Abstract
Melanopsin is the photopigment that confers photosensitivity to a subset of retinal ganglion cells (pRGCs) that regulate many non-image forming tasks such as the detection of light for circadian entrainment. Recent studies have begun to subdivide the pRGCs on the basis of morphology and function but the origin of these differences is not yet fully understood. Here we report the identification of two isoforms of melanopsin from the mouse Opn4 locus, a previously described long isoform (Opn4L) and a novel short isoform (Opn4S) which more closely resembles the sequence and structure of rat and human melanopsins. Both isoforms, Opn4L and Opn4S, are expressed in the ganglion cell layer of the retina, traffic to the plasma membrane and form a functional photopigment in vitro. Quantitative PCR revealed that Opn4S is 40 times more abundant than Opn4L. The two variants encode predicted proteins of 521 and 466 amino acids and only differ in the length of their C-terminal tails. Antibodies raised to isoform specific epitopes identified two discrete populations of melanopsin expressing RGCs, those that co-express Opn4L and Opn4S and those that express Opn4L only. Recent evidence suggests that pRGCs show a range of anatomical subtypes which may reflect the functional diversity reported for mouse Opn4-mediated light responses. The distinct isoforms of Opn4 described in this study provide a potential molecular basis for generating this diversity and it seems likely that their differential expression plays a role in generating the variety of pRGC light responses found in the mammalian retina.
Keywords: Melanopsin, Splice variant, pRGCs, circadian, GPCR, Entrainment
Introduction
The melanopsin gene (Opn4) encodes a functional photopigment involved in the mediation of non-visual photoreceptive tasks, such as circadian entrainment, pupillary constriction and masking of locomotor activity (for review see Hankins et al., 2008). Melanopsin was originally isolated in 1998 from the melanophores of Xenopus, specialized light-sensitive cells in the skin (Provencio et al., 1998). Subsequently, Provencio and colleagues (2000) isolated melanopsin from mammals and demonstrated that it is expressed in a subset of ganglion cells in the inner retina (RGCs). Several studies, using a variety of approaches, went on to demonstrate that these RGCs (1-3% of the total), are intrinsically photosensitive (pRGCs) and project to several brain areas including the suprachiasmatic nuclei (SCN), the master circadian pacemaker and the olivary pretectal nuclei (OPN) (Berson et al., 2002; Panda et al., 2002; Ruby et al., 2002; Lucas et al., 2003; Sekaran et al., 2003; Melyan et al., 2005; Panda et al., 2005; Qiu et al., 2005; Hattar et al., 2006).
An initial physiological study of the responses of pRGCs in mice revealed a diversity of melanopsin dependent light responses described as transient, sustained and repetitive (Sekaran et al., 2003). Further work, using multi-electrode array recording in the neonatal mouse retina, also demonstrated differences in functional responses based on sensitivity and latency (Tu et al., 2005). In addition it has been reported that in the primate retina there are two morphologically distinct subtypes of melanopsin RGCs, with dendrites that ramify in either the inner or outer strata of the inner plexiform layer (IPL) (Dacey et al., 2005). Similarly in the mouse retina, two types of melanopsin ganglion cells, termed M1 and M2, have been described (Hattar et al., 2006). Two further studies have extended this anatomical diversity to define three types of melanopsin expressing cells. These are M1 which have dendrites in the outer IPL, close to the inner nuclear layer (INL), M2 which have dendrites in the inner IPL, close to the ganglion cell layer (GCL), and cells that are bistratified, with dendrites in the same strata as both M1 and M2 cells (Viney et al., 2007; Schmidt et al., 2008). Recently it has been suggested that these subtypes of melanopsin expressing cells might differentially innervate retino-recipient brain areas (Hattar et al., 2006; Baver et al., 2008).
The isolation and subsequent characterisation of melanopsin revealed that the C-terminal tails of the deduced amino acid sequences from human and mouse differed significantly (Provencio et al., 2000). This observation as well as the finding that rat Opn4 is more similar to the human sequence than to mouse led us to undertake a more detailed analysis of the melanopsin gene structure. Here we report the existence of two isoforms of Opn4 (Opn4L and Opn4S), in the adult retina, generated by alternate splicing of a single melanopsin gene in the mouse genome. We go on to demonstrate that both isoforms encode functional photopigments and are differentially expressed in sub-populations of RGCs, a finding that offers an insight into their role in the mammalian retina.
Materials and Methods
Animals
Wild type mice (C3H/He; not carrying rd mutation) and Opn4−/− (tau-LacZ+/+) mice (mixed C57BL/6 and 129/SvJ background) (Hattar et al., 2002) were housed under a 12:12 LD cycle with food and water ad libitum. Animals were sacrificed at ZT 6-10, according to Schedule 1 of the UK Home Office Animals (Scientific Procedures) Act 1986. Eyes were removed and either processed for immunocytochemistry or retina dissected and snap frozen on dry ice at −80°C until required.
RNA extraction and cDNA synthesis
Retinal tissue was homogenised in 1ml TRIzol (Life Technologies) using a micropestle. Total RNA was then extracted according to the manufacturer’s instructions, resuspended in TE and stored at −80°C prior to use. 0.5mg of total RNA was DNase treated (Sigma-Aldrich) and reverse transcribed with an oligo d(T)n primer using the RetroScript kit (Ambion) according to manufacturer’s instructions.
Isolation of two isoforms of mouse Opn4
Primers were designed in exon 8 (mOpn4 8F 5′-GCTACCGCTCTACCCACC-3′) and around the predicted stop codons of the putative long and short isoforms (mOpn4 long 5′-CTACAGATGTCTGAGAGTCAC-3′, mOpn4 short 5′-CTACATCCCGAGATCCAGACT-3′). PCR was then performed under the following conditions: an initial denaturation step at 94°C for 3min, then 94°C for 30s, 56°C for 30s, 72°C for 30s for 35 cycles, followed by a final extension at 72°C for 7min. Each 25μl reaction contained 0.2mM dNTPs, 0.2μM of each primer, 1μl template cDNA, prepared as described above, and 1U of Taq polymerase (Thermoprime plus, ABgene). Using primer pairs mOpn4 8F/mOpn4 long and mOpn4 8F/mOpn4 short generated products of 425bp and 260bp respectively. These fragments were cloned into pGEM-T Easy (Promega) according to the manufacturer’s instructions and sequenced. Full-length coding sequences of both isoforms were generated using PCR with a primer to the start site of mOpn4 (mOpn4 1F 5′-ATGGACTCTCCTTCAGGA-3′) and mOpn4 long or mOpn4 short. PCR was carried out using Platinum Taq Supermix (Life Technologies) with an initial denaturation step at 94°C for 3min, then 94°C for 30s, 54°C for 30s, 72°C for 1min 30s for 35 cycles, followed by a final extension at 72°C for 7min. The products of 1566bp and 1401bp were cloned into pGEM-T Easy and sequence verified.
3′ RACE
3′ RACE ready cDNA was synthesised with the RLM-RACE kit (Ambion) using 1μg retinal RNA and the 3′ adapter primer. First round RACE was carried out with primer mOpn4 6F (5′-GGAAGATGGCCAAGGTCGCA-3′) and the 3′ RACE outer primer (5′-GCGAGCACAGAATTAATACGACT-3′) according to the manufacturer’s protocol, but briefly, PCR was carried out under the following conditions: an initial denaturation step at 94°C for 3min, then 94°C for 30s, 60°C for 30s, 72°C for 30s for 35 cycles, followed by a final extension at 72°C for 7min. 1μl of first round product was used in a nested PCR with the primers mOpn4 8F and 3′ RACE inner primer (5′-CGCGGATCCGAATTAATACGACTCACTATAGG-3′) using the same conditions. The two products obtained were cloned into pGEM-T easy and sequenced.
Quantitative PCR
Quantitative real-time PCR (qPCR), using cDNA synthesized as described above and the primer pairs mOpn4 8F/mOpn4 long, mOpn4 8F/mOpn4 short, was performed using Sybr Green I or TaqMan mastermixes on a StepOne thermal cycler (Applied Biosystems, USA). Relative quantification of transcript levels was performed as previously described (Peirson et al., 2003). Two genes were used for normalization, acidic ribosomal phosphoprotein (ARP) and β-actin, primer sequences were as previously described (Peirson et al., 2004).
