Abstract
We have generated a humanised double-reporter transgenic rat for whole body in vivo imaging of endocrine gene expression, using the human prolactin gene locus as a physiologically important endocrine model system. The approach combines the advantages of BAC recombineering to report appropriate regulation of gene expression by distant elements, with double reporter activity for the study of highly dynamic promoter regulation in vivo and ex vivo. We show first that this rat transgenic model allows quantitative in vivo imaging of gene expression in the pituitary gland, allowing the study of pulsatile dynamic activity of the prolactin promoter in normal endocrine cells in different physiological states. Using the dual reporters in combination, dramatic and unexpected changes in PRL expression were observed following inflammatory challenge. Expression of PRL was shown by RT-PCR to be driven by activation of the alternative upstream extra-pituitary promoter and flow cytometry analysis pointed at diverse immune cells expressing the reporter gene. These studies demonstrate the effective use of this type of model for molecular physiology and illustrate the potential for providing novel insight into human gene expression using a heterologous system.
Keywords: prolactin, in vivo imaging, gene expression, alternate promoter
Introduction
A major challenge in physiology is the understanding and analysis of dynamic temporal control of gene expression in living intact tissues in real time in different physiological conditions. In this study we developed transgenic rat lines using large reporter transgenes in bacterial artificial chromosomes, with the purpose of studying dynamic regulation of the important hormone prolactin, assessing gene expression in the intact animal in vivo and in living cells ex vivo. Surprisingly little is known about the timing of gene expression in living tissues and whole organisms, although previous work has indicated that in cell lines, gene transcription in individual cells behave in a highly dynamic and heterogeneous manner (1, 2).
Prolactin is a polypeptide hormone mainly produced by the lactotrope cells of the anterior pituitary gland and its major function in mammals is related to lactation (3). The prolactin gene is also expressed in humans and primates at extra-pituitary sites, such as brain, decidualised endometrium, myometrium and circulating lymphocytes (4) and more than three hundred biological functions have been attributed to this hormone, including reproduction, immuno-modulation and behaviour. The human PRL gene maps on the short arm of chromosome 6 and it is organised in five exons and four introns (4). In human decidua and in lymphocytes the PRL mRNA is longer including an extra exon (exon 1a), produced by the activity of an alternative promoter located 5.8 kb upstream of the pituitary transcription start site (4-6). A number of comparisons suggest that the human and rodent prolactin loci are significantly different. Where the human PRL locus lies in a gene desert (7) surrounded by around one megabase of non-coding DNA (http://genome.ucsc.edu), rodent prolactin (Prl) genes exist as a large family of 26 closely related paralogous genes arising from gene duplication (8). The prolactin gene families in the mouse and rat are clustered on chromosomes 13 and 17 respectively (4). Differential expression of the alternative human promoter has been suggested to compensate for the lack of expansion of the PRL gene in primates (9). No mutations have been described in the human prolactin gene, suggesting that such mutations might be lethal. This differs from studies in mouse where neither vitality nor immune competency is compromised in Prl (10) and Prlr knock-out mice (10, 11), despite many indications for a role of prolactin in the immune system (12). This is surprising given the high conservation of the PRL gene sequence in humans. Therefore, the mouse knock-out models available may not be ideal for studying the human prolactin gene, as a result of redundancy of the rodent system.
Here we describe the generation of a “humanised” double-transgenic rat that expresses luciferase and destabilised eGFP under the control of the entire human prolactin gene locus. Our strategy is based on a BAC recombineering approach to permit the inclusion of long-distance regulatory elements and to minimise site of transgene integration effects. We have previously used luciferase to image the dynamic pattern of gene expression, which is characteristic of the human PRL promoter (2, 13, 14). We show that the resulting rat double-transgenic model allows in vivo imaging and ex-vivo analysis of human PRL gene expression driven by the pituitary and also the extra-pituitary promoter, making this an ideal tool for the study of human PRL gene expression in different physiological and pathological conditions.
Results
Generation of a BAC-reporter transgenes
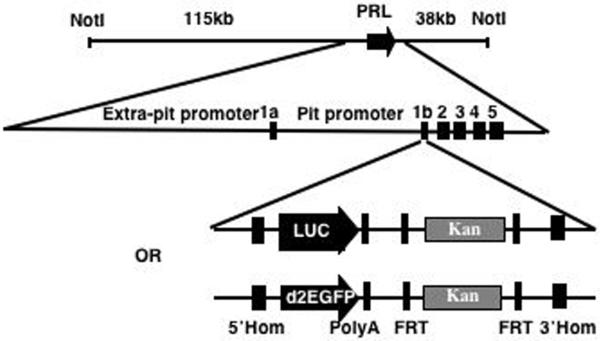
We have generated a BAC-luciferase and a BAC-d2eGFP construct by BAC recombineering (15) using BAC RP11-237G3, which spans 163kb of the human PRL genomic locus including 115kb upstream and 38kb downstream of the PRL gene (Fig. 1a). Both Photinus pyralis luciferase and destabilised eGFP (d2eGFP) were selected as reporter genes due to their short half-life, which allows for the imaging of highly dynamic gene expression patterns (2) and for their suitability for in vivo imaging (16) (17). The BAC was targeted with a linear double-strand DNA cassette containing either the luciferase or the d2eGFP gene and a Kan selectable marker flanked by FRT sites. Homologous recombination arms were designed to span the PRL gene 5′UTR and the first intron in order to substitute exon 1b with the targeting cassette (Fig. 1) (verified using Southern blot hybridisation, see supplemental figure 1b and 2b published as supplemental data on the Endocrine Society’s Journals Online web site at http://mend.endojournals.org). Exon 1b contains the translation ATG initiator and its removal prevents the production of PRL from the targeted transgene.
Figure 1.
