptional elongation [23,24]. It is worth mentioning that a role for DNA methylation in suppressing “transcriptional noise” in large genomes, i.e. initiation of spurious transcription from cryptic promoters within coding regions of genes or non-coding DNA, has been proposed and still requires more detailed investigation [25,26].
There are two models of how DNA methylation exerts its repressive effect on transcription. In the first model, CpG methylation alters binding sites of transcription factors and directly interferes with gene activation [11]. Examples validating this model include E2F, CREB and c-myc [27-29]. In the second model, methylated cytosines serve as docking sites for proteins that specifically recognize and bind to methylated CpGs and repress transcription indirectly via recruitment of corepressors that modify chromatin [11,15]. In this review we focus on the methyl-CpG binding proteins and how they translate DNA methylation patterns into functional states of chromatin.
Families of methyl-CpG binding proteins
Currently two major families of methyl-CpG binding proteins are known in vertebrates: MBDs and Kaiso-like proteins. In addition, recent studies indicate that SRA domain proteins, characterized in some detail in plants, have the ability to bind methylated DNA in non-CpG context.
MBD family proteins
Ironically, the first methyl-CpG binding protein, MeCP2, was discovered by accident by Adrian Bird and co-workers, who at the time were attempting to identify factors that bind to unmethylated DNA and would function to protect CpG islands from DNA methylation. Instead, protein factors, initially named MeCP1 and MeCP2 that bind specifically to methylated DNA were detected [30,31]. MeCP2 was purified first and represents a 53 kDa protein containing a N-terminal methyl-CpG binding domain (MBD) and a C-terminal transcriptional repression domain (TRD) [32,33]. Mammalian EST database homology searches for sequences encoding a conserved MBD domain led to the identification of four additional proteins currently known as MBD1, MBD2, MBD3 and MBD4 [34] (Figure1). Of those, MBD2 and MBD3 are closely related to each other outside the MBD domain (77% identity) and are likely to represent the ancestral MBD family founders since a homologous MBD2/3 like protein is present in invertebrates, including Drosophila where low levels of DNA methylation are detectable only in early development [5,35]. All MBD proteins, except MBD3, specifically recognize and bind to methylated DNA in vitro and in vivo [34]. Mammalian MBD3, unlike its amphibian homologue, harbours a critical mutation in the MBD domain and does not bind to methylated DNA [15,34]. The MBD family proteins, including MeCP2, are highly conserved in all vertebrates [36]. Interestingly, at least 12 MBD proteins have been identified in the plant Arabidopsis thaliana. Of these AtMBD5, AtMBD6 and AtMBD7 have been shown to bind methylated DNA in vitro [37].
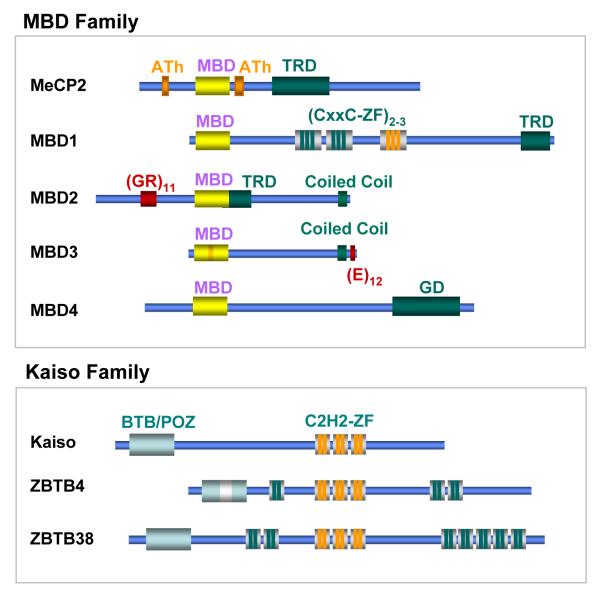
Figure 1. Families of methyl-CpG binding proteins.
MBD family proteins share a conserved MBD domain, which is required for binding to methylated DNA. MBD3 carries a mutation (shown in orange) in the MBD domain and does not bind to methylated CpGs. MeCP2 has two AT-hook motifs (ATh) which potentially could bind AT-rich DNA. These motifs are not required for high affinity binding to sequences containing a methylated CpG followed by an [A/T]≥4 run. MBD1 is characterized by two (or three in some isoforms) CxxC-type zinc fingers. The third CxxC motif (orange) binds unmethylated CpGs.
TRD indicates transcriptional repression domains mapped by functional and deletion analyses. GD indicates Glycosylase domain of MBD4 which is involved in excision of CG:TG mismatches. (GR)11 motif of MBD2 is a stretch of Glycine and Arginine residues that can be methylated by PRMT5 protein methylase [98]. (E)12 is a glutamate-rich domain. Kaiso family of proteins is characterized by three homologous C2H2 zinc finger motifs that are required for binding to methylated and in some instances unmethylated DNA. In addition, all proteins of this family carry a BTB/POZ domain likely to be involved in either homo- or heterodimerization or protein-protein interactions. ZBTB4 and ZBTB38 have additional three and seven, respectively, zinc finger motifs.
Kaiso family proteins
Kaiso, the founder protein of Kaiso-like family, was independently discovered in two different laboratories as a DNA binding factor involved in non-canonical Wnt signalling and as a protein that binds to methylated DNA [38,39]. Unlike the MBD family members, Kaiso and the two recently identified Kaiso-like proteins ZBTB4 and ZBTB38 contain a conserved POZ domain involved in protein-protein interactions and three C2H2 zinc finger motifs, two of which are essential for binding to methylated DNA [40] (Figure1). Unmethylated sequences recognized by Kaiso with high affinity have also been reported [41] raising the possibility that in some circumstances Kaiso and Kaiso-like proteins may bind to nonmethylated DNA and to methylated DNA in others.
SRA domain proteins
Recent reports suggest that another protein fold, the SRA (or YNG) domain could interpret DNA methylation [42-44]. SRA domain-containing proteins fall in two distinct families. The first one is characterised by association of the SRA domain with PHD and RING finger domains. At least 5 members of this family exist in A. thaliana including the product of the recently cloned vim1 gene [42]. So far described mammalian homologues include the Np95 protein, the closely related Np97 (or NIRF) and ICBP90 [42,44]. Recent studies suggest that Np95 plays a critical role in epigentic inheritance of DNA methylation [45,46]. The second family of SRA domain proteins is plant- specific, without obvious mammalian counterparts, and includes members of the SUVH family of SET domain histone methyltransferases in A. thaliana [43]. Interestingly, the SRA domain seems more versatile than the MBD domain in recognizing methylated DNA as in vitro it binds to methyl-cytosine at CpG, CpNpG and even at the asymmetric CpNpN sites, with a marked preference for mCpNpG [42,43].
In the rest of this review we will focus in more detail on the MBD and in part the Kaiso-like proteins in mammalian cells.
DNA binding properties of methyl-CpG binding proteins
The molecular functions of methyl-CpG binding proteins rely on their ability to recognize and bind methylated DNA. As this property is central to understanding their roles in vivo, we will review the current progress in this area in more detail.
