Abstract
To extend understanding of the genetic architecture and molecular basis of type 2 diabetes (T2D), we conducted a meta-analysis of genetic variants on the Metabochip involving 34,840 cases and 114,981 controls, overwhelmingly of European descent. We identified ten previously unreported T2D susceptibility loci, including two demonstrating sex-differentiated association. Genome-wide analyses of these data are consistent with a long tail of further common variant loci explaining much of the variation in susceptibility to T2D. Exploration of the enlarged set of susceptibility loci implicates several processes, including CREBBP-related transcription, adipocytokine signalling and cell cycle regulation, in diabetes pathogenesis.
Type 2 diabetes (T2D) is a chronic metabolic disease with multifactorial pathogenesis1. Although the genetic contribution to T2D is well recognized, the current set of 56 established susceptibility loci, identified primarily through large-scale genome-wide association studies (GWAS)2-11, captures at best 10% of familial aggregation of the disease. The characteristics (effect sizes and risk allele frequencies (RAF)) of the variants contributing to the “unexplained” genetic variance remain far from clear. At the same time, difficulties in inferring biological mechanisms from the variants of modest effect identified by GWAS have inhibited progress in defining the pathophysiological basis of disease susceptibility. One key question is whether characterization of increasing numbers of risk loci will provide evidence, at the functional level, that susceptibility involves a limited set of molecular processes.
To extend the discovery and characterization of variants influencing T2D susceptibility, we performed large-scale genotyping using the Metabochip. This custom array of 196,725 variants was designed to facilitate cost-effective follow-up of nominal associations for T2D and other metabolic and cardiovascular traits, and to enhance fine-mapping of established loci12. The T2D-nominated component of Metabochip comprises 21,774 variants, including 5,057 “replication” SNPs that capture the strongest, independent (CEU r2 < 0.2) autosomal association signals from the GWAS meta-analysis conducted by the DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium. This genome-wide meta-analysis (“DIAGRAMv3”) includes data from 12,171 cases and 56,862 controls of European descent imputed up to 2.5 million autosomal SNPs, and augments the previously published “DIAGRAMv2” meta-analysis4 with four additional GWAS (Supplementary Table 1). The T2D-nominated content of Metabochip includes a further 16,717 variants, most chosen from 1000 Genomes Project pilot data13, to fine-map 27 established susceptibility loci.
RESULTS
Study overview
Our primary investigation combined the DIAGRAMv3 (“Stage 1”) GWAS meta-analysis with a “Stage 2” meta-analysis comprising 22,669 cases and 58,119 controls genotyped with Metabochip, including 1,178 cases and 2,472 controls of Pakistani descent (PROMIS) (Online Methods and Supplementary Table 1). There was little evidence of heterogeneity in allelic effects between European- and Pakistani-descent studies in Stage 2 (Supplementary Fig. 1), so we report the combined meta-analysis including PROMIS with genomic control correction.
T2D susceptibility loci reaching genome-wide significance
Combining Stage 1 and Stage 2 meta-analyses (Supplementary Fig. 2), we identified eight new T2D susceptibility loci at genome-wide significance (P < 5 × 10−8) (Table 1, Supplementary Fig. 3 and Supplementary Table 2). By convention, we have labelled loci according to the gene nearest to the lead SNP, unless a compelling biological candidate maps nearby. The strongest signals mapped to ZMIZ1 (P = 1.0 × 10−10), ANK1 (P = 2.5 × 10−10), and the region flanking KLHDC5 (P = 6.1 × 10−10). We also observed genome-wide significant association at HMG20A (P = 4.6 × 10−9) and GRB14 (P = 1.0 × 10−8), both implicated in a recent meta-analysis of T2D in South Asians10. Neither has previously been reported in European studies, and both remain genome-wide significant after removing PROMIS from the meta-analysis (HMG20A P = 1.9 × 10−9; GRB14 P = 5.8 × 10−9). The lead SNPs from both meta-analyses are in strong linkage disequilibrium (LD) (HMG20A r2 = 0.89 and GRB14 r2 = 0.77 in CEU), and likely represent the same association signals. At the previously unreported loci, we observed nominal evidence of association (P < 0.05) in the South Asian10 and recent East Asian11 meta-analyses for the lead SNPs at MC4R and ZMIZ1 (Supplementary Table 3), with consistent directions of effect across all three ancestry groups.
Table 1.
T2D susceptibility loci achieving genome-wide significance (combined meta-analysis P < 5 × 10−8) for the first time in European descent populations
| SNP | Chr | Position (Build 36) |
Allelesa | Risk allele frequencyb | Nearby gene | Stage 1 meta-analysis: up to 12,171 cases and 56,862 controls | Stage 2 meta-analysis: up to 22,669 cases and 58,119 controls | Combined meta-analysis: up to 34,840 cases and 114,981 controls | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk | Othe r |
OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | |||||
| New loci not previously reported in any population | ||||||||||||
| rs12571751 | 10 | 80,612,637 | A | G | 0.52 | ZMIZ1 | 1.09 (1.06- 1.13) |
7.0 × 10−7 | 1.07 (1.04- 1.10) |
1.5 × 10−6 | 1.08 (1.05-1.10) | 1.0 × 10−10 |
| rs516946 | 8 | 41,638,405 | C | T | 0.76 | ANK1 | 1.10 (1.06- 1.15) |
2.1 × 10−6 | 1.08 (1.05- 1.12) |
1.1 × 10−6 | 1.09 (1.06-1.12) | 2.5 × 10−10 |
| rs10842994 | 12 | 27,856,417 | C | T | 0.80 | KLHDC5 | 1.09 (1.04- 1.13) |
3.0 × 10−4 | 1.10 (1.07- 1.14) |
2.8 × 10−8 | 1.10 (1.06-1.13) | 6.1 × 10−10 |
| rs2796441 | 9 | 83,498,768 | G | A | 0.57 | TLE1 | 1.07 (1.03- 1.12) |
4.8 × 10−4 | 1.07 (1.04- 1.10) |
3.3 × 10−7 | 1.07 (1.05-1.10) | 5.4 × 10−9 |
| rs459193 | 5 | 55,842,508 | G | A | 0.70 | ANKRD55 | 1.05 (1.01- 1.10) |
2.7 × 10−2 | 1.10 (1.06- 1.13) |
2.0 × 10−9 | 1.08 (1.05-1.11) | 6.0 × 10−9 |
| rs10401969 | 19 | 19,268,718 | C | T | 0.08 | CILP2 | 1.13 (1.05- 1.21) |
9.2 × 10−4 | 1.14 (1.08- 1.20) |
2.0 × 10−7 | 1.13 (1.09-1.18) | 7.0 × 10−9 |
| rs12970134 | 18 | 56,035,730 | A | G | 0.27 | MC4R | 1.08 (1.03- 1.12) |
2.3 × 10−4 | 1.08 (1.05- 1.11) |
2.0 × 10−6 | 1.08 (1.05-1.11) | 1.2 × 10−8 |
| rs7202877 | 16 | 73,804,746 | T | G | 0.89 | BCAR1 | 1.15 (1.07- 1.23) |
5.0 × 10−5 | 1.10 (1.05- 1.15) |
1.9 × 10−5 | 1.12 (1.07-1.16) | 3.5 × 10−8 |
| Loci not previously reported in European descent populations | ||||||||||||
| rs7177055 | 15 | 75,619,817 | A | G | 0.68 | HMG20A | 1.08 (1.04- 1.12) |
1.2 × 10−4 | 1.08 (1.05- 1.11) |
8.8 × 10−7 | 1.08 (1.05-1.10) | 4.6 × 10−9 |
| rs13389219 | 2 | 165,237,122 | C | T | 0.60 | GRB14 | 1.05 (1.01- 1.09) |
1.3 × 10−2 | 1.09 (1.06- 1.12) |
9.5 × 10−9 | 1.07 (1.05-1.10) | 1.0 × 10−8 |
Chr, chromosome; OR, odds ratio; CI, confidence interval.
Alleles are aligned to the forward strand of NCBI Build 36.
Weighted mean frequency of T2D risk allele across Stage 2 studies.
Several of these signals map to loci previously implicated in T2D-related metabolic traits (Supplementary Table 4). The lead SNP at MC4R is in strong LD with variants associated with BMI14,15 (CEU r2 = 0.80) and triglycerides16 (CEU r2 = 0.84) and is associated with waist circumference and insulin resistance17. As with FTO, the T2D-effect at MC4R is probably secondary to the BMI association. The lead SNP at GRB14 is highly correlated with variants associated with waist-hip ratio (WHR)18 and high-density lipoprotein (HDL) cholesterol16 (CEU r2 = 0.93). At CILP2, the lead SNP for T2D is also associated with triglycerides, low-density lipoprotein (LDL) and total cholesterol16. In contrast, the previously-reported association signals for haemoglobin A1C (HbA1C) levels19 near ANK1 are both independent (CEU r2 < 0.01) of the lead T2D SNP from our meta-analysis. Given the role played by rare ANK1 mutations in hereditary anemias, the HbA1C associations at this locus were assumed to be driven by abnormal erythrocyte development and/or function. However, our newly discovered independent association with T2D (in cohorts where HbA1C was not used for diagnosis) suggests that variation at this locus also has direct effects on glucose homeostasis.
