Abstract
Background
Antimicrobial peptides (AMPs) have the potential to act against multiple pathogenic targets.
Results
AMPs that maintain conformational flexibility are more potent against multiple pathogens and less haemolytic.
Conclusion
Antimicrobial action and haemolysis proceed via differing mechanisms.
Significance
The potency, selectivity and ability of AMPs to reach intracellular pathogens can be modulated using general principles.
Keywords: peptide antibiotics, CD and NMR spectroscopy, proline kinks, peptide self-association, malaria, Plasmodium falciparum, tuberculosis
We use a combination of fluorescence, circular dichroism (CD) and NMR spectroscopies, in conjunction with size exclusion chromatography, to help rationalize the relative antibacterial, antiplasmodial and cytotoxic activities of a series of proline free and proline containing model AMPs in terms of their structural properties. When compared with proline free analogues, proline containing peptides had greater activity against Gram negative bacteria, two mammalian cancer cell lines and intraerythocytic Plasmodium falciparum which they were capable of killing without causing haemolysis. In contrast, incorporation of proline did not have a consistent effect on peptide activity against Mycobacterium tuberculosis. In membrane mimicking environments, structures with high α-helix content were adopted by both proline-free and proline-containing peptides. In solution, AMPs generally adopted disordered structures unless their sequences comprised more hydrophobic amino acids or until coordinating phosphate ions were added. Proline-containing peptides resisted ordering induced by either method. The roles of the angle subtended by positively charged amino acids and the positioning of the proline residues were also investigated. Careful positioning of proline residues in AMP sequences is required to enable the peptide to resist ordering and maintain optimal antibacterial activity, while varying the angle subtended by positively charged amino acids can attenuate haemolytic potential albeit with a modest reduction in potency. Maintaining conformational flexibility improves AMP potency and selectivity towards, bacterial, plasmodial and cancerous cells while enabling the targeting of intracellular pathogens.
Linear cationic amphipathic α-helical peptides comprise a class of molecule that often have highly potent antimicrobial properties and have consequently attracted considerable attention in the development of therapeutic agents (1-4). A major goal in this field is to improve the potency of the antimicrobial peptides (AMPs) while reducing their toxicity to host cells.
The hydrophobicity and secondary structure of linear cationic amphipathic antimicrobial peptides influences their ability to both self-associate and insert into biological membranes. These properties contribute both to antimicrobial potency and host cell toxicity. Many α-helical AMPs incorporate proline or glycine residues which may enable helical sections to be interrupted, conferring conformational flexibility on the peptide. The α-helix conformation adopted by many AMPs in biological membranes, which constitute either their target of action or the main barrier to intracellular targets (5), is now regarded as a key determinant of activity (6). Therefore, the presence of helix destabilizing residues such as glycine or proline would be expected to have a critical effect. Accordingly, a number of previous studies have found that incorporating proline residues into AMPs decreases the ability of the peptide to permeabilize the cytoplasmic membrane of Escherichia coli (7) with a concomitant reduction in the antibacterial activity (7-9). Similarly, substituting helix promoting residues for glycines in the primary sequence of magainin peptides improved the antimicrobial activity considerably though a small increase in haemolicity was also detected (10).
In contrast, other studies have found that incorporating either L- or D-proline into either the hydrophobic or hydrophilic face of an amphipathic cationic α-helix-model-peptide, thus disordering the peptide secondary structure, can lead to an improvement, rather than a reduction, in antibacterial activity as well as a reduction in haemolicity (11, 12). This has been ascribed to the ability of proline-containing peptides to resist self-association and interact selectively with anionic lipids, as found in the bacterial target membranes but not in those of erythrocytes.