Cell culture
RGC-5 cells were grown in DMEM/F12 with Glutamax-I (Life Technologies) and 10% FBS and 1% (v/v) penicillin/streptomycin (Sigma). Neuro-2A cells (ECACC) were cultured in DMEM (Sigma) supplemented with 10% FBS, 2mM L-glutamine, 1% (v/v) penicillin/streptomycin. All cells were incubated in a humidified chamber at 37°C with 5% CO2, fed fresh media every 2-3 days and passaged prior to reaching confluence.
Transfection
The full-length coding regions of both isoforms of Opn4 were cloned into the expression vector pIRES2-AcGFP (BD Biosciences). Constructs were sequence verified and DNA for transfections was prepared using a plasmid Midiprep kit (Qiagen). Transfection of RGC-5 cells was performed using the Lipofectamine Plus transfection reagent (Life Technologies) according to the manufacturer’s guidelines and as previously described (Kosmaoglou and Cheetham, 2008). Transfection of Neuro-2A cells was performed using the Genejuice transfection reagent (Novagen) according to the manufacturer’s guidelines. Briefly, Neuro-2A cells were seeded at a density of 2×105 cells per 35mm Petri dish. 24h after seeding cells were incubated in RPMI media (Sigma) containing 2μg plasmid DNA and 6μl Genejuice reagent for 6h. Cells were then fed normal cell culture media and cultured for 48h prior to protein isolation and ICC.
Whole cell electrophysiology
Post transfection (24h), cells were differentiated by the addition of 20μM retinoic acid to the culture media for a further 48h in the dark. All subsequent steps were performed under dim red light. Prior to patch clamp recordings cells were perfused with extracellular saline (140mM NaCl, 4mM KCl, 1mM MgCl2, 2mM CaCl2, 5mM glucose, 10mM HEPES, pH adjusted to 7.4 with NaOH) containing 20μM 9-cis-retinal (Sigma) or 11-cis-retinal (kind gift from Rosalie Crouch) for 2h in the dark. Glass microelectrodes were made from 1.5mm diameter thin walled glass capillaries (Harvard Apparatus), with a final open pipette resistance of 3-5MΩ. Internal pipette saline contained 140mM KCl, 10mM NaCl, 1mM MgCl2, 10mM HEPES and 10mM EGTA, with osmolarity adjusted to 285 ± 5mosmol/L and pH to 7.4 with KOH. Successfully transfected cells were identified based on expression of GFP and then dark adapted for at least 1h prior to recordings, subsequent visualisation of cells was performed using infra-red light. Whole cell recordings were performed at room temperature (22-25°C) using an Axopatch 200B amplifier and PClamp9 data acquisition software (Axon Instruments) with a sampling rate of 20kHz. Whole cell currents were recorded from cells voltage clamped at holding potentials of −50mV. Access resistance during recordings was less than 20MΩ. Light stimuli were generated using a Cairn Optoscan Xenon arc source comprising a slit monochromator. Stimuli were 10s in duration with a 20nm half-bandwidth. Irradiance was measured using an optical power meter (Macam Photometrics) and converted to photon flux. The intensity of light used was 8 × 1014 photons cm−2 s−1 and is approximately 1 log unit above threshold. The magnitude of responses was defined by the peak sustained current measured using Clampfit analysis software (Axon Instruments).
Antibodies
Specific polyclonal antibodies were raised to each Opn4 isoform using different animal models (OPN4L: Rabbit; OPN4S: Goat) to enable co-localisation. An additional rabbit polyclonal antibody was raised to the amino terminus of melanopsin, which is common to both isoforms (PAS8331). Each polyclonal antibody was raised against a 15aa synthetic peptide conjugated to KLH by Harlan UK, according to their standard procedures (Short: SPQTKGHLPSLDLGM; Long: PHPHTSQFPLAFLED, N-term: MDSPSGPRVLSSLTQ, shown on Fig. 1A). All antibodies were affinity purified prior to use (Thiolink gel kit, Severn Biotech Ltd). A chicken anti-β-galactosidase antibody (ab9361, Abcam) was used for localisation of β-gal expression in tau-lacZ+/− mice. SDS PAGE Gel loading was assessed using a rabbit polyclonal anti-β-actin antibody (ab8227, Abcam, UK). Secondary antibodies: for western blotting, donkey anti-goat and anti-rabbit IgG HRP linked secondary antibodies were used (SC2304 and SC2305, Insight Biotechnology). For secondary labelling in immunofluorescence studies, Cy3 labelled donkey anti-rabbit antibody (Jackson Immunoresearch), Alexa 555 labelled goat anti-rabbit, Alexa 568 donkey anti-goat and Alexa 488 and 555 donkey anti-Rabbit, and Alexa 488 goat anti-chicken antibodies (Life Technologies) were used as stated.
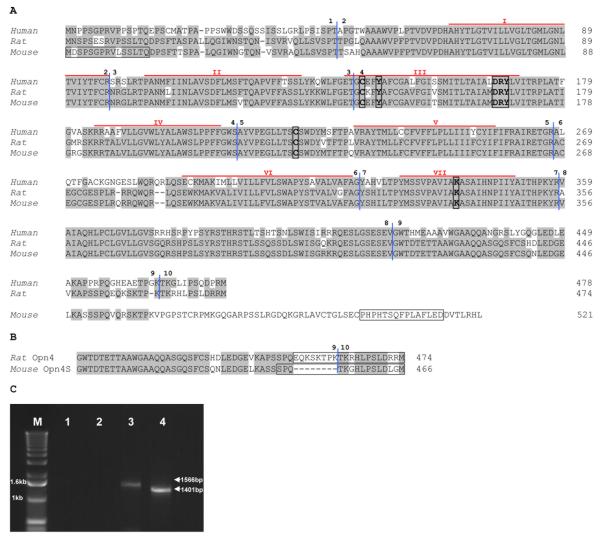
Figure 1. (A) Alignment of mouse, rat and human Opn4 deduced amino acid sequences.
Residues that are identical in two out the three sequences are shaded. The seven probable transmembrane domains are marked by red lines above the sequence and numbered using roman numerals. The characteristic features of an opsin are shown boxed: lysine (K) to form a Schiff’s base at position 337; tyrosine (Y), a possible counterion at position 145; aspartate, arginine and tyrosine (DRY) tripeptide for transducin binding at position 166–168; and cysteines (C) at positions 142 and 220 for disulphide bridge formation (numbers correspond to the mouse sequence). The intron-exon boundaries are delineated by vertical blue lines and are numbered. The epitopes of the N-terminal antibody (PAS8331) and OPN4L are shown boxed. Accession numbers: Homo sapiens NM_033282, Rattus norvegicus NM_138860, Mus musculus NM_013887. (B) Alignment of amino acids encoded by rat Opn4 exons 9 and 10 with those of the newly identified mouse Opn4S showing that the mouse isoform exon 9 is 8 amino acids shorter than the rat sequence. Boxes show the epitopes of OPN4S and the C-terminal rat antibody (see discussion for more details). (C) Amplification of Opn4L and Opn4S coding regions from adult retina cDNA. Products are 1566bp and 1401bp. M marker (1kb ladder, Life Technologies), Lane 1 no template control for Opn4L primers, Lane 2 no template control for Opn4S primers, Lane 3 Opn4L, Lane 4 Opn4S.