Diagram showing the BAC targeting strategy using two (luciferase and d2eGFP) double strand linear cassettes. 5′ Hom: 5′ homology arm; Luc: luciferase; 3′Hom: 3′ homology arm
Hormonal responses of stably transfected BAC cell lines
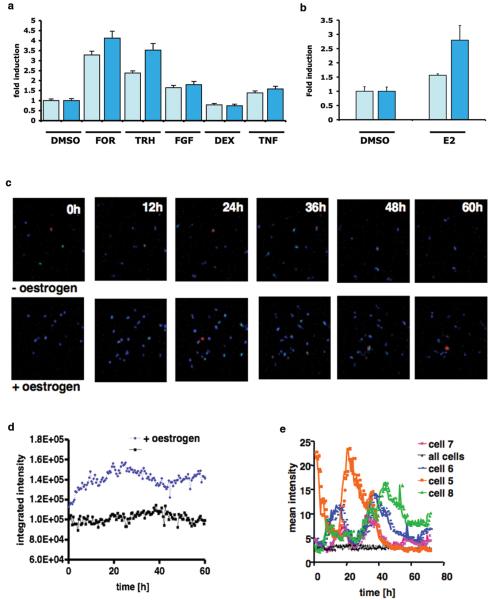
PRL-Luc BAC construct validation was performed by generating stably transfected pituitary GH3 cell lines. Eighteen recombinant clones were analysed for basal luciferase activity (see supplemental figure 3) and a subset of nine of them were challenged with a variety of well-characterised prolactin-regulating stimuli. A comparison with GH3 cells expressing luciferase under the control of 5kb of human prolactin promoter (D44 cell line) (2)) is presented in Fig. 2a. A 2.8 fold induction of luciferase activity was observed in the PRL-Luc BAC cell lines after stimulation with oestrogen compared to the 1.6 fold induction in D44 (p<0.05) (18) (Fig. 2b). Real-time luminescence imaging showed significantly greater oestrogen induction in the PRL-Luc BAC transfected GH3 cells than that observed using the 5kb prolactin promoter (Fig. 2c). Single cells revealed heterogeneous, fluctuating transcriptional activity under resting conditions (Fig. 2d and 2e), as seen previously in clonal cell lines (2), adenovirus infected (14) or micro-injected primary pituitary cells (19).
Figure 2.
Analysis of cell lines stably transfected with the PRL-Luc BAC construct a) Stimulation of BAC cell lines and D44 cells (FOR, 1μM forskolin, TRH 300 nM, FGF-2 10 ng/ml, Dex 10 nM and TNF-α 10 ng/ml) for 8h. Results are shown as fold induction ± SD. b) Oestrogen (E2) stimulation of BAC cell lines and D44 (1nM) for 24h. Results are shown as fold induction ± SD c) Cell imaging. Oestrogen was added at time 0 and images for untreated (upper panels) and treated cells (lower panels) are shown. Scale bar, 125 μm; d) Total photon counts of a whole field of cells (~20 cells) with (blue) or without (black) addition of oestrogen; e) Quantification of the transcriptional activity in single cells under resting conditions. The data are the mean intensity of individual cell areas and the mean intensity of the whole field of cells (~ 20 cells).
Generation of PRL-Luc and PRL-d2eGFP transgenic rats
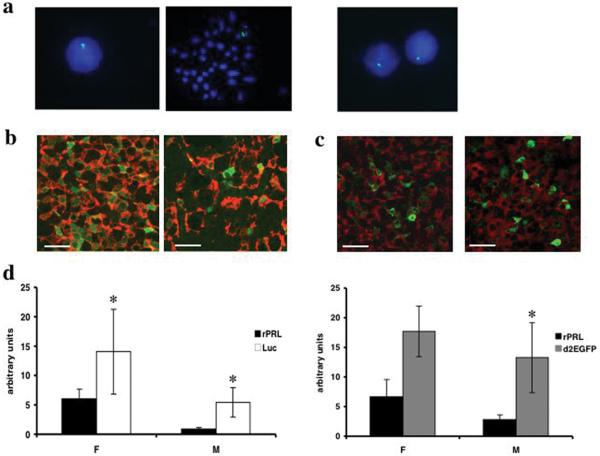
The targeted PRL-Luc and PRL-d2eGFP BAC constructs were injected into the pronucleus of Fisher344 fertilised rat oocytes. Of 64 potential founder rats for PRL-Luc construct, 5 transgenic rats were identified (PRL-Luc25, PRL-Luc34, PRL-Luc37, PRL-Luc47, PRL-Luc49) and of 26 potential founders for PRL-d2eGFP construct, 2 transgenic rats were identified by PCR and confirmed by Southern blot hybridisation (data not shown). All the lines except PRL-Luc25 and PRL-Luc34, transmitted the transgene to their progeny and showed normal growth and viability. FISH analysis of interphase and metaphase nuclei showed multiple insertion sites of the transgene in lines PRL-Luc34, PRL-Luc37 and PRL-Luc47 (see supplemental figure 4), but a single insertion site in line PRL-Luc49, PRL-d2eGFP455 (Fig. 3a) and PRL-d2eGFP485. Southern blot analysis showed that more than 155kb of the transgene was integrated into the genome of line PRL-Luc 49 in high copy number and in PRL-d2eGFP455 in low copy number. Thus, these two lines were selected for further characterisation and study. Double immuno-cytochemistry analysis shows that a good co-localisation of prolactin/reporter gene signal can be observed in male and female pituitaries. A higher density of lactotropes was observed using immunocytochemistry (ICC) throughout the female pituitary, compared to male pituitary (Fig. 3b, p=0.0005) in PRL-Luc rats. In these animals expression of luciferase was higher in females than males (Fig. 3b) and the two genes were co-expressed in 93.9% ± 5.9 of female lactotropes and in 59% ± 13.9 of male lactotropes (p<0.0001). In PRL-d2eGFP transgenic rats the two genes were co-expressed in 18.4% ± 8% of female lactotropes and in 21.6% ± 8% of male lactotropes (Fig.3c).
Figure 3.