Insights into the genetic architecture of T2D
The associated lead variants at the eight newly identified loci were common (Stage 2 RAF 0.08-0.89) and had modest effects on T2D susceptibility (allelic odds ratios (OR) 1.07-1.14). Under a multiplicative model within and between variants, the sibling relative risk attributable to lead SNPs rose from λS = 1.093 at the 55 previously described autosomal T2D loci represented on Metabochip (DUSP9 on chromosome X is not captured) to λS = 1.104 after inclusion of the eight newly discovered loci (Supplementary Table 5). Assuming a T2D population prevalence of 8%, these 63 newly discovered and established autosomal loci together account for 5.7% of variance in disease susceptibility, as calculated by transforming dichotomous disease risk onto a continuous liability scale20 (Online Methods).
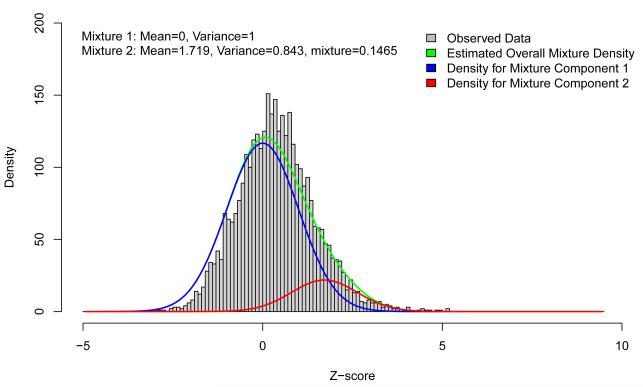
To determine the extent to which additional common variant associations contribute to the overall variance explained, we compared directional consistency in allelic effects between the two stages of the meta-analysis. Figure 1 presents the distribution of Z-scores from Stage 2, aligned to the risk allele from Stage 1, at a subset of 3,412 independent (CEU r2 < 0.05) T2D replication variants that excludes lead SNPs and possible proxies (CEU r2 ≥ 0.1) at the 63 newly discovered and established loci represented on Metabochip. The blue curve represents the expected distribution of Stage 2 Z-scores under the null hypothesis of no association. There is a clear shift in the observed distribution, corresponding to closer agreement in the direction of allelic effect than expected by chance: 2,172 (69.1%) of the 3,412 SNPs are concordant (binomial test P = 2.0 × 10−104). For comparison, we examined T2D association patterns in 2,707 independent replication SNPs for QT-interval, the trait showing weakest correlation with T2D susceptibility among those contributing to Metabochip and found far less directional consistency (54.4%, binomial test P = 3.3 × 10−6). This modest enrichment most likely reflects weak overlap of risk alleles between the two traits, since exclusion of SNPs mapping within 300 kb of directionally consistent T2D replication variants reduced this excess (52.5%, binomial test P = 0.060).
Figure 1.
Distribution of Z-scores from the Stage 2 meta-analysis, aligned to the risk allele from Stage 1. Z-scores were calculated at a subset of 3,412 independent T2D replication SNPs (CEU r2 < 0.05), excluding the 63 established and newly discovered autosomal susceptibility loci represented on Metabochip. The Z-score distribution is a mixture of: (i) the “null distribution” of SNPs having no effect on T2D (blue curve); and (ii) the “alternative distribution” of SNPs associated with the disease (red curve).
The observed distribution of Z-scores can be considered a mixture of: (i) the “null distribution” of SNPs having no effect on T2D; and (ii) the “alternative distribution” of T2D-associated SNPs (Online Methods). We estimated the features of this alternative distribution (red curve) and noted that addition of this class of SNPs significantly improved the fit to the observed Z-scores over the null model. Using simulations, based on parameter estimates from this mixture model, we estimated that 488 (95% confidence interval (CI) 456-521) of the independent replication SNPs, in addition to the 63 newly discovered and established loci, are associated with T2D susceptibility. For comparison, we undertook false-discovery rate (FDR) analysis of the 64,646 SNPs on the Metabochip selected for replication of any trait, using P-values from the combined meta-analysis (Online Methods). We observed broad agreement between combined meta-analysis P-values, FDR Q-values and the posterior probability of alternative distribution membership from the mixture model (Supplementary Fig. 4).
We were concerned that these additional, weaker association signals might reflect subtle stratification effects not eliminated by genomic control correction. However, using diverse European populations from the 1000 Genomes Project13 (Online Methods), we found no evidence that directionally-consistent T2D replication SNPs differed from other Metabochip replication SNPs with respect to FST (P = 0.88).
As expected, the estimated allelic ORs of the 488 SNPs are modest (1.01-1.11 in Stage 2), and larger samples would be required to establish association at genome-wide significance. For example, by simulating an additional 100,000 T2D cases and 100,000 controls as a “third stage” to the combined meta-analysis, we calculate that only ~37% of the 488 replication SNPs in the alternative distribution would achieve this threshold. We estimate that these variants jointly account for λS = 1.088 (95% CI 1.083-1.094), increasing the overall liability-scale variance explained to 10.7% (10.4-11.0%).
Additional sources of variation contributing to susceptibility
These estimates likely set a lower bound to the overall liability-scale variance attributable to common SNPs. The mixture model does not take account loci not represented by Metabochip T2D replication SNPs due to failures in array design or manufacture or because the association signal in DIAGRAMv3 was too weak to merit inclusion. Indeed, the latter applied to two of the genome-wide significant loci, ANKRD55 and GRB14, which were nominated for inclusion on Metabochip because of associations with WHR (ANKRD55 and GRB14), blood pressure (ANKRD55) and plasma lipid concentrations (GRB14), rather than T2D.
To estimate the contribution to the variance explained by common variants genome-wide, we undertook polygenic mixed linear modelling analyses using GCTA21,22 in two DIAGRAMv3 GWAS data sets: DGI (1,022 cases, 1,075 controls) and WTCCC (1,924 cases, 2,938 controls). The estimated liability-scale variance explained by the full set of GWAS SNPs was consistent between the two studies: 62.6% for DGI (95% CI 38.1-87.1%) and 63.9% for WTCCC (95% CI 52.1-75.8%). These results are similar to those obtained from a complementary method integrating polygenic risk score analysis and approximate Bayesian computation23 applied to the DIAGRAMv2 meta-analysis4, which estimated that ~49% of liability-scale variance was explained by common variants genome-wide. These data indicate that a substantial proportion of the variation in T2D risk is captured by common variant association signals that, individually, lie beyond unequivocal detection in single SNP analyses.
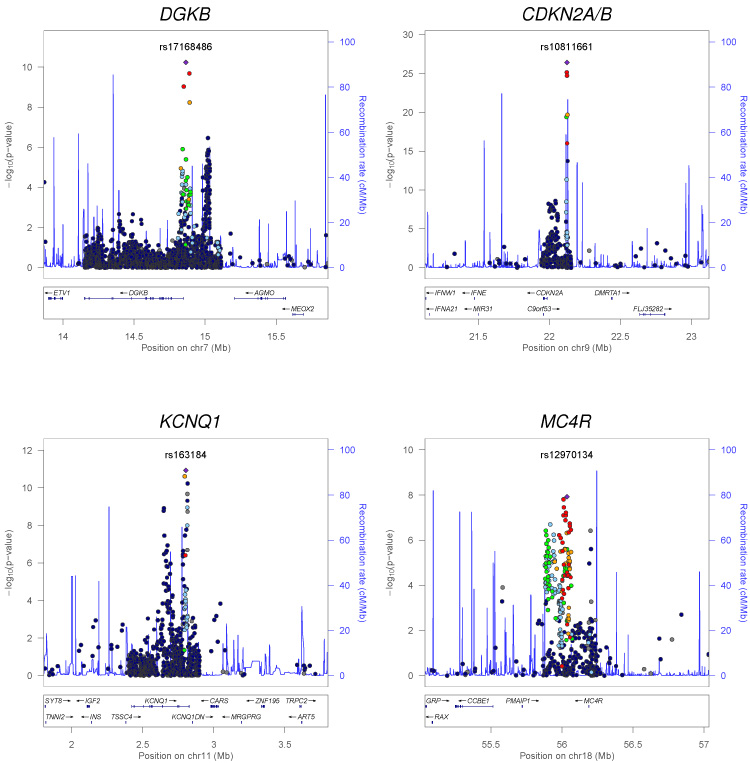
The DIAGRAMv2 meta-analysis4 had provided some evidence for loci harboring multiple independent association signals. To understand the extent to which additional variance might be attributable to multiple variants at established and newly discovered loci, we extended these analyses, focusing on the detection of independent (CEU r2 < 0.05) association signals that lie outside the recombination interval containing the lead SNP (Supplementary Table 2). We detected two loci at which multiple independent association signals attained genome-wide significance: KCNQ1 (rs163184, P = 1.2 × 10−11; rs231361, P = 1.2 × 10−9; CEU r2 = 0.01) and CDKN2A/B (rs10811661, P = 3.7 × 10−27; rs944801, P = 2.4 × 10−9; CEU r2 = 0.01) (Fig. 2). Both signals at KCNQ1 have previously been reported in East Asian and European populations4,24. However, the secondary signal at CDKN2A/B, which maps to the non-coding CDKN2B-AS1 (ANRIL) transcript, has not previously been implicated in T2D susceptibility. This signal is independent of the previously reported haplotype effect at the primary T2D signal at this locus, which is itself likely due to the phase relationships between two clades of partially correlated variants25,26. We also observed putative independent associations (P < 10−5) at DGKB (rs17168486, P = 5.9 × 10−11; rs6960043, P = 3.4 × 10−7; CEU r2 = 0.01) and MC4R (rs12970134, P = 1.2 × 10−8; rs11873305, P = 3.8 × 10−7; CEU r2 = 0.02). These results suggest that multiple independent association signals are widespread at T2D susceptibility loci. Imputation up to the more complete reference panels emerging from the 1000 Genomes Project13 and recently developed approaches that support approximate conditional analyses using meta-analysis summary level data27 will be important tools for documenting the full extent of such effects, especially where the variants map to the same recombination interval.