Western blotting
Retinal tissue samples were homogenised in 2% (w/v) SDS, 10 mM DTT in PBS with mini complete protease inhibitors (Roche) and centrifuged at 23000×g for 30min. Transiently transfected cells were centrifuged at 1000×g and the resulting cell pellets were washed with PBS and resuspended in 200μl of lysis buffer (1% (w/v) DM (Sigma), 5mM EDTA in PBS with mini complete protease inhibitors) prior to passage through a 25g needle × 10. The lysate was incubated at 4°C for 15min and then centrifuged at 23000×g at 4°C for 30min. The resulting supernatant fraction of both sample preparations was combined 1:1 with modified sample buffer without heat treatment (Saliba et al., 2002). Samples were resolved on an 8% SDS PAGE minigel and electro-transferred onto PVDF membrane (Bio-Rad). The membrane was blocked in 5% (w/v) BSA in Tris buffered saline, 1% (v/v) Tween 20 (TBST) for 1h and incubated overnight at 4°C with primary antibody (diluted in 5% (w/v) BSA in TBST). Blots were washed in TBST and incubated with HRP linked secondary antibody (Autogen Bioclear) for 1h. Following incubation, the blots were washed in TBST and developed using an ECL system (Thermo Scientific). Immunoreactivity was detected by exposure of the blots to X-ray film and subsequent development (XOgraph Imaging Systems). To assess gel loading, membranes were stripped following ECL development, by incubation at 55°C in 87.7mM Tris pH6.8, 2% (w/v) SDS, 0.1M DTT for 30min. Stripped blots were washed and blocked as before, prior to incubation with β-actin antibody.
Immunocytochemistry
Fluorescent immunolabelling was performed using standard techniques. Briefly, all slides were blocked for 1h at RT in PBS with 10% serum from the same species as the corresponding secondary antibodies. All antibodies were diluted in PBS with 2.5% serum. All wash steps were performed with PBS Tween (0.1%) for 5mins ×4. RGC-5 cells were fixed with methanol at −20°C for 20min, incubated with PAS8331 antibody (1:500) for 1h RT, followed by a Cy3 labelled secondary (1:100) for 1h RT. Slides were counterstained with DAPI (2μg/ml) in PBS for 15min then mounted with fluorescent mounting medium (DAKO). Neuro-2A cells were fixed with 4% PFA (Pierce) for 15min and permeabilised with 0.05% Triton-X in PBS for 5min RT. PAS8331, anti-Opn4L and anti-Opn4S antibodies were incubated for 1h RT diluted 1:100. Alexa labelled secondary antibodies were incubated for 1h RT diluted 1:400. Slides were mounted with anti-fade mountant with DAPI (Life Technologies). Removal, fixation and cryostat sectioning of whole mouse eyes was performed as described previously (Sekaran et al., 2007). Whole eye sections (16μm) were permeabilised with 0.2% triton-X in PBS for 20min RT. Primary antibodies were incubated for 18h at 4°C, anti-Opn4L (1:100), anti-Opn4S (1:100), anti-β-gal (1:400). Alexa labelled secondary antibodies were incubated for 1h RT diluted 1:200. For double labelling experiments slides were incubated with primary antibodies and secondary antibodies in a sequential manner; Opn4S then Opn4L or β-gal. Slides were mounted with anti-fade mountant with DAPI (Life Technologies). Fluorescent images were collected using a Carl Zeiss LSM510 confocal laser scanning microscope, excitation 405, 488 and 543nm with emission wavelengths of 420-450, 505-530 and 550-754nm for DAPI, green and red fluorescence respectively.
Results
Two isoforms of mouse Opn4 in the adult retina
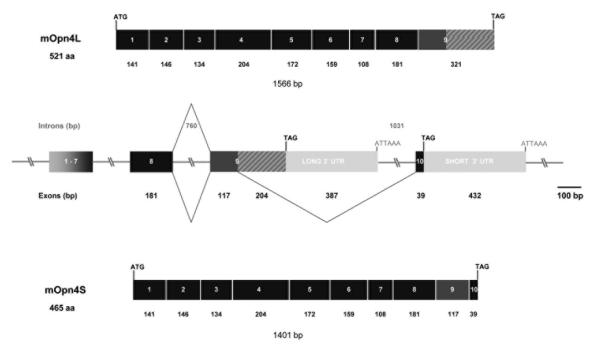
The published mouse Opn4 sequence (AF147789) is 2137bp and encodes a predicted protein of 521 amino acids containing all of the expected features of an opsin (Provencio et al., 2000). Alignment of the predicted amino acid sequences of human (NM_033282), mouse (NM_013887) and rat (NM_138860) melanopsins (Fig. 1A) shows that the mouse Opn4 sequence has a longer C-terminus than either human or rat. The human sequence consists of 10 exons spanning 11.9kb of genomic sequence on chromosome 10q23.2 (Provencio et al., 2000). Using TBLASTN searches of both the mouse and rat genomes, with the corresponding Opn4 amino acid sequence, enabled us to determine the genomic structure of these genes. The rat Opn4 gene also consists of 10 exons spanning 9.2kb on chromosome 16; however the mouse gene only has 9 exons spanning 7.8kb of genomic sequence on chromosome 14. This discrepancy led us to examine the genomic sequence of the mouse Opn4 locus in more detail. The sizes of the exons of all three genes are shown in Table 1. This comparison reveals that exon 9 in mouse is 321bp whereas exon 9 in human is 144bp and in rat 141bp. A BLAST search using the nucleotide sequence of the rat melanopsin gene (NM_138860) against the mouse genome identified a region downstream of exon 9 showing 87% identity at the nucleotide level to rat exon 10. This was the first indication that there may be two isoforms of the mouse melanopsin gene generated by alternate splicing.
Table 1.
Comparison of exon sizes of human, rat and mouse melanopsin
| Exon | Size (bp) | ||
|---|---|---|---|
|
| |||
| Human | Rat | Mouse | |
| 1a | 144 | 144 | 141 |
| 2 | 146 | 146 | 146 |
| 3 | 134 | 134 | 134 |
| 4 | 204 | 204 | 204 |
| 5 | 172 | 172 | 172 |
| 6 | 165 | 156 | 159 |
| 7 | 108 | 108 | 108 |
| 8 | 181 | 181 | 181 |
| 9 | 144 | 141 | 321 |
| 10 | 39 | 39 | |
Length from start codon
In order to determine whether this potential exon 10 is actually expressed in mouse and produces a transcript that gives rise to a short isoform similar to that seen in both human and rat, primers were designed to this novel sequence. PCR was carried out using adult mouse retina cDNA as template using a forward primer in exon 8 (mOpn4 8F) and reverse primers designed around the predicted stop codons of both isoforms (mOpn4 long and mOpn4 short). Both primer pairs generated products: 8F/long, the expected fragment of 425bp but 8F/short produced a band of 260bp compared to the predicted 284bp. Sequence analysis showed the 8F/long fragment has 100% identity to the expected sequence. However the 8F/short fragment was indeed 24bp shorter than the rat sequence. This discrepancy occurs because exon 9 in the mouse short isoform is 117bp compared to the 141bp in rat resulting in the predicted amino acid sequence being 8 amino acids shorter (Fig. 1B).
To verify this result and to confirm the presence of both isoforms in adult mouse retina, primers were designed to amplify the complete coding sequences. Using the primer pairs Opn4 1F/long and Opn4 1F/short generated products of 1566bp and 1401bp respectively using wild type retinal cDNA as template, cDNA from mice where Opn4 has been replaced with tau-lacZ (tau-lacZ+/+) (Hattar et al., 2003) was used as a negative control (Fig. 1C). These products were sequenced and comparison with the mouse genome database enabled the genomic structure to be confirmed. The first 1362 nucleotides of each clone were identical, corresponding to exons 1 to 8. A schematic diagram of the genomic structure of the gene is shown in Fig. 2 which also demonstrates how the alternate splicing occurs to generate the two isoforms, Opn4L (Genbank accession number EU303118) and Opn4S (Genbank accession number EU303117).
Figure 2. Schematic diagram of the genomic structure of mouse Opn4.
The mouse Opn4 gene consists of 10 exons that span approximately 9.6kb of genomic DNA. Exons are shown as boxes and introns as lines; all are to scale except for exons 1 to 7 and regions of intronic DNA larger than 1kb that are represented as slashed lines. Intron and exon sizes are marked. The start and stop codons in each gene are also indicated, as are the polyadenylation signals. The gene gives rise to two splice variants, the mOpn4L isoform generated by retention of intron 9 and mOpn4S by splicing to exon 10. The products generated by these two events are shown.