Molecular characterisation of BAC PRL-Luc and PRL-d2eGFP transgenic rats. a) FISH analysis of metaphase and interphase nuclei of transgenic rat spleenocytes. Left panels PRL-Luciferase line 49; right panel PRL-d2eGFP line 455 b and c) Fluorescent immuno cytochemistry of female (left) and male (right) pituitary sections. Green fluorescence: luciferase b) or d2eGFP c), red fluorescence: prolactin. Scale bar, 20 μm; d) Real-time qPCR of luciferase, d2eGFP and rat prolactin in the pituitary gland of female and male transgenic rats. (*p<0.05 rat Prl vs luciferase in males and females; n=5 female; n=4 male; * p<0.05 rat Prl vs d2eGFP in males; n=3 female; n=4 male)
Endogenous rat Prl and luciferase or d2eGFP gene expression analysis by qPCR shows a significantly higher expression of the reporter genes compared to the endogenous Prl (Fig. 3d) in both male and females. In male luciferase transgenic rats, expression of the Prl gene is significantly decreased, suggesting competition for transcription factors (transcriptional squelching; Fig.3d). Human prolactin activity was undetectable by radioimmunoassay (RIA) in the transgenic rat plasma and pituitary homogenates.
In vivo imaging of pituitary specific expression
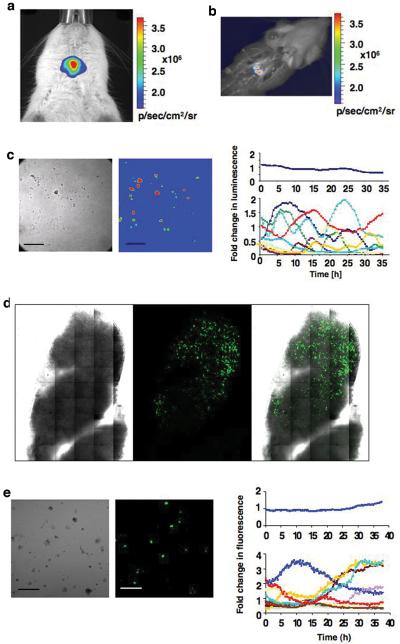
In vivo bioluminescence imaging detected strong signals, 1.33×106 ± 6.7×105 p/sec/cm2/sr in females and 5.6×105 ± 4.2×105 in males (Fig. 4a), after 30 seconds signal integration time in luciferase transgenic rats lying supine. No strong signal was detected in this position with the animal lying prone, most likely due to the optical properties of the brain and the skull (20). Specificity of the signal for the pituitary gland was assessed after the rat was sacrificed (Fig. 4b).
Figure 4.
In vivo imaging and ex vivo analysis of BAC PRL-Luc and PRL-d2eGFP transgenic rats. a) In vivo imaging of a female BAC PRL-Luc transgenic rat. The image was acquired with an IVIS Spectrum (Xenogen/ Caliper): 60s integration time, Bin (HR) 4, FOV 6.6, f1. b) Image taken after the rat has been euthanased. c) Ex vivo imaging of single cell preparations derived from PRL-Luc transgenic rat pituitary. Left: bright field image of the pituitary cells. Middle: pseudo-colour image of the same cells. Right: Quantification of the dynamic changes in luminescence from individual cells. Scale bar, 200 μm.
d) Tile scan imaging, with a 40x 1.3 fluar objective, of a pituitary thick section (400μm) from PRL-d2eGFP rat e) Ex vivo imaging of single cell preparations derived from PRL-d2eGFP rat pituitary. Left: bright field image of the pituitary cells. Middle: colour image of the same cells. Right: Quantification of the dynamic changes in fluorescence from individual cells. Scale bar, 200 μm.
Ex vivo study of pituitary single cell gene expression
Pituitaries from transgenic positive animals were harvested and the cells dispersed. Light emission could be detected in approximately one third of cells from each dispersed pituitary gland as assessed by bright field imaging. Continuous imaging and photon emission were recorded over 35 h. The cells showed a dynamic expression pattern, which was not synchronised (Fig. 4c), but similar to the responses in the BAC cell lines (Fig. 2e). A movie showing the dynamic and oscillatory pattern of expression is in supplemental movie 1. Heterogeneity in prolactin expression was also observed in d2eGFP-expressing cells dispersed from transgenic positive pituitaries (Fig. 4d and 4e).
Extra-pituitary luciferase expression
Prolactin expression in man has been demonstrated in a number of extra-pituitary sites including cells of the immune system especially in T lymphocytes and lymphoid tissues (12). Detectable levels of luciferase and d2eGFP gene expression and luciferase activity were found to be present in the thymus and spleen of transgenic rats, whereas endogenous rat Prl gene expression was absent (see supplemental figure 5). Luminescence signals were also detected in the paws and ears of male and female transgenic rats, suggesting human prolactin promoter-driven luciferase gene expression in cartilage (manuscript in preparation).
To address possible regulation of PRL promoter activity in immune-related cells, male rats were injected i.p. with either physiological saline (vehicle) or 3mg/kg of body weight of LPS. After 16 h the rats were anaesthetised, given an i.p injection of luciferin and imaged. One vehicle-treated and one LPS-treated rat were imaged simultaneously. Fig. 5a shows two representative experiments where strong multiple signals were seen in LPS-treated rats in lower and upper left abdomen, but also overlying the mediastinum (thymus and parathymic lymph nodes), the axillae (axillary/brachial lymph nodes) and posterior inter-mandibular space (mandibular lymph nodes). This pattern of light emission in LPS-treated rats suggests luciferase expression in cells resident in lymphoid tissues, but also in peritoneal exudate cells. Fainter signals could also be seen in the abdomen of vehicle-injected rats and they may be due to local and superficial LPS-independent inflammation at the site of i.p. injection of the vehicle and luciferin substrate. The vehicle treated animal in the right panel showed very low expression of pituitary luciferase (see supplemental figure 6).
Figure 5.
Extra-pituitary expression of luciferase in BAC PRL-Luc transgenic rat. a) In vivo imaging of vehicle (V) and LPS-treated rats. The rats were injected i.p (arrows indicate site of injection) either with physiological saline or with 3mg/ml LPS and imaged 16 h after the treatment. The images were acquired with an IVIS Spectrum (Xenogen/ Caliper): 30s integration time, Bin (HR) 8, FOV 19.6, f1 b) RT-PCR strategy for the analysis of luciferase expression driven by the extra-pituitary promoter. Dotted line: observed alternative splicing c) RT-PCR result. M: 100bp ladder T: thymus, SP: spleen.