Figure 2.
Regional plots of T2D susceptibility loci with evidence of multiple association signals. Each point represents a Metabochip SNP passing quality control in our combined meta-analysis, plotted with their P-value (on a -log10 scale) as a function of genomic position (NCBI Build 36). In each panel, the lead SNP is represented by the purple diamond. The color coding of all other SNPs (circles) indicates LD with the lead SNP (estimated by CEU r2 from the 1000 Genomes Project June 2010 release): red r2 ≥ 0.8; gold 0.6 ≤ r2 < 0.8; green 0.4 ≤ r2 < 0.6; cyan 0.2 ≤ r2 < 0.4; blue r2 < 0.2; grey r2 unknown. Recombination rates are estimated from the International HapMap Project and gene annotations are taken from the University of California Santa Cruz genome browser.
It has been argued that common variant association signals will often reflect unobserved causal alleles of lower frequency and greater effect size28. The fine-mapping content of Metabochip allowed us to seek empirical evidence to support this “synthetic association” hypothesis. We estimate, using 1000 Genomes Project data13 applied to HapMap CEU samples, that the array captures (CEU r2 ≥ 0.8) 89.6% of common SNPs (minor allele frequency (MAF) ≥ 5%) and 60.0% of low-frequency variants (1% ≤ MAF < 5%) across Metabochip fine-mapping regions12. This represents a substantial improvement over HapMap29,30 which, across the same regions, captures 76.8% and 32.4% of common and low-frequency variants, respectively.
Across 36 fine-mapping regions on Metabochip that contain T2D susceptibility loci (including 27 explicitly chosen by DIAGRAM), we compared the characteristics of previously reported lead SNPs (defined by GWAS and HapMap imputation) and those emerging from the Stage 2 Metabochip meta-analysis. We restricted these comparisons to Stage 2 to avoid penalizing low-frequency variants not typed or well-imputed in Stage 1. The GWAS and Metabochip lead SNPs were the same, or highly-correlated (CEU r2 > 0.8), at 20 loci (15 with CEU r2 > 0.95) (Supplementary Table 6). The low LD between GWAS and Metabochip lead SNPs at DGKB and KCNQ1 (both CEU r2 = 0.00) arises because they “switch” between independent association signals at these loci (Fig. 2). For the remaining 14 loci, there was only modest LD between the previously reported GWAS and Metabochip-defined lead SNPs (CEU r2 between 0.06 and 0.77). However, at only two loci did the lead SNP after Metabochip fine-mapping have substantially lower MAF and higher OR than the previously reported GWAS lead SNP: PROX1 (rs17712208, MAF = 0.03, OR = 1.20; rs340874, MAF = 0.48, OR = 1.06) and KLF14 (7-130116320, MAF = 0.02, OR = 1.10; rs972283, MAF = 0.48, OR = 1.01). Since coverage across Metabochip fine-mapping regions is incomplete, we cannot unequivocally exclude the presence of causal low-frequency alleles at any single locus. However, the paucity of low-frequency candidate alleles across 36 loci suggests that most causal variants at these loci are common. A contribution of even rarer causal alleles (too rare to be represented on Metabochip) is also unlikely because the substantial effect sizes required to drive common variant association signals are inconsistent with the modest familial aggregation of T2D23. This interpretation, favoring common causal alleles, is in agreement with the observed consistency of T2D risk variant associations across major ancestry groups31.
Sex-differentiated analyses
We performed sex-differentiated meta-analysis32 (Online Methods and Supplementary Figs. 5 and 6) to test for association of each SNP with T2D, allowing for heterogeneity in allelic effects between males (20,219 cases, 54,604 controls) and females (14,621 cases, 60,377 controls), thereby identifying two additional loci achieving genome-wide significance (Table 2 and Supplementary Table 7). The association signal mapping near CCND2 is most significant in males (male P = 1.1 × 10−9, female P = 0.036, heterogeneity P = 0.013), while that upstream of GIPR is most significant in females (female P = 2.2 × 10−7, male P = 0.0037, heterogeneity P = 0.057) (Supplementary Fig. 7). The lead sex-differentiated SNP in GIPR is only weakly correlated with previously reported associations with BMI15 (CEU r2 = 0.06) and two-hour glucose levels33 (CEU r2 = 0.07) (Supplementary Table 4).
Table 2.
T2D susceptibility loci with sex-differentiated evidence of association
| SNP | Chr | Position (Build 36) |
Allelesa | Risk allele frequencyb | Nearby gene | Male meta-analysis: up to 20,219 cases and 54,604 controls | Female meta-analysis: up to 14,621 cases and 60,377 controls | Sex-differentiated meta-analysis: up to 34,840 cases and 114,981 controls | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk | Othe r |
OR (95% CI) | P-value | OR (95% CI) | P-value | Association P- value |
Heterogeneity P- value |
|||||
| New loci identified through sex-differentiated meta-analysis achieving genome-wide significance (P < 5 × 10−8) | ||||||||||||
| rs11063069 | 12 | 4,244,634 | G | A | 0.21 | CCND2 | 1.12 (1.08-1.16) | 1.1 × 10−9 | 1.04 (1.00-1.09) | 3.6 × 10−2 | 9.8 × 10−10 | 1.3 × 10−2 |
| rs8108269 | 19 | 50,850,353 | G | T | 0.31 | GIPR | 1.05 (1.02-1.08) | 3.7 × 10−3 | 1.10 (1.06-1.14) | 2.2 × 10−7 | 2.1 × 10−8 | 5.7 × 10−2 |
| Other loci with nominally significant evidence (P < 0.05) of heterogeneity in allelic odds ratios between sexes | ||||||||||||
| rs163184 | 11 | 2,803,645 | G | T | 0.50 | KCNQ1 | 1.12 (1.09-1.16) | 8.5 × 10− 15 |
1.05 (1.01-1.08) | 7.8 × 10−3 | 2.4 × 10−15 | 1.3 × 10−3 |
| rs17168486 | 7 | 14,864,807 | T | C | 0.19 | DGKB | 1.15 (1.11-1.19) | 6.5 × 10− 13 |
1.06 (1.02-1.11) | 5.2 × 10−3 | 1.2 × 10−13 | 6.8 × 10−3 |
| rs3923113 | 2 | 165,210,095 | A | C | 0.63 | GRB14 | 1.05 (1.01-1.08) | 4.9 × 10−3 | 1.11 (1.08-1.15) | 1.8 × 10−9 | 2.6 × 10−10 | 8.0 × 10−3 |
| rs243088 | 2 | 60,422,249 | T | A | 0.45 | BCL11A | 1.10 (1.06-1.13) | 6.5 × 10− 10 |
1.04 (1.00-1.07) | 2.8 × 10−2 | 4.7 × 10−10 | 1.2 × 10−2 |
Chr, chromosome; OR, odds ratio; CI, confidence interval.
Alleles are aligned to the forward strand of NCBI Build 36.
Weighted mean frequency of T2D risk allele across Stage 2 studies.
The sex-differentiated analyses also revealed nominal evidence of heterogeneity (P < 0.05) at four established T2D susceptibility loci (Table 2 and Supplementary Tables 7 and 8): KCNQ1 (P = 0.0013), DGKB (P = 0.0068) and BCL11A (P = 0.012) were most significantly associated in males, and GRB14 (P = 0.0080) in females. The sex-differentiated association at GRB14 is consistent with the female-specific effect on WHR observed at this locus18. As KCNQ1 and DGKB demonstrate multiple independent associations in the sex-combined meta-analysis, we investigated whether sex differences in allelic effects were consistent across these signals (Supplementary Fig. 8). This appeared true for DGKB (rs17168486, male P = 6.5 × 10−13, female P = 0.0052; rs6960043, male P = 7.9 × 10−7, female P = 0.015), but not KCNQ1 (rs163184, male P = 8.5 × 10−15, female P = 7.8 × 10−3; rs231361, male P = 2.9 × 10−6, female P = 2.9 × 10−6).