Deletion analyses have identified the minimal region of MeCP2 responsible for the interaction with methylated CpGs [32]. Further comparison with other MBD proteins defined the MBD domain as a protein motif of about 75 amino acids [34,47]. Since the “classical” MBD was described, proteins containing MBD-like domain, including ESET/SETDB1 and TIP5, have also been identified in different species. However, as in the case of TIP5 protein, the MBD-like domains are predicted not to form specific interactions with methylated DNA and therefore may serve other functions. A unifying name of TAM domain (TIP5, ARBP, MBD) is now used to unify both canonical MBD and MBD-like domains [47]. MBD-like domains proteins are not the subject of this review.
Sequence comparison of all human MBD family proteins show the presence of 16 strictly conserved amino acids within the MBD domain. MBD3, which does not bind to methylated DNA, lacks four of these conserved residues [34]. Pairwise comparison reveals the presence of two subclasses, with the MBD domains of MBD4 and MeCP2 being more closely related to each others, while those of MBD1, MBD2 and even MBD3 forming a separate subgroup. Solution structures of MBD domains of human MeCP2 and MBD1 have been determined by NMR revealing a similar α/β sandwich fold composed of four β-strands and an α-helix [48,49] (Figure2A and B). Detailed information on how the MBD fold binds symmetrically methylated CpG was derived from the NMR structure of the MBD domain of MBD1 in complex with methylated DNA [50] (Figure2C and D).
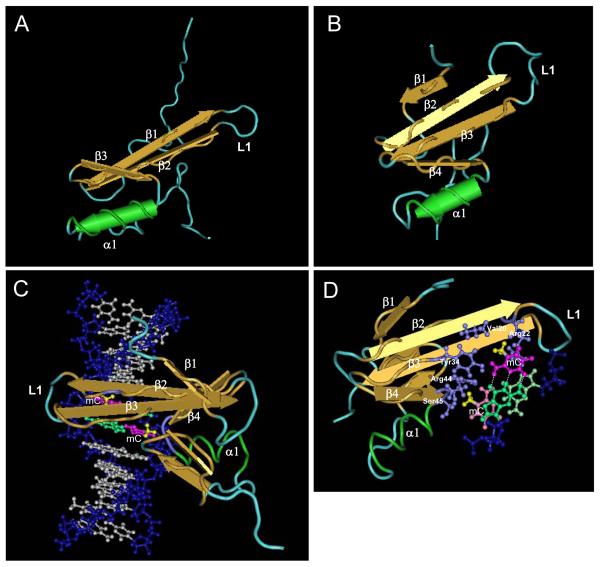
Figure 2. Interaction of the MBD domain with methylated DNA.
A. Schematic representation of the solution structure of MBD domain of MeCP2 [49]. The beta sheets (β), an alpha helix (α1q) and a long loop 1 (L1) are indicated. B. Solution structure of MBD domain of MBD1 [48]. C. MBD domain of MBD1 bound to DNA in the major groove where the methyl-groups (yellow) of methylated cytosines (magenta) are exposed towards the exterior of the double helix [50]. Residues (purple) from the two beta sheets as well as L1 and shorter loot connecting β4 with α1 are involved in interactions with the methyl-groups and the cytosine bases. G bases of the CpG pairs are colored in green. D. Top view of the MBD domain of MBD1 in contact with a pair of symmetrically methylated CpG (the top pair is indicated with brighter colours). The side chains (purple) of Valine 20 and Arginine 22 are involved in recognizing the first (top) methyl-group while those of Tyrosine 34, Arginine 44 and Serine 45 contact the second methyl-group. The figures were generated from published structures by Cn3D software.
MBD proteins interact with methylated DNA in the major groove, where the two methyl groups from the mCpG point towards the exterior of the double helix (Figure 2C). Several residues from the L1 loop, connecting the β2 and β3 strands, and the α helix respectively make several contacts with the sugar/phosphate backbone on each strand of the DNA molecule (Figure 2D). Four conserved residues (R22, Y34, R44, S45) in MBD1 are involved in recognizing the methyl-CpGs via a complex set of interactions. It appears that each side chain interacts with DNA in a somehow bivalent way, where the polar moiety of each of these residues contacts C or G base, while their hydrophobic regions stack around the methyl groups. Such of bivalent contacts from each important amino acid side chain may explain why both the CpG dinucleotide and the two methyl groups are strictly required for efficient recognition by the MBD. Subtle variations in this network might abolish binding. MBD3 for example has only three of the four conserved residues with Tyrosine (Y) to Phenylalanine (F) substitution at the equivalent position of Y34. The loss of a single hydroxyl group renders MBD3 incapable of binding to methylated DNA [34,51]. This particular arrangement of critical amino acids is likely to explain the high selectivity observed in vitro towards methylated DNA versus either hemimethylated or unmethylated CpGs. The structural data also confirm that one MBD domain can only accommodate one symmetrically methylated CpG as the MBD domain binds DNA as a monomer [32,48]. However, this does not exclude the presence of potential homo or heterodimerisation interfaces on MBD proteins, even if MeCP2 appear to be mostly monomeric in solution [52]. The only case that complicates this picture is MBD4, whose MBD domain seems capable of interacting preferentially with mCpG:TpG mismatches arising from deamination of methyl-cytosine and even with hemimethylated DNA [53,54]. The structural information available so far does not explain why MBD4, whose MBD domain is related to MeCP2 more than any other, would display an altered DNA binding specificity. However, it seems that cytosine methylation and even the MBD itself can be dispensable for MBD4 G:T mismatch-specific thymine glycosylase activity [55].
About 70 to 80% of CpG are methylated in mammalian genomes, creating a relatively high number of potential binding sites for MBD proteins [2]. Then what determines their pattern of occupancy at these sites? One possible model would be that each MBD protein randomly occupies any available methylated CpG (Figure 3A). In this scenario, the relative abundance of each MBD protein within a cell, together with the methylation density will dictate the occupancy of individual methylated sites. This random behaviour would imply high redundancy and is the principal argument to explain the relatively mild phenotypes of MBD1, MBD2 and Kaiso null mice [56-58]. In another model, one can envisage that other factors may influence the distribution of MBD proteins within a cell nucleus, making it non-uniform and non-random, with each MBD protein occupying unique sites in the genome (Figure 3B). This model would predict that a subset of genes would be affected by the loss of one MBD protein but not other. Examples of genes missexpressed in the absence of specific MBD proteins are becoming more abundant and the phenotypes of MBD deficient mice, although subtle, are markedly different [56,58-61].
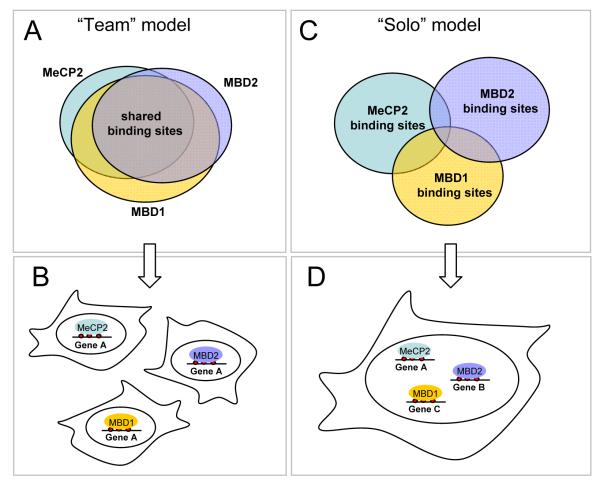
Figure 3. Models of how MBD proteins recognize methylated DNA.
