Abstract
Forkhead transcription factors of the O class (FOXOs) are important targets of the PI3-kinase/Akt pathway, and are key regulators of the cell cycle, apoptosis and response to oxidative stress. FOXOs have been shown to have tumour suppressor function and are important for stem cell maintenance.
We have performed a detailed analysis of the transcriptional programme induced in response to FOXO3a activation. We observed that FOXO3a activation results in the repression of a large number of nuclear-encoded genes with mitochondrial function. Repression of these genes was mediated by FOXO3a-dependent inhibition of c-Myc. FOXO3a activation also caused a reduction in mitochondrial DNA copy number, expression of mitochondrial proteins, respiratory complexes and mitochondrial respiratory activity.
FOXO3a has been previously implicated in the detoxification of reactive oxygen species (ROS) through induction of manganese-containing superoxide dismutase (SOD2). We observed that reduction in ROS levels following FOXO3a activation was independent of SOD2, but required c-Myc inhibition. Hypoxia increases ROS production from the mitochondria, which is required for stabilisation of the hypoxia inducible factor 1α (HIF-1α). FOXO3a activation blocked the hypoxia-dependent increase in ROS and prevented HIF-1α stabilisation. Our data suggests that FOXO factors regulate mitochondrial activity through inhibition of c-Myc function and alter the hypoxia response.
Keywords: FOXO, c-Myc, Mitochondrial biogenesis, HIF-1α, Reactive oxygen species
INTRODUCTION
FOXO proteins form a subgroup of the larger family of forkhead-box-containing transcription factors, which are identified by the winged-helix structure of their DNA binding domain [1]. The mammalian genome encodes four different FOXO proteins (FOXO1, FOXO3a, FOXO4 and FOXO6). Phosphorylation of FOXO proteins by Akt results in their exclusion from the nucleus and enhanced degradation [2, 3]. Importantly, FOXO factors have emerged as tumour suppressors in several systems [4]. FOXO factors regulate the expression of genes involved in the inhibition of cell cycle progression and induction of apoptosis [5]. FOXO also regulates detoxification of reactive oxygen species (ROS) through upregulation of Manganese-containing superoxide dismutase (SOD2) [6].
Mitochondria are central hubs for cellular bioenergetics and are an important source of ROS within mammalian cells. ROS are produced by the respiratory complexes located in the inner mitochondrial membrane. Mitochondrial ROS can cause oxidative damage to the mitochondrial DNA (mtDNA), proteins and lipids, but are also involved in signalling from the mitochondria to the cytoplasm. The regulation of mitochondrial function is crucial for the survival of both normal and cancer cells [7].
The genome of mitochondria in mammalian cells encodes only 13 proteins. The majority of proteins required for the maintenance of the structure and function of mitochondria are encoded by the nuclear genome. The expression of these genes is regulated by a network of transcription factors, including the nuclear respiratory factors 1 and 2 (NRF1 and NRF2) and the estrogen related receptor α (ERRα). These are bound to and activated by cofactors, peroxisome proliferator-activated receptor gamma coactivator 1α and 1β (PGC1α̣□□□̣PGC1β) and PGC-related 1 (PRC) [8]. In addition, the transcription factor c-Myc emerged as an important regulator of mitochondrial gene expression [9]. Interestingly, the hypoxia inducible factor 1α (HIF-1α) downregulates mitochondrial biogenesis via inhibiting c-Myc as part of the cellular adaptation to hypoxia [10].
We have shown previously that FOXO3a induces the expression of c-Myc antagonists of the Mad/Mxd family. In particular, FOXO3a drives the expression of Mxi1 by binding to several conserved Daf-16 binding elements (DBEs) within the first intron of the gene [11]. Induction of Mad/Mxd proteins is required for efficient inhibition of c-Myc-dependent gene expression and cell cycle arrest following FOXO3a activation by inhibiting c-Myc [11].
Here we present a comprehensive analysis of the transcriptional response to FOXO3a which revealed the repression of a large number of nuclear encoded genes through the inhibition of c-Myc function. We show that through this signalling arm, FOXO3a activation alters mitochondrial activity and reduces cellular ROS production, independently of SOD2 activation. Regulation of mitochondrial structure and function could be an important role of FOXO factors in regulating ROS production and affect cellular adaptation to hypoxia.
RESULTS
FOXO3a activation causes downregulation of mitochondrial gene expression
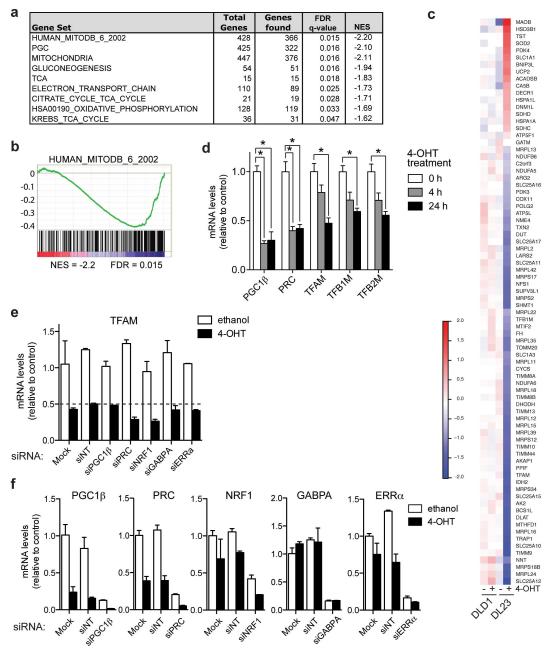
In order to characterise the complete transcriptional response to FOXO3a activation, we used DLD-1 colon cancer cells expressing a 4-hydroxytamoxifen (4-OHT) inducible version of FOXO3a in which the three Akt phosphorylation sites have been replaced with alanine (FOXO3a.A3-ER), thereby rendering its activity completely dependent on the presence of agonist. This cell line (termed DL23) has previously been used to determine the role of FOXO3a in cell cycle regulation [12]. We identified over 2,700 genes with significantly altered expression levels following FOXO3a activation in these cells, including many known FOXO target genes (supplementary Table 1). To characterise these changes in gene expression, we compared our results to publicly available gene sets (Molecular Signature Database, Broad Institute) using Gene Set Enrichment Analysis (GSEA). Among the most significantly enriched genes sets were signatures previously associated with mitochondrial functions, including a curated collection of genes with mitochondrial function (HUMAN_MITODB_6_2002), PGC-target genes, tricarboxylic acid (TCA) cycle enzymes and genes involved in electron transport chain and oxidative phosphorylation (Figure 1a and b). These gene sets show a strong association with genes downregulated following FOXO activation, as indicated by the negative enrichment score (NES). As predicted by the GSEA, most genes within the HUMAN_MITOCB_6_2002 gene set are downregulated following FOXO3a activation, but are unaffected by 4-OHT treatment in the parental line (DLD-1) (Figure 1c). Among the upregulated genes within this gene set are SOD2 and pyruvate decarboxylase kinase 4 (PDK4), two previously identified FOXO target genes [6, 13].
Figure 1. FOXO3a downregulates mitochondrial genes independently of regulatory transcription factors.