Here, in order to reconcile these differing views of the effects of proline induced conformational flexibility on antibacterial potency, we test the hypothesis that the ability to maintain a disordered structure in certain environments is crucial to AMP potency against a range of bacterial and eukaryotic pathogens and consider the importance of the positioning of the proline residue. We performed a detailed study of the structure and conformation of a series of proline-free and proline-containing model peptides. We find that the properties conferred on AMPs by proline residues depend strongly on the properties of the proline-free template peptide as well as the positioning of the proline residue in the primary sequence. Using CD spectroscopy supported by fluorescence spectroscopy, size exclusion chromatography and NMR diffusion measurements, we show that in solution, analogously to melittin (13), ordering of structure, that may be related to peptide self-association, of model peptides can be induced through the addition of phosphate anion at fixed pH or through increasing the hydrophobicity of the peptides by incorporating phenylalanine residues at the N-terminus. Proline containing peptides resisted this process whether induced by either phosphate anions or increased hydrophobicity. The relationship between the structural ordering of the peptides and their activities against eukaryotic cell targets was notable. Proline containing peptides were more active against both mammalian cancer cells and the malaria parasite Plasmodium falciparum while having substantially reduced haemolytic potential and differential effects against Mycobacterium tuberculosis.
Some small but significant differences in activity against E. coli and Pseudomonas aeruginosa were observed within the series of proline containing peptides and were ascribed to the effects of the differing positions of the proline residues. Investigating this further, we have obtained high resolution structures of three proline containing analogues in the presence of the anionic detergent sodium dodecyl sulphate (SDS) using NMR spectroscopic methods. The structures reveal that the position of the single proline in the primary sequence of the model peptide has a considerable effect on the ability of the peptide to adopt an α-helix conformation in a membrane mimicking environment.
MATERIALS AND METHODS
Peptides
Peptides comprising L-amino acids (Table 1) were purchased from either EZBiolab (Carmel, IN) or Pepceuticals Ltd (Nottingham, UK) as desalted grade. Peptides comprising D-amino acids were synthesized using standard manual Fmoc solid-state chemistry. HPLC purification was performed using methanol/water or acetonitrile/water gradients and the identity of the product confirmed by matrix assisted laser desorption ionization mass spectrometry. Peptides were lyophilized from 10% acetic acid to remove the trifluoroacetic acid counter ion.
Table 1.
| Peptide | Sequence | (H)1 | (H)2 | (μH)1* |
|---|---|---|---|---|
| D-LAK120 | KKLALLALKKWLLALKKLALLALKK-NH2 | 1.26 | −0.05 | 1.28 |
| D-LAK120-P13 | KKLALLALKKWLPALKKLALLALKK-NH2 | 0.87 | −0.07 | 1.66 |
| D-LAK120-A | KKLALALAKKWLALAKKLALALAKK-NH2 | −0.02 | −0.08 | 2.28 |
| D-LAK120-AP13 | KKLALALAKKWLPLAKKLALALAKK-NH2 | 0.00 | −0.10 | 2.25 |
| D-LAK120-H | KKLALHALKKWLHALKKLAHLALKK-NH2 | −0.35 | −0.16 | 2.80 |
| D-LAK120-HP13 | KKALAHALKKWLPALKKLAHALAKK-NH2 | −1.07 | −0.17 | 3.81 |
| D-LAK80-HP13 | KKALAKALKHWLPALHKLAKALAKK-NH2 | −1.07 | −0.17 | 4.02 |
| D-LAK160-HP13 | KKALKHALAKWLPALKALAHKLAKK-NH2 | −1.07 | −0.17 | 3.40 |
