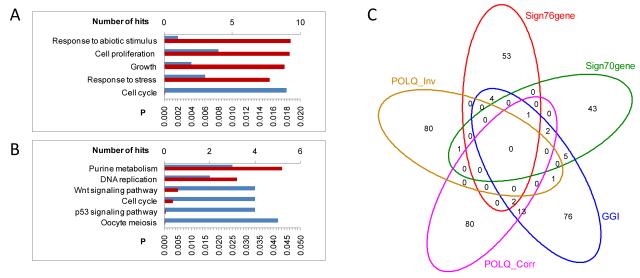
Figure 3.
Pathway analysis and overlap with prognostic signatures of POLQ co-expressed genes. Seed-clustering was used in 1015 breast cancer samples to identify genes whose expression was co- and inversely associated with POLQ expression. A) Over-represented KEGG pathways and B) GO Biological processes amongst genes co-expressed with POLQ. The number of genes in each pathway is shown in blue, top x-axis, and a hypergeometric test p-value (FDR adjustment for multiple testing) is shown in red, bottom axis. C) Venn-diagram showing the overlap of genes whose expression is co- (POLQ_Corr) and inversely (POLQ_Inv) associated with expression of POLQ with the Genomic Grade Index Signature (GGI) [25], the 76-gene signature (Sign76gene) [16], and the 70-genes signature (Sign70genes) [24].