DLD-1 colon cancer cells expressing inducible FOXO3a.A3.ER (DL23 cells) were treated with 100 nM 4-hydroxy-tamoxifen (4-OHT) to induce activation of FOXO3a. The expression profile from these cells following FOXO3a activation was analysed using Affymetrix exon microarrays. (a) Geneset enrichment analysis (GSEA) was used to determine gene sets significantly regulated by FOXO3a. Selected mitochondrial genesets are shown. NES = Normalised Enrichment Score; q-values = FDR-adjusted p-value. (b) Enrichment plot of the top mitochondrial gene set (MitoDB_2002) found to be significantly associated with downregulation by FOXO3a. (c) Heatmap showing the expression profile of genes significantly regulated by FOXO3a in DL23 cells that are also present in the HUMAN_MITODB_6_2002 gene set. Values show fold change over the median across all samples. (d) DL23 cells were treated with 4-OHT or solvent for 4 or 24 h. Expression of PGC1β, PRC, TFAM. TFB1M and TFB2M was determined by qRT-PCR. (e) DL23 cells were transfected with the indicated siRNAs or mock treated for 72 h and stimulated with 4-OHT for the final 24 h (NT = non-targeting control). Expression of the indicated genes after 24 h of solvent (white bars) or 4-OHT treatment (black bars) was determined by qRT-PCR. Data shown are representative of 3 independent experiments. (f) Silencing of PGC1β, PRC, NRF1, GABPA and ERRα was confirmed by qRT-PCR. Data shown are representative of 3 independent experiments. All data are shown as mean ± SEM. * indicates statistical significance as determined by Student’s t-test (P < 0.05, n=4).
We next investigated whether FOXO3a altered the expression of coactivators known to be involved in the regulation of nuclear-encoded mitochondrial genes (PGC1β and PRC) or the mitochondrial transcription factors A, B1 and B2 (TFAM, TFB1M and TFB2M), which regulate the expression of genes encoded by the mitochondrial genome and are required for maintenance of mtDNA [8]. We found that FOXO3a activation resulted in rapid and pronounced downregulation of PGC1β and PRC expression (Figure 1d). Furthermore, the expression of TFAM, TFB1M and TFB2M was significantly reduced upon FOXO3a activation, albeit with slower kinetics (Figure 1d). Expression of PGC1α was not detected in DL23 cells (data not shown). Activation of endogenous FOXOs by treatment of DLD-1 cells with the phosphatidylinositol 3-kinase (PI3-kinase) inhibitors LY-294002 or PI-103 also resulted in downregulation of PGC1β, PRC and TFAM, while direct FOXO target genes SOD2 and Mxi1 were upregulated (Supplemental Figure S1).
We then investigated whether PGC1β or PRC are involved in the regulation of mitochondrial gene expression by FOXO3a. Silencing of PGC1β or PRC did not alter TFAM expression in the presence or absence of FOXO3a activation (Figure 1e). Furthermore, silencing of NRF1, ERRα or GABPA (DNA-binding subunit of NRF2) did not affect inhibition of TFAM expression following FOXO3a activation (Figure 1e). Efficient ablation of genes by siRNA was confirmed by qRT-PCR (Figure 1f). We also investigated the effect of FOXO3a activation on the activity of a reporter construct carrying proximal sequences from the promoter of the human TFAM gene, containing binding sites for NRF1, NRF2 and SP1 [14]. Supplementary Figure S2b shows that this reporter was unaffected by FOXO3a activation. Together, these results indicate that the repression of mitochondrial gene expression by FOXO3a is independent of the PGC/NRF-regulatory network.
The Mad/Mxd family of transcriptional repressors contribute to the inhibition of mitochondrial genes by FOXO3a
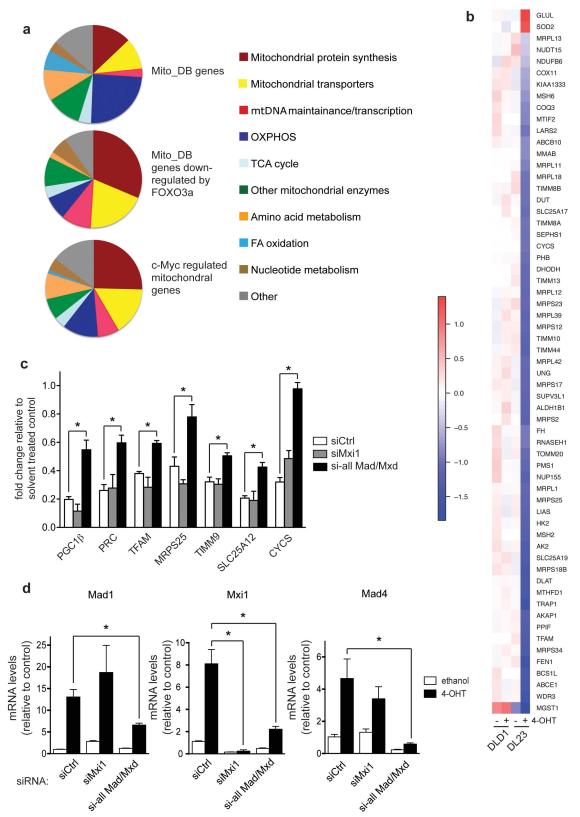
c-Myc induces the expression of a number of mitochondrial genes, including TFAM, by directly binding to their promoters [9]. In order to investigate whether the inhibition of mitochondrial gene expression following FOXO3a activation is mediated by c-Myc inhibition, we compared the functional classification of FOXO3a regulated mitochondrial genes with published mitochondrial target genes of c-Myc [9]. Three functional categories are over-represented in the Myc-regulated gene expression signature and the genes downregulated following FOXO3a activation: mitochondrial protein synthesis, mitochondrial transporters and mitochondrial DNA maintenance and transcription (Figure 2a). Indeed, the majority of c-Myc-regulated mitochondrial genes identified in the study by Li et al. were repressed following FOXO3a activation in our dataset (Figure 2b).
Figure 2. Induction of the Mad/Mxd family contributes to the downregulation of mitochondrial regulators by FOXO3a.
(a) Charts showing the functional classification of genes present in the MitoDB gene set (upper), MitoDB genes that are downregulated by FOXO3a (middle) and previously identified c-Myc regulated mitochondrial genes [9] that are downregulated by FOXO3a activation (lower). (b) Heatmap showing the expression profile of genes found to be significantly regulated by FOXO3a in DL23 cells which have been previously identified to be c-Myc regulated mitochondrial genes [9]. (c) Silencing of Mad/Mxd proteins partially rescues repression of mitochondrial genes. DL23 cells were transfected with 100 nM of control (siCtrl, white bars), siRNAs targeting Mxi1 (grey bars) or pools of siRNAs targeting all Mad/Mxd genes (Mad1, Mxi1, Mad3 and Mad4) (black bars) for 72 h and treated with 4-OHT for the final 24 h. Expression levels of the indicated mitochondrial genes were determined by qRT-PCR. Values represent mRNA expression levels of 4-OHT treated cells relative to solvent treated controls. (d) Silencing of Mad1, Mxi1 and Mad4 was confirmed by qRT-PCR.
All data are shown as mean ± SEM. * indicates statistical significance as determined by Student’s t-test (P < 0.05, n=4).
We have shown that FOXO3a inhibits c-Myc-dependent gene expression by inducing the expression of members of the Mad/Mxd family of c-Myc antagonists [11]. We therefore asked whether induction of these antagonists is also involved in the inhibition of mitochondrial genes by FOXO3a. Silencing of Mxi1 alone had no effect on the expression of a panel of mitochondrial genes. However, combined silencing of all four Mad/Mxd genes (Mad1, Mxi1, Mad3 and Mad4) resulted in a partial yet significant inhibition of FOXO3a dependent downregulation (Figure 2c and d). This indicates that induction of these factors does indeed contribute to inhibition of mitochondrial gene expression by FOXO3a.
