Abstract
Depletion of POLQ (DNA polymerase theta) has recently been shown to render tumour cells more sensitive to radiotherapy whilst having little or no effect on normal tissues. This finding led us to investigate whether tumours that overexpress POLQ are associated with an adverse outcome. We therefore correlated the clinical outcomes of two retrospective series of patients with early breast cancer with the expression levels of POLQ, as determined by microarray gene expression analysis. We found that a significant number of tumours overexpressed POLQ and that overexpression was correlated with ER negative disease (p=0.047) and high tumour grade (p=0.004), both of which are associated with poor clinical outcomes. POLQ overexpression was associated with poor relapse free survival rates on both univariate (HR 5.80; 95% CI, 2.220 to 15.159; p<0.001) and multivariate analysis (HR 8.086; 95% CI 2.340 to 27.948 p=0.001). Analysis of other published clinical series confirmed that POLQ overexpression is associated with adverse clinical outcomes. The poor prognosis associated with POLQ is independent of other clinical or pathological features. The mechanism that causes this adverse outcome remains to be elucidated but may in part arise from resistance to adjuvant treatment. These findings, combined with the limited normal tissue expression of POLQ, make it a very appealing target for possible clinical exploitation.
Keywords: Translational research, POLQ, breast cancer, prognosis, radiotherapy
Introduction
POLQ (DNA Polymerase Theta) is a member of the A family of DNA polymerases, which, unusually for this class of polymerases, synthesizes DNA with very low fidelity [1, 2]. The precise physiological functions of this protein are currently unclear. It has previously been suggested that mice deficient in POLQ had a substantially decreased frequency of mutations in immunoglobulin genes [3, 4]. However a recent study found that mutation types and frequencies were similar in wild type, POLQ−/−, POLH−/−, and POLQ−/− POLH−/− mice [5]. Accordingly this group suggested that POLQ does not have a significant role in the hypermutation pathway.
It has been suggested that POLQ has a role in base excision repair (BER) but this also remains unresolved. It has previously been shown in the DT40 chicken B cell lymphoma line, that POLQ/POLβ mutants had significantly higher sensitivity to methyl methanesulfonate than either single mutant. Extracts obtained from this cell line were used to show that POLQ mutant cells have markedly reduced single nucleotide BER capacity in vitro and that this reduction was of a similar magnitude to cells deficient in POLβ [6]. These findings led to the suggestion that POLQ and POLβ cooperate in BER.
Recent biochemical analysis has shown that cloned full-length human POLQ as well as a C-terminal fragment of POLQ, have 5′-deoxyribose phosphate (5′-dRP) lyase activity. The full-length protein and the C-terminal fragment were shown to have BER activity in vitro [7]. Although these findings have been used to support the argument that POLQ may have a role in BER in vivo, it should be noted that the rate of 5′-dRP lyase activity of POLQ is approximately 40 fold slower than that of POLβ. Cells with deficiencies in the BER pathway have been shown to have increased sensitivity to temozolomide [8]. Since cells depleted of POLQ do not show hypersensitivity to this drug, it has been questioned as to whether POLQ has any physiologically significant role in BER [9].
We have recently published a siRNA screen that aimed to identify molecular determinants of tumour radiosensitivity [9]. This study demonstrated that POLQ siRNA transfection resulted in radiosensitisation of a panel of tumour cell lines but had little or no effect on normal tissue lines. These differences reflect previous work showing significant disparity in expression between normal tissues and tumour cells [10]. Normal tissue expression appears to be mainly limited to lymphoid tissues such as the fetal liver, thymus, and bone marrow. However POLQ is known to be overexpressed in a large proportion of tumours derived from patients with colon, lung, and gastric cancer.
In view of the in vitro evidence linking POLQ expression to tumour cell radioresistance, we hypothesised that POLQ overexpression may increase the likelihood of treatment failure in cancer patients, and therefore confer an adverse clinical prognosis.
We therefore correlated the clinical outcomes of two series of breast cancer patients (n=279 in total) with the expression levels of POLQ as determined by microarray gene expression analysis. We also analysed the pathways associated with POLQ expression in vivo by data-mining gene expression data from published breast cancer studies (n=1015 samples). To the best of our knowledge this is the first study to demonstrate that POLQ is overexpressed in breast cancer, that its overexpression confers a significant adverse prognosis, and that it is associated with key cancer pathways.