To assess whether this in vivo expression was reflecting alternative promoter usage in immune-related organs, an RT-PCR approach was designed to amplify luciferase cDNA in thymus and spleen of transgenic rats, using two sets of primers. The first set of primers amplified a portion of cDNA including exon 1a, the extra-pituitary 5′UTR (21) and the pituitary 5′ UTR. The second set of primers amplified a portion of the luciferase gene as well (Fig. 5b). Amplification was successful in both tissues with the two sets of primers, demonstrating luciferase expression driven by the extra-pituitary promoter, but a band was obtained that was 148bp larger than the expected size and corresponded to the most abundant amplicon obtained in addition to the expected fragment (Fig. 5c). Sequence analysis allowed for the precise mapping of the alternative splice acceptor site to position -246 relative to the luciferase translation start site (equivalent to the human prolactin ATG position). These data suggest activation of the human alternative prolactin promoter upon immune challenge.
In vivo imaging and ex vivo analysis of prolactin mediated LPS response in a double-transgenic rat
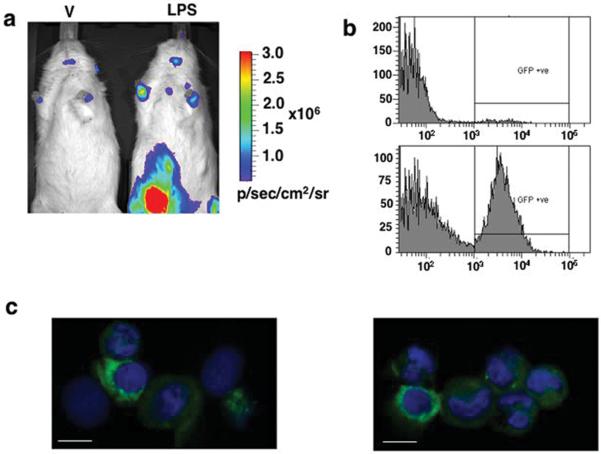
Double-transgenic PRL-Luc/d2eGFP rats were generated by mating from single transgenics. Male double-transgenic rats were injected i.p. with either physiological saline (vehicle) or 3mg/kg of body weight of LPS. After 16 h the rats were anaesthetised, given an i.p injection of luciferin and imaged using the IVIS Spectrum. One vehicle-treated and one LPS-treated rat were imaged simultaneously. Figure 6a shows bioluminescence imaging of the PRL-mediated immune response as was also observed in the PRL-Luc transgenic animals (Fig 5a). Furthermore, this model offers the option for fluorescence-based analysis of the cellular response to the LPS challenge. Fluorescent positive cells were observed in the peritoneal exudate of vehicle and LPS treated rats in dramatic different proportions (Fig 6b) by flow cytometry. Confocal imaging of peritoneal exudate cells suggested the expression of the d2eGFP protein by different populations of immune cells, including monocytes/macrophages and granulocytes (Fig 6c).
Figure 6.
In vivo and ex vivo imaging of reporter genes in LPS treated PRL-Luc/d2eGFP double transgenic rats. a) In vivo imaging of vehicle (V) and LPS-treated rats. The rats were injected i.p either with physiological saline or with 3mg/ml LPS and imaged 16 h after the treatment. The images were acquired with an IVIS Spectrum (Xenogen/ Caliper): 30s integration time, Bin (HR) 8, FOV 19.6, f1. b) Flow cytometric analysis of d2eGFP positive cells from the peritoneal exudate of a vehicle and a LPS treated double transgenic rats. c) Confocal imaging of cytocentrifuge preparation of cells from the peritoneal exudate of LPS treated double-transgenic rats. Scale bar, 10μm.
Discussion
We have used a BAC recombineering approach to generate reporter transgenic rats that express the luciferase and the d2eGFP gene under the control of both the pituitary and extra-pituitary specific human prolactin gene promoters. Whole body in vivo imaging of human PRL promoter activity in these models readily reveals physiological pituitary expression and striking evidence of alternative promoter activation after immune challenge.
The human PRL gene is surrounded by highly conserved non-coding sequences that span over one megabase of genomic DNA. These sequences are likely to contain functional regulatory elements and long-distance enhancers (22, 23). We selected a BAC clone spanning 163kb of the human prolactin locus and engineered it to insert either the luciferase or the destabilised eGFP genes into the human PRL exon 1b, thus removing expression of human prolactin. BAC transgenesis offers several advantages over the use of small transgenes. These constructs encompass large stretches of flanking DNA and behave very similarly to the endogenous gene as effects due to the position of insertion can be practically nullified (24). They are also readily modified by BAC recombineering (25), and can be used for generating transgenic animals and for transfecting mammalian cell lines (26). To validate the PRL-Luc BAC construct, stably transfected pituitary GH3-based cell lines were generated. They showed dramatic heterogeneity in prolactin transcription at single cell level even under resting conditions, confirming a dynamic pattern of gene regulation (2, 13, 14, 19).
The BAC cell lines showed a response to a variety of stimuli, such as forskolin, TRH, FGF-2 and TNF-α. We have recently demonstrated that TNF-α is a stimulator of the human prolactin promoter and its effect is mediated by NF-κB signalling pathway (27). Interestingly, the stimulation of several BAC cell lines with E2 elicited larger luciferase emission (18) than D44 cells which express luciferase under the control of a shorter human prolactin promoter (2). These data suggest the presence of further oestrogen-responsive regulatory elements in the BAC sequence, in addition to the one identified at -1189bp relative to the transcription start site.
Although cell studies are very valuable for looking at molecular mechanisms of gene regulation, in vivo imaging strategies are very important tools to understand biological processes as they occur in living animals. We have generated 4 lines of BAC PRL-Luc and two lines of BAC PRL-d2eGFP transgenics but selected luciferase line 49 and eGFP line 455 for further studies due to the integrity of the BAC construct at a single insertion point in the rat genome. Immunocytochemistry results and real-time qPCR taken together show that endogenous rat prolactin and the reporter genes (luciferase and d2eGFP) are expressed in lactotropes cells at good levels. Given the high transgene copy number in the PRL-Luc49 line, male transgenic rats showed a marked reduction in endogenous rat prolactin mRNA compared to the luciferase mRNA. Both pituitaries and plasma of transgenic rats were tested for production of human prolactin and results were negative. Having excluded negative feedback by human prolactin on endogenous rat prolactin production, one could speculate that given the sexual dimorphism in the expression of the pituitary specific transcription factor Pit-1 (28), transcriptional squelching could occur in male pituitaries, but to at a lesser extent in female pituitaries. Indeed, silencing of gene expression has been reported for large transgenes when present in higher copy numbers (8-14 copies) (29). The reduced production of endogenous rat PRL in male animals could account for the highly significant reduced number of lactotropes in luciferase transgenic male pituitaries. In PRL-d2eGFP animals the low copy number of the transgene, is consistent with a normal expression of the endogenous rat PRL as shown by the qPCR. A similar, but low proportion of male and female lactotropes co-express the two genes; this could be possibly due to different half-lives of the proteins, differential expression of the human and rat prolactin promoters, or stochastic expression of individual prolactin loci under condition of low stimulation and in different lactotropes subtypes in the pituitary.