FOXO3a inhibits mitochondrial gene expression by reducing c-Myc stability
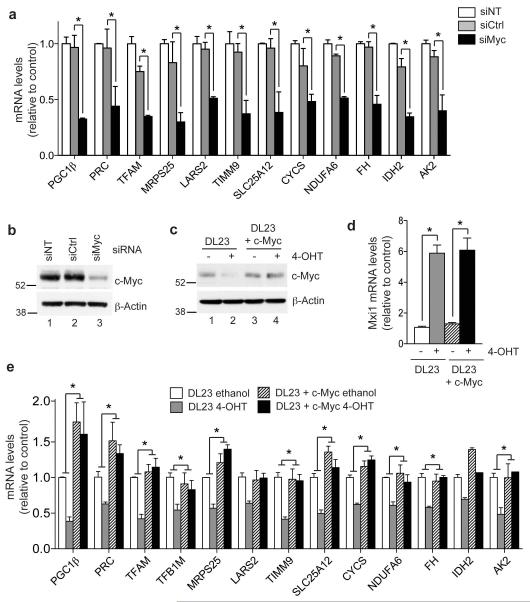
FOXO3a activation results in a considerable reduction of c-Myc protein levels ([11] and Figure 3c, lanes 1 and 2). We therefore investigated whether ablation of c-Myc induces changes in the expression of mitochondrial genes, similar to those observed following FOXO3a activation. The majority of genes in a panel representative of the functional categories identified in Figure 2a were downregulated following silencing of c-Myc in parental DLD-1 cells (Figure 3a). We then asked whether restoration of c-Myc protein levels would prevent the FOXO3a-dependent downregulation of mitochondrial genes. Expression of c-Myc in DL23 cells using a retroviral vector restored c-Myc protein to similar levels observed prior to FOXO3a activation (Figure 3c) and did not alter the induction of direct transcriptional targets, such as Mxi1, by FOXO3a (Figure 3d). However, c-Myc expression fully restored the expression of most mitochondrial genes within this panel in the presence of active FOXO3a, except IDH2, indicating that inhibition of c-Myc is vital for the inhibition of these genes (Figure 3e).
Figure 3. Downregulation of mitochondrial genes by FOXO3a is mediated by inhibition of c-Myc.
(a) Parental DLD-1 cells were transfected with 100 nM of either non-targeting (siNT), control (siCtrl) or c-Myc (siMyc) siRNA for 48 h. Expression levels of the indicated mitochondrial genes was determined by qRT-PCR. (b) Silencing of c-Myc was confirmed by immunoblotting lysates from cells transfected in parallel. β-actin is shown as a loading control. (c) DL23 cells were infected with pWZL-Blast-c-Myc and selected for 96 h. Cells were then treated with 4-OHT or solvent for 24 h. Expression of c-Myc was determined by immunoblotting. β-actin is shown as a loading control. (d) Expression levels of Mxi1 following FOXO3a activation in DL23 parental cells or DL23 cells expressing c-Myc (DL23+c-Myc) was determined by qRT-PCR. (e) Expression of the indicated mitochondrial genes in DL23 (white and grey bars) or DL23+c-Myc (hatched and black bars) was determined by qRT-PCR. Statistical analysis of the effect of 4-OHT treatment in DL23 or DL23+c-Myc cells was performed by comparing the fold change relative to ethanol treated control in each cell line.
All data are shown as mean ± SEM. * indicates statistical significance as determined by Student’s t-test (P < 0.05, n=3).
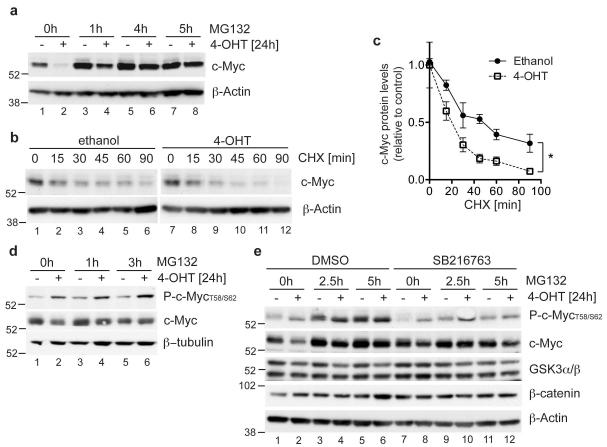
Since the loss of c-Myc protein following FOXO3a activation was not associated with a significant reduction in c-Myc mRNA levels (Supplementary Figure S3), we examined the effect of FOXO3a activation on c-Myc protein stability. c-Myc is regulated by phosphorylation-dependent ubiquitination and proteasomal degradation [15]. Addition of the proteasome inhibitor MG132 prevented the loss of c-Myc protein following FOXO3a activation (Figure 4a). Furthermore, FOXO3a activation decreased the stability of c-Myc protein in the presence of cycloheximide (Figure 4b and c). The recognition of c-Myc by the Fbw7 ubiquitin ligase requires phosphorylation of threonine 58 by GSK3 [16, 17], as well as a priming phosphorylation on serine 62 [18]. We used antibodies raised against a c-Myc-derived peptide carrying phosphates on threonine 58 and serine 62. Previous reports have shown that this serum detects c-Myc protein phosphorylated at threonine 58 under the conditions used by us [19]. FOXO3a activation increased the recognition of c-Myc by this serum indicating enhanced phosphorylation of the protein at threonine 58 (Figure 4d, lanes 1 and 2). Inhibition of c-Myc degradation by MG132 increased the levels of phosphorylated c-Myc in the presence of FOXO3a (Figure 4d, lanes 3-6). Furthermore, inhibition of GSK3 by SB216763 resulted in a reduction in c-Myc phosphorylation (Figure 4e, lanes 9-12) and prevented the reduction in c-Myc protein following FOXO3a activation (Figure 4e, compare lanes 1 and 2 with 7 and 8). Levels of β-catenin, another target of GSK3-dependent phosphorylation and degradation [20], were not affected by FOXO3a activation suggesting that FOXO3a does not alter GSK3 activity itself, but may instead regulate the priming phosphorylation.
Figure 4. FOXO3a induces GSK-3 dependent phosphorylation and proteasomal degradation of c-Myc.
(a) DL23 cells were induced with 4-OHT or solvent for 24 h. 25 μM MG132 was added for the final 1, 4 or 5 h, as indicated. c-Myc protein levels were determined by immunoblotting. β-actin is shown as loading control. (b) DL23 cells were treated with 4-OHT or solvent for 15 minutes before addition of 2 μg/ml cycloheximide (CHX). Cells were lysed after 15, 30, 45, 60 or 90 minutes of cycloheximide treatment. c-Myc protein levels were determined by immunoblotting. β-actin is shown as a loading control. (c) Quantitation of c-Myc protein levels in the presence of cycloheximide in DL23 cells treated with 4-OHT or solvent from three independent experiments. Data are shown as mean ± SEM. * indicates statistical significance as determined by Student’s t-test (P < 0.05, n=3). (d) DL23 cells were treated with 4-OHT or solvent for 24 h. 25 μM MG132 was added for the final 1, 3 or 5 h, as indicated. Levels of phosphorylated and total c-Myc were determined by immunoblotting. β-tubulin is shown as a loading control. (e) DL23 cells were treated with 4-OHT or solvent for 24 h in the presence of DMSO or 5 μM of the GSK3 inhibitor SB216763. 25 μM MG132 was added for the final 1, 2.5 or 5 h, as indicated. Levels of GSK3 α/β, β-catenin, and phosphorylated and total c-Myc were determined by immunoblotting. β-actin is shown as a loading control.
