Abstract
Considerable non-allelic heterogeneity for autosomal recessively inherited Charcot-Marie-Tooth (ARCMT) disease has challenged molecular testing and often requires a large amount of work in terms of DNA sequencing and data interpretation or remains unpractical. This study tested the value of SNP array-based whole-genome homozygosity mapping as a first step in the molecular genetic diagnosis of sporadic or ARCMT in patients from inbred families or outbred populations with the ancestors originating from the same geographic area. Using 10 K 2.0 and 250 K Nsp Affymetrix SNP arrays, 15 (63%) of 24 CMT patients received an accurate genetic diagnosis. We used our Java-based script eHoPASA CMT—easy Homozygosity Profiling of SNP arrays for CMT patients to display the location of homozygous regions and their extent of marker count and base-pairs throughout the whole genome. CMT4C was the most common genetic subtype with mutations detected in SH3TC2, one (p.E632Kfs13X) appearing to be a novel founder mutation. A sporadic patient with severe CMT was homozygous for the c.250G > C (p.G84R) HSPB1 mutation which has previously been reported to cause autosomal dominant dHMN. Two distantly related CMT1 patients with early disease onset were found to carry a novel homozygous mutation in MFN2 (p.N131S). We conclude that SNP array-based homozygosity mapping is a fast, powerful, and economic tool to guide molecular genetic testing in ARCMT and in selected sporadic CMT patients.
Keywords: Autosomal recessive CMT, Homozygosity mapping, SNP array, Hereditary neuropathies, Gene
Introduction
Charcot-Marie-Tooth (CMT) disease is a common group of inherited neuropathies with autosomal dominant (ADCMT), autosomal recessive (ARCMT) or X-linked inheritance. It has been shown that ARCMT, also known as CMT type 4 (CMT4), is genetically heterogeneous with at least 14 genes identified so far [1] (Supplementary Table 1). CMT4 is usually characterized by an early disease onset and a more severe course of the disease compared to ADCMT [2, 3]. The underlying pathology of the peripheral nerves has been shown to be due to demyelinating or axonal nerve damage or a combination of both [1]. However, the resulting phenotypes are generally very similar making it often impossible to distinguish the different genetic subtypes on clinical and electrophysiological grounds. Given the large number of genes involved in the pathogenesis of CMT and that all Mendelian modes of inheritance might apply in a sporadic case, molecular testing often requires a considerable effort in terms of cost and time, thus becoming a challenge for clinicians and human geneticists.
Recent molecular genetic studies have shown considerable progress in the identification of recessive disease-causing genes using positional cloning by homozygosity (equal autozygosity) mapping that tests the assumption that a homozygous mutation in a recessive disease is identical by descent (IBD) by segregating twice to the affected person from a common ancestor through both the maternal and paternal line [4, 5]. The value of single nucleotide polymorphism (SNP) array-based homozygosty mapping in the identification of a recessive disease gene for both inbred and outbred populations has been demonstrated [4,6]. We here show that whole genome SNP homozygosity mapping and subsequent DNA sequencing of a known candidate gene provide a fast, cost-effective, and powerful approach to identify the disease locus in autosomal recessive and in selected sporadic CMT patients and can guide molecular genetic testing.
Materials and methods
Patients
The present study included a total of 24 unrelated index probands with sporadic or ARCMT neuropathy who had received a detailed neurological and neurophysiological examination. The study was performed with written informed consent and was approved by the local Ethical Committee of the Medical University of Graz, Austria.
Based on the family history the patients were subdivided into three groups:
Group 1 (G1)
Includes 6 probands (G1-1 to G1-6) diagnosed as CMT4 whose parents are first cousins.
Group 2 (G2)
Includes 11 probands (G2-7 to G2-17) with sporadic or ARCMT whoes ancestors originate from the same or a near geographical area making distant relationship of the parents possible.
Group 3 (G3 control group)
Includes 7 patients (G3-18 to G3-24) with genetically confirmed CMT or related neuropathies caused by homozygous mutations [7, 8]. In these individuals SNP homozygosity mapping was carried out to retrospectively test for homozygous areas. For G3-18 to G3-21 consanguinity of the parents was known. Group 3 also includes a patient with a mutation in the HSN2 gene which has been published previously [8].