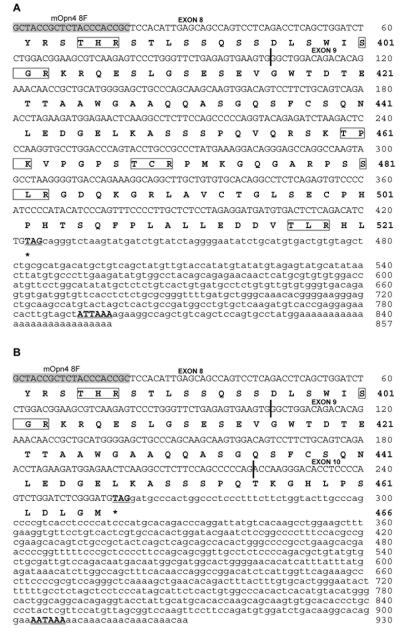
The final verification of the presence of both isoforms in adult mouse retina was achieved using 3′ RACE which generated two fragments of 857bp and 930bp. Sequence analysis showed that the 857bp fragment, corresponding to the long isoform (Opn4L), consisted of 425bp of coding sequence (104bp of exon 8 and 321bp of exon 9) and 432bp of 3′ UTR contiguous to exon 9. This is shown schematically in Fig. 2 and the sequence shown in Fig. 3A. This fragment encodes the C-terminus originally reported by Provencio et al. (Provencio et al., 2000). The second fragment of 930bp, corresponding to the short isoform (Opn4S) is composed of 260bp of coding sequence split across 3 exons (104bp of exon 8, 117bp of exon 9 and 39bp of exon 10). The remaining 670bp of 3′ UTR is contiguous to the newly identified exon 10 shown in Figs. 2 and 3B. Both clones contained polyadenylation signals (ATTAAA in the long 3′ UTR and AATAAA in the short 3′ UTR) shown boxed in Figs. 3A and 3B. Taken together, these data confirm the presence of two isoforms, Opn4L (EU303118) and Opn4S (EU303117), in the adult mouse retina. We also have preliminary RT-PCR evidence for a long isoform of human melanopsin (OPN4) indicating that a similar splicing mechanism may be present in the human retina (Pires and Halford unpublished data).
Figure 3. 3′ RACE products.
3′ RACE with the mouse specific primer mOpn4 8F generated two fragments of 857bp and 930bp. The nucleotide sequence of each fragment is shown with the deduced amino acid sequence below. Exon boundaries are delineated with vertical blue lines, potential polyadenylation signals are underlined and potential protein kinase C sites are boxed. (A) The 857bp fragment consists of 425bp of coding sequence (104bp of exon8 and 321bp of exon 9) and 432bp of 3′ UTR contiguous to exon 9 and corresponds to Opn4L. (B) The 930bp fragment is composed of 260bp of coding sequence split across 3 exons (104bp of exon 8, 117bp of exon 9 and 39bp of exon 10). The remaining 670bp of 3′ UTR is contiguous to the newly identified exon 10. This product corresponds to Opn4S.
Quantitative PCR was then used to determine the relative abundance of each isoform. The specificity of the Opn4L and Opn4S primers was determined by melting curve analysis (Supplementary Fig. 1A) as well as by amplification of a dilution series (standard curve) based upon Opn4L and S cloned into the vector pIRES-AcGFP (Supplementary Fig. 1B). Both isoforms exhibited comparable amplification efficiency and were detectable in the mouse retina although the Opn4S transcript was present at levels approximately 40× more than Opn4L (Supplementary Fig. 1C).
Opn4L and Opn4S encode predicted proteins of 521 and 466 amino acids respectively, with the first 454aa being identical. Both contain all the characteristic features of an opsin and only differ in the lengths of their C-terminal tails (Fig. 1A and B). Prediction of putative post-translational modification sites was performed using the PREDICTPROTEIN program (http://www.predictprotein.org/submit.html) (Rost et al., 2004), which revealed the presence of three potential N-glycosylation sites (aa positions 30, 34 and 87), three casein kinase II (aa positions 140, 411 and 418) and one cAMP phosphorylation site (at position 183) in Opn4L and Opn4S. Both isoforms also have six potential protein kinase C phosphorylation sites (aa positions 36, 182, 264, 381, 385, 401) but Opn4L has 4 more potential sites in its longer C-terminal tail (aa positions 460, 468, 481 and 517) (marked on Fig. 3A and B).
Sub-cellular localisation
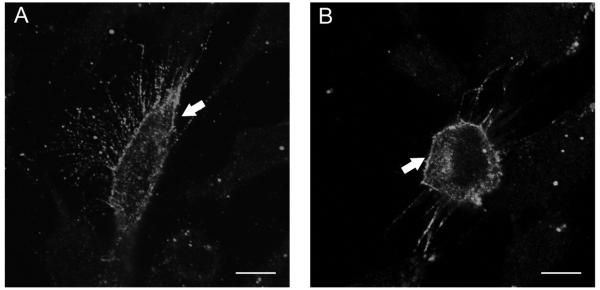
To test potential differences in processing between the Opn4L and Opn4S, both isoforms were expressed in the rat retinal ganglion cell line RGC-5. RGC-5 cells have been reported to have many of the properties of retinal ganglion cells (Krishnamoorthy et al., 2001) and should therefore process these isoforms similarly to retinal ganglion cells in vivo. RGC-5s did not express detectable levels of endogenous Opn4, but immunofluorescent labelling of transfected cells with an antibody raised against the N-terminus region of melanopsin, present in both isoforms, revealed that both Opn4L and Opn4S were trafficked predominantly to the plasma membrane in approximately 90% of cells analyzed (Fig. 4). No staining of non-transfected cells was observed with PAS8331, secondary antibody only controls were also negative. No differences were observed in the processing of these isoforms in RGC-5 cells, or the mouse neuronal cell line, Neuro-2A (Supplementary Fig. 3).
Figure 4. Opn4 localisation in RGC-5 cells.
RGC-5 cells were transfected with Opn4L (A) and Opn4S (B) and fixed after 24 hours. Melanopsin expression was detected using a rabbit anti-Opn4 antibody targeted to the N-terminus of the protein (PA8331) and immunofluorescence signal was observed mainly on the plasma membrane (arrowed). Scale bar 10μm.
Opn4L and Opn4S both encode a functional sensory photopigment
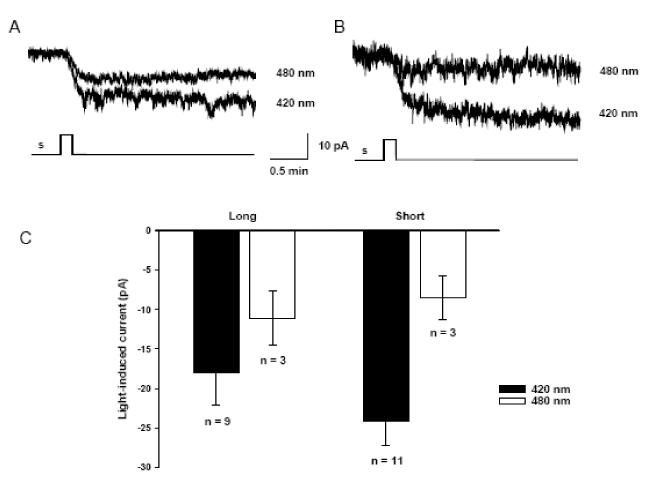
In order to directly determine whether both isoforms encode a functional sensory pigment, we utilised the Neuro-2A heterologous expression system previously used to examine melanopsin (Melyan et al., 2005; Bellingham et al., 2006). Neuro-2A cells were transiently transfected with either Opn4L or Opn4S in the expression vector pIRES2-AcGFP which also contains a GFP reporter gene. Whole cell patch clamp recordings from fluorescent cells revealed light-dependent inward currents in cells transfected with either Opn4L or Opn4S, in the presence but not the absence of 9-cis-retinal or 11-cis-retinal chromophore (Fig. 5). Increasing the duration of light stimuli elicited inward currents of increasing amplitude (Supplementary Fig. 2). For both Opn4L and Opn4S transfected cells, the light evoked responses show only limited recovery over periods of 10-30min following stimulation. Responses were absent from cells transfected with GFP alone (data not shown).
Figure 5. Heterologous expression of Opn4L and Opn4S suggests that both variants can form a sensory photopigment.