Materials and Methods
Ethics Statement
Informed consent was obtained and all clinical investigations were conducted according to the ethical standards and principles expressed in the Declaration of Helsinki. Ethical approval was obtained from the local research ethics committee.
Patient Details
Individual tumour samples were obtained from retrospective series of patients with early primary breast cancer who were treated in Oxford, UK, between 1989 and 1998. Patients received adjuvant chemotherapy and/or adjuvant hormone therapy, or no adjuvant treatment. Tamoxifen was used as endocrine therapy for 5 years in estrogen receptor (ER) positive patients. Patients who were ≥50 years of age, with lymph node positive tumors, or ER– and/or a primary tumor >3 cm in diameter, received adjuvant cyclophosphamide, methotrexate, and 5-fluorouracil (CMF) for six cycles, in a three weekly intravenous regimen. Patients ≥50 years of age with ER–, lymph node–positive tumors also received CMF. Two series of 152 (Series 1) and 127 (Series 2) samples respectively were analysed. Series 1 has been described previously [11]; this series had completed 7 years of follow-up for all but 4 patients, and the median follow-up time for patients leaving the study alive and without a relapse was 12 years. Series 2 is part of a published series [12]; the published cohort had 93 cases in common with Series 1, these have been excluded from this study so that Series 1 and 2 have no overlapping cases. Series 2 had completed 10 year of follow-up apart from one case. Patient demographic details of Series 1 and 2 as analysed in this study are summarised in supplementary table 1.
RNA extraction and gene expression profiling
Total RNA was isolated by Trizol method (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. mRNA expression was measured using Affymetrix U133 arrays for Series 1 and Illumina Human RefSeq-8 arrays (Illumina inc., San Diego, CA, USA) for Series 2. RNA was amplified using Ambion Illumina Amplification Kit. Methods for both protocols have been previously described [12, 13]. Affymetrix data were pre-processed using gcrma [14]; signal from Illumina arrays was background subtracted with local background subtraction (BeadStudio). Data from both series were quantile normalized in Bioconductor (www.bioconductor.org) and logged (base 2). The target sequence of the probes that corresponded to POLQ expression in Affymetrix and Illumina arrays are shown in supplementary table 2. Two additional published datasets of patients with early breast cancer were accessed to validate the findings observed in the Oxford datasets [15, 16].
Published clinical series
NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) was searched for gene expression studies in cancer, published in peer-reviewed journals, where microarrays were performed on frozen material extracted before treatment with either chemotherapy, radiotherapy or endocrine treatment. Five data sets [11, 15, 17] of 1015 samples in total (supplementary table 3) were selected that used latest generation Affymetrix 3′ array platforms (Affymetrix U133 and plus2, www.affymetrix.com). All handling and processing of the downloaded data was performed as previously described [18].
Data-mining of gene expression data
Seed-clustering with bootstrap resampling was applied as previously described [18] to obtain genes co- and inversely expressed with POLQ in the 1015 published breast cancer samples. In short, the two probesets targeting POLQ (supplementary table 2) were chosen as initial seeds. Transcripts on the arrays showing significant association (Spearman Rank Test, Bonferroni multiple test correction) with each seed after bootstrap resampling of the breast cancer samples were considered. Amongst these, transcripts showing a concordant association with both seeds that was significantly higher than observed by random simulation were selected as POLQ co-/inversely expressed genes. A pathway enrichment analysis was thus performed using GeneCodis2 [19] to study the Gene Ontology classes and the KEGG pathways which are over-represented in POLQ co-/inversely expressed genes.
Survival analysis
Endpoints were relapse free survival for Series 1; and distant-relapse free survival and recurrence free survival as defined by the STEEP criteria [20] for Series 2. Endpoints as published were considered for the other datasets. Univariate and multivariate analysis was performed. Cox multivariate models were reduced using stepwise backward likelihood selection. In univariate analyses, expression of POLQ and other genes was considered either as binary variable, with median expression as binary cut-off, or as continuous variable, ranked and normalised between 0 and 1. In multivariate analysis the latter was always considered.