SNP array genotyping and identification of homozygous regions
In order to define the molecular genetic basis of the disease in the CMT patients, a search for regions of putative homozygosity-by-descent was initiated by conducting a whole genome scan using Affymetrix SNP arrays. Genomic DNA samples from all patients of group 1 and group 3 (G1-1–6; G3-18–24), from G2-7–10, G2-13, G2-15, G2-17, and from a healthy control were hybridized to Gene-Chip® Human Mapping 10 K 2.0 SNP arrays (Affymetrix, Santa Clara, CA, USA) using the protocols recommended by the manufacturer. Samples from patients G1-1–4, G2-7–14, G2-16, G3-18, G3-23, G3-24, and from a healthy control were also hybridized to GeneChip® Human Mapping 250 K Nsp SNP arrays subsequently. Genotypes of both array types were called using GeneChip Genotyping Analysis Software (Version 4.0) and default thresholds. The overall call rate of each array succeeded 95%.
Both the 10 K 2.0 and the 250 K Nsp SNP results of each patient were further analyzed by using our self-developed Java-based tool (eHoPASA-CMT—Easy Homozygosity Profiling of Affymetrix SNP arrays for CMT patients) which had been designed to quickly extract genome wide regions of homozygosity (“homozygosity plotting”) that may harbor the disease gene without any linkage computations [9]. The software accepts tab-delimited SNP array genotype data and generates result files for scaled homozygosity profile display. Single occurrences of genotyping failures or undetermined genotypes (“no calls”) are being ignored, when at least seven neighboring genotypes on each side are homozygous [10]. For the homozygosity regions plotting we arbitrarily defined a homozygous region on the occurrence of at least eight or more consecutive homozygous SNPs (hSNPs, 10 K 2.0 array) and 20 hSNPs (250 K Nsp array) by the user-selectable parameter in the program. Homozygosity profiles along the entire human genome were plotted using the R statistical software package [11]. In addition to other software programs [10] our tool was adapted to quickly inspect and visualize homozygosity profiles at the ARCMT loci. In the next step the resulted homozygosity profiles were further mapped against ARCMT loci using only homozygosity regions with at least 15 consecutive homozygous SNPs (hSNPs, 10 K 2.0 array) and 40 hSNPs (250 K Nsp array) to discover the regions of homozygosity within these loci. Subsequently only CMT genes located within these regions were sequenced (Supplementary Table 2). All analyses are based on human reference sequence hg18 (NCBI Build 36 Version 1) genome assembly annotations. The eHoPASA-CMT software itself and R programming code for plotting of the homozygosity profiles are freely available for download (http://ehopasa.sourceforge.net).
Mutation analysis
All exons and exon–intron boundaries of ganglioside-induced differentiation-associated protein 1 (GDAP1), SH3 domain and tetratricopeptide repeats 2 (SH3TC2), heat shock 27 kDa protein 1 (HSPB1), early growth response 2 (EGR2), neurofilament protein, light chain (NEFL), lamin A/C (LMNA) and mitofusin 2 (MFN2) were screened for mutations by direct sequencing (ABI 3730 Genetic Analyzer, Applied Biosystems, Foster City, CA, USA) following standard methods. Frataxin (FXN) was analyzed using standard methods [12].
Results
Patients
Detailed clinical and neurophysiological findings in the 24 CMT patients (6 females, 18 males) aged 7–59 years (mean 24 years) with disease onset ranging from birth to 25 years (mean 7 years) are shown in Supplementary Table 3. In ten patients the phenotype was complicated and included additional proximal weakness, deafness, mild retardation, ataxia, nephropathy, hyperhydrosis, and hyperextensible skin. NCS of the median or ulnar motor nerve were known in 21 of 24 probands and confirmed the presence of a demyelinating (14), axonal (4) or intermediate neuropathy (3) in the patients.
Homozygosity plotting identifies the individual ARCMT locus in the majority of cases
Low- and high-density (10 and 250 K) array-based homozygosity plotting revealed up to four regions of homoyzgosity at ARCMT loci, with a higher number and larger extent of homozygosity in individuals from group 1 as compared to individuals from group 2 and the control group (group 3); see Supplementary Table 2. As expected, high-density arrays revealed a higher resolution of homozygosity across the genome at ARCMT loci. Using our criteria to define a homozygous region disease-causing mutations at homozygous ARCMT loci were subsequently identified in 5/6 patients from group 1, in 4/11 patients from group 2, and 6 of 7 patients from group 3 (Supplementary Table 2).