FOXO3a activation alters mitochondrial function
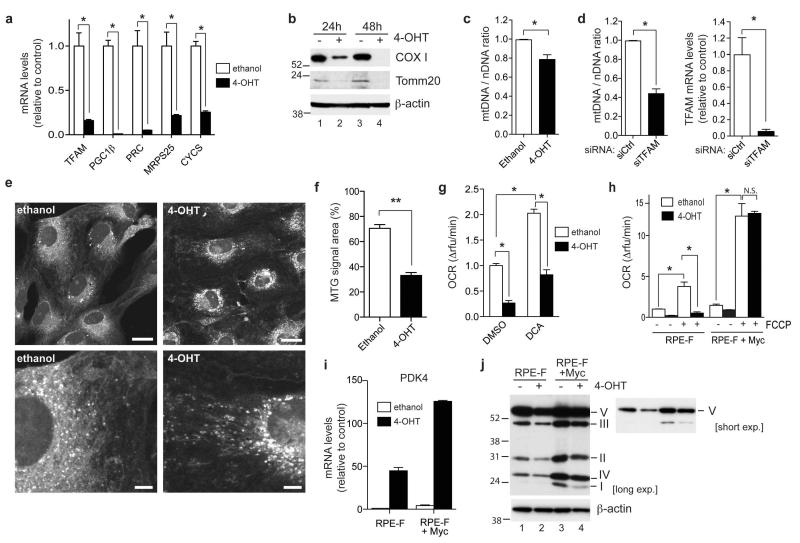
Many cancer cell lines show defects in their respiratory activity due to oncogenic signalling or mutational alterations within the mitochondrial genome [21]. We therefore used a non-malignant, immortalised epithelial cell line (RPE-hTERT) to further investigate the effect of FOXO3a on mitochondrial function. FOXO3a.A3-ER was introduced into these cells by retroviral transduction to generate a stable cell line (termed RPE-F). Activation of FOXO3a following 4-OHT treatment was confirmed by analysing the induction of p27KIP1 and Mxi1-SRα (data not shown). Similar to the results in DL23 cells, FOXO3a activation resulted in substantial downregulation of mitochondrial genes in these cells (Figure 5a). We next investigated whether FOXO3a activation induced changes in the levels of mitochondrial proteins, mitochondrial DNA copy number and mitochondrial mass. FOXO3a activation lowered the expression of Tomm20, a mitochondrial transporter, and cytochrome c oxidase 1 (COX1), a gene encoded by the mitochondrial genome (Figure 5b). FOXO3a activation also resulted in a moderate but significant reduction of the ratio of mitochondrial to nuclear DNA (mtDNA/nDNA) indicative of a reduction in mtDNA copy number (Figure 5c). Silencing of TFAM, which is known to regulate mtDNA replication, caused a 50% reduction in the mtDNA copy number (Figure 5d). The reduction of mtDNA copy number observed following FOXO3a activation is consistent with the levels of TFAM downregulation (Figure 5a). We also used Mitotracker green (MTG) staining to examine effects on mitochondrial mass. Although a substantial increase in total MTG fluorescence could be detected following resveratrol treatment, a known activator of mitochondrial biogenesis [22], no significant change was observed in response to FOXO3a activation in these cells (Supplementary Figure S4). Confocal microscopy of MTG stained cells revealed that FOXO3a activation induced a change in mitochondrial appearance from a filamentous network to more punctate structures (Figure 5e). Image analysis showed that the total area of MTG fluorescence was reduced (Figure 5f), suggesting that FOXO3a activation causes a contraction of the mitochondrial network.
Figure 5. Regulation of mitochondrial function by FOXO3a.
(a) RPE.FOXO3a.A3.ER (RPE-F) cells were treated with solvent (white bars) or 100 nM 4-OHT (black bars) for 24 h. Expression of the indicated mitochondrial genes was determined by qRT-PCR. (b) RPE-F cells were treated with 4-OHT of solvent for 24 or 48 h. Expression levels of Cox I and Tomm20 were determined by immunoblotting. β-actin is shown as a loading control. (c) Ratio of mitochondrial DNA (mtDNA) to nuclear DNA (nDNA) was determined by qPCR using DNA from RPE-F cells treated with solvent (white bars) or 4-OHT (black bars) for 48h. (d) RPE-F cells were transfected with 100 nM of either control (white bars) or a pool of siRNAs targeting TFAM (black bars) for 72 h. Ratio of mtDNA to nDNA was determined by qPCR. Silencing of TFAM was confirmed by qRT-PCR. (e) RPE-F cells were treated with ethanol or 4OHT for 48 h, fixed in formaldehyde, stained using 100 nM Mitotracker green and visualised using confocal microscopy. Scale bar, upper panels 20 μm, lower panels 5 μm. (f) Quantification of area occupied by Mitotracker green (MTG) stain. The area occupied by MTG in each cell was measured from confocal images using Metamorph software in 20 cells from 2 independent experiments. (g) Oxygen consumption was measured in RPE-F cells treated with solvent (white bars) or 4-OHT (black bars) for 48 h. 50 mM dichloroacetate (DCA) or solvent (DMSO) was added for the final 24 h before measurement. (h) Oxygen consumption rates were measured in RPE-F or RPE-F+c-Myc cells following 48 h of solvent (white bars) or 4-OHT (black bars). 2 μM FCCP was added to determine mitochondrial capacity. (i) RPE-F or RPE-F+c-Myc cells were treated with 24 h of solvent (white bars) or 4-OHT (black bars). Expression of PDK4 was determined by qRT-PCR. (j) RPE-F or RPE-F+c-Myc cells were treated with solvent or 4-OHT for 48 h. Total cell lysates were used to detect OXPHOS complexes by immunoblotting. β-actin is shown as a loading control.
All data are shown as mean ± SEM. * indicates statistical significance as determined by Student’s t-test (P < 0.05, n=3). ** indicates statistical significance as determined by Student’s t-test (p=1×10−8, n=20). N.S. = non-significant.
We next assessed mitochondrial respiratory activity by determining oxygen consumption rates (OCR). FOXO3a activation resulted in a significant decrease in OCR (Figure 5f). Although FOXO3a activation caused a substantial increase in the expression of PDK4 (Figure 5i, left part), the FOXO3a-dependent decrease in OCR was still observed when cells were incubated with dichloroacetate (DCA), an inhibitor of PDK activity (Figure 5g). This suggests that the inhibition of mitochondrial activity by FOXO3a is at least in part independent of PDK4. Furthermore, FOXO3a activation caused a strong reduction in the OCR in the presence of the uncoupling agent FCCP indicative of lower mitochondrial maximal capacity, providing additional evidence for a PDK4-independent effect on mitochondrial activity (Figure 5h, left part). FOXO3a activation also resulted in a decrease in the amounts of mitochondrial respiratory complexes (Figure 5j, lanes 1 and 2).
We next investigated the role of c-Myc in the regulation of mitochondrial activity by FOXO3a. Expression of c-Myc in RPE-F cells caused a moderate increase in basal oxygen consumption but this was still reduced by FOXO3a activation (Figure 5h, left part). This reduction could be due to the enhanced expression of PDK4 observed in these cells following 4-OHT treatment (Figure 5i). c-Myc expression resulted in a substantial increase in mitochondrial maximal capacity (FCCP induced OCR) and blocked the ability of FOXO3a to reduce capacity (Figure 5h). c-Myc also increased the amount of mitochondrial respiratory complexes both in the presence and absence of FOXO3a activity (Figure 5i).
It has been shown that the mitochondrial fission machinery is involved in mitochondrial remodelling during FOXO3a-dependent muscle atrophy [23]. However, we did not observe induction in the fission regulators FIS1 or DRP1 upon FOXO3a activation (Supplemental Figure S5a). Furthermore, inhibition of mitochondrial fission using the DRP1 inhibitor Mdivi-1 did not prevent the downregulation of mitochondrial activity or capacity by FOXO3a (Supplemental Figure S5b). Although we cannot exclude that mitochondrial fission contributes to the morphological changes induced by FOXO3a activation, it seems unlikely that it is responsible for the alterations in mitochondrial activity observed here.
FOXO3a activation reduces levels of reactive oxygen species independently of SOD2
The mitochondrial respiratory chain is a major source of cellular ROS. Superoxide is produced by complexes I and III and is released into the mitochondrial matrix where it can be converted into hydrogen peroxide and subsequently into water by SOD2 and catalase (CAT). Complex III can also release superoxide into the intermembrane space, thereby affecting cytoplasmic signalling processes [24].