Representative whole cell patch-clamp recording from Neuro-2A cells transfected with Opn4L (A) or Opn4S (B), in the presence of 9-cis-retinal. Monochromatic light stimuli 420nm and 480nm (presented for 10s 8 × 1014 photons cm−2 s−1) evoke a stimulus dependent inward current. Holding potential −50mV. No differences in amplitude of responses, kinetics or spectral sensitivity were observed between Opn4L and Opn4S.
In all aspects, the light-evoked currents observed in the cells transfected with either Opn4L and Opn4S are consistent with those obtained with human melanopsin in this expression system (Melyan et al., 2005) and suggest that both isoforms can form a functional retinaldehyde-dependent sensory photopigment. In the Neuro-2A heterologous expression system, we could find no significant differences in the light-evoked responses of either isoform with regards to response amplitude, kinetics, or spectral sensitivity.
Opn4L and Opn4S are differentially expressed in the mammalian retina
Isoform specific antibodies were raised against peptides corresponding to the differing C-terminal regions of Opn4L and Opn4S (shown in Fig.1). The specificity of these antibodies was confirmed by immunolabelling of Neuro-2A cells transiently transfected with Opn4L or Opn4S. Both antibodies were found to be isoform specific, with the anti-Opn4L antibody only labelling Opn4L transfected cells, and the anti-Opn4S antibody only labelling Opn4S transfected cells. In each case labelling was only observed for GFP positive cells with no labelling observed in neighbouring untransfected cells (Supplementary Fig. 3) or non-transfected controls (data not shown). The antibodies showed equal levels of staining in cells expressing similar levels of the GFP transfection reporter. As expected the N-terminal melanopsin antibody labelled cells transfected with both Opn4L and Opn4S (Supplementary Fig. 3).
Western blotting of transiently transfected cells and adult retinal tissue was undertaken with the isoform specific antibodies. Immunoreactive proteins were detected at ~55 and 60kDa by the short and long isoform specific antibodies respectively (Fig. 6A and B). These masses are comparable to the calculated molecular weights of 51kDa for Opn4S and 57kDa for Opn4L. The additional immunoreactive proteins observed with the Opn4L antibody, in both transiently transfected cells and retinal samples, could result from differential post-translational modification of the protein such as glycosylation or phosphorylation, which is currently under investigation. An alternative explanation is that the expression of Opn4L at high concentrations in mammalian cells results in differential processing of the protein, as suggested with melanopsin from other species (Terakita et al., 2008). Western blot analysis confirmed a higher concentration of Opn4S than Opn4L when compared to β-actin loading controls (Fig. 6A and B). The higher expression of Opn4S correlates with the quantitative PCR analysis which demonstrated that the Opn4S transcript is 40 times more abundant in the mouse retina than the Opn4L transcript.
Figure 6. Western blot analysis of isoform specific expression of melanopsin in retina.
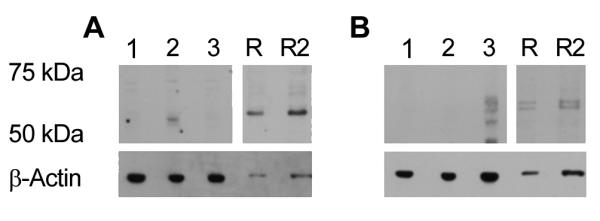
(A) Western blot analysis using Opn4S antibody, 1: Wild type Neuro-2A cells; 2: Opn4S transfected cells; 3: Opn4L transfected cells R: Tau-lacZ+/− retina single load; R2: Tau-lacZ+/− retina double load. (B) Western blot analysis using Opn4L antibody, gel loading as previous figure. β-actin was used to confirm equal loading of the gel.
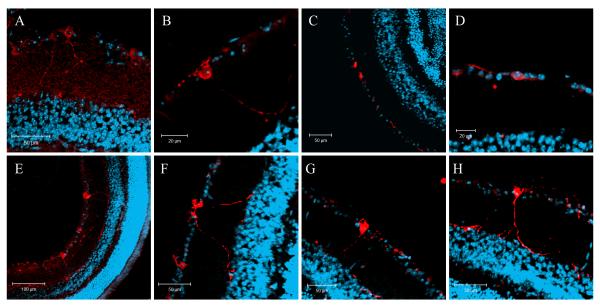
Initially, immunolabelling of wild type mouse eye sections was performed using either the anti-Opn4L or the anti-Opn4S antibody alone (single labelling). These experiments confirmed the expression of both long and short isoforms of melanopsin in a sub-set of RGCs within the mouse retina (Fig. 7). The majority of Opn4L positive cells had relatively low levels of staining and cellular processes were often difficult to visualise. In contrast, Opn4S positive cells appeared brightly labelled and cellular processes were more easily observed. When visible, the processes of Opn4L positive cells revealed a number of sub-populations. Opn4L positive cells were observed with processes in the outer IPL close to the INL or were found to be bi-stratified with processes located both in the inner (ON) and outer (OFF) layers of the IPL (Fig. 7A and B). A second population of Opn4L positive cells were identified whose processes were confined to the inner layer of the IPL (Fig. 7C and D). Opn4S positive cells had processes localised to the outer IPL or were bi-stratified with processes in both the inner and outer layers of the IPL (Fig. 7E-H). Cells with processes confined only to the inner layer of the IPL were absent in Opn4S single labelling experiments.
Figure 7. Single immunolabelling of the mouse retina with anti-Opn4L and anti-Opn4S antibodies.
Single labelling of whole eye sections with anti-Opn4L (A-D) and anti-Opn4S (E-H) antibodies (red) and DAPI counterstain (blue) shows that both isoforms are expressed in a subset of RGCs. Higher levels of labelling were observed for Opn4S compared to Opn4L. When visible, the majority of Opn4L cells have dendrites localised near the inner nuclear layer (INL) or are bi-stratified with processes in the INL and ganglion cell layer. A number of Opn4L cells were also identified whose processes were confined to the vicinity of the ganglion cell layer. All Opn4S positive cells have dendrites located in the INL and are often bi-stratified with processes also seen in the ganglion cell layer.
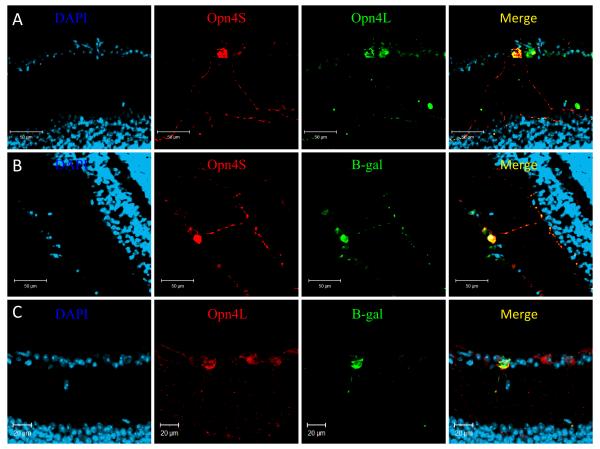
Subsequent double labelling using both anti-Opn4L and anti-Opn4S confirmed a differential pattern of expression for each isoform. The majority of melanopsin positive RGC cells (~70%) were found to express both the long and short isoforms of melanopsin, with processes clearly evident in the outer IPL (Fig. 8A). In these cells the intensity of Opn4S labelling was markedly higher than that of Opn4L. A smaller population of pRGCs (~30%) were found to express only the Opn4L isoform, and when visible, had processes confined to the inner layer of the IPL (Fig. 8A). Co-labelling experiments using an N-terminal antibody, which recognises both isoforms, and the anti-Opn4S antibody again showed a high degree of co-localisation, with ~70% of all cells labelled with the N-terminal melanopsin antibody also labelling with Opn4S (data not shown).
Figure 8. Immunolabelling of the mouse retina shows a differential pattern of expression for Opn4L and Opn4S.
A) Double labelling with Opn4L (green) and Opn4S (red) identified two subsets of pRGCs, those expressing both Opn4L and Opn4S and a second subset of cells expressing only Opn4L. B) Double labelling with β-gal (green) and Opn4S (red) shows a 100% overlap of expression, with all cells positive for both β-gal and Opn4S. C) Labelling with β-gal (green) and Opn4L (red) reveals a sub-set of Opn4L positive cells that lack detectable β-gal expression. For all images DAPI = blue.