Mutations in the SH3TC2 gene
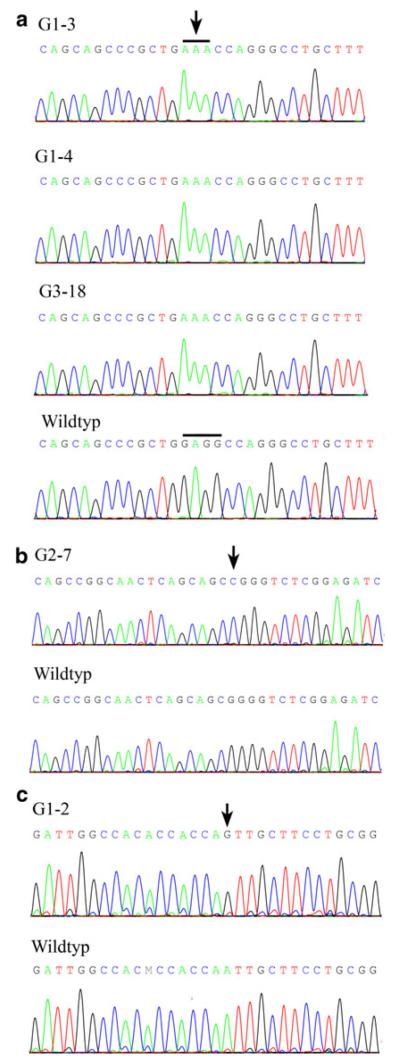
In three Turkish CMT1 patients (G1-3, G1-4 and G3-18) without known relationship, we identified a novel mutation (c.1894_1897 delGAGGinsAAA) in the SH3TC2 gene (Fig. 1a). Patients G1-3 and G1-4 were homozygous at the SH3TC2 locus but haplotypes obtained from the 10 K 2.0 SNP array were different. In individual G3-19 homozygosity at this locus was even absent when using the 10 K 2.0 SNP array. However, using 250 K Nsp arrays revealed a 2.5 Mb interval in all three patients including 284 identical hSNPs suggesting a common haplotype. Patient G2-8 and G2-15 from Austria with severe early onset CMT1 demonstrated significant homozygosity at the SH3TC2 locus (Supplementary Table 2) which guided initial genetic testing of the SH3TC2 gene. Both patients carried the known c.2860 C > T (p.R954X) mutation in a homozygous state. In the Turkish patient G2-17 with early onset CMT1 and scoliosis homozygosity at the SH3TC2 locus also suggested subsequent screening of the SH3TC2 gene that revealed the c.1285 G > T (p.E429X) mutation.
Fig. 1.
Detection of ARCMT mutations. a Partial sequence chromatogrom displaying SH3TC2 mutations c.1894_1897 del-GAGGinsAAA (p.E632Kfs13X, exon 11, arrow) identified in three patients. b Homozygous HSPB1 c.250G > C (p.G84R, exon 1) mutation (arrow). c Homozygous MFN2 c.392A > G, p.N131S (exon 3) mutation (arrow)
Mutations in the HSPB1 gene
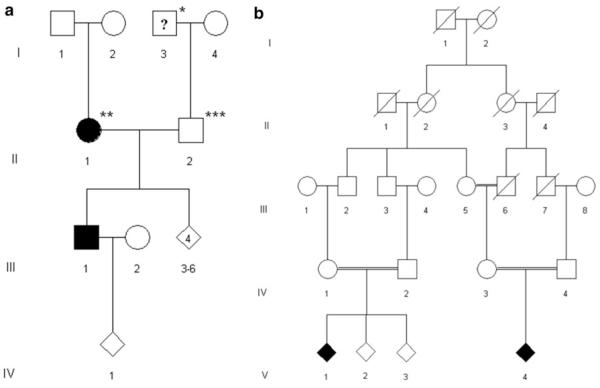
Patient G2-7, whose parents originated from the same geographic area, was found to be homozygous at the HSPB1 locus (Supplementary Table 2, Fig. 2a). This proband exhibited an early onset severe CMT phenotype. Median NCS were reduced to 35 m/s. Sensory symptoms were not reported and sensory NCS were within the normal range. There was no family history except for prominent foot deformity in the paternal grandfather. Direct sequencing of HSPB1 detected a homozygous mutation (c.250G > C; p.G84R; Fig. 1b) which has previously been reported to segregate with autosomal dominant CMT [13]. Subsequent examination of the patient’s father revealed absent achilles tendon reflexes and reduced patellar and upper limb tendon reflexes. Muscle strength and tone were normal and vibration sense was preserved including vibration at the hallux. Motor and sensory NCS were in the normal range in upper and lower limbs. The mother did not complain of any symptoms and neurological examination was normal except for reduced achilles tendon reflexes. However, motor NCS were severely slowed and reduced in amplitudes in the lower limbs (left tibial nerve: 27 m/s; 1.5 mV; right tibial nerve: 31 m/s, 1.4 mV). NCS of the sural nerve revealed reduced amplitudes and mild conduction slowing (left side: 7 μV; 40 m/s; right side: 8.9 μV; 41 m/s). NCS in the upper limbs were in the normal range except for mild slowing of distal motor latencies in the absence of clinical signs of a carpal tunnel syndrome. Both parents were heterozygous for the HSPB1 c.250G > C; p.G84R mutation.