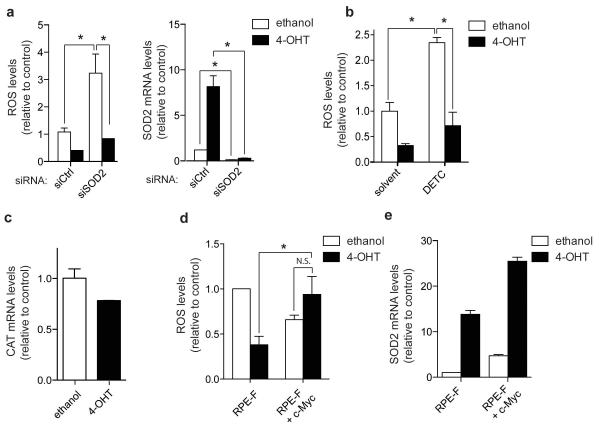
We found that FOXO3a activation caused a significant reduction in the amount of ROS in RPE-F cells (Figure 6a). FOXO3a increases resistance to oxidative stress caused by glucose depletion in quiescent fibroblasts by inducing the expression of SOD2 [6]. Although silencing of SOD2 significantly increased ROS levels in the cell line used here, FOXO3a activation was still able to cause a strong reduction in ROS levels in the absence of SOD2 (Figures 6a). Furthermore, inhibition of SOD2 using the inhibitor diethyldithiocarbamate (DETC) did not prevent the reduction in ROS levels following FOXO3a activation (Figure 6b). Catalase expression was not induced by FOXO3a activation in these cells (Figure 6c).
Figure 6. FOXO3a dependent reduction in ROS is independent of SOD2 and is rescued by re-expression of c-Myc.
(a) RPE-F cells were transfected with 100 nM of either controls (siCtrl) or siRNAs targeting SOD2 (siSOD2) for 72 h and treated with 4-OHT or solvent for the final 48 h. ROS levels were determined by measuring fluorescence of CM-H2DCFDA by flow cytometry. Expression levels of SOD2 were determined by qRT-PCR. (b) RPE-F cells were treated with 4-OHT or solvent for 48 h before treatment with 15 mM of the SOD2 inhibitor DETC for 90 mins. ROS levels were determined by measuring fluorescence of CM-H2DCFDA by flow cytometry. (c) RPE-F cells were induced with 4-OHT or solvent for 24 h. Expression of catalase (CAT) was determined by qRT-PCR. Data shown are representative of 3 independent experiments. (d) RPE-F or RPE-F+c-Myc cells were treated with 4-OHT or solvent for 48 h. ROS levels were determined by measuring fluorescence of CM-H2DCFDA by flow cytometry. (e) Expression levels of SOD2 mRNA in RPE-F and RPE-F+c-Myc cells treated with 4-OHT or solvent for 48 h were determined by qRT-PCR. Data shown are representative of 3 independent experiments.
All data are shown as mean ± SEM. * indicates statistical significance as determined by Student’s t-test (P < 0.05, n=3). N.S. = non-significant.
Since the regulation of mitochondrial genes by FOXO3a is dependent on c-Myc inhibition, we asked whether inhibition of c-Myc function is also required for the reduction in ROS levels by FOXO3a. Importantly, expression of c-Myc in RPE-F cells prevented the downregulation of ROS levels following FOXO3a activation (Figure 6d). In the c-Myc expressing cells, FOXO3a still induces SOD2, although SOD2 itself is upregulated by c-Myc expression (Figure 6e). Together, these data indicate that FOXO3a inhibits ROS levels through inhibition of c-Myc rather than through SOD2 upregulation.
Endogenous FOXO3a affects expression of mitochondrial genes in hypoxic cells
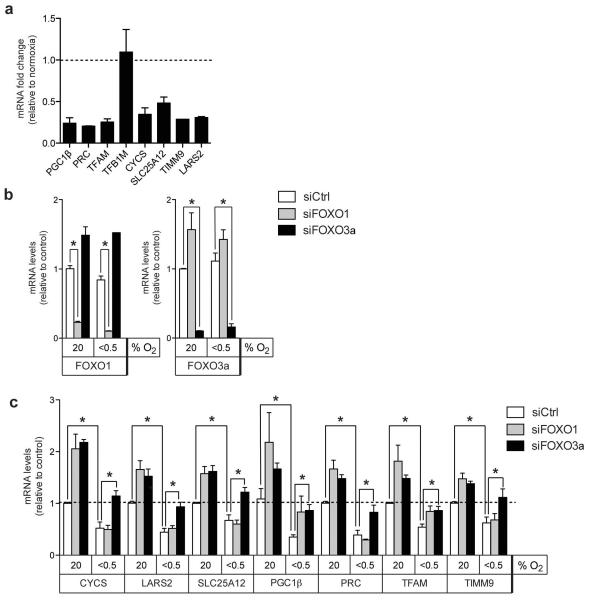
Inhibition of mitochondrial biogenesis is an important metabolic adaptation to hypoxia. We therefore investigated whether mitochondrial FOXO3a target genes are regulated by hypoxic conditions. Apart from TFB1M, all mitochondrial FOXO3a target genes tested showed a significant reduction in mRNA levels after 24 h of hypoxia (<0.5% oxygen) in parental RPE-hTERT cells (Figure 7a). It has been reported that FOXO3a is induced in response to hypoxic stress [25]. However, we did not observe significant changes in FOXO1 or FOXO3a mRNA levels in hypoxia in the cells used here (Figure 7b). While silencing of FOXO1 or FOXO3a increased expression of all mitochondrial genes in normoxic cells, hypoxia-dependent downregulation was mainly affected by depletion of FOXO3a (Figure 7c). In contrast, silencing of FOXO1 only affected the expression of PGC1β and TFAM in hypoxic cells (Figure 7c), suggesting that FOXO3a is responsible for the inhibition of mitochondrial genes by hypoxia.
Figure 7. Endogenous FOXO1 and FOXO3a are involved in the downregulation of mitochondrial genes in hypoxic cells.
(a) Parental RPE-hTERT cells were cultured in normoxic (20% O2) or hypoxic (<0.5% O2) conditions for 24 h. Expression of mitochondrial target genes was determined by qRT-PCR. (b) RPE-hTERT cells were transfected with siRNAs targeting FOXO1 or FOXO3a. 72 h post-transfection, cells were placed in normoxic or hypoxic conditions for 24 h. Expression of FOXO1 and FOXO3a was determined by qRT-PCR. (c) Expression of mitochondrial target genes in the same RNA samples as those in (b) was determined by qRT-PCR.
All data are shown as mean ± SEM * indicates statistical significance as determined by Student’s t-test (P < 0.05, n=3).
FOXO factors regulate ROS and HIF-1α levels in hypoxia
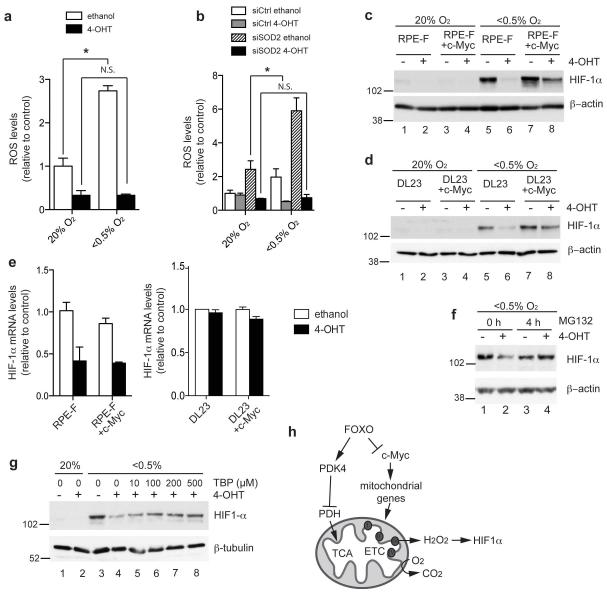
Mitochondrial ROS production is increased in hypoxia, and ROS produced through the electron transport chain have a key role in the adaptation to hypoxic conditions [26]. We therefore investigated whether FOXO3a activation modulates ROS levels during the hypoxia response. Activation of FOXO3a blocks the increase in ROS levels in cells exposed to hypoxia (Figure 8a). This activity is independent of SOD2 induction since silencing of SOD2 caused a 3-fold increase in ROS levels in hypoxic cells but did not affect the ability of FOXO3a to reduce ROS levels in either normoxic or hypoxic conditions (Figure 8b).