Results
POLQ is overexpressed in breast cancer compared to normal breast tissue
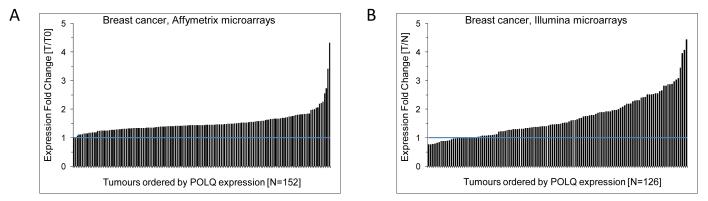
In order to assess POLQ expression, we identified two independent gene expression datasets that were obtained using arrays from different manufacturers. Series 1 and 2 were obtained using Affymetrix and Illumina arrays respectively. POLQ expression was normalised to the lowest level of tumour expression in the Affymetrix series, and to a panel of normal breast tissue samples for the Illumina series. POLQ expression is upregulated in a large proportion of breast tumour samples (Fig 1).
Figure 1.
POLQ expression in breast cancer. A) Breast cancer samples, Series 1, described in this study (N=152). No normal breast tissue samples were available for Series 1 so POLQ data were normalised to the sample with the lowest expression of POLQ (named T0). Expression fold change (FC) between all other tumours and T0 is shown for POLQ (207746_at). Expression is measured by Affymetrix array and quantile normalized. B) Breast cancer samples, Series 2, (N=127) described in this study. The FC between POLQ (ILMN_1450687) expression in each tumour and the median expression of 10 normal pools is shown. Expression is measured by Illumina arrays and quantile normalized.
POLQ overexpression is independently associated with significantly worse relapse free survival (RFS) rates
The samples from Series 1 were divided into the top and bottom 50th centiles and a univariate analysis of the differences in RFS was conducted (Fig 2A). POLQ overexpression was associated with a markedly increased risk of disease relapse (HR 5.80; 95% CI, 2.220 to 15.159; p<0.001). We then correlated the level of POLQ expression with multiple pathological and demographic features such as patient age, tumour grade and tumour size. We found that POLQ overexpression correlated with both ER negative disease (p=0.047) and high tumour grade (p=0.004) (Fig 2B). As both of these features are recognised as being associated with poor clinical outcomes [21-23], we performed a multivariate analysis which showed that POLQ expression confers a poor prognosis which is independent of any other clinical features (HR 8.086; 95% CI 2.340 to 27.948; p=0.001). The multivariate models contained POLQ as continuous variable, ranked and normalised between 0 and 1, and the following clinical features; ER status, lymph node status, patient age, tumour grade, tumour size. To confirm the validity of this finding we performed further univariate and multivariate analyses on Series 2 and the two additional datasets previously described (supplementary table 4). In total, three of the four datasets analysed demonstrated that POLQ overexpression was strongly associated with significantly worse survival outcomes (Fig 2C).
Figure 2.
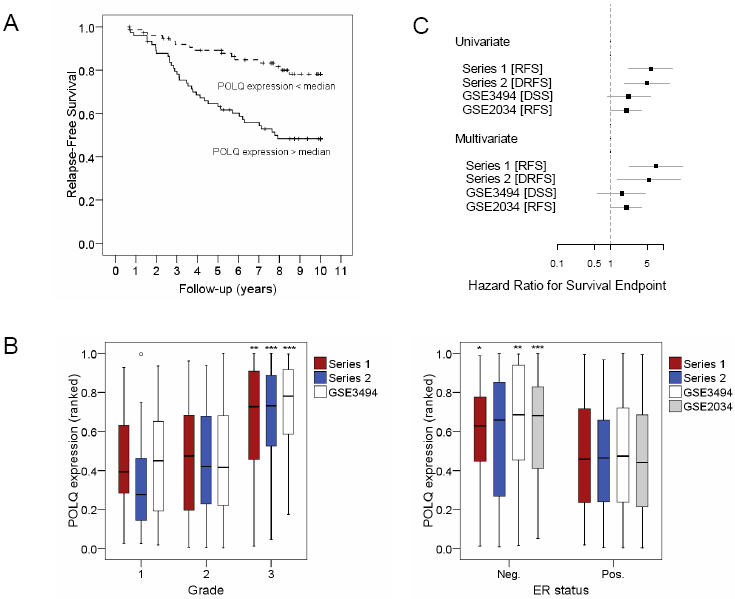
POLQ expression is prognostic in breast cancer independently from clinico-pathological variables. A) Univariate analysis in 152 breast cancers (Series 1). POLQ expression is divided in two groups by median value. B) POLQ expression is associated with tumour grade (left) and ER status (right) in Series 1 and 2 described in this study (Affymetrix and Illumina arrays respectively) and two published series (Affymetrix arrays, see Methods), although grade information was not available for GSE2034. Boxes summarize the median, quartiles and extreme values of POLQ expression in the different categories. One outlier is shown (circle), defined as case with values between 1.5-3 box lengths from the edge of the box. Mann-Whitney and Spearman Rank Association significance levels for the null hypotheses of POLQ expression not varying with ER and Grade respectively, are indicated on the highest category of each plot:*=p<0.05, **=p<0.01, ***=p<0.001. C) Forest plot of POLQ Hazard Ratio for Survival Endpoints in univariate and multivariate analysis in the 2 series described in this study and 2 published datasets (GEO Ids shown). Dots represent Hazard Ratios of POLQ expression and grey bars the 95% confidence intervals. Dot dimensions are proportional to dataset size. The expression of POLQ is entered in this model as a continuous ranked variable, normalised between 0 (lowest rank) and 1 (highest rank). RFS= Recurrence Free Survival, DRFS=Distant Relapse Free Survival, DSS= Disease Specific Survival.