A. The “team” model indicates a non-discriminative binding of MBD proteins to any accessible methylated CpG. Thus most binding sites will be potentially shared and the overall pattern of occupancy will be governed by the concentration of each MBD protein. B. The “team” model would predict redundancy of MBD proteins where the repression of any methylated gene, for example gene A, could be achieved by any methyl-CpG binding protein. C. The “solo” model implies specificity of binding where some MBD proteins discriminate between binding sites either by recognition of bases adjacent to the mCpG or other nearby DNA motifs. Therefore most of the binding sites will not be shared.
D. The “solo” model would imply that each MBD protein by occupying a distinct set of binding sites would regulate a specific subset of genes.
In support of the second model, a recent study demonstrates that in primary human fibroblasts, MBD1, MBD2 and MeCP2 do not share binding sites in vivo, at least at the number of genomic sequences examined [62]. Morpholino-mediated depletion of MeCP2 and MBD2 suggested the existence of a mechanism dictating preference of MeCP2 but not MBD2 for a subset of methylated sites in vivo [62]. Whether this selective binding is retained in cancer cells, which tend to accumulate aberrant DNA methylation patterns, is unclear [63]. Specific targeting of MBD proteins, observed in primary cells, may be achieved via interactions with binding partners (see below) including other DNA binding activities which may facilitate targeting of MBDs to chromatin or DNA at specific loci. Experimental evidence for recruitment via partner proteins is currently missing. Another possibility, which has been validated to some extent, is that the various members of the MBD family display different DNA binding specificity, meaning that they recognize and bind to more complex sequences than a single methylated CpG.
Recent in vitro experiment showed that, unlike MBD2, MeCP2 requires a run of four or more A/T base pairs adjacent to methylated CpG for high affinity binding [62]. Furthermore [A/T]≥4 runs are present at MeCP2 target sequences identified in vivo [62]. These findings constitute the first example where the enhanced binding specificity towards a particular set of methylated sequences allows discriminative binding site occupancy of an MBD protein. Whether this is the case for other MBD proteins remains to be determined. However, as MeCP2 and MBD1 contain additional DNA binding domains, it is possible that a single methylated CpG is not sufficient to support high affinity binding of an MBD protein to DNA. Early studies on MeCP2 detected a potential second DNA binding activity independent of the MBD domain and sequence analyses identified the presence of two AT-hooks [31,62,64] (Figure1). The AT-hook motif is capable of interacting with the minor groove of AT rich DNA and has been characterized in high mobility group proteins such as HMGA1 [65]. However, the AT hooks are frequently present in conjunction with other functional DNA or chromatin binding domains [66], and in the case of MeCP2 their functionality remains to be determined. Surprisingly, the AT hooks are not required for selective binding of MeCP2 to CpG followed by [A/T]≥4 run [62]. However, these motifs may interact with other stretches of A/T-rich DNA in cis or trans. Additionally, a role of the C-terminus of MeCP2 in helping binding to DNA, matrix attachment regions and nucleosome has also been reported [67-70]. Whether multiple DNA and chromatin binding interfaces play a role in MeCP2 function requires further studies.
On the other hand, MBD1 protein carries a second functional DNA binding motif separate from the MBD. Depending on the isoform, MBD1 can have two or three zinc finger motifs defined by 8 conserved cysteines, the CxxC zinc finger [71,72]. However, each copy is not strictly equivalent to one another, as they display primary sequence differences that would alter their biochemical properties. The most C terminal zinc finger (usually referred as CxxC3), which we will consider as a canonical CxxC motif, is also present in various other proteins, including DNMT1, CpG binding protein CGBP, H3K4 histone methylase MLL and H3K36 histone demethylases of the Jumonji family JHDM1A and JHDM1B [71,73,74]. This canonical version of the CxxC zinc finger has been shown to bind non methylated CpGs in vitro in the case of MBD1, MLL CGBP and JHDM1B [71,74-76]. The two other CxxC motifs of MBD1 lack a conserved glutamine residue and a KFGG motif, characteristic of all DNA binding CxxC zinc fingers, and as a consequence are unable to bind DNA [71]. The role of these divergent CxxC zinc fingers is unclear but they might be involved in protein-protein interactions [61].
In reporter gene assays, MBD1 represses transcription from CpG rich unmethylated promoters in a CxxC3 domain-dependant manner [71,72]. This suggests that this domain could be as efficient as the MBD for targeting MBD1 to DNA in vivo and therefore, MBD1 may play a role in silencing certain unmethylated CpG island promoters. However, the CxxC3 domain by itself does not provide enough sequence specificity to discriminate between different CpG islands, which are defined by their high CpG content. A mechanism(s) that would account for the specific targeting of MBD1, and other CxxC containing protein, to specific DNA loci remains to be uncovered. An attractive hypothesis would be that MBD1 requires each of its two DNA binding domains for efficient binding at specific loci in vivo. As each domain interacts with a very short sequence (2 nucleotides) compared to classical DNA binding transcription factors, one can speculate that the use of two separate DNA binding domains might enhance the specificity for particular sequences in vivo. Another possibility is that, similar to MeCP2 which requires a methylated CpG followed by an [A/T]≥4 run, each DNA binding domain of MBD1 recognizes a more complex sequence than currently known. Whether MBD1 binds DNA through an independent use of its two DNA binding domains, or they collaborate with each other to target efficiently MBD1 to specific loci will be an intriguing question to answer.
It might appear surprising that a methyl-CpG binding protein carries a domain that allows it to bind unmethylated CpGs. However, this ability to interact with methylated and unmethylated DNA is not a unique feature of MBD1. The identification of Kaiso showed that the MBD domain is not the only protein fold able to recognize DNA methylation, as Kaiso and the Kaiso-like proteins ZBTB4 and ZBTB38 use a set of C2H2 zinc fingers to bind methylated DNA [39,40]. Early studies suggested that Kaiso requires at least two mCpGs for efficient binding, while ZBTB4 and ZBTB38 seem to interact with a single mCpG [39,40]. In vitro studies also show that Kaiso interacts specifically with unmethylated consensus sequence, the Kaiso Binding Site (KBS:TCCTGCNA), which is present at promoters of Wnt target genes [77,78]. Interestingly, only zinc fingers 2 and 3 of Kaiso are necessary and sufficient for binding to either type of sequences in vitro [41]. The ability to bind unmethylated DNA is shared by ZBTB4, but surprisingly not by ZBTB38. High resolution structural information may help to explain how the zinc-fingers of Kaiso and Kaiso-like proteins interact with methylated and unmethylated DNA. Such structural studies may facilitate the design of specific point mutations which would allow uncoupling of mCpG and KBS binding activities and clear cut discrimination between the functions of Kaiso and ZBTB4 that rely on their interaction with either methylated or unmethylated DNA.
In summary, several lines of evidence suggest that methyl-CpG binding proteins recognize more complex sequences than a single methylated CpG, thus favouring a gene or locus specific role for each member of the MBD and Kaiso-like families. As MBD proteins are widely expressed in different tissues and constitute relatively abundant chromosomal proteins, it has been suggested that they may also exert functions unrelated to recognition of methylated DNA. Although binding of MBD proteins to other nucleic acids such as RNA and cruciform DNA structures in vitro has been reported [79,80], evidence in vivo for the most part firmly supports the function of MBD proteins in reading DNA methylation patterns at specific loci. One likely explanation for the existence of two (and maybe more) families of divergent methyl-CpG binding proteins, with members that display different sequence specificity towards methylated DNA, could be the evolutionary adaptation of pre-existing nucleic acid binding motifs for binding to methylated DNA.