The bioluminescent transgenic rats are valuable tools for studying gene expression in vivo, because in spite of the intracranial location of the pituitary gland, high quality images were obtained with short integration times (30-60 sec) in both males and females. While the signal resolution was low when imaged through intact tissue (and a precise localisation of the light output could not be determined), the removal of the skull and the brain revealed pituitary specific localisation of luciferase expression. Studies on cultured transgenic pituitary cells revealed striking fluctuations in promoter activity, and more detailed ex-vivo studies will be important to further characterise these transcription profiles.
Extra-pituitary expression of prolactin promoter activity was dramatically visualised in BAC transgenic animals when the animals were challenged with an i.p. injection of LPS, and we confirmed that this arose from alternate promoter usage in vivo. LPS is the major virulence factor of Gram-negative bacteria and great progress has been made in the elucidation of LPS recognition and signalling in mammalian cells (30). Recognition of LPS by macrophages leads to the rapid activation of an intracellular signalling pathway, which results in the release of pro-inflammatory mediators and the recruitment of humoral and cellular components of the immune system. In vivo imaging of male luciferase transgenic and luciferase/d2eGFP double-transgenic rats, 16 h after the injection of LPS, showed luciferase signal in the abdominal area and also in the mediastinum, mandibular and axillary lymph nodes. The double transgenic rat poses a real advantage in the study of the involvement of PRL in the immune response by combining in vivo imaging with ex vivo analysis. Flow cytometry analysis has revealed a population of eGFP-expressing cells in the peritoneal exudate of LPS treated rats, which has been confirmed by confocal imaging with the suggestion that diverse cells types express d2eGFP. These data suggest that these rat models will therefore allow the in vivo and ex vivo study of the role of human prolactin in immune related processes, clarifying its function in autocrine and paracrine immuno-modulation. Moreover, we have developed a unique approach for monitoring inflammation processes in vivo in real-time, and this could facilitate the screening of drugs with anti-inflammatory potential. Despite the recognition of PRL as an immune cytokine, its synthesis by the immune system is very low and resting cells normally require activation to express PRL (12). Expression of luciferase and d2eGFP was observed by qPCR in thymus and spleen of un-stimulated male and female transgenic rats, but endogenous rat Prl gene expression was undetectable. This observation confirms the conflicting evidence regarding PRL gene expression in the rodent immune system, where it could be both weak and transient. Conversely, stronger evidence is available for human prolactin gene expression in T lymphocytes and peripheral blood lymphocytes (PBL) (12) as confirmed here by the expression of luciferase in thymus and spleen of un-stimulated rats. We have analysed gene expression in vivo and ex vivo in two rat models with independent insertion sites of the BAC transgene and with different copy numbers and appreciated luciferase activity and fluorescence emission in cells of the immune system in both models; therefore, these observations suggest that the transgenic rat models we have generated are humanised model organisms that could be very valuable for studying human prolactin gene regulation in vivo under physiological and pathological conditions; further studies are now required to characterise the reporter gene expression in leukocytes.
Interestingly, the extra-pituitary mRNA transcribed in thymus and spleen is 148bp longer than the previously described and most common extra-pituitary mRNAs (5, 6). However, it is of note that a less abundant human decidual PRL cDNA has been reported with the same structure (21).
In conclusion, we have generated humanised reporter-gene transgenic rats for whole body imaging and ex vivo study of human prolactin gene expression. These models combine the advantages of BAC recombineering for appropriate regulation of gene expression and of reporter gene activity for the study of highly dynamic promoter regulation in vivo and ex vivo in different tissues. Moreover, it offers the benefit of a heterologous system for the characterisation of human-specific prolactin gene expression in a rat. To our knowledge, these are the first BAC transgenic rats for in vivo imaging of gene expression. Given the limitations of the prolactin and prolactin receptor knock-out mouse for the understanding of human prolactin gene function, these transgenic rats provide the first in vivo model to accurately study the physiological regulation of the human prolactin gene.
Methods
Transgene construction
BAC RP11-237G3 (CHORI, http://www.chori.org/) was selected. For the generation of the luciferase recombination cassette, a 5′ homology arm (234-bp) was amplified using primers #1 and #2 from human BAC RP11-237G3 and using primers #3 and #4 (see supplemental table 1) for the 3′ (227-bp) homology arm. The homology arms were designed into the human prolactin 5′UTR and the first intron, so to replace the starting ATG of the human prolactin gene (in exon 1b) with the starting codon of the reporter gene. The FRT-Neo-FRT cassette was PCR-amplified from the pIGCN21 (15) plasmid (kindly provided by Neal Copeland). The PCR fragments were sub-cloned into pGL3-promoter plasmid (see supplemental figure 1a). The luciferase/kanamycin targeting cassette was released from the pGL3-promoter plasmid by a HindIII/SalI digestion, gel purified and used for targeting RP11-273G3 BAC by recombineering (15) in EL250 bacterial strain according to the protocol available at http://recombineering.ncifcrf.gov/Protocol.asp. In a second step of recombination the kanamycin gene was removed after arabinose induction of the cells.
Recombinant PRL-Luc BAC clones were screened by southern blot hybridisation after EcoRI digestion (see supplemental figure 1b). For the generation of the d2eGFP recombination cassette, the pGL3-promoter plasmid containing the luciferase/kanamycin cassette was targeted with a double-strand linear cassette containing d2eGFP together with a tetracycline gene flanked by lox-P sites. The luciferase gene was substituted by the d2eGFP/tetracycline cassette by homologous recombination in EL350 cells (15). In the second step of recombination the tetracycline gene was removed after arabinose induction (see supplemental figure 2).