Further double-labelling experiments were then performed on eye sections from tau-lacZ+/− mice (Hattar et al., 2002), using each isoform specific antibody with a chicken anti-β-galactosidase antibody. When both anti-Opn4S and anti-β-gal antibodies were used together all the immunopositive cells were co-labelled (Fig. 8B). Whereas staining with anti-Opn4L and anti-β-gal revealed sub-populations of cells, one expressing both Opn4L and β-gal, and a second subset of Opn4L positive cells lacking β-gal expression (Fig. 8C). For all antibodies, controls including omission of primary antibody, and absorption of antibody with immunizing peptides were negative.
Discussion
The isolation of both human and mouse melanopsin was originally described in 2000 (Provencio et al., 2002) and highlighted that the predicted C-terminal tail of the mouse protein was 46 amino acids longer than that of the predicted human protein. This was not thought to be relevant as humans and mice diverged approximately 65 million years ago. However, the subsequent isolation of the rat Opn4 gene (Hattar et al., 2002) revealed the unexpected finding that the predicted C-terminal tail of the rat protein was more similar to that of human than mouse (Fig. 1A). The additional observation that the human melanopsin gene consists of 10 exons and the mouse only of 9 (Provencio et al., 2000) and the demonstration in a recent study that the Opn4 gene in the Australian marsupial, the fat-tailed dunnart, also consisted of 10 exons (Pires et al., 2007) prompted us to undertake a comprehensive examination of the mouse gene. This analysis revealed that the mouse Opn4 gene also consists of 10 exons spanning approximately 9kb of genomic DNA and that transcription at this locus generates 2 isoforms, Opn4L and Opn4S, by alternate splicing (Fig. 2). Both Opn4 isoforms are expressed in adult mouse retina and are restricted to a subset of retinal ganglion cells. However, quantitative PCR shows that the short isoform, Opn4S, is expressed at levels 40× greater than the long isoform, Opn4L (Supplementary Fig. 1). Whether this result is due to higher de novo expression or greater mRNA stability remains to be determined.
Melanopsin belongs to the opsin superfamily of G-protein coupled receptors (GPCRs), which function through the activation of a guanine nucleotide binding protein (G-protein) and an effector enzyme. Opsins consist of seven α–helical transmembrane regions which form a bundle within the membrane creating a hollow cavity on the extracellular side that serves as a binding site for the chromophore, retinal (Palczewski et al., 2000). The predicted proteins encoded by Opn4L (521aa) and Opn4S (466aa) are identical for the first 454 amino acids only differing in the length of their C-terminal tails. Both isoforms encode a seven transmembrane domain protein and contain all of the key features expected to be present in an opsin (see Fig. 1). However, in order to test whether the newly described Opn4S is also functional, the full-length coding sequences of both isoforms were cloned into a mammalian expression vector. Transient transfection of Opn4L and Opn4S into the rat retinal ganglion cell line, RGC-5, and the mouse neuronal cell line, Neuro-2A, showed that both were trafficked predominantly to the plasma membrane and that there was no difference in the processing of these isoforms (Fig. 4 and Supplementary Fig. 3). Similarly, no functional differences were observed for Opn4L and Opn4S following expression in the Neuro-2A expression system used previously to examine both human and chicken melanopsin (Melyan et al., 2005; Bellingham et al., 2006). Both Opn4L and Opn4S transfected cells demonstrated light-dependent inward currents only in the presence of retinaldehyde chromophore. There were no differences in amplitude, kinetics or spectral sensitivity of responses recorded from the two forms. However in this heterologous expression system the opsin is acting through a non-native G-protein signalling cascade. We therefore cannot assume that the light response in native pRGCs will necessarily be equivalent for Opn4L and Opn4S. Indeed differences in the C-terminal motifs in the two isoforms may result in coupling to quite discrete transduction cascades within the native cell environment. Until recently GPCRs were thought to be a homogeneous family with the majority (>90%) being intronless. However, it is now clear that a small sub-set undergo alternate splicing mostly at the C-terminus. Little is known about the functional roles of these splice variants but differences relating to ligand binding, signalling efficiency, constitutive activity and desensitization have been reported (for review see Minneman, 2001).
It was previously assumed that the vertebrates had a single melanopsin gene but the study by Bellingham and colleagues described the identification of two distinct melanopsin genes in chicken, Opn4m and Opn4x (Bellingham et al., 2006). They also demonstrated that the Opn4x gene has been lost from the mammalian lineage. The alternate splicing described in this study may therefore generate functional diversity required in the mammals. However, there is now evidence that C-terminal splice variants are expressed from both chicken genes (Torii et al., 2007) so this may be another example of the reduction in photosensory capability seen in the mammals.
Immunolabelling of mouse eye sections with isoform specific antibodies demonstrates that both Opn4L and Opn4S are expressed within the mouse retina. Double immunolabelling using Opn4L and Opn4S antibodies together revealed distinct sub-populations of melanopsin expressing cells. These observations are consistent with previous reports that have described sub-classes of melanopsin ganglion cells that can be characterised based on the nature of their dendrites, termed M1 (type I), M2 (type II) or bi-stratified (type III) (Hattar et al., 2006; Viney et al., 2007; Schmidt et al., 2008). The majority of melanopsin positive cells (~70%) were labelled with both Opn4S and Opn4L antibodies, with higher levels of Opn4S labelling observed compared to Opn4L. These cells stratified in the OFF layer of the IPL near the border of the INL (M1 or type I) or were bistratified with processes evident in both the OFF layer and ON layer of the IPL (type III). The remaining cells (~30%) were labelled weakly for Opn4L only and their processes stratified in the ON layer of the IPL near to the GCL and resemble those previously described as M2 (type II).
A number of previous studies using heterozygous tau-lacZ+/− mice have distinguished M1 and M2 type cells based on morphology and differential staining with N-terminal melanopsin and β-gal antibodies (Provencio et al., 2002; Hattar et al., 2006; Baver et al., 2008). M1 type cells stain with both the N-terminal melanopsin and β-gal antibody, whereas M2 type cells only stain with the melanopsin antibody. This unexpected finding has been attributed to a low level of expression of the β-galactosidase gene that is undetectable in M2 cells. In this study double-labelling of tau-lacZ+/− mice with anti-Opn4S or anti-Opn4L and a β-gal antibody revealed a 100% overlap in expression of Opn4S and β-gal, with β-gal expression absent from a subset of Opn4L positive cells. Based on the morphology of their processes and the co-expression of β-gal, the Opn4S positive pRGCs identified in our study would again seem to correspond to those previously described as M1 (type I) or bi-stratified (type III). Morphological characterisation of the ‘Opn4L only’ cells identified in our studies is more difficult due to the low levels of staining typically observed for these cells. However, in examples where processes are clearly visible for Opn4L only cells they stratify in the ON layer of the IPL and resemble those of M2 (type II) cells. This conclusion is supported by the observation that M2 type cells are only detected using the Opn4L antibody, and are absent from single labelling experiments using the Opn4S antibody. The characteristically low levels of melanopsin staining and the lack of detectable β-gal in these cells further indicate that they are M2 cells.