Establishment of silenced chromatin by methyl-CpG binding proteins
Early studies have demonstrated that DNA methylation has no major impact on gene expression until the DNA template is assembled into chromatin [81,82]. Once the MBD proteins were discovered and shown to function as methylation-dependent transcriptional repressors, the work in several laboratories has focused on identification of co-repressor complexes associated with these proteins. Currently, it is established that MBD proteins cooperate with histone deacetylases and histone methylase activities that modify chromatin and prevent productive initiation of transcription [15] (Figure 4).
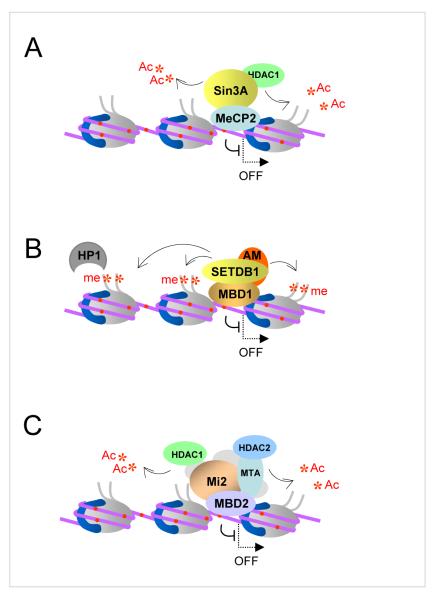
Figure 4. Complexes of MBDs with co-repressor proteins.
A. MeCP2 co-immunoprecipitates with Sin3A/HDAC1 complex from most cell types. HDAC1 removes acetyl groups (*Ac) from histone tails and is responsible for MeCP2-mediated repression. Histone tails free of acetylation can be modified by histone methylase activity associated with MeCP2 to generate heterochromatin. B. MBD1 interacts with SETDB1 histone methylase and SETDB1 co-factor AM/MCAF1. SETDB1 methylates K9 of histone H3 tail to generate silenced chromatin. H3K9me3 is further recognized by heterochromatin protein HP1. C. MBD2 participates in a large protein complex known as NuRD, which includes HDAC1 and HDAC2, which, similar to MeCP2 associated complex, deacetylates histone tails to generate transcriptionally silent chromatin. Methylated CpGs are shown as red dots.
Protein complexes containing MeCP2 and MBD2 have been purified by biochemical fractionation and found to contain class I histone deacetylase activities HDAC1 and HDAC2 [33,83,84]. MeCP2 was co-purified with Sin3A/HDAC2 complex from Xenopus oocyte extract and a number of studies have shown than the interactions of MeCP2 with Sin3A and HDAC2 are conserved in mammalian cells and are essential for MeCP2-mediated repression [33,83]. A number of other proteins directly or indirectly interacting with MeCP2 have been found including DNMT1, CoREST, NCoR/SMRT, c-SKI, histone H3 lysine 9 methylase activity, RNA splicing factors and chromatin remodelling activities such as ATRX and Brahma (Brm1)-related SWI/SNF complex [85-92]. Nevertheless, the composition of a well defined MeCP2 co-repressor complex that would be present in all cell types or tissues remains elusive. Purification of MeCP2 from mammalian sources, including brain where this protein is most abundant, has produced conflicting results, ranging from complete lack of stable association of MeCP2 with either Sin3A, or any other proteins in nuclear extracts [52,93], to the identification of high molecular weight MeCP2 containing complexes [89,91,94]. Some of these discrepancies could be explained by the use of different biochemical methods that may vary in sensitivity of detection of MeCP2-associated proteins. It is also possible that most of the interactions of MeCP2 with partner proteins are either relatively unstable or cell type- and/or locus- specific [94]. In addition, these findings raise the possibility that DNA-bound MeCP2 may interact with partner proteins differently compared to the unbound form of MeCP2, which behaves as an unusually elongated monomeric molecule in solution [52]. Perhaps affinity tagging of the endogenous MeCP2 protein and tandem purification of the MeCP2 containing complexes from a variety of tissues may provide some interesting insights.
MBD2 and MBD3 co-purify with a large protein complex known as NuRD (Nucleosome Remodelling and Histone Deacetylation) which contains chromatin remodelling ATPase Mi-2, HDAC1 and HDAC2 histone deacetylases as well as other proteins [84,95]. The NuRD complex exists in several forms and may or may not contain MBD proteins. Initially, it was suggested that MBD2 and MBD3 together associate with NuRD [84,96,97], but recent affinity tag purifications of MBD2 and MBD3 complexes from mammalian cells showed that NuRD associates either with MBD2 or MBD3 but never with both proteins [98]. These different MBD complexes have probably no or very little functional overlap since MBD3 null mice die early during embryogenesis while MBD2-deficient animals are viable and fertile [56]. As MBD3 does not bind to methylated DNA, these findings indicate that only a proportion of NuRD complexes would be recruited to methylated DNA and participate in methylation-dependent transcriptional repression. It appears that association with ATP-dependent chromatin remodelling activities is a common feature of MeCP2- and MBD2-associated protein complexes. Although MeCP2 and MBD2 are likely to be responsible for the initial recruitment of these complexes to chromatin assembled on methylated DNA, studies in vitro and in vivo suggest that chromatin remodelling activities further facilitate binding of MBD proteins to methylated sites that are not readily accessible on nucleosomal templates and by doing so stimulate MBD-mediated gene repression [89,99].
MBD1 protein in most assays behaves as a histone deacetylation-independent transcriptional repressor [100]. At least two histone H3 lysine 9 (H3K9) methylase activities SETDB1 and SUV39H have been found associated with MBD1 as well as the heterochromatin protein HP1 [61,101]. In addition the C-terminus of MBD1 binds a SETDB1 co-factor AM/MCAF which stimulates SETDB1 activity to allow more efficient di- and trimethylation of H3K9 [102,103]. Furthermore, it was shown that during S-phase of the cell cycle MBD1/SETDB1 complex can be displaced from methylated DNA by progressing replication forks to allow the formation of a transient complex with p150 subunit of chromatin assembly factor CAF-1 [61]. As a result of this interaction, the MBD1-bound SETDB1 methylates H3K9 of the H3/H4 dimers associated with CAF-1. Thus the S-phase specific MBD1 complex facilitates post-replicative maintenance of the repressive H3K9 chromatin modification on methylated daughter DNA strands [61]. This mechanism provides a plausible explanation of how silenced chromatin can be heritably transmitted through DNA replication and cell division in synchrony with DNA methylation. The function of MBD1 in transcriptional repression and maintenance of H3K9 methylation is negatively regulated by conjugation of SUMO1 [104]. Two E3 SUMO-ligases PIAS1 and PIAS3 sumoylate MBD1 in human cells and compete against SETDB1 for interaction with MBD1. SETDB1 can bind MBD1-SUMO1 in vitro but not in vivo suggesting that there could be a specific binding partner(s) for sumoylated MBD1, which disrupt the formation of the MBD1/SETDB1 complex [104]. Identification of factors that bind MBD1-SUMO1 but not MBD1 will be essential for the mechanistic understanding of how the function of MBD1 might be regulated in response to physiological stimuli. Intriguingly, conjugation of SUMO2/3 to MBD1 has also been reported and, unlike SUMO1, seems to stimulate transcriptional repression by MBD1 [105]. Therefore it is possible that these two modifications recruit different binding partners to MBD1. Given that a number of proteins were found to associate with MBD1, biochemical purification of MBD1 complex(es) may help to determine whether MBD1 stably associates with a set of co-repressor proteins or, like MeCP2, could cooperate with many different nuclear factors.