Figure 8. FOXO3a activation prevents ROS induction in hypoxia independently of SOD2 induction and blocks HIF-1α stabilisation.
(a) RPE-F cells were pre-treated with solvent (white bars) or 4-OHT (black bars) for 6 h and placed in normoxic (20% O2) or hypoxic (<0.5% O2) conditions for a further 48 h. ROS levels were determined by measuring fluorescence of CM-H2DCFDA by flow cytometry. (b) RPE-F cells were transfected with 100 nM of either control (siCtrl) or siRNAs targeting SOD2 (siSOD2) before being treated with 4-OHT or solvent in either normoxic or hypoxic conditions for 48 h. ROS levels were determined by measuring fluorescence of CM-H2DCFDA by flow cytometry. (c) RPE-F or RPE-F+Myc cells were treated with 4-OHT or solvent for 24 h and then placed in normoxic or hypoxic conditions for a further 24 h. Expression of HIF-1α was determined by immunoblotting. β-actin is shown as loading control. (d) DL23 or DL23+c-Myc cells were treated and analysed as in (c). (e) RPE-F and RPE-F+c-Myc and DL23 and DL23-c-Myc cells were treated with 4-OHT or solvent for 24 h. HIF-1α mRNA levels were determined by qRT-PCR. Data shown are representative of 3 independent experiments. (f) DL23 cells were treated with 4-OHT or solvent for 24 h and placed in hypoxic conditions for a further 24 h. 25 μM MG132 was added for the final 4 h. Expression of HIF-1α was determined by immunoblotting. β-actin is shown as loading control. (g) DL23 cells were treated with 4-OHT or solvent for 24 h and then placed in normoxia or hypoxia for a further 24 h. Boluses of tert-Butyl hydroperoxide (TBP), were applied at the indicated concentrations every 20 mins for the final 2 h. Expression of HIF-1α was determined by immunoblotting. β-tubulin is shown as loading control. (h) Diagram of FOXO action on mitochondrial activity. Induction of PDK4 by FOXO3a reduces TCA cycle activity and respiration. Inhibition of c-Myc function by FOXO3a lowers mitochondrial gene expression, reduces the formation of ROS and blocks stabilisation of HIF-1α. (TCA = tricarboxylic acid cycle; ETC = electron transport chain)
All data are shown as mean ± SEM. * indicates statistical significance as determined by Student’s t-test (P < 0.05, n=3). N.S. = non-significant.
One role of mitochondrial ROS production in hypoxic conditions is the regulation of the stability of hypoxia inducible factor 1α (HIF-1α). Several studies have shown that ROS produced by complex III is required for stabilisation of HIF-1α in hypoxia [27-30]. Complex III-derived ROS can be released into the cytoplasm and putatively decrease the activity of prolyl hydroxylases (PHDs), thereby preventing ubiquitination and degradation of HIF-1α by the Von Hippel Lindau (VHL) protein [26, 31]. We therefore investigated the effect of FOXO3a activation on HIF-1α stability in hypoxic RPE-F and DL23 cells. Figures 8c and d show that FOXO3a activation abolished HIF-1α induction by hypoxia. Interestingly, this activity of FOXO3a is dependent on inhibition of c-Myc, since re-expression of c-Myc restored HIF-1α expression in hypoxic cells (Figure 8c and d, lanes 7 and 8).
Surprisingly, FOXO3a activation had differential effects on HIF-1α mRNA in the 2 cell lines studied. FOXO3a activation led to a substantial reduction in HIF-1α mRNA in RPE-F cells but this was not observed in DL23 cells (Figure 8e and Supplementary Figure S6). Due to the reduction in HIF1-α mRNA following FOXO3a activation in RPE-F cells, inhibition of PHDs or silencing of VHL did not rescue the loss of HIF-1α in these cells (Supplementary Figure S7). However, downregulation of HIF-1α mRNA in RPE-F cells was not dependent on c-Myc inhibition since this was not affected by c-Myc re-expression (Figure 8e). The ability of c-Myc to rescue HIF-1α protein levels following FOXO3a activation without affecting its mRNA suggests that FOXO3a regulates the stability of HIF-1α protein through c-Myc inhibition. Indeed, in DL23 cells where there is no effect of FOXO3a on mRNA levels, treatment with the proteasome inhibitor MG132 blocked the downregulation of HIF-1α protein levels (Figure 8f). Furthermore, in this cell line, addition of an exogenous oxidant tert-Butyl hydroperoxide (TBP) to increase ROS levels, rescued the loss of HIF-1α protein levels following FOXO3a activation under hypoxia in a dose-dependent manner (Figure 8g).
Overall, our data suggest that FOXO3a can reduce ROS levels in hypoxic cells independently of SOD2 induction. Furthermore, activation of FOXO3a reduces the accumulation of HIF-1α in hypoxic cells and this is overcome by re-expression of c-Myc.
DISCUSSION
In this study, we have analysed the global transcriptional response to FOXO3a activation and identified a novel connection between FOXO3a and mitochondrial function. Surprisingly, the repression of mitochondrial target genes by FOXO3a was independent of the established transcriptional networks involving NRF1, NRF2 or ERRα. A previous study has shown that insulin like growth factor (IGF) induces expression of nuclear-encoded mitochondrial genes and mitochondrial biogenesis in Schwann cells through activation of ERRα [32]. We found that inhibition of mitochondrial gene expression by FOXO3a was mediated through inhibition of the c-Myc proto-oncogene. c-Myc has a well-established role in regulating mitochondrial gene expression. Activation of c-Myc drives expression of nuclear-encoded mitochondrial genes in B-lymphocytes [9]. It has been suggested that activation of mitochondrial gene expression by c-Myc involves the induction of PGC1β [10]. However, it is likely that c-Myc can also directly regulate a number of nuclear-encoded mitochondrial NRF1 target genes by binding to E-boxes embedded within the NRF1 DNA-binding motif [33].
Several previous studies have indicated that FOXO factors can antagonise c-Myc function [11, 34, 35] and FOXOs intersect with c-Myc function on multiple levels. We show here that FOXO3a inhibits mitochondrial gene expression by inducing the expression of Mad/Mxd proteins and by modulating the stability of the c-Myc protein. The reduction in c-Myc stability following FOXO3a activation could be an acute mechanism to rapidly downregulate the expression of mitochondrial genes while the induction of Mxd/Mxd proteins may ensure their sustained inhibition.
Consistent with the inhibition of mitochondrial gene expression, FOXO3a activation reduced mtDNA copy number, caused a decrease in the expression of mitochondrial proteins and lowered the levels of respiratory complexes. FOXO3a activation also reduced respiration both in coupled and uncoupled conditions. It is likely that induction of PDK4 contributes to the inhibition of basal oxygen consumption by blocking the entry of pyruvate into the TCA cycle. However, FOXO3a activation reduced oxygen consumption also in the presence of DCA, an inhibitor of PDK4. Interestingly, re-expression of c-Myc did not alter the effect of FOXO3a activation on basal oxygen consumption but prevented the reduction in mitochondrial capacity. Thus FOXO3a has multiple effects on mitochondrial function: it reduces mitochondrial capacity through inhibition of c-Myc and lowers the entry of pyruvate into the mitochondrial metabolism by induction of PDK4 (Figure 8h).