Fig. 2.
Pedigree. a Pedigree of the family of patient G2-7 (III/1) carrying the HSPB1 mutation c.250G > C, p.G84R. * This individual was reported having had prominent pes cavus. ** Clinically normal, but severe slowing of NCV in the LL. *** Clinically normal, but reduced tendon reflexes. b Pedigree of the family of patient G1-2 (V/1) carrying the MFN2 mutation c.392A > G, p.N131S
Mutations in the MFN2 gene
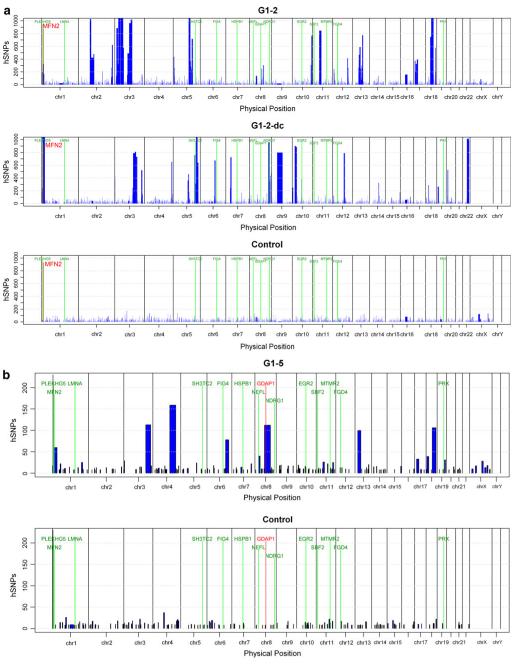
Patient G1-2 with severe and typical early onset CMT1 was initially excluded for mutations in the common CMT1 genes (PMP22, MPZ, GJB1, LITAF, GDAP1). Subsequent analysis of the 250 K Nsp SNP array revealed significant homozygosity at the PLEKHG5/MFN2 locus (Supplementary Table 2, Fig. 3a). While mutations in PLEKHG5 were excluded initially, we identified the c.392A > G, (p.N131S) mutation in MFN2. This mutation is not listed in databases (Ensemble, NCBI, CMT mutation database) and has not been reported in the 1,000 genomes project (http://www.1000genomes.org). The parents of this patient were first cousins and were normal on clinical and elecropyhsiological examination (Fig. 2b). The same homozygous mutation was confirmed in a distantly related individual exhibiting the same phenotype. Comparing homozygosity profiles of both patients revealed only one overlapping homozygous area at the MFN2 locus on chromosome 1 (Figs. 3a, 1c).
Fig. 3.
Homozygosity profiles. Homozygosity profiles of selected individuals. The intervals of homozygosity are shown as (blue blocks) and are plotted across the genome for each individual. The chromosomal positions of the homozygous intervals are indicated on the x-axis, the length of each block represents the size of the homozygous interval in basepairs (bp). The y-axis shows the number of homozygous SNPs (hSNPs). The physical positions of the known ARCMT genes are included and shown in green color. a High-density (250 K Nsp) array-derived homozygosity profiles of G1-2 and a distantly related cousin (G1-2-dc) (MFN2 marked in red) compared to a control individual with no parental consanguinity. b Low-density (10 K 2.0) array-derived homozygosity profiles for probands G1-5, carrying distinct GDAP1 mutations each (marked in red) and for a control sample
Mutations in the GDAP1 gene
Patient G1-5 with axonal early onset CMT showed an extended homozygous area of 112 hSNPs using the 10 K 2.0 array along the GDAP1 locus (Supplementary Table 2, Fig. 3b) and direct sequencing of GDAP1 revealed the known homozygous c.482G > A, (p.R161H) mutation. The two further GDAP1 mutations (c.174_176del3ins-TGTG, p.Leu58LeufsX4, and c.458 C > T, p.P153L) have been published previously [7].
Mutations in the FXN gene
Patient G1-6 who had been previously diagnosed with CMT2 showed no homozygosity at any CMT locus. However, because mild retardation and mild ataxia were known in this patient we also looked for homozygous regions at the AR ataxia loci and identified a convincing area of 71 hSNPs at the FXN locus using the 10 K 2.0 array. We thus tested for Friedreich ataxia and detected a biallelic trinucleotide repeat expansion.
Mutations in the NDRG1, PRX and HSN2 gene
Patient G3-22 and G3-23 with severe CMT 1 and partial hearing loss had no history of parental consanguinity. Both patients who were already known to carry the NDRG1 founder mutation (c.442 C > T, p.R148X) [14], showed convincing homozygous areas at the NDRG1 locus (Supplementary Table 2) when using the 250 K Nsp Array. The homozygous PRX mutation (c.2098 delG, p.A700PfsX17) and the HSN2 mutation (c.550 C > T, p.Q184X) have been published previously [7, 8]. Also, at these loci the probands were homozygous at the corresponding loci (Supplementary Table 2).