Similar to MBDs, Kaiso-like proteins function as HDAC-dependent transcriptional repressors. From HeLa cell nuclear extracts, Kaiso co-purifies with NCoR (Nuclear receptor Co-Repressor) complex containing histone deacetylase HDAC3 and this association is required for silencing of methylated MTA2 promoter [106]. Depletion of Kaiso in Xenopus embryos results in derepression of methylated genes before the midblastula transition [107]. However, Xenopus Kaiso is also involved in a non-canonical Wnt pathway where its function is controlled through an association with p120-catenin and is essential for regulation of target genes in a methylation-independent manner [77,78]. Complexes containing Kaiso-like proteins have not yet been purified but ZBTB38 protein was shown to interact with several histone deacetylases activities and co-repressor CtBP [108].
Taken together, methyl-CpG binding proteins represent an important class of chromosomal proteins which associate with multiple protein partners to modify surrounding chromatin and silence transcription, providing a functional link between DNA methylation and chromatin remodelling and modification.
MBD proteins and chromatin architecture
Independently from the establishment of transcriptionally inactive chromatin via cooperation with corepressor proteins, a more direct role of MBD proteins in organization of higher order chromatin structure has also been proposed. Earlier studies have shown that MeCP2 forms discrete complexes with nucleosomes assembled on methylated DNA, can displace histone H1 from pre-assembled chromatin and in addition is able to interact with the nucleosome core via its C-terminus [67,109]. More recent studies have reported that purified recombinant MeCP2 when added to nucleosomal arrays in vitro causes chromatin compaction, which has been attributed to additional interactions between MeCP2 and DNA or chromatin in cis or trans via a domain(s) different from the MBD [68,69].
In vivo, in mouse cells, MeCP2 as well as other MBD and Kaiso-like proteins localize to condensed pericentric heterochromatin regions known as chromocenters (Figure 5A-D). These chromatin domains are also enriched in histone H3 trimethylated at lysine 9 and heterochromatin proteins HP1α and HP1β [110,111]. During myogenic differentiation of mouse C2C12 cells the pericentric heterochromatin domains undergo reorganization and cluster into smaller number of larger chromocenters [112]. These events are accompanied by an increase in methylation of major satellite DNA and accumulation of MeCP2 and MBD2 proteins in the nuclei of terminally differentiated muscle cells (myotubes). Interestingly, overexpression of MeCP2 and MBD2 in C2C12 myoblasts in the absence of differentiation also induces aggregation of chromocenters indicating that these proteins may be directly involved in reorganization of heterochromatin architecture [112]. As overexpression of MBD domain of MeCP2 is sufficient to cause fusion of chromocenters, these events are unlikely to involve co-repressor HDAC complexes or other proteins interacting with the C-terminal region of MeCP2. The mechanism by which MBD proteins induce aggregation of chromocenters is unclear although it has been suggested that it may be caused by oligomerization of MBD proteins and formation of DNA-MBD-MBD-DNA structures or multiple interactions between MBD protein and DNA as mentioned above [69]. Given that neither MeCP2 nor MBD2 nor their MBD domains form dimers or oligomers in vitro, the second interpretation seems more plausible. However, the chromocenters in mouse cells do not undergo decompaction or visible reorganization in the absence of MeCP2 or MBD2, and the differentiation of muscle tissues in MeCP2 and MBD2 null animals seems to proceed normally [56,113]. Therefore the functional significance of MBD proteins localizing to pericentric heterochromatin domains is yet to be established. Intriguingly, a recent study reports aggregation of chromocenters in mouse ES cells null for Dnmt3a/3b where major satellite DNA is mostly unmethylated [114]. It is possible that mouse cells respond to any drastic changes in components of constitutive heterochromatin by dynamic rearrangement of chromocenters.
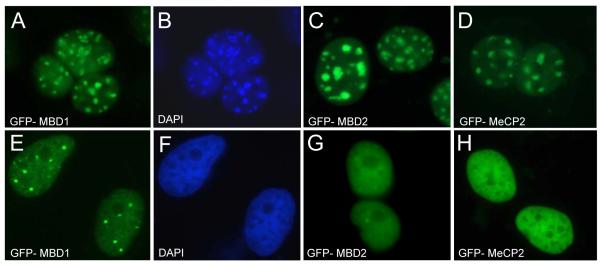
Figure 5. Nuclear localization of MBD proteins in mouse and human cells.
A, C, D. Mouse 3T3 cells transfected with plasmids expressing human GFP-tagged MBD proteins display identical localization that coincides with the heterochromatic chromocenters B. The chromocenters in mouse cells (the same cells as in A) as visualized by staining with DAPI. E, G and H. GFP-tagged MBD proteins transfected into human HeLa cells show localization specific to each protein. F. The DNA of same cells as shown in E was also stained with DAPI. Note that pericentric heterochromatin does not form chromocenters in human cells.
In human cells, where the pericentric heterochromatin does not cluster into specific domains that can be visualized by DNA-staining dyes, each MBD protein displays distinct localization (Figure 5E-H). In most human cell types MeCP2 is diffusely distributed throughout the nucleus, while MBD2 and MBD1 form several bright foci on a weaker background of diffuse distribution throughout the nucleus [101,104]. The nature and functional significance of MBD1 and MBD2 foci are currently unclear. In the case of MBD1, these structures seem to be enriched in heterochromatin-specific histone modifications and proteins such H3K9 methylases SUV39H and SETDB1, and HP1 [105]. It would be of interest to determine whether genes silenced by DNA methylation are recruited to MBD foci. Additional studies by chromatin conformation capture (3C) analyzes may provide some essential information regarding these structures.
The role of MBD proteins in human disease
It is becoming clear that mutations in proteins involved in establishment of DNA methylation patterns as well as DNA methylation effectors, such as MBD proteins, lead to complex human disease phenotypes. For example, mutations in the de novo DNA methyltransferase DNMT3B result in immunodeficiency/centromeric instability/facial anomalies (ICF) syndrome and mutations in the MeCP2 gene, located on the X-chromosome, cause one of the most common forms of mental retardation in females known as Rett syndrome [115,116]. In addition, genome-wide loss of DNA methylation and aberrant methylation of CpG island promoters of genes controlling restricted cellular growth are considered important epigenetic hallmarks of cancer [13]. Despite the functional importance of MBD proteins in recognizing DNA methylation, the means by which MBD proteins mediate the physiological functions of DNA methylation in normal tissues remain for the most part unclear. However, genetic analyses of mice null for specific methyl-CpG binding proteins have allowed their role in disease processes to be investigated in detail.