Understanding the biology of T2D susceptibility loci
For most T2D susceptibility loci, the underlying causal variants and the genes through which they act are yet to be identified, and the pathophysiological processes mediating disease risk remain unclear. We applied a variety of approaches to the newly discovered and established T2D susceptibility loci, and in some cases to putative loci with more modest evidence of association, to identify mechanisms involved in disease pathogenesis.
Physiological analyses
As noted earlier, lead SNPs at several newly identified loci are in strong LD with variants associated with other T2D-related metabolic traits. To gain a more complete picture of patterns of trait overlap, we first assessed the effect of T2D risk alleles on glycemic traits in European-descent meta-analyses from the MAGIC Investigators (Online Methods). Fasting glucose associations were analyzed for up to 133,010 non-diabetic individuals with GWAS and/or Metabochip data34. In addition to the nine loci previously reported (MTNR1B, DGKB, ADCY5, PROX1, GCK, GCKR, TCF7L2, SLC30A8 and C2CD4A)4,5, four more T2D association signals were genome-wide significant for fasting glucose: CDKN2A/B (P = 5.7 × 10−18), ARAP1 (P = 1.2 × 10−10), IGF2BP2 (P = 1.8 × 10−8) and CDKAL1 (P = 2.0 × 10−8) (Supplementary Table 9). The ZBED3 locus also attained genome-wide significance with fasting glucose after adjustment for BMI (P = 1.2 × 10−8). In contrast, lead T2D SNPs at 27 of the newly discovered and established loci showed no evidence of association with fasting glucose (P > 0.05), despite sample sizes ranging from 38,424 to 132,999 individuals (Supplementary Table 10 and Supplementary Fig. 9). Lead T2D SNPs at the remaining 24 loci were nominally associated with fasting glucose (P < 0.05), all with directionally consistent effects. These data extend previous reports indicating that the genetic landscape of pathological and physiological variation in glycemia is only partially overlapping, and are consistent with reciprocal analyses reported in the companion MAGIC paper34.
Second, we extended our previous analysis4 of the physiological consequences of T2D risk alleles to include the newly identified loci. We used the published MAGIC meta-analysis (up to 37,037 non-diabetic individuals) of HOMA indices of beta-cell function and insulin sensitivity5 as these traits were not included in the enlarged Metabochip study34. The risk allele at ANK1 has features (nominally significant reduction in HOMA-B) indicating a primary effect on beta-cell function, whereas those at GRB14 and AKNRD55 are characteristic of loci acting primarily through insulin resistance (increased HOMA-IR) (Supplementary Fig. 10 and Supplementary Table 10). The results for GRB14 are consistent with its broad impact on insulin-resistance related traits (described below), while at AKNRD55, these analyses point to MAP3K1, encoding MEK kinase, a key component of the insulin-signalling pathway, as the stand-out local candidate.
Next, we examined the effect of T2D risk alleles on anthropometric and lipid traits using data from the GIANT Consortium (up to 119,600 individuals after excluding data from T2D case series)15 and the Global Lipids Genetics Consortium (up to 100,184 individuals)16 (Online Methods and Supplementary Tables 11 and 12). The only lead SNP to demonstrate convincing evidence of association (P < 10−5) with adiposity was at MC4R. The lead SNPs at MC4R and GRB14 show the same pattern of lipid associations (P < 10−5): reduced HDL and raised triglycerides. In contrast, the lipid associations at CILP2 and GIPR ran counter to expected epidemiological correlations: T2D risk alleles were associated with reduced triglyceride levels at both loci, and at CILP2 with reduced LDL and total cholesterol.
Finally, we noticed that the lead T2D SNP at the BCAR1 locus is genome-wide significant for type 1 diabetes (T1D)35, although risk is conferred by the opposite alleles. Across 37 T1D susceptibility loci (Supplementary Fig. 11), we observed nominal evidence (P < 0.05) of association to T2D at six. For three of these (BCAR1, GLIS3 and RAD51L1), the T1D risk allele was protective for T2D, while at the others (C6orf173, COBL and C10orf59), the effects were coincident. These data indicate that rates of diagnostic misclassification among T2D cases in our study are low, and also highlight interesting points of overlap in the processes involved in risk of, and protection from, these two major forms of diabetes.
Mapping potential causal transcripts and variants
The T2D-association signals emerging from the present meta-analysis map to regions containing many transcripts and potential functional variants. To identify promising regional transcripts, we examined expression quantitative trait locus (eQTL) data from a variety of tissues (Online Methods and Supplementary Note). At six of the newly discovered loci, the lead T2D SNP showed strong cis-eQTL associations and was highly correlated (CEU r2 > 0.8) with the lead cis-eQTL SNP (Supplementary Table 13). These “coincident” eQTL implicate GRB14 (omental fat), ANK1 (omental and subcutaneous fat, liver and prefrontal cortex), KLHDC5 (blood, T cells and CD4+ lymphocytes), BCAR1 (blood), ATP13A1 (at the CILP2 locus, blood and monocytes), HMG20A (liver) and LINGO1 (also at the HMG20A locus, adipose tissue). For those loci (GRB14, ANK1 and BCAR1) for which individual-level expression data for the appropriate tissues were available36, we confirmed signal coincidence by conditional analyses (Online Methods and Supplementary Table 14).
We used 1000 Genomes Project data13 to search for non-synonymous variants in strong LD (CEU r2 > 0.8) with lead SNPs at the newly discovered loci (Online Methods). The only candidate allele uncovered was a non-synonymous variant in exon 6 of TM6SF2 (19-19379549, CEU r2 = 0.98 with rs10401969) at the CILP2 locus. This change is predicted by SIFT37 to have no appreciable effect on protein function.
Pathway and protein-protein interaction analyses
To extend previous efforts to define pathways and networks involved in T2D pathogenesis4, we combined meta-analysis data with protein-protein interactions (PPI), semantic relationships within the published literature and annotated pathways (Fig. 3). For these analyses, we generated a “primary” list of 77 transcripts mapping nearest to lead SNPs at T2D susceptibility loci or implicated in monogenic diabetes38 (Online Methods and Supplementary Table 15).
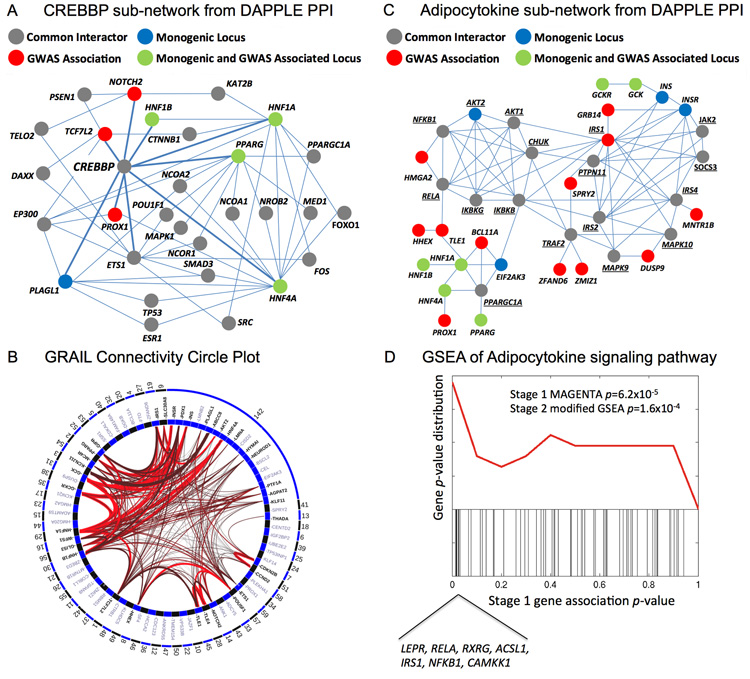
Figure 3.
Functional analyses. (a,b) Protein-protein interaction (PPI) sub-network for CREBBP and adipocytokine interactions. All direct interactions and common interactors between direct connections were extracted from the larger network of 314 proteins defined in the DAPPLE network analysis. Genes in the network are circles (nodes), colored according to the statistical relationship with T2D: common interactors between GWAS identified or monogenic loci are depicted as grey, monogenic loci (only) in blue, GWAS identified loci (only) in red, and loci with GWAS association and implicated by monogenic forms of diabetes are shown in green. Each interaction defined in the inWEB network is depicted by a line (edge) between nodes. (c) GRAIL circle plot of locus connectivity. Each locus is plotted in a circle where significant connections (P < 0.05) based on PubMed abstracts are drawn spanning the circle. Conservatively, we treated all monogenic loci (region 142) as a single locus by which connectivity is assessed. The strongest connections (P < 0.001) are colored in bright red. (d) GSEA of associations in the adipocytokine signaling pathway. The black bars represent the Stage 1 meta-analysis P-values of 63 autosomal genes in the Adipocytokine Signaling pathways (KEGG). A density plot of the black bars is depicted in the top panel (red line). The replicating genes in the leading edge of the GSEA are listed. The Stage 2 modified GSEA P = 1.6×10−4 was calculated based on both the primary and secondary transcripts using the LD locus definition.