Clustering analysis identifies genes co-expressed with POLQ with functions in key cancer pathways
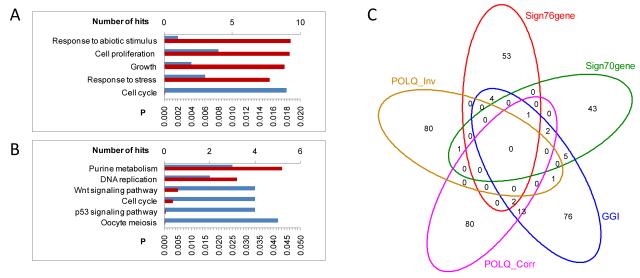
In order to identify genes which were co-expressed with POLQ, a seed-clustering analysis was performed on gene expression data obtained from five different breast cancer data sets (details of datasets in supplementary table 3). This identified a total of 97 genes that were strongly associated with POLQ overexpression in breast cancer (supplementary table 5). Pathway analysis of these genes showed that genes co-expressed with POLQ are involved in several pathways that have been associated with cancer development and progression such as cell cycle progression, p53 signalling, Wnt signalling and DNA replication (Fig 3A and 3B).
Figure 3.
Pathway analysis and overlap with prognostic signatures of POLQ co-expressed genes. Seed-clustering was used in 1015 breast cancer samples to identify genes whose expression was co- and inversely associated with POLQ expression. A) Over-represented KEGG pathways and B) GO Biological processes amongst genes co-expressed with POLQ. The number of genes in each pathway is shown in blue, top x-axis, and a hypergeometric test p-value (FDR adjustment for multiple testing) is shown in red, bottom axis. C) Venn-diagram showing the overlap of genes whose expression is co- (POLQ_Corr) and inversely (POLQ_Inv) associated with expression of POLQ with the Genomic Grade Index Signature (GGI) [25], the 76-gene signature (Sign76gene) [16], and the 70-genes signature (Sign70genes) [24].
Genes co-expressed with POLQ overlap with several genes that comprise the Gene expression Grade Index (GGI)
Previous studies such as the ‘70-gene’ expression signature [24] have identified groups of genes that form expression profiles which correlate with clinical outcome. Although POLQ expression has not previously been shown to be independently associated with clinical outcome, it is interesting to note that POLQ is included in both the GGI [25], and the ‘76-gene’ signature [16]. The correlation between POLQ expression and tumour grade and prognosis (Fig. 2) led us to assess whether genes that are co-expressed with POLQ are included in these validated gene expression signatures (Fig. 3C). Eighteen of the genes that are significantly co-expressed with POLQ (supplementary table 5) are components of the GGI index (Table 1). The large number of genes that overlap between these two groups may account for the clinical correlation between POLQ expression and high tumour grade.
Table 1.