Our results strongly suggest that expression of Opn4S is restricted to β-gal positive cells and is absent from M2 type cells. These findings are corroborated in a recent study by Baver and colleagues who reported that an antibody raised against the C-terminus of the rat Opn4 sequence selectively labels the β-gal positive subset of pRGCs in tau-lacZ+/− mice (Baver et al., 2008). The epitope recognised by this antibody is not present in the previously reported mouse Opn4 sequence (termed Opn4L here) (Provencio et al., 2000) but is present in the novel Opn4S sequence described here. Hence the data presented by Baver et al., confirms that Opn4S expression is absent in M2 type cells (Baver et al., 2008). Based on morphology and co-expression of the β-gal reporter, our studies suggest that 70% of pRGCs are Opn4S positive cells (M1 and bistratified), with the remaining 30% being M2 type cells, These results are in contrast with previous studies which have suggested that between 40-50% of pRGCs are M2 (Viney et al., 2007; Baver et al., 2008; Schmidt et al., 2008). A possible explanation for this disparity is that due to the relatively low expression of Opn4L observed in the Opn4S negative cells we may have failed to detect a number of these cells. Alternative explanations could include age-related differences in the mice used for our studies and circadian variation in expression (Gonzalez-Menendez et al., 2009).
pRGCs are known to receive light information from rods and/or cones (Altimus et al., 2008; Guler et al., 2008) via synaptic contacts with bipolar and amacrine cells (Belenky et al., 2003; Viney et al., 2007; Wong et al., 2007). Processes of pRGCs located in the OFF layer of the IPL are known to receive input from OFF bipolar cells and are also closely associated with dopaminergic amacrine cells (Belenky et al., 2003; Ostergaard et al., 2007; Viney et al., 2007; Vugler et al., 2007; Wong et al., 2007; Zhang et al., 2008), whereas cells with processes in the ON layer of the IPL receive inputs from ON bipolar cells (Wong et al., 2007). This difference in retinal architecture offers a clear mechanism to differentially regulate the functions of distinct pRGC sub-types. However it has also been reported that M1 type cells (that have processes in the OFF layer of the IPL) may receive direct (Wong et al., 2007; Hoshi et al., 2009) or indirect (Pickard et al., 2009) excitatory inputs from the ON bipolar cells, although the mechanisms involved are not entirely clear.
The precise natures of the synaptic contacts influencing the cells identified in this study have not been fully characterised. We have confirmed that both Opn4L and Opn4S positive processes in the OFF layer of the IPL interact closely with processes of dopaminergic amacrine cells (data not shown) but have not fully characterised the nature of bipolar cell contacts to either the Opn4S (and Opn4L) positive M1 type cells, or to the Opn4L only M2 type cells. However, our data identify an intriguing possibility that may have functional relevance. As pRGCs with processes in the OFF layer of the IPL (M1) express predominantly Opn4S (in addition to lower levels of Opn4L), and pRGCs that receive exclusively ON inputs (M2 cells) express only Opn4L, it would seem that the dominant isoform of melanopsin in cells influenced by the OFF pathway is Opn4S, whereas the dominant isoform in cells receiving exclusively ON inputs is Opn4L.
In addition to differences in cellular morphology and stratification of dendrites, a number of previous studies have identified sub-sets of pRGCs based on functional differences. At least three different type of response have been observed during Ca2+ imaging (Sekaran et al., 2003) and MEA recordings of mouse retina explants (Tu et al., 2005). These studies did not correlate these different responses with the morphologically distinct sub-sets of pRGCs. However, more recently Schmidt and Kofuji have reported functional differences in light responses recorded from M1 and M2 type pRGCs using dual whole cell electrophysiology (Schmidt and Kofuji, 2009). M1 cells were shown to be significantly more sensitive to light than M2 cells. In addition, the responses of M1 cells were significantly larger and consisted of a rapid time inactivating component and smaller sustained component. In contrast, M2 cells exhibited markedly smaller responses which showed little signs of time dependant inactivation. This study suggests that the difference in light sensitivity between M1 and M2 cells may be influenced by levels of melanopsin expression, differences in resting membrane potential, input resistance and ion channel expression. Our data now offer a potential molecular explanation for the functional differences observed between sub-populations of pRGCs based on the differential expression of Opn4L and Opn4S within these cells. The different C-terminal regions may potentially facilitate differential interactions with intracellular signalling molecules, and in turn convey functional differences to Opn4L and Opn4S expressing cells.
This study details the identification of two distinct isoforms of melanopsin in the mouse retina, a previously described long isoform and a new novel short isoform which more closely resembles the sequences of rat and human melanopsin. Both isoforms are capable of forming functional pigments in vitro and show a differential pattern of expression in sub-populations of pRGCs. Our results strongly indicate that Opn4S is the most abundant isoform in the mouse retina, although Opn4L is actually expressed in all pRGCs with expression of Opn4S restricted to M1 (type I) and bi-stratified (type III) cells. These findings provide further insight into the complexity of melanopsin signalling pathways and offer a potential mechanism to explain the functional diversity observed between different subsets of pRGCs. The application of RNAi based techniques is now needed to investigate the specific contributions of Opn4L and Opn4S isoforms to the different functional responses observed from individual pRGCs, but also to evaluate the contribution of each isoform to more complex behavioural responses such as pupillary light responses and circadian entrainment.
Supplementary Material
Acknowledgements
This work was supported by a Wellcome Trust programme grant (GR069714MA) to RGF, a BBSRC grant to MWH (BB/E021671/1) and by a D.Phil. GABBA Studentship to S.S.P. from Fundacão para a Ciência e Tecnologia (FCT), Portugal. Steven Hughes is funded by F. Hoffmann-La Roche. MK was funded by Fight for Sight. JB is grateful to the NIHR Manchester Biomedical Research Centre (BRC) and Manchester Academic Health Science Centre (MAHSC). We are grateful to Rosalie Crouch for providing the 11-cis-retinal, King-Wai Yau & Samer Hattar for the Opn4 mice and Neeraj Agarwal (Fort Worth) for providing the RGC-5 cells and Francesca Cordeiro and Li Guo for advice on culture conditions. We would also like to thank Sumathi Sekaran for useful discussions.
References
- Altimus CM, Guler AD, Villa KL, McNeill DS, Legates TA, Hattar S. Rods-cones and melanopsin detect light and dark to modulate sleep independent of image formation. Proc Natl Acad Sci U S A. 2008;105:19998–20003. doi: 10.1073/pnas.0808312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baver SB, Pickard GE, Sollars PJ. Two types of melanopsin retinal ganglion cell differentially innervate the hypothalamic suprachiasmatic nucleus and the olivary pretectal nucleus. Eur J Neurosci. 2008;27:1763–1770. doi: 10.1111/j.1460-9568.2008.06149.x. [DOI] [PubMed] [Google Scholar]
- Belenky MA, Smeraski CA, Provencio I, Sollars PJ, Pickard GE. Melanopsin retinal ganglion cells receive bipolar and amacrine cell synapses. J Comp Neurol. 2003;460:380–393. doi: 10.1002/cne.10652. [DOI] [PubMed] [Google Scholar]
- Bellingham J, Chaurasia SS, Melyan Z, Liu C, Cameron MA, Tarttelin EE, Iuvone PM, Hankins MW, Tosini G, Lucas RJ. Evolution of melanopsin photoreceptors: discovery and characterization of a new melanopsin in nonmammalian vertebrates. PLoS Biol. 2006;4:e254. doi: 10.1371/journal.pbio.0040254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson DM, Dunn FA, Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Science. 2002;295:1070–1073. doi: 10.1126/science.1067262. [DOI] [PubMed] [Google Scholar]