Using a refined database of high-confidence PPI39,40, we constructed a network of 314 proteins from these 77 transcripts using DAPPLE41. We detected an excess of physical interactions in the network, both direct (between the associated transcripts themselves, P < 10−4) and indirect (via 237 shared interactors not on the list of associated transcripts, P = 0.0070). There was no evidence that this set of shared interactors was enriched for T2D-associated variants. Some interactions, such as those between the potassium channel encoding genes KCNJ11 and ABCC8, are expected, while other sub-networks are of greater novelty. For example, the transcriptional co-activator protein CREBBP, implicated in the coupling of chromatin remodelling to transcription factor recognition, does not map to any T2D susceptibility locus. However, it is the most connected gene for protein-level interactions (P < 0.005) in the PPI network, interacting with nine primary transcripts, eight implicated in monogenic diabetes or mapping to established T2D susceptibility loci (HNF1A, HNF1B, HNF4A, PLAGL1, TCF7L2, PPARG, PROX1 and NOTCH2) and one from a locus with a strong, but not genome-wide significant, association (ETS1, lead SNP rs7931302, P = 3.8 × 10−7). Other shared interactors identified through these analyses included SERTAD1, FOXO1, PPARGC1A, GRB10 and MAFA. Several of these play roles in the transcriptional regulation of diabetes-relevant tissues, and some also interact with CREBBP. We used a pre-defined set of 1,814 genes encoding “DNA-binding proteins” (Online Methods) to show that: (i) T2D signals are highly enriched for transcription factors (21 of 71 primary transcripts listed within the HGNC catalog, compared to 1,793 of 19,162, P = 2.3 × 10−6); and (ii) transcription factors within T2D loci are enriched for interaction with CREBBP (taking the 1,164 listed in the protein interaction database, 9 of 21 compared with 127 of 1,143, P = 2.7 × 10−4). These data suggest that modulation of CREBBP-binding transcription factors plays an important role in T2D susceptibility.
The same set of 77 primary transcripts showed modest evidence of excess connectivity (P = 0.020 by permutation) using text-mining approaches42 (Online Methods). When we used this set of 77 genes as a “seed” to query a list of 77 “secondary” transcripts (nearest to lead SNPs with posterior probability of T2D-association >75% from the mixture model) (Supplementary Table 15), we found significant connections (P < 0.001) between the primary associated transcripts and four other genes: LEPR (leptin obesity pathways), MYC (cell-cycle pathway), GATA6 (pancreas development pathway) and DLL4 (Notch signalling target).
We also tested for enrichment of GWAS associated transcripts in pathway data. To retain power, we focused on 16 biological hypotheses chosen for assumed relevance to T2D pathogenesis4,43-45 (Supplementary Note). We used a two-step modified gene-set enrichment analysis (GSEA) approach applied sequentially to Stage 1 (using MAGENTA46) and Stage 2 meta-analyses (Online Methods and Supplementary Table 16). Of the 16 biological hypotheses tested, two demonstrated reproducible enrichment of T2D associations. The strongest enrichment was observed for a broader set of primary and secondary transcripts mapping to T2D-associated loci in the adipocytokine signalling pathway (MAGENTA P = 6.2 × 10−5; modified GSEA P = 1.6 × 10−4). This gene set includes the adiponectin, leptin and TNF-alpha signalling pathways previously implicated in the development of insulin resistance47, but for which genome-wide significant common variant associations with T2D susceptibility have not been previously reported. This analysis highlighted eight genes in this pathway most likely to be causal for T2D susceptibility: IRS1, LEPR, RELA, RXRG, ACSL1, NFKB1, CAMKK1 and a monogenic diabetes gene AKT2. Members of this pathway were also strongly represented (17 out of 314) in the DAPPLE PPI network (P = 7.5 × 10−14). Modest but robust enrichment was also observed for genes influencing cell cycle, in particular regulators of the G1 phase during mitosis (MAGENTA P = 2.0 × 10−4; modified GSEA P = 3.0 × 10−3). The majority of genes driving these cell-cycle enrichments were cyclin-dependent kinase (CDK) inhibitors (CDKN2A/B, CDKN1C and CDKN2C) and cyclins that activate CDKs (CCNE2, CCND2 and CCNA2). Many of these regulate CDK4 or CDK6, which are known to play a role in pancreatic beta-cell proliferation48,49. We saw no evidence of enrichment for other processes implicated in T2D pathogenesis, including amyloid formation, ER stress and insulin signalling.
DISCUSSION
We have expanded T2D association analysis to almost 150,000 individuals. In so doing, we have added another 10 loci to the list of confirmed common variant signals: for several of these, we have identified strong positional candidates based on expression data and known biology. The data support the view that much of the overall variance in T2D susceptibility can be attributed to the impact of a large number of common causal variants, most of very modest effect. While such a model poses challenges for accumulating genome-wide significant evidence of association at a specific variant, it does suggest that genetic profiling based on the entirety of sequence variation has the potential to provide useful risk stratification for T2D.
If common causal alleles explain a substantial component of T2D susceptibility, the contribution of rare and low-frequency risk variants may be less than is often assumed: re-sequencing studies will soon provide empirical data to address this question. In particular, it will be important to determine whether, as the number of susceptibility loci increases, there is evidence that the pathophysiological mechanisms implicated by human genetics coalesce around a limited set of core pathways and networks. Our data suggest that this may be the case, with a variety of analytical approaches pointing to cell cycle regulation, adipocytokine signalling and CREBBP-related transcription factor activity as key processes involved in T2D pathogenesis.
ONLINE METHODS
Stage 1 meta-analysis
The Stage 1 meta-analysis consisted of 12,171 T2D cases and 56,862 controls across 12 GWAS from European descent populations (Supplementary Table 1). Samples were typed with a range of GWAS genotyping products. Sample and SNP quality control (QC) were undertaken within each study. Each GWAS was then imputed at up to 2.5 million SNPs using CEU samples from Phase II of the International HapMap Project28. Each SNP with MAF >1% passing QC was tested for association with T2D under an additive model after adjustment for study-specific covariates, including indicators of population structure. The results of each GWAS were corrected for residual population structure using the genomic control inflation factor50 and were combined via fixed-effects inverse-variance weighted meta-analysis. The results of the Stage 1 meta-analysis were subsequently corrected by genomic control (λGC = 1.10).
Stage 2 meta-analysis
The Stage 2 meta-analysis consisted of 21,491 T2D cases and 55,647 controls across 25 studies from European descent populations and 1,178 T2D cases and 2,472 controls from one study of Pakistani descent (PROMIS) (Supplementary Table 1). All samples were genotyped with Metabochip. Sample and SNP QC were undertaken within each study. Each SNP with MAF >1% passing QC was tested for association with T2D under an additive model after adjustment for study-specific covariates. We would expect inflation in association signals across the content of Metabochip, even in the absence of population structure, because it has been designed to be enriched for T2D and other T2D-related metabolic trait loci. The results of each study were thus corrected for residual population structure using the genomic control inflation factor obtained from a subset of 3,598 independent “QT-interval” SNPs (CEU r2 < 0.05), which were not expected to be associated with T2D. The Stage 2 meta-analysis was performed in two steps: (i) combine all studies of European descent; and (ii) add the PROMIS study. In both steps, the results of each study were combined via fixed-effects inverse-variance weighted meta-analysis. The results of the Stage 2 European meta-analysis were corrected by “QT-interval” genomic control (λQT = 1.19), but this adjustment was not then necessary after the addition of PROMIS (λQT = 0.99 was less than 1). Heterogeneity in allelic effects between European descent studies and subsequently between the European meta-analysis and PROMIS was assessed by means of Cochran’s Q-statistic51.
Combined meta-analysis
The results of the Stage 1 and Stage 2 meta-analyses were combined for all Metabochip SNPs via fixed-effects inverse-variance weighted meta-analysis. The combined meta-analysis consisted of 34,840 cases and 114,981 controls. This was performed in two steps: (i) combine Stage 1 meta-analysis with European descent Stage 2 meta-analysis; and (ii) add the PROMIS study. The results of the combined European meta-analysis was corrected by “QT-interval” genomic control (λQT = 1.13), but this adjustment was not necessary after the addition of PROMIS (λQT = 0.98 was less than 1) (Supplementary Fig. 12). Heterogeneity in allelic effects between the Stage 1 and Stage 2 meta-analyses was assessed by means of Cochran’s Q-statistic.
Look-up of meta-analysis results for lead SNPs in GWAS of South and East Asian descent
We obtained summary statistics (RAFs, association P-values, allelic ORs and 95% CIs) for lead SNPs at the newly discovered loci in meta-analyses of T2D GWAS in: (i) 5,561 cases and 14,458 controls of South Asian descent10, excluding 1,958 overlapping samples from PROMIS that were also included in our study, comprising 568,976 directly genotyped autosomal SNPs; and (ii) 6,952 cases and 11,865 controls of East Asian descent11, comprising 2,626,356 directly genotyped and imputed autosomal SNPs. For each SNP, summary statistics were aligned to the risk allele in our primarily European descent meta-analysis.