Overlap between the Genomic Grade Index (GGI) signature [25] and transcripts co- or inversely associated with POLQ in seed-clustering of 1015 breast cancer samples
| Symbol | GGI grades |
Accession Number |
Gene ID | Full name/description |
|---|---|---|---|---|
| Transcripts co-expressed with POLQ | ||||
| AURKA | G3 | NM_003158 | 6790 | aurora kinase A |
| CCNB2 | G3 | NM_004701 | 9133 | cyclin B2 |
| CCNE2 | G3 | NM_004702 | 9134 | cyclin E2 |
| CDKN3 | G3 | AF213033 | 1033 | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) |
| CEP55 | G3 | NM_018131 | 55165 | centrosomal protein 55kDa |
| ESPL1 | G3 | NM_012291 | 9700 | extra spindle pole bodies homolog 1 (S. cerevisiae) |
| ESPL1 | G3 | D79987 | 9700 | extra spindle pole bodies homolog 1 (S. cerevisiae) |
| GTSE1 | G3 | NM_016426 | 51512 | G-2 and S-phase expressed 1 |
| KIFC1 | G3 | BC000712 | 3833 | kinesin family member C1 |
| LMNB1 | G3 | NM_005573 | 4001 | lamin B1 |
| MCM2 | G3 | NM_004526 | 4171 | MCM2 minichromosome maintenance deficient 2, mitotin (S. cerevisiae) |
| MELK | G3 | NM_014791 | 9833 | maternal embryonic leucine zipper kinase |
| MYBL2 | G3 | NM_002466 | 4605 | v-myb myeloblastosis viral oncogene homolog (avian)-like 2 |
| NA | G3 | BE966236 | NA | NA |
| NCAPG | G3 | NM_022346 | 64151 | non-SMC condensin I complex, subunit G |
| POLQ | G3 | NM_006596 | 10721 | polymerase (DNA directed), theta |
| PRC1 | G3 | NM_003981 | 9055 | protein regulator of cytokinesis 1 |
| RRM2 | G3 | BC001886 | 6241 | ribonucleotide reductase M2 polypeptide |
| TIMELESS | G3 | NM_003920 | 8914 | timeless homolog (Drosophila) |
| TRIP13 | G3 | NM_004237 | 9319 | thyroid hormone receptor interactor 13 |
| Transcripts whose expression is inversely associated with POLQ expression | ||||
| CX3CR | G1 | U20350 | 1524 | chemokine (C-X3-C motif) receptor 1 |
G1 and G3 are the sets of genes with increased expression in histologic grade 1 and 3 tumors, respectively.
POLQ overexpresssion confers a poor prognosis that is independent of published prognostic signatures
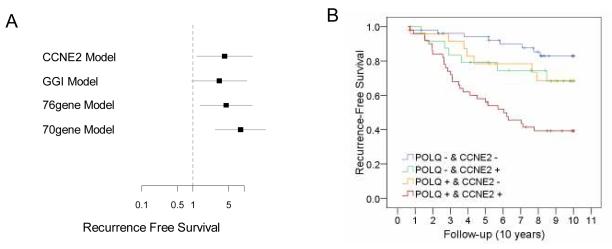
As POLQ has several genes in common with the GGI signature, and is itself part of the GGI and ‘76 gene’ signatures, we assessed whether POLQ expression remained an independent predictor of relapse when these signatures were included in a multivariate analysis of the data from Series 1 (Fig 4A and supplementary table 6). POLQ expression remained a strong, independent predictor of disease relapse after statistical consideration of these validated expression profiles and reinforces the close association between POLQ expression and adverse outcome.
Figure 4.
POLQ expression shows prognostic potential in multivariate models including clinical variables, published signatures and CCNE2. A) Forest plot of POLQ Hazard Ratio for Recurrence Free Survival in multivariate analysis of Series 1. Dots represent Hazard Ratios (dimensions are proportional to dataset size) and grey bars the 95% confidence intervals. In each analysis, a multivariate model including POLQ expression, all significant clinical variables, CCNE2 expression, and published signature scores (GGI, 76-gene or 70-gene signature) is derived. The expression of POLQ, signature scores and CCNE2 are entered in these models as continuous ranked variables, normalised between 0 (lowest rank) and 1 (highest rank). See methods for more details. B) Kaplan-Meier plots of Series 1 data. POLQ and CCNE2 expression divided by median value (− indicates below median, + above median). A Helmert contrasts analysis demonstrated that tumours overexpressing both POLQ and CCNE2 were associated with worse outcomes than the average of the other groups (HR 3.26; 95% CI 1.88 to 5.66; p<0.001)
The poor prognosis associated with POLQ expression is independent of Cyclin E expression
CCNE2 (cyclin E) is the only gene that is a component of all three expression signatures and which is also co-expressed with POLQ. As cyclin E overexpression has been identified as being independently associated with an adverse outcome in breast cancer patients [26], we considered whether the adverse prognosis associated with POLQ expression may simply be due to the observation that CCNE2 is often co-expressed with POLQ. We therefore performed a multivariate analysis of the data from Series 1 that included CCNE2 expression and found that POLQ and CCNE2 were both independently associated with an increase in RFS (Fig 4A). It is notable that tumours that overexpress both POLQ and CCNE2 confer an extremely poor prognosis relative to the other groups (HR 3.26; 95% CI 1.88 to 5.66; p<0.001) (Fig 4B). Tumours that do not overexpress either gene are associated with a good prognosis, and those that overexpress only one of the genes are associated with an intermediate prognosis. This data suggests that the biological mechanisms by which POLQ and CCNE2 confer a poor prognosis might be independent of each other. These results could not be confirmed in the other datasets considered, where POLQ lost significance after inclusion of CCNE2. However it should be noted that Series 1 is the only one in which patients did not receive systemic chemotherapy which could potentially distort prognostically important factors.