Rett syndrome (RTT) is a late onset (6 months after birth) severe autism spectrum disorder that affects 1 in 10,000 girls and is caused almost exclusively by mutations in the MeCP2 gene [116]. Due to the random X chromosome inactivation, RTT patients are usually mosaic in the expression of the wild type and the mutant copy of the gene and show abnormal neuronal morphology but not neuronal death [117]. Conditional deletions and neuron-specific expression of MeCP2 in mice have shown that Rett phenotype is caused by MeCP2 deficiency in postmitotic mature neurons [118,119]. Mice null for MeCP2 protein have been generated and shown to recapitulate the most essential features of the human Rett Syndrome [113,119]. MeCP2-deficient males, unlike humans, survive postnatally and develop the symptoms of the disease at ~6 weeks and die after ~11 weeks. MeCP2-deficient females have milder phenotype and therefore survive longer and are fertile [113,119]. Recent reports have challenged previous views that Rett Syndrome is a neurodevelopmental disorder. Thus re-expression of the MeCP2 gene in Mecp2lox-Stop/y mice with progressing disease phenotype is sufficient to reverse the neurological symptoms of RTT [120]. These experiments clearly suggest that MeCP2-deficient neurons develop functionally normal and are not irreversibly damaged by the absence of MeCP2. Important molecular cues, presumably DNA methylation, that allow MeCP2 to function in mature neurons are established appropriately in the absence of MeCP2. These experiments convincingly demonstrate the principle of reversibility of Rett syndrome and are consistent with the hypothesis that MeCP2 is required to stabilize and maintain the state of mature neurons. Nevertheless, it is yet unclear whether MeCP2 function in the brain involves maintenance of specific chromatin conformation or regulation of a few key genes. Target genes for MeCP2-mediated repression are still sparse and microarray studies have not detected major changes in gene expression in the brain of MeCP2-null animals [121-123]. Perhaps, detailed studies of MeCP2 function in specifically in differentiated neurons will help to determine why this protein is so crucial for their integrity.
Substantial evidence in vivo suggests that MBD proteins contribute to transcriptional repression at methylated gene promoters, especially in tumours where many promoter-associated CpG islands are aberrantly methylated [13,36,63]. In mice that carry heterozygous mutant allele of the Apc gene (Apc+/min), a spontaneous inactivation of the wild type allele often occurs in the intestine and leads to development of adenomas and subsequent death of these animals at an age of about 180 days [124]. However, Apc+/min mice lacking Mbd2 or Kaiso proteins show reduced incidence of intestinal tumours and significantly improved survival compared to Apc+/min littermates with wild type Mbd2 or Kaiso alleles [36,57]. Antisense oligonucleotide knock down of MBD2 in xenographs has also been shown to suppress tumour growth validating MBD2 as a potential target for anti-cancer therapy [125]. It will be interesting to determine whether mice null for MBD1 protein are resistant to development of tumours when crossed onto Apc+/min background. On the other hand, Mbd4-deficient Apc+/min mice display accelerated tumour formation consistent with the proposed role of MBD4 in suppression of CpG mutability and tumorigenesis in vivo [126]. Taken together, these genetic studies clearly indicate that methyl-CpG binding proteins contribute to development of cancer phenotypes in mouse models.
Studies in human cancers have identified a number of aberrantly methylated promoters of tumour suppressor genes that are bound by MBD proteins. Candidate gene approach and genome-wide studies using CpG-island microarrays showed that a significant proportion of methylated promoters are bound by a single MBD protein, most often MBD2 [63,127]. However, unlike primary human cells, about half of all methylated promoters seem to be occupied by more than one MBD protein [63]. Whether cooperation between MBD proteins is required for stable silencing of densely methylated CpG islands in cancer cells is yet to be established.
Conclusions and future directions
Considerable amount of work over the last years have contributed to understanding of the role of DNA methylation in mammalian development and human disease. Nevertheless, many questions remain unresolved, including most fundamental ones of how the patterns of DNA methylation are established in vertebrate genomes and what causes dramatic changes of these patterns in pathological states such as cancer. Studies on proteins that bind methylated DNA have demonstrated that these proteins act as important effectors of DNA methylation and are involved in establishment and maintenance of transcriptionally silenced chromatin and, perhaps, higher order chromatin structure. Recent data have provided evidence that methyl-CpG binding proteins do not share binding sites in vivo and can recognize methylated DNA within specific sequence context. Perhaps, in some cases the binding selectivity could be attributed to structural properties of the domain that interacts with methylated DNA. In others, cooperation between the methyl-CpG binding domain and other DNA binding motifs could be essential for discrimination between different methylated loci and targeting distinct subsets of genes. Further work in vitro and in vivo, structural studies of these proteins in complex with appropriate DNA sequences and genome-wide analyses of their binding profiles will provide additional essential information and will facilitate more detailed mechanistic understanding of their complex functions.
Acknowledgements
We apologize to those researchers whose work could not be cited or was cited indirectly due to space limitations. Work from our laboratory is supported by Cancer Research UK, the Wellcome Trust and EMBO Young Investigators programme. T.C. is an EMBO long-term fellow.
References
- [1].Wilson GG, Murray NE. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- [2].Hutchins AS, Mullen AC, Lee HW, Sykes KJ, High FA, Hendrich BD, Bird AP, Reiner SL. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol Cell. 2002;10:81–91. doi: 10.1016/s1097-2765(02)00564-6. [DOI] [PubMed] [Google Scholar]
- [3].Selker EU. Genome defense and DNA methylation in Neurospora. Cold Spring Harb Symp Quant Biol. 2004;69:119–24. doi: 10.1101/sqb.2004.69.119. [DOI] [PubMed] [Google Scholar]
- [4].Vanyushin BF. DNA methylation in plants. Curr Top Microbiol Immunol. 2006;301:67–122. doi: 10.1007/3-540-31390-7_4. [DOI] [PubMed] [Google Scholar]
- [5].Lyko F, Ramsahoye BH, Jaenisch R. DNA methylation in Drosophila melanogaster. Nature. 2000;408:538–40. doi: 10.1038/35046205. [DOI] [PubMed] [Google Scholar]
- [6].Zhu H, Geiman TM, Xi S, Jiang Q, Schmidtmann A, Chen T, Li E, Muegge K. Lsh is involved in de novo methylation of DNA. Embo J. 2006;25:335–45. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- [8].Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–73. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- [9].Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- [10].Pfeifer GP. Mutagenesis at methylated CpG sequences. Curr Top Microbiol Immunol. 2006;301:259–81. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- [11].Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- [12].Cross SH, Bird AP. CpG islands and genes. Curr Opin Genet Dev. 1995;5:309–14. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- [13].Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- [14].Costello JF, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–8. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- [15].Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–4. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- [16].Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- [17].Meehan RR, Stancheva I. DNA methylation and control of gene expression in vertebrate development. Essays Biochem. 2001;37:59–70. doi: 10.1042/bse0370059. [DOI] [PubMed] [Google Scholar]
- [18].Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–40. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- [19].Jackson-Grusby L, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–9. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- [20].Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- [21].Stancheva I, Meehan RR. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev. 2000;14:313–27. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [22].Stancheva I, El-Maarri O, Walter J, Niveleau A, Meehan RR. DNA methylation at promoter regions regulates the timing of gene activation in Xenopus laevis embryos. Dev Biol. 2002;243:155–65. doi: 10.1006/dbio.2001.0560. [DOI] [PubMed] [Google Scholar]