Calculation of sibling relative risk and liability-scale variance explained
Assuming a multiplicative model (within and between variants), the contribution to the sibling relative risk of a set of N SNPs is given by
where pj and ψj denote the RAF and corresponding allelic OR at the jth SNP52. Assuming disease prevalence, K, the liability-scale variance20 explained by these SNPs is given by
In this expression, T=φ−1(1-K), T1=φ−1(1-λSK), and ω=z/K, where z is the height of the standard Gaussian density at T.
Z-score mixture modelling
We considered the distribution of Z-scores from the Stage 2 meta-analysis, aligned to the risk allele from Stage 1, at a subset of 3,412 independent T2D replication variants (CEU r2 < 0.05), excluding lead SNPs and proxies (CEU r2 ≥ 0.1) at the 63 established and newly discovered susceptibility loci on Metabochip. The Stage 2 Z-scores were modelled as a mixture of two Gaussian distributions: (i) with mean zero and unit variance (i.e. under the null hypothesis of no association); and (ii) with unknown mean (greater than zero) and variance (i.e. under the alternative hypothesis). The mean and variance of the alternative distribution, and the mixing proportion, were estimated using an expectation-maximization algorithm.
We estimated the posterior probability that each of the 3,412 independent replication SNPs is truly associated with T2D from the mixture distribution. We approximated the contribution of these SNPs to λS by simulation from the mixture distribution. For each simulated replicate, we selected “causal” variants at random from these SNPs according to their posterior probability of association. Over 1,000 replicates, we approximated the mean and 95% CI for: (i) the number of “causal” variants among the 3,412 independent replication SNPs; and (ii) the contribution to λS, using estimated RAFs and allelic ORs from the Stage 2 meta-analysis. For each replicate, we also generated a hypothetical third stage to the study consisting of 100,000 T2D cases and 100,000 controls. For each “causal” variant, we generated association summary statistics (Z-score aligned to the risk allele from Stage 1) according to the RAF and allelic OR from our Stage 2 meta-analysis.
Assessment of allele frequency variation across European populations
We calculated F-statistics (FST) across European populations using data from the 1000 Genomes Project (CEU, TSI, FIN, GBR and IBS)13 for the subset of SNPs selected for replication on Metabochip. FST was calculated by comparing mean heterozygosity across all populations to the mean within each sub-population, weighted by the number of contributing chromosomes from each sub-population. We compared FST for the subset of T2D replication SNPs that were directionally consistent between Stage 1 and Stage 2 meta-analyses with all Metabochip replication SNPs (up to 65,345 SNPs), using the Kolmogorov-Smirnov test.
False-discovery rate (FDR) analysis
We undertook FDR analysis53 of 64,646 Metabochip replication SNPs using combined meta-analysis P-values. From this analysis, we observed , consistent with an excess of true positives in this set. We compared these P-values with FDR Q-values and posterior probabilities of membership to the alternative distribution from the mixture model (Supplementary Fig. 4) at the set of 2,172 T2D replication SNPs with concordant direct of allelic effect in both stages of the meta-analysis, after exclusion of 11 AT/GC SNPs with obvious strand orientation misalignments. FDR analysis also indicated an excess of expected true positives in this set of SNPs, even at relatively consistent thresholds (for example, we expect one false positive and 66 true positives at a Q-value of 0.014).
Sex-differentiated meta-analysis
The Stage 1, Stage 2 and combined meta-analyses described above were repeated for males and females separately with correction for population structure within each sex (Supplementary Fig. 13). The male-specific meta-analysis consisted of 20,219 cases and 54,604 controls, while the female-specific meta-analysis consisted of 14,621 cases and 60,377 controls. The sex-specific meta-analyses were then combined to conduct a sex-differentiated test of association and a test of heterogeneity in allelic effects between males and females32.
Physiological analyses
We obtained summary statistics (association P-values and Z-scores for direction of effect or allelic effects and standard errors) for lead T2D SNPs in GWAS meta-analyses of metabolic traits in European descent populations. Summary statistics were aligned to the T2D risk allele from the combined meta-analysis. We obtained summary statistics for lead SNPs in all newly discovered and established loci for glycemic traits in non-diabetic individuals from the MAGIC Investigators5,34. For fasting glucose and fasting insulin, the meta-analysis comprised up to 133,010 individuals, genotyped with GWAS arrays and imputed on up to ~2.5 million SNPs, or genotyped with Metabochip. We also considered surrogate estimates of beta-cell function (HOMA-B) and insulin resistance (HOMA-IR) derived by homeostasis model assessment in up to 38,238 individuals (from GWAS meta-analysis only since these traits were not investigated in the enlarged MAGIC Metabochip study). We obtained summary statistics for lead SNPs in the newly discovered T2D loci (also including GRB14 and HMG20A) for BMI in up to 119,600 individuals from the GIANT Consortium15. To eliminate potential bias in BMI allelic effect estimates at T2D susceptibility loci54, we restricted our attention to meta-analysis of population-based studies not ascertained for disease status for ~2.8 million directly genotyped and/or imputed SNPs. We obtained summary statistics for the same SNPs for plasma lipid concentrations from the Global Lipids Genetics Consortium16. This meta-analysis comprised ~2.6 million directly genotyped and/or imputed SNPs assessed for association to plasma concentrations of: total cholesterol (up to 100,184 individuals); LDL (up to 95,454 individuals); HDL (up to 99,900 individuals); and triglycerides (up to 96,598 individuals).
We also examined T2D association summary statistics at lead SNPs for 37 established T1D susceptibility loci. For each of these SNPs, we reported the allelic OR (aligned to the T2D risk-allele) and P-values in: (i) our Stage 1 T2D meta-analysis; and (ii) a GWAS meta-analysis of 7,514 T1D cases and 9,045 population controls from European descent populations from the Type 1 Diabetes Genetics Consortium35.
Expression analyses
We identified proxies (CEU r2 > 0.8) for each lead T2D SNP in our newly discovered loci (also including GRB14 and HMG20A). We interrogated public databases and unpublished resources for cis-eQTL expression with these SNPs in multiple tissues (details of these resources are summarized in the Supplementary Note). The collated results from these resources met study-specific criteria for statistical significance for association with transcript expression. For each transcript associated with a lead T2D SNP (or proxy), we identified the lead cis-eQTL SNP, and then estimated LD between them using 1000 Genomes Project data to assess coincidence of the signals.
We subsequently tested for association of each lead T2D SNP with the expression of flanking transcripts (within a 1 Mb window) in 603 subcutaneous adipose tissue samples and 745 peripheral blood samples from individuals from the Icelandic population, genotyped using the Illumina HumanHap 300 Bead Array, and imputed up to ~2.5M SNPs36. We modelled the log-average expression ratio of two fluorphores as a function of the allele count (expected allele count for imputed SNPs) in a linear regression framework, with adjustment for age and sex (and differential cell count for blood samples) as covariates. All P-values were also adjusted for the relatedness between individuals by simulating genotypes through the corresponding Icelandic genealogy55. We also identified the most strongly associated cis-eQTL SNP for each flanking transcript. We then performed a conditional test of association of the transcript with the cis-eQTL SNP within the same linear regression framework, with additional adjustment for the lead T2D SNP as a covariate. The conditional analyses determine whether the cis-eQTL SNP association with the transcript can be explained by the lead T2D SNP.
We searched the 1000 Genomes Project data (Phase I interim release) for non-synonymous variants in strong LD (CEU r2 > 0.8) with lead T2D SNPs in the newly discovered loci (also including GRB14 and HMG20A). Identified non-synonymous variants were subsequently interrogated for likely downstream functional consequences using SIFT37.
Pathway, text mining and PPI analyses
We generated two lists of transcripts on the basis of the results of the sex-combined and sex-differentiated meta-analyses. The “primary” list included: (i) the nearest transcript to the lead SNP at 41 previously reported common variant loci identified in European descent populations; (ii) the nearest transcript to the lead SNP at the ten newly identified loci (P < 5 × 10−8) from the sex-combined meta-analysis, including GRB14 and HMG20A; (iii) the nearest transcript to the lead SNP at both novel signals (P < 5 × 10−8) from the sex-differentiated meta-analysis; (iv) the nearest transcript to the lead SNP at six additional loci with the strongest evidence of association (P < 5 × 10−7) from the sex-combined meta-analysis; and (v) 18 genes implicated in monogenic forms of diabetes38, not already overlapping other loci included in the list. The “secondary” list incorporated the nearest transcript to the lead SNP at 77 additional loci with posterior probability of association of at least 75% from the mixture model, not already included in the primary list.
We tested the hypothesis that a PPI network built from the 77 primary transcripts was significantly enriched for physical interaction over and above that expected by chance using DAPPLE41. To build networks, DAPPLE uses a refined database of high-confidence interactions39,40, which emphasizes confidence of interaction over completeness, with the result that not all proteins are represented. We considered two categories of interactions: direct (i.e. between the associated transcripts themselves) and indirect (i.e. via common interactors that were not among the associated transcripts). We assessed the significance of the enrichment of physical interactions by permutation. Subsequently, we used the network as a “seeds” to query against the 77 secondary transcripts.