Discussion
The identification of at least 14 genes involved in the pathogenesis of ARCMT has challenged successful molecular genetic testing. In our series of 24 index patients with sporadic or ARCMT disease, 15 (63%) received an accurate genetic diagnosis by application of SNP array-based whole-genome homozygosity plotting used as the first step of the genetic analysis. Our newly-developed software programe considering the physical positions of the known ARCMT loci enables fast and simple inspection of homozygosity at ARCMT loci, and displays in detail the extent of homozygosity throughout the genome.
Because our patients originated from consanguineous families or their ancestors came from the same geographic area, homozygosity for a disease-causing mutation IBD was likely or possible. In the majority of all probands born to consanguineous parents or in those who were already known to carry rare homozygous mutations, comparison of the low-density (10 K 2.0) array and the high-density 250 K Nsp array data showed that the 10 K 2.0 SNP array was sufficient to pinpoint the ARCMT locus harboring the mutation. However, in patients of groups 2 and 3 with possible distant parental relationship application of the 10 K 2.0 SNP array enabled a genetic diagnosis in only 4 of 15 patients, whereas 3 further patients were diagnosed following 250 K Nsp array data analysis. Proband G1-1 showed homozygosity at the NEFL locus only but mutations in this gene could be excluded by direct sequencing suggesting that another dominant or a hitherto unknown ARCMT gene might be responsible for the disease. Also, smaller deletions or duplications in NEFL cannot be ruled out definitely as has been shown previously [15, 16]. Based on data obtained here we conclude, that homozygous mutations known ARCMT genes can be excluded with high probability in patients who do not show extended homozygosity at known loci. Moreover, using SNP array analysis may also be straightforward in the detection of large homozygous deletions. Patient G1-6, finally diagnosed with Friedreich ataxia, highlights the value of SNP-array based whole-genome homozygosity mapping in the differential diagnosis of ARCMT.
Our data obtained in patients from groups 2 and 3 are consistent with the findings of recent studies demonstrating that high-density SNP array-based homozygosity mapping can successfully be applied to non-consanguineous single individuals with autosomal recessive diseases [4, 6, 9]. Homozygous disease-causing mutations could be detected in single cases of outbred populations within homozygous areas often larger than 2 Mb in size [4]. Another study explored the potential of estimating individual autozygosity from data on runs of homozygosity (ROHs) in different European populations and showed that ROHs measuring up to 4 Mb are common in outbred populations [17].
In six of our CMT patients mutations in the SH3TC2 gene were identified including a novel alteration which may be traced back to a common founder. As described previously [18], CMT4C is one of the most common genetic ARCMT subtypes followed by CMT4A caused by mutations in GDAP1 [19] which were found in three of our patients.
Of special interest is the homozygous c.250G > C (p.G84R) mutation (Fig. 1b) in the small heat shock protein 27 gene (HSPB1) in patient G2-7. HSPB1 has originally been associated with autosomal dominant HMSN (CMT2F/CMT2L) and dHMN [20], but a recent study demonstrated that distinct HSPB1 mutations may also act in a recessive way to cause dHMN [13]. The same study also reported four novel heterozygous mutations in HSPB1 including the c.250G > C (p.G84R) mutation in a patient with a dHMN phenotype. The patient included in our study did not report any neuromuscular problems in his sibs, parents or grandparents except for considerable foot deformity in the paternal grandfather. However, subsequent examination of the patient’s parents heterozygous for this HSPB1 mutation detected a subclinical phenotype. As shown for other genes involved in the pathogenesis of CMT, like GDAP1, NEFL, MFN2, PMP22, EGR2, and HSPB1 mutations should also be considered in both ADCMT and ARCMT patients [21-28].
In summary, this study identified a novel, presumably founder mutation in SH3TC2, a novel homozygous mutation in MFN2, and the rare event of severe CMT caused by a homozygous HSPB1 mutation. We suggest that SNP array-based homozygosity plotting without linkage analyses is a simple, practical, and inexpensive screening method that should be initially applied to ARCMT patients with known parental consanguinity and can also be used for a small and selected number of patients with sporadic CMT syndrome.
Acknowledgments
We are grateful for the participation of the patients and families in this study. This work was supported by the Austrian Science Fond (FWF, P19455-B05), the Land Steiermark (GZ: A3-12G 114-06/1) and the Oesterreichische Nationalbank (ÖNB, project 13010).