- [23].Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–9. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- [24].Suzuki MM, Kerr AR, De Sousa D, Bird A. CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res. 2007;17:625–31. doi: 10.1101/gr.6163007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bird AP. Gene number, noise reduction and biological complexity. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- [26].Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–13. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- [27].Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc Natl Acad Sci U S A. 2000;97:6481–6. doi: 10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186–9. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- [29].Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–9. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- [30].Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- [31].Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–14. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- [32].Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–92. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–9. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- [34].Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–47. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Marhold J, Kramer K, Kremmer E, Lyko F. The Drosophila MBD2/3 protein mediates interactions between the MI-2 chromatin complex and CpT/A-methylated DNA. Development. 2004;131:6033–9. doi: 10.1242/dev.01531. [DOI] [PubMed] [Google Scholar]
- [36].Sansom OJ, Berger J, Bishop SM, Hendrich B, Bird A, Clarke AR. Deficiency of Mbd2 suppresses intestinal tumorigenesis. Nat Genet. 2003;34:145–7. doi: 10.1038/ng1155. [DOI] [PubMed] [Google Scholar]
- [37].Zemach A, Grafi G. Characterization of Arabidopsis thaliana methyl-CpG-binding domain (MBD) proteins. Plant J. 2003;34:565–72. doi: 10.1046/j.1365-313x.2003.01756.x. [DOI] [PubMed] [Google Scholar]
- [38].Daniel JM, Reynolds AB. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol. 1999;19:3614–23. doi: 10.1128/mcb.19.5.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Prokhortchouk A, Hendrich B, Jorgensen H, Ruzov A, Wilm M, Georgiev G, Bird A, Prokhortchouk E. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–8. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Filion GJ, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, Defossez PA. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol Cell Biol. 2006;26:169–81. doi: 10.1128/MCB.26.1.169-181.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Daniel JM, Spring CM, Crawford HC, Reynolds AB, Baig A. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 2002;30:2911–9. doi: 10.1093/nar/gkf398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Woo HR, Pontes O, Pikaard CS, Richards EJ. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 2007;21:267–77. doi: 10.1101/gad.1512007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007;17:379–84. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–10. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- [45].Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–4. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- [46].Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007 doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- [47].Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269–77. doi: 10.1016/S0168-9525(03)00080-5. [DOI] [PubMed] [Google Scholar]
- [48].Ohki I, Shimotake N, Fujita N, Nakao M, Shirakawa M. Solution structure of the methyl-CpG-binding domain of the methylation-dependent transcriptional repressor MBD1. Embo J. 1999;18:6653–61. doi: 10.1093/emboj/18.23.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wakefield RI, Smith BO, Nan X, Free A, Soteriou A, Uhrin D, Bird AP, Barlow PN. The solution structure of the domain from MeCP2 that binds to methylated DNA. J Mol Biol. 1999;291:1055–65. doi: 10.1006/jmbi.1999.3023. [DOI] [PubMed] [Google Scholar]
- [50].Ohki I, Shimotake N, Fujita N, Jee J, Ikegami T, Nakao M, Shirakawa M. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell. 2001;105:487–97. doi: 10.1016/s0092-8674(01)00324-5. [DOI] [PubMed] [Google Scholar]
- [51].Saito M, Ishikawa F. The mCpG-binding domain of human MBD3 does not bind to mCpG but interacts with NuRD/Mi2 components HDAC1 and MTA2. J Biol Chem. 2002;277:35434–9. doi: 10.1074/jbc.M203455200. [DOI] [PubMed] [Google Scholar]
- [52].Klose RJ, Bird AP. MeCP2 behaves as an elongated monomer that does not stably associate with the Sin3a chromatin remodeling complex. J Biol Chem. 2004;279:46490–6. doi: 10.1074/jbc.M408284200. [DOI] [PubMed] [Google Scholar]
- [53].Bellacosa A, et al. MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1. Proc Natl Acad Sci U S A. 1999;96:3969–74. doi: 10.1073/pnas.96.7.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–4. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- [55].Petronzelli F, et al. Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase. J Biol Chem. 2000;275:32422–9. doi: 10.1074/jbc.M004535200. [DOI] [PubMed] [Google Scholar]
- [56].Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–23. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Prokhortchouk A, et al. Kaiso-deficient mice show resistance to intestinal cancer. Mol Cell Biol. 2006;26:199–208. doi: 10.1128/MCB.26.1.199-208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhao X, et al. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc Natl Acad Sci U S A. 2003;100:6777–82. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Barr H, Hermann A, Berger J, Tsai HH, Adie K, Prokhortchouk A, Hendrich B, Bird A. Mbd2 contributes to DNA methylation-directed repression of the Xist gene. Mol Cell Biol. 2007;27:3750–7. doi: 10.1128/MCB.02204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Berger J, Sansom O, Clarke A, Bird A. MBD2 is required for correct spatial gene expression in the gut. Mol Cell Biol. 2007;27:4049–57. doi: 10.1128/MCB.02023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- [62].Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol Cell. 2005;19:667–78. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- [63].Ballestar E, et al. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. Embo J. 2003;22:6335–45. doi: 10.1093/emboj/cdg604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–92. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Reeves R, Nissen MS. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990;265:8573–82. [PubMed] [Google Scholar]
- [66].Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–21. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Chandler SP, Guschin D, Landsberger N, Wolffe AP. The methyl-CpG binding transcriptional repressor MeCP2 stably associates with nucleosomal DNA. Biochemistry. 1999;38:7008–18. doi: 10.1021/bi990224y. [DOI] [PubMed] [Google Scholar]
- [68].Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J Biol Chem. 2003;278:32181–8. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- [69].Nikitina T, Shi X, Ghosh RP, Horowitz-Scherer RA, Hansen JC, Woodcock CL. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol Cell Biol. 2007;27:864–77. doi: 10.1128/MCB.01593-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Weitzel JM, Buhrmester H, Stratling WH. Chicken MAR-binding protein ARBP is homologous to rat methyl-CpG-binding protein MeCP2. Mol Cell Biol. 1997;17:5656–66. doi: 10.1128/mcb.17.9.5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Jorgensen HF, Ben-Porath I, Bird AP. Mbd1 is recruited to both methylated and nonmethylated CpGs via distinct DNA binding domains. Mol Cell Biol. 2004;24:3387–95. doi: 10.1128/MCB.24.8.3387-3395.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Fujita N, Takebayashi S, Okumura K, Kudo S, Chiba T, Saya H, Nakao M. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol Cell Biol. 1999;19:6415–26. doi: 10.1128/mcb.19.9.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- [74].Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem. 2005;280:41725–31. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- [75].Lee JH, Skalnik DG. CpG-binding protein is a nuclear matrix- and euchromatin-associated protein localized to nuclear speckles containing human trithorax. Identification of nuclear matrix targeting signals. J Biol Chem. 2002;277:42259–67. doi: 10.1074/jbc.M205054200. [DOI] [PubMed] [Google Scholar]
- [76].Birke M, Schreiner S, Garcia-Cuellar MP, Mahr K, Titgemeyer F, Slany RK. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res. 2002;30:958–65. doi: 10.1093/nar/30.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Daniel JM. Dancing in and out of the nucleus: p120(ctn) and the transcription factor Kaiso. Biochim Biophys Acta. 2007;1773:59–68. doi: 10.1016/j.bbamcr.2006.08.052. [DOI] [PubMed] [Google Scholar]