We used GRAIL to highlight genes from T2D susceptibility loci using similarity of text in PubMed abstracts or in gene-ontology associated codes42. To reduce confounding by published T2D GWAS analyses, we restricted our analysis to abstracts published prior to December 2006. We first tested for enrichment of connectivity in the list of 77 primary transcripts (treating the 18 monogenic loci as a single locus to reduce confounding), and assessed significance via permutation4. These gene sets were then used as the “seed” against which the list of 77 secondary transcripts was queried for connectivity.
We employed a two-step GSEA strategy to test for enrichment of transcripts in T2D susceptibility loci within pathways pertaining to 16 biological hypotheses related to disease pathogenesis (full details of these hypotheses are presented in the Supplementary Note). In the first step, we applied MAGENTA46 to the Stage 1 meta-analysis. Genes in each pathway were scored on the basis of the most significant “local” SNP association using -110 kb/+40 kb boundaries. The 95th percentile of association P-values from all genes in the genome was used to determine the enrichment cut-off. In the second “replication” step, nominally significant gene sets from step one (MAGENTA P < 0.05) were tested for enrichment of T2D association signals in the Stage 2 meta-analysis. To account for the bias in the Metabochip design to SNPs nominally associated with T2D and related metabolic traits, we employed a modified GSEA approach. We tested for enrichment among a broader set of primary or primary and secondary transcripts within LD regions defined by r2 > 0.5 on either side of the lead SNP, extended to the nearest recombination hotspot and then an additional 50 kb (if there was no gene within the LD region, we used the nearest transcript). For robustness testing, we also examined enrichment in the nearest gene to the lead SNPs. The modified GSEA P-value was computed as the fraction of randomly sampled sets of loci, matched for number and local gene density to our primary and secondary lists, which have the same or more significant hyper-geometric probability than that of the T2D loci. For the “null” set, we used 1,600 LD-pruned Metabochip T2D replication SNPs with the lowest posterior probability of association (<5%) from the mixture model. To control for potential confounders, we applied the modified GSEA approach to two negative control lists: (i) loci defined by the lowest ranked independent T2D replication SNPs from our Stage 2 meta-analysis; and (ii) loci for QT-interval on the basis of our Stage 2 meta-analysis for independent replication SNPs for this trait, excluding those within our primary and secondary lists of T2D susceptibility loci and those near monogenic diabetes genes.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this study was provided by: Academy of Finland (77299, 102318, 110413, 118065, 123885, 124243, 129680, 129293, 129494, 136895, 139635, 141005, 213506, 251217); Agence Nationale de la Recherche (France); American Diabetes Association (7-08-MN-OK); Association Française des Diabétiques; Association de Langue Française pour l’Etude du Diabète et des Maladies Métaboliques (France); Association Diabète Risque Vasculaire (France); BDA Research (UK); British Heart Foundation (RG/98002; RG2008/08); Cancer Research UK; Central Norway Health Authority; Central Finland Hospital District; Center for Inherited Disease Research (CIDR) (USA) ; Chief Scientist Office, Scotland (CZB/4/672); City of Kuopio (Finland); City of Leutkirch (Germany); Dept of Health (UK); Deutsche Forschungsgemeinschaft (ER1 55/6-2); Diabetes UK; Doris Duke Charitable Foundation (USA); Estonian Government SF0180142s0; European Commission: ENGAGE (HEALTH-F4-2007-201413); EXGENESIS (LSHM-CT-2004-005272); 245536; QLG1-CT-2002-00896; 2004310); European Commission (Marie Curie: FP7-PEOPLE-2010-IEF); European Regional Development Fund; Faculty of Medicine, Norwegian University of Science and Technology; Finnish Diabetes Association; Finnish Diabetes Research Foundation; Finnish Foundation for Cardiovascular Research; Finnish Heart Association; Finnish Medical Society; Folkhälsan Research Foundation (Finland); Food Standards Agency (UK); Foundation for Life and Health in Finland; Federal Ministry of Education and Research (BMBF) (Germany); Federal Ministry of Health (Germany); General Secretary of Research and Technology (Greece); German Center for Diabetes Research (DZD); German Research Council (GRK 1041); Great Wine Estates of the Margaret River region of Western Australia; Groupe d’Etude des Maladies Métaboliques et Systémiques (France); Harvard Medical School (USA); Heinz Nixdorf Foundation (Germany); Helmholtz Zentrum München-Research Center for Environment and Health (Germany); Helsinki University Central Hospital Research Foundation (Finland); IngaBritt and Arne Lundberg’s Research Foundation (Sweden) (grant nr. 359); Ministry of Health (Ricerca Corrente) (Italy); Karolinska Institutet (Sweden); Knut and Alice Wallenberg Foundation (Sweden) (KAW 2009.0243); Kuopio University Hospital (Finland); Municipal Heath Care Center and Hospital, Jakobstad, Finland; Ministry of Social Affairs and Health (Finland); Ministry of Education and Culture (Finland) (627;2004-2011); Ministry of Innovation, Science, Research and Technology of the state North Rhine-Westphalia (Germany); Medical Research Council (UK) (G0000649,G0601261); MRC-GSK pilot programme grant (UK); Munich Center of Health Sciences (MC Health) (Germany); National Genome Research Network (NGFN) (Germany); NHLBI (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, HHSN268201100012C, R01HL087641, R01HL59367, R01HL086694, N01-HC-25195, N02-HL-6-4278); NHGRI (U01HG004402, N01-HG-65403); National Institutes for Health (USA) (HHSN268200625226C, UL1RR025005, U01HG004399, 1R21NS064908, 1Z01-HG000024, AG028555, AG08724, AG04563, AG10175, AG08861, CA055075); NIDDK (DK062370, DK058845, DK072193, DK078616, DK080140, DK073490); Närpes Health Care Foundation (Finland); National Health Screening Service of Norway; National Institute of Health Research (UK); National Institute for Health and Welfare (Finland) ; Nord-Trøndelag County Council (Norway); Nordic Center of Excellence in Disease Genetics; Norwegian Institute of Public Health; Norwegian Research Council; Novo Nordisk Fonden (Denmark); Ollqvist Foundation (Sweden); Oxford NIHR Biomedical Research Centre (UK); Paavo Nurmi Foundation (Finland); Päivikki and Sakari Sohlberg Foundation (Finland); Perklén Foundation (Sweden); Pfizer; Pirkanmaa Hospital District (Finland); Programme National de Recherche sur le Diabéte (France); Programme Hospitalier de Recherche Clinique (French Ministry of Health); Region of Nord Pas De Calais (Contrat de Projets état-Région) (France); Research into Ageing (UK); Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center; Royal Swedish Academy of Sciences; Sarstedt AG & Co. (Germany); Signe and Ane Gyllenberg Foundation (Sweden); Slottery Machine Association (Finland); Social Insurance Institution of Finland (4/26/2010); South OstroBothnia Hospital District (Finland); State of Baden-Württemberg, Germany; Stockholm County Council (560183, 562183; Stroke Association (UK); Swedish Research Council (8691;09533; 2009-1039; Dnr 521-2010-3490, Dnr 521-2007-4037, Dnr 521-2008-2974, Dnr 825-2010-5983; Dnr 349-2008-6589); Swedish Cultural Foundation in Finland; Swedish Diabetes Foundation; Swedish Heart-Lung Foundation; Swedish Foundation for Strategic Research; Swedish Society of Medicine; Swedish Research Council; Swedish Research Council for Infrastructures; The Sigrid Juselius Foundation (Finland); Torsten and Ragnar Söderberg Foundation (Sweden) (MT33/09); University Hospital Essen (Germany); University of Tromsø (Norway); Uppsala University (Sweden); Uppsala University Hospital (Sweden); Wellcome Trust (GR072960; 076113, 077016, 081682, 083948, 083270, 084711, 086596, 090367, 090532, 098051). A more detailed set of acknowledgements is provided in the Supplementary Note.
Footnotes
AUTHOR CONTRIBUTIONS Writing group: A.P.M., B.F.V., T.M.T., T. Ferreira, A.V.S., V. Steinthorsdottir, R.J.S., H.K., H.G., A. Mahajan, I.P., M.B., M.I.M.
GWAS re-analysis: A.P.M., B.F.V., A.V.S., V. Steinthorsdottir, H.G., I.P., C.D., C.M.L., N.W.R., L.J.S., S.W., S. Raychaudhuri, H. Chen, C.F., C. Liu, D.R., D.J.C., W.H.L.K., M. Li, C.M.C., P.K., Q.S., R.M.v.D., H.M.S., P.S.C., A. Kong, N.R., G.T., R.B., L.L.B., N.B., G.C., C.J.G., C. Guiducci, C.H., W.R., N.K., C. Sigurðsson, B.T., H. Campbell, C.v.D., A.G.U., A. Hofman, E.S., G.R.A., K.R.O., E.Z., B.B., C.N.A.P., V. Lyssenko, T. Tuomi, B.I., D.J.H., L.Q., M.R., J.F.W., F.S.C., K.L.M., R.N.B., J. Tuomilehto, S.C., P. Froguel, T.I., A.D.M., T.M.F., A.T.H., E.B., P.M.N., U.T., L.C.G., K. Stefansson, F.H., J.S.P., J. Dupuis, J.B.M., D.A., M.B., M.I.M.