Footnotes
C. Fischer and S. Trajanoski contributed equally to this work.
Electronic supplementary material The online version of this article (doi:10.1007/s00415-011-6213-8) contains supplementary material, which is available to authorized users.
Conflict of interest None.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Carina Fischer, Center for Medical Research, Medical University of Graz, Graz, Austria.
Slave Trajanoski, Center for Medical Research, Medical University of Graz, Graz, Austria.
Lea Papić, Department of Internal Medicine, Division of Endocrinology and Metabolism, Medical University of Graz, Stiftingtalstraße 24, 8010 Graz, Austria.
Christian Windpassinger, Institute of Human Genetics, Medical University of Graz, Graz, Austria.
Günther Bernert, Preyersches Kinderspital, Vienna, Austria.
Michael Freilinger, Department of Pediatrics and Adolescent Medicine, Medical University Vienna, Vienna, Austria.
Maria Schabhüttl, Department of Internal Medicine, Division of Endocrinology and Metabolism, Medical University of Graz, Stiftingtalstraße 24, 8010 Graz, Austria.
Mine Arslan-Kirchner, Institute of Human Genetics, Medizinische Hochschule Hannover, Hannover, Germany.
Poupak Javaher-Haghighi, Institute of Human Genetics, Medizinische Hochschule Hannover, Hannover, Germany; Amedes MVZ Hamburg, Hamburg, Germany.
Barbara Plecko, Department of Neuropediatrics, Medical University of Graz, Graz, Austria.
Jan Senderek, Institute of Human Genetics, RWTH Aachen University, Aachen, Germany; Institute of Neuropathology, RWTH Aachen University, Aachen, Germany.
Christian Rauscher, Department of Neuropediatrics, Christian Doppler Klinik, Salzburg, Austria.
Wolfgang N. Löscher, Department of Neurology, Innsbruck Medical University, Innsbruck, Austria
Thomas R. Pieber, Department of Internal Medicine, Division of Endocrinology and Metabolism, Medical University of Graz, Stiftingtalstraße 24, 8010 Graz, Austria
Andreas R. Janecke, Department of Pediatrics II, Innsbruck Medical University, Innsbruck, Austria
Michaela Auer-Grumbach, Department of Internal Medicine, Division of Endocrinology and Metabolism, Medical University of Graz, Stiftingtalstraße 24, 8010 Graz, Austria.
References
- 1.Reilly MM, Shy ME. Diagnosis and new treatments in genetic neuropathies. J Neurol Neurosurg Psychiatry. 2009;80(12):1304–1314. doi: 10.1136/jnnp.2008.158295. [DOI] [PubMed] [Google Scholar]
- 2.Dubourg O, Azzedine H, Verny C, Durosier G, Birouk N, Gouider R, Salih M, Bouhouche A, Thiam A, Grid D, Mayer M, Ruberg M, Tazir M, Brice A, LeGuern E. Autosomal-recessive forms of demyelinating Charcot-Marie-Tooth disease. Neuromolecular Med. 2006;8(1-2):75–86. doi: 10.1385/nmm:8:1-2:75. [DOI] [PubMed] [Google Scholar]
- 3.Bernard R, De Sandre-Giovannoli A, Delague V, Lévy N. Molecular genetics of autosomal-recessive axonal Charcot-Marie-Tooth neuropathies. Neuromolecular Med. 2006;8(1-2):87–106. doi: 10.1385/nmm:8:1-2:87. [DOI] [PubMed] [Google Scholar]
- 4.Hildebrandt F, Heeringa SF, Rüschendorf F, Attanasio M, Nürnberg G, Becker C, Seelow D, Huebner N, Chernin G, Vlangos CN, Zhou W, O’Toole JF, Hoskins BE, Wolf MT, Hinkes BG, Chaib H, Ashraf S, Schoeb DS, Ovunc B, Allen SJ, Vega-Warner V, Wise E, Harville HM, Lyons RH, Washburn J, Macdonald J, Nürnberg P, Otto EA. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 2009;5:e1000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr IM, Szymanska K, Sheridan E, Markham AF, Bonthron DT, Johnson CA. Shadow autozygosity mapping by linkage exclusion (SAMPLE): a simple strategy to identify the genetic basis of lethal autosomal recessive disorders. Hum Mutat. 2009;30:1642–1649. doi: 10.1002/humu.21105. [DOI] [PubMed] [Google Scholar]