- [78].Park JI, et al. Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev Cell. 2005;8:843–54. doi: 10.1016/j.devcel.2005.04.010. [DOI] [PubMed] [Google Scholar]
- [79].Galvao TC, Thomas JO. Structure-specific binding of MeCP2 to four-way junction DNA through its methyl CpG-binding domain. Nucleic Acids Res. 2005;33:6603–9. doi: 10.1093/nar/gki971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Jeffery L, Nakielny S. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J Biol Chem. 2004;279:49479–87. doi: 10.1074/jbc.M409070200. [DOI] [PubMed] [Google Scholar]
- [81].Buschhausen G, Wittig B, Graessmann M, Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci U S A. 1987;84:1177–81. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol. 1997;7:157–65. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- [83].Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–91. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- [84].Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–35. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J Biol Chem. 2003;278:4806–12. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- [86].Lunyak VV, et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–52. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- [87].Stancheva I, Collins AL, Van den Veyver IB, Zoghbi H, Meehan RR. A mutant form of MeCP2 protein associated with human Rett syndrome cannot be displaced from methylated DNA by notch in Xenopus embryos. Mol Cell. 2003;12:425–35. doi: 10.1016/s1097-2765(03)00276-4. [DOI] [PubMed] [Google Scholar]
- [88].Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–40. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- [89].Harikrishnan KN, et al. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat Genet. 2005;37:254–64. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- [90].Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, Shu X, Kriaucionis S, Bird A. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc Natl Acad Sci U S A. 2007;104:2709–14. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Young JI, et al. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci U S A. 2005;102:17551–8. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kokura K, et al. The Ski protein family is required for MeCP2-mediated transcriptional repression. J Biol Chem. 2001;276:34115–21. doi: 10.1074/jbc.M105747200. [DOI] [PubMed] [Google Scholar]
- [93].Hu K, Nan X, Bird A, Wang W. Testing for association between MeCP2 and the brahma-associated SWI/SNF chromatin-remodeling complex. Nat Genet. 2006;38:962–4. doi: 10.1038/ng0906-962. author reply 964-7. [DOI] [PubMed] [Google Scholar]
- [94].Harikrishnan KN, et al. Reply to “Testing for association between MeCP2 and the brahma-associated SWI/SNF chromatin-remodeling complex”. Nat Genet. 2006;38:964–967. doi: 10.1038/ng0906-962. [DOI] [PubMed] [Google Scholar]
- [95].Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–6. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- [96].Ng HH, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- [97].Mahajan MC, Narlikar GJ, Boyapaty G, Kingston RE, Weissman SM. Heterogeneous nuclear ribonucleoprotein C1/C2, MeCP1, and SWI/SNF form a chromatin remodeling complex at the beta-globin locus control region. Proc Natl Acad Sci U S A. 2005;102:15012–7. doi: 10.1073/pnas.0507596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, Stunnenberg HG. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006;26:843–51. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001;15:827–32. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ng HH, Jeppesen P, Bird A. Active repression of methylated genes by the chromosomal protein MBD1. Mol Cell Biol. 2000;20:1394–406. doi: 10.1128/mcb.20.4.1394-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Fujita N, Watanabe S, Ichimura T, Tsuruzoe S, Shinkai Y, Tachibana M, Chiba T, Nakao M. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J Biol Chem. 2003;278:24132–8. doi: 10.1074/jbc.M302283200. [DOI] [PubMed] [Google Scholar]
- [102].Fujita N, Watanabe S, Ichimura T, Ohkuma Y, Chiba T, Saya H, Nakao M. MCAF mediates MBD1-dependent transcriptional repression. Mol Cell Biol. 2003;23:2834–43. doi: 10.1128/MCB.23.8.2834-2843.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wang H, et al. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol Cell. 2003;12:475–87. doi: 10.1016/j.molcel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- [104].Lyst MJ, Nan X, Stancheva I. Regulation of MBD1-mediated transcriptional repression by SUMO and PIAS proteins. Embo J. 2006;25:5317–28. doi: 10.1038/sj.emboj.7601404. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [105].Uchimura Y, Ichimura T, Uwada J, Tachibana T, Sugahara S, Nakao M, Saitoh H. Involvement of SUMO modification in MBD1- and MCAF1-mediated heterochromatin formation. J Biol Chem. 2006;281:23180–90. doi: 10.1074/jbc.M602280200. [DOI] [PubMed] [Google Scholar]
- [106].Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell. 2003;12:723–34. doi: 10.1016/j.molcel.2003.08.008. [DOI] [PubMed] [Google Scholar]
- [107].Ruzov A, Dunican DS, Prokhortchouk A, Pennings S, Stancheva I, Prokhortchouk E, Meehan RR. Kaiso is a genome-wide repressor of transcription that is essential for amphibian development. Development. 2004;131:6185–94. doi: 10.1242/dev.01549. [DOI] [PubMed] [Google Scholar]
- [108].Sasai N, Matsuda E, Sarashina E, Ishida Y, Kawaichi M. Identification of a novel BTB-zinc finger transcriptional repressor, CIBZ, that interacts with CtBP corepressor. Genes Cells. 2005;10:871–85. doi: 10.1111/j.1365-2443.2005.00885.x. [DOI] [PubMed] [Google Scholar]
- [109].Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–81. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- [110].Maison C, et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–34. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- [111].Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- [112].Brero A, Easwaran HP, Nowak D, Grunewald I, Cremer T, Leonhardt H, Cardoso MC. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J Cell Biol. 2005;169:733–43. doi: 10.1083/jcb.200502062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–6. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- [114].Gilbert N, Thomson I, Boyle S, Allan J, Ramsahoye B, Bickmore WA. DNA methylation affects nuclear organization, histone modifications, and linker histone binding but not chromatin compaction. J Cell Biol. 2007;177:401–11. doi: 10.1083/jcb.200607133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96:14412–7. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–8. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- [117].Armstrong D, Dunn JK, Antalffy B, Trivedi R. Selective dendritic alterations in the cortex of Rett syndrome. J Neuropathol Exp Neurol. 1995;54:195–201. doi: 10.1097/00005072-199503000-00006. [DOI] [PubMed] [Google Scholar]
- [118].Luikenhuis S, Giacometti E, Beard CF, Jaenisch R. Expression of MeCP2 in postmitotic neurons rescues Rett syndrome in mice. Proc Natl Acad Sci U S A. 2004;101:6033–8. doi: 10.1073/pnas.0401626101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–31. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- [120].Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–7. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Tudor M, Akbarian S, Chen RZ, Jaenisch R. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci U S A. 2002;99:15536–41. doi: 10.1073/pnas.242566899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Kriaucionis S, Paterson A, Curtis J, Guy J, Macleod N, Bird A. Gene expression analysis exposes mitochondrial abnormalities in a mouse model of Rett syndrome. Mol Cell Biol. 2006;26:5033–42. doi: 10.1128/MCB.01665-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- [124].Moser AR, Luongo C, Gould KA, McNeley MK, Shoemaker AR, Dove WF. ApcMin: a mouse model for intestinal and mammary tumorigenesis. Eur J Cancer. 1995;31A:1061–4. doi: 10.1016/0959-8049(95)00181-h. [DOI] [PubMed] [Google Scholar]
- [125].Campbell PM, Bovenzi V, Szyf M. Methylated DNA-binding protein 2 antisense inhibitors suppress tumourigenesis of human cancer cell lines in vitro and in vivo. Carcinogenesis. 2004;25:499–507. doi: 10.1093/carcin/bgh045. [DOI] [PubMed] [Google Scholar]
- [126].Millar CB, et al. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002;297:403–5. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- [127].Lopez-Serra L, Ballestar E, Fraga MF, Alaminos M, Setien F, Esteller M. A Profile of Methyl-CpG Binding Domain Protein Occupancy of Hypermethylated Promoter CpG Islands of Tumor Suppressor Genes in Human Cancer. Cancer Res. 2006;66:8342–6. doi: 10.1158/0008-5472.CAN-06-1932. [DOI] [PubMed] [Google Scholar]