The new d2eGFP/kanamycin cassette was then released from the pGL3-promoter plasmid by a HindIII/SalI digestion, gel purified and used for targeting RP11-273G3 BAC by recombineering (15) as described above.
Cell culture and generation of stable transfected BAC cell lines
The construction of the GH3/hPRL-luc (D44) cell line was described previously(2). Cells were maintained in DMEM with pyruvate/glutamax (Life Technologies Inc) and 10% FCS (Harlan Sera-Lab).
For the generation of the PRL-Luc BAC cell line, GH3 cells were (107 cells/ml) were mixed with 30 μg of BAC-DNA (after Kanamycin being substituted by neomycin in the BAC construct) and transfected using an Easyjet Plus (Equibio) electroporation system (250V, 600μF). Stable transfectant clones were selected and re-cloned using G418 (500 μg/ml). The clones were screened for luciferase expression and responses to stimuli (17β-estradiol 10nM, forskolin 1μM, FGF-2 10ng/ml, TRH 300nM, dexamethasone 10nM, TNF-α 10ng/ml).
Production of transgenic rats
Recombined RP11-237G3 BAC inserts were freed by a NotI digestion and purified by preparative pulse field gel electrophoresis, β-agarase treatment (New England Biolabs), and dialysis against injection buffer (10 mM Tris-HCl, pH 7.5 and 0.1 mM EDTA with 100 mM NaCl)(31). BAC DNA (1 μg/ml) was microinjected into the pronucleus of Fisher 344 rats (Harlan,UK) zygotes. Five transgenic PRL-Luc positive founders were identified by PCR using primers #5 and #6 (S Table 1) and southern blot hybridisation. Primers #14 and #15 (see supplemental table 1) were used instead to identify PRL-d2eGFP positive founders by PCR. Animal studies were undertaken under UK Home Office Licence, following review by local ethics committee. All rats were housed individually, given free access to water and standard commercial rat chow (Special Diet Service, Witham, Essex UK), and maintained under controlled conditions of temperature (21±1°C) and humidity (50±10%), under a 12 h light/dark cycle. Female rats were treated with 0.04 mg of LHRH (Sigma-Aldrich) at day -3 to synchronise their estrus cycle before real-time qPCR.
Cell imaging
Either collagenase type I dispersed pituitaries or GH3 BAC-transfected cells (105) were cultured in 10% foetal calf serum, and then serum starved for 24 h before imaging. Luciferin (1mM; Bio-Synth) was added at least 10 h before the start of the experiment, and the cells were transferred to a Zeiss (Welwyn Garden City) Axiovert 100M in a dark room. The cells were maintained at 37° C with 5% CO2-95% air. Bright-field images were taken before and after luminescence imaging. Luminescence images were obtained using either a Fluar 20x, 0.75 NA objective and captured using a photon-counting charge-coupled device camera (Orca II; Hamamatsu Photonics, Figure 2) or a Fluar 10x, 0.5 NA objective using a photon-counting Hamamatsu 2-stage VIM intensified camera (Figure 4). Sequential images, each integrated over 30 min, were taken using 4 × 4 binning and analysed using Kinetic Imaging software AQM6 (Andor, Belfast, UK). Regions of interest were drawn around each single cell, and total photon counts for individual cell areas obtained from each image.
Mean intensity data were collected after the average instrument noise (corrected for the number of pixels being used) was subtracted from the luminescence signal.
FISH
Rat spleen cells were cultured in RPMI 1640 medium/10% foetal calf serum with LPS (O111:B4, Sigma-Aldrich) 25μg/ml, Concanavalin A (Sigma-Aldrich) 5μg/ml and β-Mercaptoethanol 0.5%. After 48 h, colcemid (0.25 μg/ml) was added for 60 min. Standard techniques for hypotonic treatment, methanol/acetic acid fixation, and slide preparation were used (32).
FISH was performed as previously described (33) using biotin 16-dUTP (Roche) labelled PRL-Luc BAC DNA a 1:500 dilution of an avidin-FITC (Vector Laboratories) antibody, followed by 30° min incubation with an anti-avidin biotin antibody (1:100) (Vector Laboratories) and a second incubation with the avidin-FITC antibody. Slides were finally mounted in Vectashield (Vector Laboratories) containing DAPI counter-stain.
Immunocytochemistry
Immunofluorescent staining was performed as described previously (34) using the following antibodies: rabbit anti-ovine prolactin (MCNR51) (35), mouse anti-luciferase (P.pyralis) (Invitrogen/Zymed) and a monoclonal antibody against GFP (Aequorea victoria) (Clontech) were used at 1:4000 and 1:50 and 1:100 respectively. All tissues were blocked using goat serum (NGS; Diagnostics Scotland) diluted 1:5 in PBS (phosphate buffered saline, pH7.4) containing 5% bovine serum albumin (BSA) prior to the addition of the primary antibody. All primary antibodies were incubated overnight at 4°C. For the direct detection of prolactin in PRL-Luc rats, goat anti-rabbit Alexa fluor 488 (1:200, Molecular Probes) was used. However, indirect detection using biotinylated goat anti-mouse IgG (1:500 Vector Laboratories) followed by Streptavidin Alexa 546 (1:200; Molecular Probes) was used to reveal luciferase. For the detection of prolactin in PRL-d2eGFP rats a goat anti-rabbit peroxidase (1:200, Dako) was used followed by tyramide Cy3 (1:50, PerkinElmer). However, antigen retrieval and indirect detection with goat anti-mouse biotinylated (1:200, Dako) followed by avidin Alexa 488 (1:200, Invitrogen) was performed for detecting eGFP. Finally, sections were mounted using Permafluor fluorescent mounting medium then examined using a Zeiss LSM510 confocal (Carl Zeiss). For each section, 4 fields were examined at random and the number of bi-hormonal prolactin/luciferase and prolactin-only cells were determined using Image-Pro Plus analysis software (Media Cybernetics UK) (n=8, total number of field counted for each sex).