Discussion
We have recently demonstrated that tumour cells depleted of POLQ are rendered more sensitive to radiotherapy and that its limited expression in normal tissues made POLQ a potentially exploitable clinical target [9]. In this study we have demonstrated that POLQ is frequently upregulated in breast cancers. Although POLQ overexpression has previously been demonstrated in lung, gastric and colorectal cancers [10], to the best of our knowledge, this has not previously been shown in breast cancer.
In this current study we have demonstrated strong associations between POLQ expression and the presence of other individual factors such as tumour grade and ER negative disease that are known to confer an adverse prognosis. We have also demonstrated that POLQ overexpression is associated with markedly increased rates of disease relapse, and using multivariate analysis, that these increased failure rates are independent of its association with features like tumour grade and ER status.
The mechanisms by which POLQ overexpression causes these adverse outcomes are not presently clear. POLQ associated radioresistance is likely to contribute to these findings and further work is required to assess whether POLQ expression increases the tumour cell resistance to the cytotoxic and endocrine treatments typically used to treat breast cancers. The co-expression of POLQ with genes linked to pathways associated with tumour progression, as well as several genes that are contained within the gene expression grade index, suggests that POLQ overexpression promotes a more aggressive phenotype, increasing the likelihood of disease recurrence.
The clinical significance of tumour expression of POLQ has not previously been examined in detail. A previous study in colorectal cancer correlated the expression levels of genes involved in DNA replication with clinical outcomes in 74 patients with colorectal cancer [27]. Although POLQ was not independently associated with adverse outcome, its co-overexpression with at least three other genes involved in DNA replication ‘firing’ (from among CDC45, CDC6, CDT1, SLD5, MCM2, and MCM7) was associated with a worse overall survival. The overall significance of POLQ on this finding is not clear since MCM7 overexpression was shown to independently be associated with adverse survival rates. This group suggested that the expression of these genes could produce a more aggressive tumour phenotype by contributing to ‘replication stress’. As POLQ is known to repair DNA damage in an error-prone fashion [1, 2], it would seem likely that the poor prognosis that we have described in this study is partially due to POLQ contributing to increased replication stress and genomic instability.
To the best of our knowledge this is the first study to demonstrate an adverse association with POLQ expression in patients with breast cancer. In recent years, attempts have been made to identify gene expression signatures that are capable of predicting patient outcomes with greater accuracy than is currently achievable in routine clinical practice. It is possible that specific gene expression profiles could identify the likelihood of response to individual therapies, enabling clinicians to refine the adjuvant therapy offered to individual patients. The GGI signature [25] identified 97 genes with differential expression between low and high grade breast carcinomas. This signature enabled a more accurate and refined assessment of the risk of disease recurrence in patients with intermediate grade disease. Subsequent studies have confirmed the ability of the GGI signature to accurately predict disease relapse [13, 28]. A separate expression profile has been created to more accurately identify patients at risk of developing metastatic disease [16]. This study used tumours derived from patients who did not receive adjuvant systemic therapy, thereby eliminating potentially confounding predictive factors occurring as a result of systemic treatment. The resulting ‘76 gene’ signature was shown to predict both distant failure as well as overall survival. Further studies have reinforced the prognostic accuracy of this gene signature [29, 30]. A third gene expression profile utilising a 70 gene signature has also been shown to predict clinical outcome [24] and has also been subsequently validated [31]. The prognostic effect of POLQ expression on its own has not previously been assessed, but it is interesting to note that POLQ is a component of both the GGI and the ‘76 gene’ expression profiles. Given the large differences that we have shown in relapse rates on the basis of POLQ expression, and that these differences are maintained on multivariate analyses that include these signatures, it is possible that POLQ may be amongst the most important determinants within these signatures.