- Dacey DM, Liao HW, Peterson BB, Robinson FR, Smith VC, Pokorny J, Yau KW, Gamlin PD. Melanopsin-expressing ganglion cells in primate retina signal colour and irradiance and project to the LGN. Nature. 2005;433:749–754. doi: 10.1038/nature03387. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Menendez I, Contreras F, Cernuda-Cernuda R, Garcia-Fernandez JM. Daily rhythm of melanopsin-expressing cells in the mouse retina. Front Cell Neurosci. 2009;3:3. doi: 10.3389/neuro.03.003.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guler AD, Ecker JL, Lall GS, Haq S, Altimus CM, Liao HW, Barnard AR, Cahill H, Badea TC, Zhao H, Hankins MW, Berson DM, Lucas RJ, Yau KW, Hattar S. Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature. 2008;453:102–105. doi: 10.1038/nature06829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankins MW, Peirson SN, Foster RG. Melanopsin: an exciting photopigment. Trends Neurosci. 2008;31:27–36. doi: 10.1016/j.tins.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science. 2002;295:1065–1070. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S, Kumar M, Park A, Tong P, Tung J, Yau KW, Berson DM. Central projections of melanopsin-expressing retinal ganglion cells in the mouse. J Comp Neurol. 2006;497:326–349. doi: 10.1002/cne.20970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S, Lucas RJ, Mrosovsky N, Thompson S, Douglas RH, Hankins MW, Lem J, Biel M, Hofmann F, Foster RG, Yau KW. Melanopsin and rod-cone photoreceptive systems account for all major accessory visual functions in mice. Nature. 2003;424:76–81. doi: 10.1038/nature01761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi H, Liu WL, Massey SC, Mills SL. ON inputs to the OFF layer: bipolar cells that break the stratification rules of the retina. J Neurosci. 2009;29:8875–8883. doi: 10.1523/JNEUROSCI.0912-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmaoglou M, Cheetham ME. Calnexin is not essential for mammalian rod opsin biogenesis. Mol Vis. 2008;14:2466–2474. [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy RR, Agarwal P, Prasanna G, Vopat K, Lambert W, Sheedlo HJ, Pang IH, Shade D, Wordinger RJ, Yorio T, Clark AF, Agarwal N. Characterization of a transformed rat retinal ganglion cell line. Brain Res Mol Brain Res. 2001;86:1–12. doi: 10.1016/s0169-328x(00)00224-2. [DOI] [PubMed] [Google Scholar]
- Lucas RJ, Hattar S, Takao M, Berson DM, Foster RG, Yau KW. Diminished pupillary light reflex at high irradiances in melanopsin-knockout mice. Science. 2003;299:245–247. doi: 10.1126/science.1077293. [DOI] [PubMed] [Google Scholar]
- Melyan Z, Tarttelin EE, Bellingham J, Lucas RJ, Hankins MW. Addition of human melanopsin renders mammalian cells photoresponsive. Nature. 2005;433:741–745. doi: 10.1038/nature03344. [DOI] [PubMed] [Google Scholar]
- Minneman KP. Splice variants of G protein-coupled receptors. Mol Interv. 2001;1:108–116. [PubMed] [Google Scholar]
- Ostergaard J, Hannibal J, Fahrenkrug J. Synaptic contact between melanopsin-containing retinal ganglion cells and rod bipolar cells. Invest Ophthalmol Vis Sci. 2007;48:3812–3820. doi: 10.1167/iovs.06-1322. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Panda S, Nayak SK, Campo B, Walker JR, Hogenesch JB, Jegla T. Illumination of the melanopsin signaling pathway. Science. 2005;307:600–604. doi: 10.1126/science.1105121. [DOI] [PubMed] [Google Scholar]
- Panda S, Sato TK, Castrucci AM, Rollag MD, DeGrip WJ, Hogenesch JB, Provencio I, Kay SA. Melanopsin (Opn4) requirement for normal light-induced circadian phase shifting. Science. 2002;298:2213–2216. doi: 10.1126/science.1076848. [DOI] [PubMed] [Google Scholar]
- Peirson SN, Butler JN, Foster RG. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:e73. doi: 10.1093/nar/gng073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirson SN, Bovee-Geurts PH, Lupi D, Jeffery G, DeGrip WJ, Foster RG. Expression of the candidate circadian photopigment melanopsin (Opn4) in the mouse retinal pigment epithelium. Brain Res Mol Brain Res. 2004;123:132–135. doi: 10.1016/j.molbrainres.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Pickard GE, Baver SB, Ogilvie MD, Sollars PJ. Light-induced fos expression in intrinsically photosensitive retinal ganglion cells in melanopsin knockout (opn4) mice. PLoS One. 2009;4:e4984. doi: 10.1371/journal.pone.0004984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires SS, Shand J, Bellingham J, Arrese C, Turton M, Peirson S, Foster RG, Halford S. Isolation and characterization of melanopsin (Opn4) from the Australian marsupial Sminthopsis crassicaudata (fat-tailed dunnart) Proc Biol Sci. 2007;274:2791–2799. doi: 10.1098/rspb.2007.0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencio I, Rollag MD, Castrucci AM. Photoreceptive net in the mammalian retina. This mesh of cells may explain how some blind mice can still tell day from night. Nature. 2002;415:493. doi: 10.1038/415493a. [DOI] [PubMed] [Google Scholar]
- Provencio I, Jiang G, De Grip WJ, Hayes WP, Rollag MD. Melanopsin: An opsin in melanophores, brain, and eye. Proc Natl Acad Sci U S A. 1998;95:340–345. doi: 10.1073/pnas.95.1.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencio I, Rodriguez IR, Jiang G, Hayes WP, Moreira EF, Rollag MD. A novel human opsin in the inner retina. J Neurosci. 2000;20:600–605. doi: 10.1523/JNEUROSCI.20-02-00600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Kumbalasiri T, Carlson SM, Wong KY, Krishna V, Provencio I, Berson DM. Induction of photosensitivity by heterologous expression of melanopsin. Nature. 2005;433:745–749. doi: 10.1038/nature03345. [DOI] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby NF, Brennan TJ, Xie X, Cao V, Franken P, Heller HC, O’Hara BF. Role of melanopsin in circadian responses to light. Science. 2002;298:2211–2213. doi: 10.1126/science.1076701. [DOI] [PubMed] [Google Scholar]
- Saliba RS, Munro PM, Luthert PJ, Cheetham ME. The cellular fate of mutant rhodopsin: quality control, degradation and aggresome formation. J Cell Sci. 2002;115:2907–2918. doi: 10.1242/jcs.115.14.2907. [DOI] [PubMed] [Google Scholar]
- Schmidt TM, Kofuji P. Functional and morphological differences among intrinsically photosensitive retinal ganglion cells. J Neurosci. 2009;29:476–482. doi: 10.1523/JNEUROSCI.4117-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TM, Taniguchi K, Kofuji P. Intrinsic and extrinsic light responses in melanopsin-expressing ganglion cells during mouse development. J Neurophysiol. 2008;100:371–384. doi: 10.1152/jn.00062.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekaran S, Foster RG, Lucas RJ, Hankins MW. Calcium imaging reveals a network of intrinsically light-sensitive inner-retinal neurons. Curr Biol. 2003;13:1290–1298. doi: 10.1016/s0960-9822(03)00510-4. [DOI] [PubMed] [Google Scholar]
- Sekaran S, Lall GS, Ralphs KL, Wolstenholme AJ, Lucas RJ, Foster RG, Hankins MW. 2-Aminoethoxydiphenylborane is an acute inhibitor of directly photosensitive retinal ganglion cell activity in vitro and in vivo. J Neurosci. 2007;27:3981–3986. doi: 10.1523/JNEUROSCI.4716-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakita A, Tsukamoto H, Koyanagi M, Sugahara M, Yamashita T, Shichida Y. Expression and comparative characterization of Gq-coupled invertebrate visual pigments and melanopsin. J Neurochem. 2008;105:883–890. doi: 10.1111/j.1471-4159.2007.05184.x. [DOI] [PubMed] [Google Scholar]
- Torii M, Kojima D, Okano T, Nakamura A, Terakita A, Shichida Y, Wada A, Fukada Y. Two isoforms of chicken melanopsins show blue light sensitivity. FEBS Lett. 2007;581:5327–5331. doi: 10.1016/j.febslet.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Tu DC, Zhang D, Demas J, Slutsky EB, Provencio I, Holy TE, Van Gelder RN. Physiologic diversity and development of intrinsically photosensitive retinal ganglion cells. Neuron. 2005;48:987–999. doi: 10.1016/j.neuron.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Viney TJ, Balint K, Hillier D, Siegert S, Boldogkoi Z, Enquist LW, Meister M, Cepko CL, Roska B. Local retinal circuits of melanopsin-containing ganglion cells identified by transsynaptic viral tracing. Curr Biol. 2007;17:981–988. doi: 10.1016/j.cub.2007.04.058. [DOI] [PubMed] [Google Scholar]
- Vugler AA, Redgrave P, Semo M, Lawrence J, Greenwood J, Coffey PJ. Dopamine neurones form a discrete plexus with melanopsin cells in normal and degenerating retina. Exp Neurol. 2007;205:26–35. doi: 10.1016/j.expneurol.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Wong KY, Dunn FA, Graham DM, Berson DM. Synaptic influences on rat ganglion-cell photoreceptors. J Physiol. 2007;582:279–296. doi: 10.1113/jphysiol.2007.133751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DQ, Wong KY, Sollars PJ, Berson DM, Pickard GE, McMahon DG. Intraretinal signaling by ganglion cell photoreceptors to dopaminergic amacrine neurons. Proc Natl Acad Sci U S A. 2008;105:14181–14186. doi: 10.1073/pnas.0803893105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.