We found that the reduction in cellular ROS levels by FOXO3a activation was independent of SOD2 but could be reversed by re-expression of c-Myc. It has been shown that acute activation of c-Myc regulates glycolysis and oxidative phosphorylation to enable rapid cell cycle entry and the absence of c-Myc causes mitochondrial dysfunction [36]. The opposing effects of FOXO factors and c-Myc on mitochondrial metabolism and gene expression is likely to represent a switch between two distinct cellular states. c-Myc promotes mitochondrial production of energy and metabolites for macromolecule synthesis during cell cycle entry, while FOXO factors reduce mitochondrial output in order to prevent excess ROS production. Limiting mitochondrial ROS production is particularly important during conditions of oxygen deprivation. However, ROS can also act as important mediators of cellular signalling. In particular, mitochondria-derived ROS is required to promote the accumulation of HIF-1α in hypoxia [27]. We found that FOXO3a activation prevented the increase in ROS levels in hypoxic cells independently of SOD2 expression. Interestingly, FOXO3a activation prevented hypoxic accumulation of HIF-1α which was restored by re-expression of c-Myc. Previous studies showed that FOXO3a is induced in response to hypoxia and can inhibit HIF1α-dependent gene expression by directly binding to HIF-1α [37] or through induction of the negative transcriptional cofactor CITED2 [25]. However, studies in neural stem cells from foxo3a−/− mice revealed that FOXO3a is required for the expression of hypoxia-dependent genes [38]. Decreasing mitochondrial function is an important role of HIF-1α in the response to hypoxia. By regulating mitochondrial gene expression, FOXO3a may replace some of the function of HIF-1α in the cellular adaptation to hypoxia.
In conclusion, our study reveals a novel role of FOXO factors in regulating mitochondrial activity through the repression of c-Myc function. Mitochondrial ROS signalling is known to contribute to Ras-dependent transformation and tumourigenesis [39]. The regulation of mitochondrial gene expression and mitochondrial activity could therefore be an important feature of the role of FOXO factors as tumour suppressors.
MATERIALS AND METHODS
Plasmids and Reagents
pBABEpuro-HA-FKHR-L1.A3-ER has been described previously [12]. pWZL-Blast-c-Myc was constructed by inserting the cDNA of c-Myc into pWLZ-Blast. pBABE-Blast was obtained from M. Murillo (CRUK LRI, London). pGL3-basic mtTFA-luc WT was kindly supplied by Prof. R. Scarpulla (Northwestern Medical School, Chicago).
MG132 and PI-103 were obtained from Calbiochem. LY-294002 was from Cell Signaling. Cycloheximide, the GSK3 inhibitor SB216763, Resveratrol, FCCP, DMOG, 4-hydroxy-tamoxifen (4-OHT), the DRP-1 inhibitor Mdivi-1, the SOD2 inhibitor sodium diethyldithiocarbamate (DETC) and tert-Butyl hydroperoxide (TBP) were obtained from Sigma.
Cell culture
DL23 [12] and parental DLD-1 cells were grown in RPMI 1640 supplemented with 10% foetal calf serum. 4-OHT was dissolved in ethanol and used at a final concentration of 100 nM. RPE-hTERT cells (Clontech) and derivatives were grown in DMEM/HAMS F12 (1:1) medium supplemented with 10% FCS, glutamine and sodium bicarbonate. pBABEpuro-HA-FKHR-L1.A3-ER (FOXO3a.A3-ER) retroviruses were packaged in Phoenix cells and used to infect RPE-hTERT cells. Infected cells were selected using puromycin, and clone F12 was chosen for further study.
To create c-Myc expressing cells, pWZL-Blast-c-Myc retroviruses were packaged in Pheonix cells and used to infect DL23 or RPE-hTERT.FOXO3a.A3-ER cells. Infected cells were selected using blasticidin. All experiments were performed using early passages.
Hypoxia was generated using an InVivo2 500 hypoxic work station (Ruskinn).
Immunoblotting
Cells were lysed in Triton buffer (1% Triton X-100, 50 mM Tris pH 7.5, 300 mM NaCl, 1 mM EGTA and protease inhibitor cocktail, Roche). Plates were incubated for 20 min on ice, and lysates were cleared by centrifugation. Lysates were separated by sodium dodecyl sulphate (SDS)-polyacrylamide gel electrophoresis and transferred onto an Immobilon membrane (Millipore). Proteins were detected by immunoblotting with ECL detection. The following antibodies were used: anti-c-Myc 9E10 (CRT), anti-phospho-c-Myc (Cell Signaling), anti-GSK3α/β (Cell Signaling), anti-β-catenin (Cell Signaling), anti-CoxI antibody (Invitrogen), anti-Tom20 (Santa Cruz), MitoProfile Total OXPHOS Human WB Antibody Cocktail (Mitosciences), anti-HIF-1α (BD Biosciences), anti-β-tubulin-horseradish peroxidase (Abcam) and anti-β-actin-horseradish peroxidase (Abcam). Secondary antibodies used: Donkey anti-rabbit IgG-horseradish peroxidase (Amersham) and sheep anti-mouse IgG-horseradish peroxidase (Amersham). The intensities of Immunoblot bands from three independent experiments were quantified using Image J software and normalized to the intensities of the loading control from each experiment.
RNA preparation and array analysis
Total RNA was extracted using RNeasy kits (QIAGEN). Gene expression analysis was performed by the CRUK GeneChip service (Patterson Institute) using Affymetrix human Exon Arrays. Data analysis was performed within LIMMA, Bioconductor. Signal estimates were generated using RMA. FOXO3a regulated genes were selected using a false discovery rate of 0.05. Annotations were derived from Affymetrix release HuEx-1_0-st-v2.na24.hg18.
Gene Set Enrichment Analysis (GSEA) was performed using gene sets from the Molecular Signatures Database for curated gene sets (MSigDB-C2 v2, BROAD Institute) [40]. In order to avoid false positives due to multiple testing in GSEA, the false discovery rate (FDR) is used to adjust the p-value to give the q-value. A q-value of <0.05 is statistically significant.
Gene silencing
For transient silencing, DL23 or RPE-hTERT.FOXO3a.A3-ER cells were transfected with a total of 100 nM of small interfering RNA (siRNA) oligonucleotide pools (siGenome, Dharmacon) using DharmaFECT 3 or DharmaFECT 1 reagents, respectively (Dharmacon) in serum-free medium (optiMEM I; Invitrogen) for a total time of either 48, 72 h or 96 h, as indicated. For experiments with 72 h or 96 h of silencing, cells were split 24 or 48 h post-transfection respectively and incubated for a further 48 h. To silence all members of the Mad/Mxd family simultaneously, a mixture of siRNA pools for each Mad/Mxd (Mad1, Mxd1, Mad3 and Mad4) family was transfected (each pool present at 25 nM).
The following siRNA oligonucleotides were used: siGenome (Dharmacon) Pools for PGC1β (PPARGC1B), PRC (PPRC1), NRF1, GABPA, ERRα (ESRRA), Mad1 (Mxd1), Mxi1, Mad3 (Mxd3), Mad4 (Mxd4), c-Myc (MYC), SOD2, FOXO1 and FOXO3, TFAM, VHL, Control 4 and non-targeting control (RISC free).
Reverse transcription quantitative PCR
Total RNA (1 to 5 μg) was used for first-strand cDNA synthesis using oligo-dT primers and SuperScript II reverse transcriptase (Invitrogen). Real-time PCR was performed with SYBR Green PCR master mix (Qiagen) in 96-well plates using the Chromo 4 system (MJ Research) or ABI 7900HT (Applied Biosystems). All primers used are QuantiTect Primer Assays (Qiagen). All reactions are performed at least in duplicate. The relative amount of mRNA was calculated using the comparative CT method after normalisation to β-actin or B2M.
Reporter assays
DL23 cells were transiently transfected using Lipofectamine Plus reagent (Invitrogen) in serum-free medium (optiMEM I; Invitrogen). Cells were harvested in passive lysis buffer (Promega) and luciferase activity was determined using the luciferase dual reporter assay kit (Promega) and luminescence was read on an Envison Multilabel Plate Reader (Perkin Elmer). Activity of firefly luciferase was normalised to the activity obtained from a cotransfected expression construct for renilla luciferase.