RT-PCR and qPCR
Total mRNA was extracted from rat pituitaries, thymus (parathymic lymph nodes) and spleen using the TRIzol reagent (Invitrogen). First strand cDNA was synthesised using random octamers (8-mers) and Superscript II (Invitrogen) on 2μg total RNA.
Prolactin specific primers and probe were #7, #8 and #9; luciferase specific primers and probe were #10 #11 and #12; d2EGFP specific primers and probes were #16, #17 and #18 (see supplemental table 1). RT-PCR for the detection of extra-pituitary mRNA was performed using forward primer 98 (6) in prolactin exon 1a and reverse primers #13 in prolactin 5′UTR and #11 in the luciferase gene (supplemental Table 1).
Real-time quantitative PCR (qPCR) was performed as described previously (36) on a LightCycler 480 System (Roche). Each sample was analysed in triplicate along with no template controls. The data were normalised to 18S gene expression.
In vivo studies
Adult male and female rats from line PRL-Luc49 and from double-transgenics line PRL-Luc/d2EGFP were analysed. PRL-Luc transgene expression was monitored in vivo using the approach of biophotonic imaging (16).
Photons were detected non-invasively using a sensitive in vivo imaging system (IVIS Spectrum, Caliper/Xenogen). Rats were anaesthetised with isoflurane and injected with luciferin (Biosynth) (150 mg/kg i.p. injection) 10 min before imaging. Images were taken using the following parameters: exposure time 30-60 sec, binning 4-8, no filter, f-stop 1, variable FOV 6.6-26. Images were analysed by using Living Image Software (Caliper/Xenogen).
Adult PRL-Luc transgenic rats were treated with either 3mg/kg LPS O127:B8 (Sigma-Aldrich) or physiological saline by intra-peritoneal (i.p.) injection and imaged 16 h later.
Flow cytometry analysis
Peritoneal exudate was recovered from double-transgenic rats in PBS/Heparin (5U/ml) solution. Red blood cells were lysed with a 1mM NH4HCO3 and 114 mM NH4Cl solution. The cells were then fixed in 2%formaldehyde in PBS solution and analysed with a FACSVantage SE (Becton Dickinson) apparatus.
Statistical Analysis
Results for real-time PCR are shown as arbitrary units ± s.d. and for the immunocytochemistry as mean percentage ± s.d. Statistical significance for both of these assays was determined by non-parametric two-tailed Mann-Whitney test. Results were considered significant at p<0.05.
Supplementary Material
Acknowledgements
The authors wish to thank Sharp M., Dransfield I., Rossi A. and Jacqui Horn for useful discussion, Salter D., Ramage L. for access to human samples, Featherstone K. for preparation of thick pituitary sections, Boyle S., for performing the FISH analysis and Dunbar D. for the statistical analysis.
This work was funded by a Wellcome Trust Programme Grant (# 067252). SS is the recipient of a Wellcome Trust Intermediate Fellowship (CVRI), JJM is the recipient of a Wellcome Trust PRF, CVH was for part of this work the recipient of the Professor John Glover Memorial Fellowship, ADA held an MRC PhD studentship. ASM and JRM are funded by the Medical Research Council UK. Cell imaging instrumentation was funded by BBSRC, Carl Zeiss and Hamamatsu Photonics. We also acknowledge the support of the Wellcome Trust Functional Genomics Initiative and the British Heart Foundation Centre for Research Excellence (CoRE).
References
- 1.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 2.Takasuka N, White MRH, Wood CD, Robertson WR, Davis JRE. Dynamic changes in prolactin promoter activation in individual living lactotrophic cells. Endocrinology. 1998;139:1361–1368. doi: 10.1210/endo.139.3.5826. [DOI] [PubMed] [Google Scholar]
- 3.Bole-Feysot C, Goffin V, Edery M, Binart N, Kelly PA. Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocr Rev. 1998;19:225–268. doi: 10.1210/edrv.19.3.0334. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Jonathan N, Lapensee CR, Lapensee EW. What Can We Learn from Rodents about Prolactin in Humans? Endocr Rev. 2008;29:1–41. doi: 10.1210/er.2007-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berwaer M, Martial JA, Davis JRE. Characterization of an up-stream promoter directing extrapituitary expression of the human prolactin gene. Mol Endocrinol. 1994;8:635–642. doi: 10.1210/mend.8.5.8058071. [DOI] [PubMed] [Google Scholar]
- 6.Gellersen B, Kempf R, Telgmann R, DiMattia GE. Nonpituitary human prolactin gene transcription is independent of Pit-1 and differentially controlled in lymphocytes and in endometrial stroma. Mol Endocrinol. 1994;8:356–373. doi: 10.1210/mend.8.3.8015553. [DOI] [PubMed] [Google Scholar]
- 7.Venter JC. A part of the human genome sequence. Science. 2003;299:1183–1184. doi: 10.1126/science.299.5610.1183. [DOI] [PubMed] [Google Scholar]
- 8.Soares MJ. The prolactin and growth hormone families: pregnancy-specific hormones/cytokines at the maternal-fetal interface. Reprod Biol Endocrinol. 2004;2:51. doi: 10.1186/1477-7827-2-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerlo S, Davis JRE, Mager DL, Kooijman R. Prolactin in man: a tale of two promoters. Bioessays. 2006;28:1051–1055. doi: 10.1002/bies.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horseman ND, Zhao W, Montecino-Rodriguez E, Tanaka M, Nakashima K, Engle SJ, Smith F, Markoff E, Dorshkind K. Defective mammopoiesis, but normal hematopoiesis, in mice with a targeted disruption of the prolactin gene. Embo J. 1997;16:6926–6935. doi: 10.1093/emboj/16.23.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouchard B, Ormandy CJ, Di Santo JP, Kelly PA. Immune system development and function in prolactin receptor-deficient mice. J Immunol. 1999;163:576–582. [PubMed] [Google Scholar]