- 6.Collin RW, van den Born LI, Klevering BJ, de Castro-Miró M, Littink KW, Arimadyo K, Azam M, Yazar V, Zonneveld MN, Paun CC, Siemiatkowska AM, Strom TM, Hehir-Kwa JY, Kroes HY, de Faber JT, van Schooneveld M, Heckenlively JR, Hoyng CB, den Hollander AI, Cremers FP. High-resolution homozygosity mapping is a powerful tool to detect novel mutations causative for autosomal recessive RP in the Dutch population. Invest Ophthalmol Vis Sci. 2011;52(2):2227–2239. doi: 10.1167/iovs.10-6185. doi:10.1167/iovs.10-6185. [DOI] [PubMed] [Google Scholar]
- 7.Auer-Grumbach M, Fischer C, Papić L, John E, Plecko B, Bittner RE, Bernert G, Pieber TR, Miltenberger G, Schwarz R, Windpassinger C, Grill F, Timmerman V, Speicher MR, Janecke AR. Two novel mutations in the GDAP1 and PRX genes in early onset Charcot-Marie-Tooth syndrome. Neuropediatrics. 2008;39(1):33–38. doi: 10.1055/s-2008-1077085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coen K, Pareyson D, Auer-Grumbach M, Buyse G, Goemans N, Claeys KG, Verpoorten N, Laurà M, Scaioli V, Salmhofer W, Pieber TR, Nelis E, De Jonghe P, Timmerman V. Novel mutations in the HSN2 gene causing hereditary sensory and autonomic neuropathy type II. Neurology. 2006;66(5):748–751. doi: 10.1212/01.wnl.0000201191.57519.47. [DOI] [PubMed] [Google Scholar]
- 9.Papić L, Fischer D, Trajanoski S, Höftberger R, Fischer C, Ströbel T, Schmidt WM, Bittner RE, Schabhüttl M, Gruber K, Pieber TR, Janecke AR, Auer-Grumbach M. SNP-array based whole genome homozygosity mapping: a quick and powerful tool to achieve an accurate diagnosis in LGMD2 patients. Eur J Med Genet. 2011;54(3):214–219. doi: 10.1016/j.ejmg.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. Homozygosity mapper—an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkp369. doi:10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: 2009. http://www.R-project.org. [Google Scholar]
- 12.Campuzano V, Montermini L, Moltò MD, Pianese L, Cossée M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Cañizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271(5254):1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 13.Houlden H, Laura M, Wavrant-De Vrièze F, Blake J, Wood N, Reilly MM. Mutations in the HSP27 (HSPB1) gene cause dominant, recessive, and sporadic distal HMN/CMT type 2. Neurology. 2008;71(21):1660–1668. doi: 10.1212/01.wnl.0000319696.14225.67. [DOI] [PubMed] [Google Scholar]
- 14.Kalaydjieva L, Gresham D, Gooding R, Heather L, Baas F, de Jonge R, Blechschmidt K, Angelicheva D, Chandler D, Worsley P, Rosenthal A, King RH, Thomas PK. N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy-Lom. Am J Hum Genet. 2000;67(1):47–58. doi: 10.1086/302978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boone PM, Bacino CA, Shaw CA, Eng PA, Hixson PM, Pursley AN, Kang SH, Yang Y, Wiszniewska J, Nowakowska BA, Del Gaudio D, Xia Z, Simpson-Patel G, Immken LL, Gibson JB, Tsai AC, Bowers JA, Reimschisel TE, Schaaf CP, Potocki L, Scaglia F, Gambin T, Sykulski M, Bartnik M, Derwinska K, Wisniowiecka-Kowalnik B, Lalani SR, Probst FJ, Bi W, Beaudet AL, Patel A, Lupski JR, Cheung SW, Stankiewicz P. Detection of clinically relevant exonic copy-number changes by array CGH. Hum Mutat. 2010;31(12):1326–1342. doi: 10.1002/humu.21360. doi:10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osaka H, Hamanoue H, Yamamoto R, Nezu A, Sasaki M, Saitsu H, Kurosawa K, Shimbo H, Matsumoto N, Inoue K. Disrupted SOX10 regulation of GJC2 transcription causes Pelizaeus-Merzbacher-like disease. Ann Neurol. 2010;68(2):250–254. doi: 10.1002/ana.22022. [DOI] [PubMed] [Google Scholar]