Metabochip design: B.F.V., H.M.K., G.R.A., D.A., M.B., M.I.M.
Metabochip samples: P.A., M.A., R.B., G.C., A.S.F.D., M.D., T. Forsen, B.G., C.H., A.B.H, A. James, A. Jonsson, W.R., J. Kravic, K.L., E.L., S. Männistö, B.M., L.R., J. Saramies, B.S., S. Shah, G. Sigurðsson G, A. Silveira, G. Steinbach, B.T., F.V., R.W., D.Z., M.D.T., N.G.F., J.G.E., B.B., C.N.A.P., V. Lyssenko, T.T., B.I., A.R.S., M.R., I.B., J.B., K. Hovingh, J.F.P., J.F.W., R.R., T.A.L., L.L., G.D., I.N., N.L.P., K. Khaw, N.J.W., S.M.K., T.E.S., T.W., E.K., J. Saltevo, M. Laakso, J. Kuusisto, A. Metspalu, F.S.C., K.L.M., R.N.B., J. Tuomilehto, B.O.B., C. Gieger, K. Hveem, S.C., P. Froguel, D.B., E. Tremoli, S.E. Humphries, D.S., J. Danesh, E.I., S. Ripatti, V. Salomaa, R.E., K.H.J., S. Moebus, A.P., T.I., U.dF., A. Hamsten, A.D.M., P.J.D., T.M.F., A.T.H., O.M., S. Kathiresan, P.M.N., P.D., U.T., L.C.G., K. Stefansson, D.A., M.B., M.I.M.
Metabochip genotyping: L.L.B., J.C., A.T.C., S.E., E.E., G.G.B, C.J.G., C. Guiducci, J.H., N.K., K. Krjutškov, C. Langford, S.L., G.M., T.W.M., M.P., J. Trakalo, W.W., A. Syvänen, L.P., M.M.N.
Metabochip analysis: A.P.M., B.F.V., T.M.T., T. Ferreira, A.V.S., V. Steinsthorsdottir, R.J.S., H.K., H.G., A. Mahajan, I.P., T.E., R.M.F., S. Kanoni, L.K., A. Kumar, V. Lagou, J.L., C.M.L., M.M., S. Pechlivanis, N.W.R., L.J.S., S.W., L.Y., H.M.S., P.S.C., K.F., P. Fontanillas, O.L.H., S.E. Hunt, A.U.J., A. Kong, R.L., J.M., J.R.B.P., C.G.P.P., S. Potter, E.R., N.R., S. Sivapalaratnam, S. Stančáková, K. Stirrups, G.T., E. Tikkanen, A.R.W., K.G.
Core and additional analyses: A.P.M., B.F.V., T.M.T., T. Ferreira, A.V.S., V. Steinsthorsdottir, R.J.S., H.K., H.G., A. Mahajan, I.P., E.J.R., S. Raychaudhuri, A.D.J., A.S.D., R.J.F.L., S.V., V.E., M.B., M.I.M.
Consortium management: A.P.M., B.F.V., T.M.T., H.G., C. Langenberg, J.C.F., H. Campbell, C.v.D., G.R.A., K.R.O., E.Z., C.N.A.P., V. Lyssenko, A.R.S., I.B., J.F.W., K.L.M., C. Gieger, S.C., P. Froguel, E.I., T.I., A.D.M., T.M.F., A.T.H., U.T., L.C.G., K. Stefansson, F.H., J.S.P., J.B.M., D.A., M.B., M.I.M.
COMPETING FINANCIAL INTERESTS Valgerdur Steinthorsdottir, Gudmar Thorleifsson, Unnur Thorsteinsdottir and Kari Stefansson are employees at deCODE genetics, a biotechnology company that provides genetic testing services, and own stock/stock options in the company. Jose Florez received consulting honoraria from Novartis, Lilly and Pfizer. Inês Barroso and spouse own stock in Glaxosmithkline and Incyte Ltd.
REFERENCES
- 1.Stumvoll M, et al. Type 2 diabetes: principles of pathogenesis and therapy. Lancet. 2005;365:1333–1346. doi: 10.1016/S0140-6736(05)61032-X. [DOI] [PubMed] [Google Scholar]
- 2.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identified additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kong A, et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi L, et al. Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum. Mol. Genet. 2010;19:2706–2715. doi: 10.1093/hmg/ddq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai F-J, et al. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet. 2010;6:e1000847. doi: 10.1371/journal.pgen.1000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shu XO, et al. Identification of new genetic risk variants for type 2 diabetes. PLoS Genet. 2010;6:e1001127. doi: 10.1371/journal.pgen.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamauchi T, et al. A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nat. Genet. 2010;42:864–868. doi: 10.1038/ng.660. [DOI] [PubMed] [Google Scholar]
- 10.Kooner JS, et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat. Genet. 2011;43:984–989. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho YS, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in East Asians. Nat. Genet. 2012;44:67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voight BF, et al. The Metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. doi: 10.1371/journal.pgen.1002793. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loos RJF, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chambers JC, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat. Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 18.Heid IM, et al. Meta-analysis identifies 12 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soranzo N, et al. Common variants at 10 genomic loci influence Hemoglobin A1C levels via glycaemic and nonglycaemic pathways. Diabetes. 2010;59:3229–3239. doi: 10.2337/db10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wray NR, et al. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet. 2010;6:e1000864. doi: 10.1371/journal.pgen.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SH, et al. Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stahl EA, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat. Genet. 2012;44:483–489. doi: 10.1038/ng.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unoki H, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat. Genet. 2008;40:1098–1102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 25.Zeggini E, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shea J, et al. Comparing strategies to fine-map the association of common SNPs at chromosome 9p21 with type 2 diabetes and myocardial infarction. Nat. Genet. 2011;43:801–805. doi: 10.1038/ng.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dickson SP, et al. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The International HapMap Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waters KM, et al. Consistent association of type 2 diabetes risk variants found in Europeans in diverse racial and ethnic groups. PLoS Genet. 2010;6:e1001078. doi: 10.1371/journal.pgen.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magi R, et al. Meta-analysis of sex-specific genome-wide association studies. Genet. Epidemiol. 2010;34:846–853. doi: 10.1002/gepi.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saxena R, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat. Genet. 2010;42:142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott RA, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat. Genet. doi: 10.1038/ng.2385. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 37.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doria A, et al. The emerging genetic architecture of type 2 diabetes. Cell Metab. 2008;8:186–200. doi: 10.1016/j.cmet.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 40.Lage K, et al. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc. Natl. Acad. Sci. USA. 2008;105:20870–20875. doi: 10.1073/pnas.0810772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossin EJ, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;13:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muoio DM, Newgard CB. Mechanisms of disease: molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat. Rev. Mol. Cell. Biol. 2008;9:193–205. doi: 10.1038/nrm2327. [DOI] [PubMed] [Google Scholar]
- 44.Gangwisch JE. Epidemiological evidence for the links between sleep, circadian rhythms and metabolism. Obesity Rev. 2009;10:37–45. doi: 10.1111/j.1467-789X.2009.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boucher BJ. Vitamin D insufficiency and diabetes risks. Current Drug Targets. 2011;12:61–87. doi: 10.2174/138945011793591653. [DOI] [PubMed] [Google Scholar]
- 46.Segre AV, et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;12:e1001058. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pittas AG, et al. Adipocytokines and insulin resistance. J. Clin. Endocrinol. Metab. 2004;89:447–452. doi: 10.1210/jc.2003-031005. [DOI] [PubMed] [Google Scholar]
- 48.Rane SG, et al. Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in beta-islet cell hyperplasia. Nat. Genet. 1999;22:44–52. doi: 10.1038/8751. [DOI] [PubMed] [Google Scholar]
- 49.Fiaschi-Taesch NM, et al. Induction of beta-cell proliferation and engraftment using a single G1/S regulatory molecule, cdk6. Diabetes. 2010;59:1926–1936. doi: 10.2337/db09-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 51.Ioannidis J, et al. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin S, et al. Exhaustive allelic transmission disequilibrium tests as a new approach to genome-wide association studies. Nat. Genet. 2004;36:1181–1188. doi: 10.1038/ng1457. [DOI] [PubMed] [Google Scholar]
- 53.Storey JD, Tibshirani R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol. Biol. 2003;224:149–157. doi: 10.1385/1-59259-364-X:149. [DOI] [PubMed] [Google Scholar]
- 54.Stolerman ES, et al. TCF7L2 variants are associated with increased proinsulin/insulin ratios but not obesity traits in the Framingham Heart Study. Diabetologia. 2009;52:614–620. doi: 10.1007/s00125-009-1266-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stefansson H, et al. A common inversion under selection in Europeans. Nat. Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.