Pathway analysis identified several genes, including Cyclin E, that were frequently co-expressed with POLQ. Cyclin E over expression has been identified as being associated with an adverse outcome in breast cancer patients [26]. It is the only gene that is a component of all three gene expression signatures and which is also frequently co-expressed with POLQ. Cyclin E binds to cyclin-dependent kinase-2 (cdk-2), permitting the transition from G1 to S-phase [32]. Increased cyclin E induces enhanced cdk-2 activity, accelerating G1/S transition [33]. There is substantial evidence to suggest that CCNE overexpression confers a poor prognosis in breast cancer. A recent meta-analysis of 12 independent studies involving 2,534 patients, demonstrated that the combined HR estimate for overall survival and breast cancer specific survival was 2.98 (95% CI, 1.85–4.78) and 2.86 (95% CI, 1.85–4.41) in univariate and multivariate analysis, respectively [34]. Although there is ongoing debate as to which fragments of cyclin E are important in predicting outcome [35], the evidence supporting its use in routine clinical assessment have led for calls for large scale clinical trials [34]. In this study we have again confirmed that cyclin E overexpression was associated with a poor clinical prognosis on multivariate analysis. In addition we have shown that tumours expressing both POLQ and CCNE2 are associated with an extremely poor outcome. This suggests that these genes confer a poor prognosis through separate mechanisms. Larger studies are required to investigate whether the risk of relapse from tumours overexpressing cyclin E could be better assessed if further stratified by POLQ expression levels.
Independently of its association with other known poor pathological features, POLQ overexpression is associated with increased relapse rates. This is the first study to demonstrate that POLQ overexpression is associated with an extremely poor outcome in breast cancer on both univariate and multivariate analysis.
We believe that the poor prognosis associated with POLQ expression, the known radiosensitivity induced by its depletion, and its highly limited normal tissue expression makes POLQ an extremely appealing target for clinical exploitation.
Supplementary Material
Acknowledgements
This work was supported by grants from Cancer Research UK, the Medical Research Council and the NIHR Biomedical Research Centre, Oxford. GH was supported by a Cancer Research UK/Royal College of Radiologists Clinical Research Fellowship.
Footnotes
Conflict of Interest The Authors have no conflicts of interest to declare.
References
- 1.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger AJ, Iii, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase theta plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masuda K, Ouchida R, Hikida M, Nakayama M, Ohara O, Kurosaki T, O-Wang J. Absence of DNA polymerase theta results in decreased somatic hypermutation frequency and altered mutation patterns in Ig genes. DNA Repair (Amst) 2006;5:1384–1391. doi: 10.1016/j.dnarep.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Martomo SA, Saribasak H, Yokoi M, Hanaoka F, Gearhart PJ. Reevaluation of the role of DNA polymerase theta in somatic hypermutation of immunoglobulin genes. DNA Repair (Amst) 2008;7:1603–1608. doi: 10.1016/j.dnarep.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, Lan L, Seki M, Wood RD, Arakawa H, Buerstedde JM, Hochegger H, Okada T, Hiraoka M, Takeda S. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH. Human DNA polymerase theta possesses 5′-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009;37:1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trivedi RN, Almeida KH, Fornsaglio JL, Schamus S, Sobol RW. The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer Res. 2005;65:6394–6400. doi: 10.1158/0008-5472.CAN-05-0715. [DOI] [PubMed] [Google Scholar]
- 9.Higgins GS, Prevo R, Lee YF, Helleday T, Muschel RJ, Taylor S, Yoshimura M, Hickson ID, Bernhard EJ, McKenna WG. A small interfering RNA screen of genes involved in DNA repair identifies tumor-specific radiosensitization by POLQ knockdown. Cancer Res. 2010;70:2984–2993. doi: 10.1158/0008-5472.CAN-09-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawamura K, Bahar R, Seimiya M, Chiyo M, Wada A, Okada S, Hatano M, Tokuhisa T, Kimura H, Watanabe S, Honda I, Sakiyama S, Tagawa M, O-Wang J. DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int J Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- 11.Loi S, Haibe-Kains B, Desmedt C, Wirapati P, Lallemand F, Tutt AM, Gillet C, Ellis P, Ryder K, Reid JF, Daidone MG, Pierotti MA, Berns EM, Jansen MP, Foekens JA, Delorenzi M, Bontempi G, Piccart MJ, Sotiriou C. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 13.Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, Ellis P, Harris A, Bergh J, Foekens JA, Klijn JG, Larsimont D, Buyse M, Bontempi G, Delorenzi M, Piccart MJ, Sotiriou C. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 14.Wu ZIR, Gentleman R, Martinez-Murillo F, Spencer F. A Model-Based Background Adjustment for Oligonucleotide Expression Arrays. Journal of the American Statistical Association. 2004;99:909–917. [Google Scholar]