- 12.Matera L. Endocrine, paracrine and autocrine actions of prolactin on immune cells. Life Sci. 1996;59:599–614. doi: 10.1016/0024-3205(96)00225-1. [DOI] [PubMed] [Google Scholar]
- 13.McFerran DW, Stirland JA, Norris AJ, Khan RA, Takasuka N, Seymour ZC, Gill MS, Robertson WR, Loudon AS, Davis JRE, White MRH. Persistent synchronized oscillations in prolactin gene promoter activity in living pituitary cells. Endocrinology. 2001;142:3255–3260. doi: 10.1210/endo.142.7.8252. [DOI] [PubMed] [Google Scholar]
- 14.Stirland JA, Seymour ZC, Windeatt S, Norris AJ, Stanley P, Castro MG, Loudon AS, White MRH, Davis JRE. Real-time imaging of gene promoter activity using an adenoviral reporter construct demonstrates transcriptional dynamics in normal anterior pituitary cells. J Endocrinol. 2003;178:61–69. doi: 10.1677/joe.0.1780061. [DOI] [PubMed] [Google Scholar]
- 15.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 16.Contag CH, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Qu R, Ogura M, Shibata T, Harada H, Hiraoka M. Real-time imaging of hypoxia-inducible factor-1 activity in tumor xenografts. J Radiat Res. 2005;46:93–102. doi: 10.1269/jrr.46.93. [DOI] [PubMed] [Google Scholar]
- 18.Adamson AD, Friedrichsen S, Semprini S, Harper CV, Mullins JJ, White MRH, Davis JRE. Human Prolactin Gene Promoter Regulation by Estrogen Convergence with TNF{alpha} Signaling. Endocrinology. 2008;149:687–694. doi: 10.1210/en.2007-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shorte SL, Leclerc GM, Vazquez-Martinez R, Leaumont DC, Faught WJ, Frawley LS, Boockfor FR. PRL gene expression in individual living mammotropes displays distinct functional pulses that oscillate in a noncircadian temporal pattern. Endocrinology. 2002;143:1126–1133. doi: 10.1210/endo.143.3.8682. [DOI] [PubMed] [Google Scholar]
- 20.Cheong WF, Prahl SA, Welch AJ. A review of the optical properties of biological tissues. IEEE J Quantum Electron. 1990;26:2166–2185. [Google Scholar]
- 21.DiMattia GE, Gellersen B, Duckworth ML, Friesen HG. Human prolactin gene expression. The use of an alternative noncoding exon in decidua and the IM-9-P3 lymphoblast cell line. J Biol Chem. 1990;265:16412–16421. [PubMed] [Google Scholar]
- 22.Dubchak I, Brudno M, Loots GG, Pachter L, Mayor C, Rubin EM, Frazer KA. Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 2000;10:1304–1306. doi: 10.1101/gr.142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tautz D. Evolution of transcriptional regulation. Curr Opin Genet Dev. 2000;10:575–579. doi: 10.1016/s0959-437x(00)00130-1. [DOI] [PubMed] [Google Scholar]
- 24.Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 2001;10:83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- 25.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 26.Illenye S, Heintz NH. Functional analysis of bacterial artificial chromosomes in mammalian cells: mouse Cdc6 is associated with the mitotic spindle apparatus. Genomics. 2004;83:66–75. doi: 10.1016/s0888-7543(03)00205-2. [DOI] [PubMed] [Google Scholar]
- 27.Friedrichsen S, Harper CV, Semprini S, Wilding M, Adamson AD, Spiller DG, Nelson G, Mullins JJ, White MRH, Davis JRE. Tumor necrosis factor-alpha activates the human prolactin gene promoter via nuclear factor-kappaB signaling. Endocrinology. 2006;147:773–781. doi: 10.1210/en.2005-0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez-Parra S, Chowen JA, Garcia Segura LM, Argente J. Ontogeny of pituitary transcription factor-1 (Pit-1), growth hormone (GH) and prolactin (PRL) mRNA levels in male and female rats and the differential expression of Pit-1 in lactotrophs and somatotrophs. J Neuroendocrinol. 1996;8:211–225. doi: 10.1046/j.1365-2826.1996.04526.x. [DOI] [PubMed] [Google Scholar]
- 29.Li S, Hammer RE, George-Raizen JB, Meyers KC, Garrard WT. High-level rearrangement and transcription of yeast artificial chromosome-based mouse Ig kappa transgenes containing distal regions of the contig. J Immunol. 2000;164:812–824. doi: 10.4049/jimmunol.164.2.812. [DOI] [PubMed] [Google Scholar]
- 30.Rietschel ET, Kirikae T, Schade FU, Mamat U, Schmidt G, Loppnow H, Ulmer AJ, Zahringer U, Seydel U, Di Padova F, et al. Bacterial endotoxin: molecular relationships of structure to activity and function. Faseb J. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- 31.Mullins LJ, Kotelevtseva N, Boyd AC, Mullins JJ. Efficient Cre-lox linearisation of BACs: applications to physical mapping and generation of transgenic animals. Nucleic Acids Res. 1997;25:2539–2540. doi: 10.1093/nar/25.12.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rooney DE, Czepulkowski BH. Human cytogenetics: a practical approach. IRL; Oxford: 1986. [Google Scholar]
- 33.Newsome PN, Johannessen I, Boyle S, Dalakas E, McAulay KA, Samuel K, Rae F, Forrester L, Turner ML, Hayes PC, Harrison DJ, W.A B, Plevris JN. Human cord blood-derived cells can differentiate into hepatocytes in the mouse liver with no evidence of cellular fusion. Gastroenterology. 2003;124:1891–1900. doi: 10.1016/s0016-5085(03)00401-3. [DOI] [PubMed] [Google Scholar]
- 34.Pope C, McNeilly JR, Coutts S, Millar M, Anderson RA, McNeilly AS. Gonadotrope and thyrotrope development in the human and mouse anterior pituitary gland. Dev Biol. 2006;297:172–181. doi: 10.1016/j.ydbio.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 35.Davis JRE, McVerry J, Windeatt S, Castro MG, Lowenstein PR, Lincoln GA, McNeilly AS. Cell-type specific adenoviral transgene expression in the intact ovine pituitary gland after steriotaxic delivery: an in vivo system for long-term multiple parameter evaluation of human pituitary gene therapy. Endocrinology. 2001;142:795–801. doi: 10.1210/endo.142.2.7963. [DOI] [PubMed] [Google Scholar]
- 36.Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.