Measurement of Oxygen Consumption Rate (OCR)
RPE-hTERT.FOXO3a.A3-ER cells were stimulated with 4-OHT for 48 h. Viable cells were counted using a Vi-Cell (Beckman Coulter). 1×106 viable cells were plated in each well of an Oxygen Biosensor Plate (BD Biosciences) in media in the presence or absence of 2 μM FCCP (Sigma). Fluorescence was measured with an Envision Multilabel Plate Reader (Perkin Elmer) with an excitation wavelength of 485 nm and emission wavelength of 633 nm every 2 minutes for at least 90 minutes. Fluorescence increases as oxygen is depleted from the media. The oxygen consumption rate (OCR: Δrfu/min) was calculated as the rate of increase in fluorescence per minute from the linear part of the curve.
Mitochondrial DNA ratio
Total DNA was extracted using DNeasy kits (Qiagen). qPCR was performed to determine the relative amounts of an amplicon within a mitochondrial gene (cytochrome b) to that of a nuclear gene (β-actin). The primer sequences used are: cytochrome b 5′: GCGTCCTTGCCCTATTACTATC; cytochrome b 3′: CTTACTGGTTGTCCTCCGATTC; β-actin 5′: ACCCACACTGTGCCCATCTAC; β-actin 3′: TCGGTGAGGATCTTCATGAGGTA.
Mitotracker staining for flow cytometry
RPE-hTERT.FOXO3a.A3.ER cells were stimulated with 4-OHT for 48 h prior to fixation with 3.7% formaldehyde for 15 minutes at 37°C. Cells were incubated with 100 nM Mitotracker Green in PBS (Molecular Probes) for 10 minutes at room temperature, according to the manufacturer’s instructions for staining of fixed cells. Incubation with Mitotracker Green was carried out after fixation in order to determine mitochondrial mass independently of mitochondrial activity. Cells were washed twice in PBS and analysed with an LSRII-SORP (Becton Dickinson) flow cytometer. DAPI was used to gate out dead cells.
Confocal microscopy
RPE-hTERT.FOXO3a.A3.ER cells were grown on coverslips, stimulated with 4-OHT for 48 h and fixed with 3.7% formaldehyde for 15 minutes at 37°C. Cells were incubated with 100 nM mitotracker green for 10 minutes at room temperature, according to the manufacturer’s instructions for staining of fixed cells. Images were acquired on a Zeiss 510 confocal microscope using a 60x 1.4 oil immersion objective using the same instrument settings for each image. A projection of the Z-stack is shown. The area of cells occupied by Mitotracker Green was quantified using Metamorph software with the threshold tool set at an identical threshold value for the 2 conditions.
Determination of ROS levels
RPE-hTERT.FOXO3a.A3.ER cells were treated as indicated then incubated with 10 μM CM-H2DCFDA (Molecular Probes) for 30 minutes in media at 37°C. Cells were trypsinised, washed with PBS and analysed with an LSRII-SORP flow cytometer (Becton Dickinson). DAPI was used to gate out dead cells. Median values were taken following subtraction of background fluorescence from solvent (DMSO)-treated controls.
Statistical analysis
Student’s t tests assuming a two-tailed distribution and equal variance were performed for statistical analysis.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the LRI Research Services, particularly the FACS laboratory, Equipment Park and Light Microscopy for technical assistance. We would also like to thank Beatrice Griffiths for help with tissue culture and Julian Downward for critical discussions. This work was funded by Cancer Research UK.
ABBREVIATIONS
- FOXO3a
forkhead-box protein O3a
- GSEA
Gene Set Enrichment Analysis
- GSK3
Glycogen synthase kinase)
- HIF-1α
hypoxia inducible factor 1 alpha
- mtDNA
mitochondrial DNA
- OCR
oxygen consumption rate
- ROS
reactive oxygen species
- SEM
Standard Error of the Mean
- SOD2
Manganese-containing superoxide dismutase
- PGC
Peroxisome proliferator-activated receptor gamma
- 4-OHT
4-hydroxytamoxifen
REFERENCES
- 1.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27(16) doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 2.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96(6):857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 3.Kops GJ, et al. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398(6728):630–4. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 4.Dansen TB, Burgering BM. Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Biol. 2008;18(9):421–9. doi: 10.1016/j.tcb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 5.van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8(6):440–50. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- 6.Kops GJ, et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419(6904):316–21. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 7.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 8.Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25(14):6225–34. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11(5):407–20. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Delpuech O, et al. Induction of Mxi1-SRalpha by FOXO3a Contributes to Repression of Myc-Dependent Gene Expression. Mol Cell Biol. 2007;27(13):4917–30. doi: 10.1128/MCB.01789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kops GJ, et al. Control of cell cycle exit and entry by protein kinase B-regulated forkhead transcription factors. Mol Cell Biol. 2002;22(7):2025–36. doi: 10.1128/MCB.22.7.2025-2036.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furuyama T, et al. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem J. 2003;375(Pt 2):365–71. doi: 10.1042/BJ20030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci U S A. 1994;91(4):1309–13. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vervoorts J, Luscher-Firzlaff J, Luscher B. The ins and outs of MYC regulation by posttranslational mechanisms. J Biol Chem. 2006;281(46):34725–9. doi: 10.1074/jbc.R600017200. [DOI] [PubMed] [Google Scholar]
- 16.Welcker M, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101(24):9085–90. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yada M, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23(10):2116–25. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeh E, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6(4):308–18. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, et al. Pim kinase-dependent inhibition of c-Myc degradation. Oncogene. 2008;27(35):4809–19. doi: 10.1038/onc.2008.123. [DOI] [PubMed] [Google Scholar]
- 20.Aberle H, et al. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16(13):3797–804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carew JS, Huang P. Mitochondrial defects in cancer. Mol Cancer. 2002;1:9. doi: 10.1186/1476-4598-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127(6):1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 23.Romanello V, et al. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J. 2010;29(10):1774–85. doi: 10.1038/emboj.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem Sci. 2010;35(9):505–13. doi: 10.1016/j.tibs.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bakker WJ, Harris IS, Mak TW. FOXO3a is activated in response to hypoxic stress and inhibits HIF1-induced apoptosis via regulation of CITED2. Mol Cell. 2007;28(6):941–53. doi: 10.1016/j.molcel.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 26.Klimova T, Chandel NS. Mitochondrial complex III regulates hypoxic activation of HIF. Cell Death Differ. 2008;15(4):660–6. doi: 10.1038/sj.cdd.4402307. [DOI] [PubMed] [Google Scholar]
- 27.Chandel NS, et al. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J Biol Chem. 2000;275(33):25130–8. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 28.Guzy RD, et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1(6):401–8. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Mansfield KD, et al. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell Metab. 2005;1(6):393–9. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunelle JK, et al. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1(6):409–14. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Bell EL, et al. The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. J Cell Biol. 2007;177(6):1029–36. doi: 10.1083/jcb.200609074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Echave P, et al. Extracellular growth factors and mitogens cooperate to drive mitochondrial biogenesis. J Cell Sci. 2009;122(Pt 24):4516–25. doi: 10.1242/jcs.049734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Lee JH, Iyer VR. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One. 2008;3(3):e1798. doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouchard C, et al. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. Embo J. 2004;23(14):2830–40. doi: 10.1038/sj.emboj.7600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gan B, et al. FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell. 2010;18(5):472–84. doi: 10.1016/j.ccr.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrish F, et al. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle. 2008;7(8):1054–66. doi: 10.4161/cc.7.8.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emerling BM, et al. PTEN regulates p300-dependent hypoxia-inducible factor 1 transcriptional activity through Forkhead transcription factor 3a (FOXO3a) Proc Natl Acad Sci U S A. 2008;105(7):2622–7. doi: 10.1073/pnas.0706790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renault VM, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5(5):527–39. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107(19):8788–93. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.