- 15.Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, Bergh J. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 17.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 18.Buffa FM, Harris AL, West CM, Miller CJ. Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br J Cancer. 2010;102:428–435. doi: 10.1038/sj.bjc.6605450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37:W317–322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudis CA, Barlow WE, Costantino JP, Gray RJ, Pritchard KI, Chapman JA, Sparano JA, Hunsberger S, Enos RA, Gelber RD, Zujewski JA. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J Clin Oncol. 2007;25:2127–2132. doi: 10.1200/JCO.2006.10.3523. [DOI] [PubMed] [Google Scholar]
- 21.Parl FF, Schmidt BP, Dupont WD, Wagner RK. Prognostic significance of estrogen receptor status in breast cancer in relation to tumor stage, axillary node metastasis, and histopathologic grading. Cancer. 1984;54:2237–2242. doi: 10.1002/1097-0142(19841115)54:10<2237::aid-cncr2820541029>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 22.Kinne DW, Butler JA, Kimmel M, Flehinger BJ, Menendez-Botet C, Schwartz M. Estrogen receptor protein of breast cancer in patients with positive nodes. High recurrence rates in the postmenopausal estrogen receptor-negative group. Arch Surg. 1987;122:1303–1306. doi: 10.1001/archsurg.1987.01400230089016. [DOI] [PubMed] [Google Scholar]
- 23.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 24.van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 25.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, Desmedt C, Larsimont D, Cardoso F, Peterse H, Nuyten D, Buyse M, Van de Vijver MJ, Bergh J, Piccart M, Delorenzi M. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen NH, Arnerlov C, Emdin SO, Landberg G. Cyclin E overexpression, a negative prognostic factor in breast cancer with strong correlation to oestrogen receptor status. Br J Cancer. 1996;74:874–880. doi: 10.1038/bjc.1996.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pillaire MJ, Selves J, Gordien K, Gourraud PA, Gentil C, Danjoux M, Do C, Negre V, Bieth A, Guimbaud R, Trouche D, Pasero P, Mechali M, Hoffmann JS, Cazaux C. A ‘DNA replication’ signature of progression and negative outcome in colorectal cancer. Oncogene. 29:876–887. doi: 10.1038/onc.2009.378. [DOI] [PubMed] [Google Scholar]
- 28.Desmedt C, Giobbie-Hurder A, Neven P, Paridaens R, Christiaens MR, Smeets A, Lallemand F, Haibe-Kains B, Viale G, Gelber RD, Piccart M, Sotiriou C. The Gene expression Grade Index: a potential predictor of relapse for endocrine-treated breast cancer patients in the BIG 1-98 trial. BMC Med Genomics. 2009;2:40. doi: 10.1186/1755-8794-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foekens JA, Atkins D, Zhang Y, Sweep FC, Harbeck N, Paradiso A, Cufer T, Sieuwerts AM, Talantov D, Span PN, Tjan-Heijnen VC, Zito AF, Specht K, Hoefler H, Golouh R, Schittulli F, Schmitt M, Beex LV, Klijn JG, Wang Y. Multicenter validation of a gene expression-based prognostic signature in lymph node-negative primary breast cancer. J Clin Oncol. 2006;24:1665–1671. doi: 10.1200/JCO.2005.03.9115. [DOI] [PubMed] [Google Scholar]
- 30.Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y, d’Assignies MS, Bergh J, Lidereau R, Ellis P, Harris AL, Klijn JG, Foekens JA, Cardoso F, Piccart MJ, Buyse M, Sotiriou C. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13:3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 31.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 32.Koff A, Giordano A, Desai D, Yamashita K, Harper JW, Elledge S, Nishimoto T, Morgan DO, Franza BR, Roberts JM. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 33.Resnitzky D, Gossen M, Bujard H, Reed SI. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Shao ZM. Cyclin e expression and prognosis in breast cancer patients: a meta-analysis of published studies. Cancer Invest. 2006;24:581–587. doi: 10.1080/07357900600894799. [DOI] [PubMed] [Google Scholar]
- 35.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC., Jr. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.