- 17.McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, Smolej-Narancic N, Janicijevic B, Polasek O, Tenesa A, Macleod AK, Farrington SM, Rudan P, Hayward C, Vitart V, Rudan I, Wild SH, Dunlop MG, Wright AF, Campbell H, Wilson JF. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senderek J, Bergmann C, Stendel C, Kirfel J, Verpoorten N, De Jonghe P, Timmerman V, Chrast R, Verheijen MH, Lemke G, Battaloglu E, Parman Y, Erdem S, Tan E, Topaloglu H, Hahn A, Müller-Felber W, Rizzuto N, Fabrizi GM, Stuhrmann M, Rudnik-Schöneborn S, Züchner S, Michael Schröder J, Buchheim E, Straub V, Klepper J, Huehne K, Rautenstrauss B, Büttner R, Nelis E, Zerres K. Mutations in a gene encoding a novel SH3/TPR domain protein cause autosomal recessive Charcot-Marie-Tooth type 4C neuropathy. Am J Hum Genet. 2003;73(5):1106–1119. doi: 10.1086/379525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuesta A, Pedrola L, Sevilla T, García-Planells J, Chumillas MJ, Mayordomo F, LeGuern E, Marín I, Vílchez JJ, Palau F. The gene encoding ganglioside-induced differentiation-associated protein 1 is mutated in axonal Charcot-Marie-Tooth type 4A disease. Nat Genet. 2002;30(1):22–25. doi: 10.1038/ng798. [DOI] [PubMed] [Google Scholar]
- 20.Evgrafov OV, Mersiyanova I, Irobi J, Van Den Bosch L, Dierick I, Leung CL, Schagina O, Verpoorten N, Van Impe K, Fedotov V, Dadali E, Auer-Grumbach M, Windpassinger C, Wagner K, Mitrovic Z, Hilton-Jones D, Talbot K, Martin JJ, Vasserman N, Tverskaya S, Polyakov A, Liem RK, Gettemans J, Robberecht W, De Jonghe P, Timmerman V. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet. 2004;36(6):602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- 21.Yum SW, Zhang J, Mo K, Li J, Scherer SS. A novel recessive Nefl mutation causes a severe, early-onset axonal neuropathy. Ann Neurol. 2009;66:759–770. doi: 10.1002/ana.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholson GA, Magdelaine C, Zhu D, Grew S, Ryan MM, Sturtz F, Vallat JM, Ouvrier RA. Severe early-onset axonal neuropathy with homozygous and compound heterozygous MFN2 mutations. Neurology. 2008;70(19):1678–1681. doi: 10.1212/01.wnl.0000311275.89032.22. [DOI] [PubMed] [Google Scholar]
- 23.Chung KW, Kim SM, Sunwoo IN, Cho SY, Hwang SJ, Kim J, Kang SH, Park KD, Choi KG, Choi IS, Choi BO. A novel GDAP1 Q218E mutation in autosomal dominant Charcot-Marie-Tooth disease. J Hum Genet. 2008;53(4):360–364. doi: 10.1007/s10038-008-0249-3. [DOI] [PubMed] [Google Scholar]
- 24.Sivera R, Espinós C, Vílchez JJ, Mas F, Martínez-Rubio D, Chumillas MJ, Mayordomo F, Muelas N, Bataller L, Palau F, Sevilla T. Phenotypical features of the p.R120 W mutation in the GDAP1 gene causing autosomal dominant Charcot-Marie-Tooth disease. J Peripher Nerv Syst. 2010;15(4):334–344. doi: 10.1111/j.1529-8027.2010.00286.x. doi:10.1111/j.1529-8027.2010.00286. [DOI] [PubMed] [Google Scholar]
- 25.Baxter RV, Ben Othmane K, Rochelle JM, Stajich JE, Hulette C, Dew-Knight S, Hentati F, Ben Hamida M, Bel S, Stenger JE, Gilbert JR, Pericak-Vance MA, Vance JM. Ganglioside-induced differentiation-associated protein-1 is mutant in Charcot-Marie-Tooth disease type 4A/8q21. Nat Genet. 2002;30(1):21–22. doi: 10.1038/ng796. [DOI] [PubMed] [Google Scholar]
- 26.Shy ME, Scavina MT, Clark A, Krajewski KM, Li J, Kamholz J, Kolodny E, Szigeti K, Fischer RA, Saifi GM, Scherer SS, Lupski JR. T118M PMP22 mutation causes partial loss of function and HNPP-like neuropathy. Ann Neurol. 2006;59(2):358–364. doi: 10.1002/ana.20777. [DOI] [PubMed] [Google Scholar]
- 27.Roa BB, Garcia CA, Pentao L, Killian JM, Trask BJ, Suter U, Snipes GJ, Ortiz-Lopez R, Shooter EM, Patel PI, Lupski JR. Evidence for a recessive PMP22 point mutation in Charcot-Marie-Tooth disease type 1A. Nat Genet. 1993;5(2):189–194. doi: 10.1038/ng1093-189. [DOI] [PubMed] [Google Scholar]
- 28.Warner LE, Mancias P, Butler IJ, McDonald CM, Keppen L, Koob KG, Lupski JR. Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat Genet. 1998;18(4):382–384. doi: 10.1038/ng0498-382. [DOI] [PubMed] [Google Scholar]