| LAK80 | KKLAKALKLLALLWLKLAKALKKA-NH2 | 0.46 | −0.09 | 3.73 |
| LAK80-P7 | KKLAKAPKLLALLWLKLAKALKKA-NH2 | 0.05 | −0.11 | 3.46 |
| LAK80-P10 | KKLAKALKLPALLWLKLAKALKKA-NH2 | 0.05 | −0.11 | 3.33 |
| LAK80-P12 | KKLAKALKLLAPLWLKLAKALKKA-NH2 | 0.05 | −0.11 | 4.14 |
| LAK120 | KKLALALKKLALLWKKLALALKKA-NH2 | 0.46 | −0.09 | 3.02 |
| LAK120-P7 | KKLALAPKKLALLWKKLALALKKA-NH2 | 0.05 | −0.11 | 2.76 |
| LAK120-P10 | KKLALALKKPALLWKKLALALKKA-NH2 | 0.05 | −0.11 | 2.62 |
| LAK120-P12 | KKLALALKKLAPLWKKLALALKKA-NH2 | 0.05 | −0.11 | 3.43 |
| LAK160 | KKLKLALAKLALLWKALALKLKKA-NH2 | 0.46 | −0.09 | 2.59 |
| LAK160-P7 | KKLKLAPAKLALLWKALALKLKKA-NH2 | 0.05 | −0.11 | 2.34 |
| LAK160-P10 | KKLKLALAKPALLWKALALKLKKA-NH2 | 0.05 | −0.11 | 2.18 |
| LAK160-P12 | KKLKLALAKLAPLWKALALKLKKA-NH2 | 0.05 | −0.11 | 3.00 |
| LAK80-F1 | FKKLAKALKLLALLALKLAKALKKA-NH2 | 0.41 | −0.06 | 3.36 |
| LAK80-F2 | FFKKLAKALKLLALLALKLAKALKKA-NH2 | 0.78 | −0.04 | 3.55 |
| LAK80-F2-P9 | FFKKLAKAPKLLALLALKLAKALKKA-NH2 | 0.40 | −0.06 | 3.30 |
Comparison of physical features of peptides used in this study. Hydrophobicity (H) and mean hydrophobic moment (μH) are shown according to the Combined Consensus scale1 or Eisenberg scale2 and were calculated using the HydroMCalc Java applet made available by Alex Tossi (http://www.bbcm.univ.trieste.it/~tossi/HydroCalc/HydroMCalc.html). All peptides contain eight lysine residues and are amidated at the C-terminus conferring a nominal charge of +9 at neutral pH.
Mean hydrophobic moment assuming formation of ideal α-helix.
Broth micro-dilution assay
The activities of the peptides against two strains of E. coli were assessed in planktonic suspension in polypropylene 96 well plates (Greiner Bio-one, Frickhausen, Germany) according to a modified broth dilution assay (14). E. coli (NCTC 9001), P. aeruginosa (PAO1) and E. coli TOP10 were gifts from K.D Bruce and C. Junkes. E. coli (NCTC 9001), P. aeruginosa (PAO1) or competent E. coli TOP10 were grown without shaking in 50 ml Mueller-Hinton (MH) broth at 37°C. Peptides were tested in duplicates with two rows allocated for each peptide. In each of columns 2-11, 50 μl of MH broth was added under sterile conditions. In the first row, 50 μl of 256 μg/ml stock peptide solutions, prepared in distilled water, were added and then the broth from the second row was pipetted into the first row and thoroughly mixed before being deposited again in the second row. This process was repeated throughout the tray providing a twofold dilution of peptide with each row. Bacteria with an A620 of 0.001 were then added to each well in volumes of 50 μl giving a further twofold dilution and a final volume of 100 μl per well. The final column was used either as sterility control (100 μl broth) or negative control (no peptide). Plates were incubated overnight at 37°C and the A620 read. Growth curves prepared from duplicates were fitted to determine the peptide concentration required to inhibit growth by 50% (MIC50). The MIC50 quoted for each peptide (Table 2) is an average value from at least two independent repeats.
Table 2.
| Peptide |
E. coli (NCTC 9001) MIC50 |
P. aeruginosa (PAO1) MIC50 |
E. coli (TOP10) MIC50 |
P. falciparum (3D7) EC50 |
P. falciparum (C10) EC50 |
A549 EC50 |
RAW264.7 EC50 |
Haemolysis HC50 |
|---|---|---|---|---|---|---|---|---|
| D-LAK120 | 1.48 ± 0.53 | 2.71 ± 1.07 | 0.62 ± 0.15 | 1.95 ± 0.08 | n.d. | n.d. | n.d. | 4.06 ± 1.30 |
| D-LAK120-P13 | 1.54 ± 0.97 | 1.42 ± 0.41 | 0.58 ± 0.18 | 0.88 ± 0.08 | 2.18 ± 0.41 | 2.44 ± 0.43 | 3.80 ± 0.34 | 11.66 ± 0.03 |
| D-LAK120-A | 1.23 ± 0.47 | 1.83 ± 0.63 | 0.38 ± 0.21 | 1.45 ± 0.12 | 2.13 ± 0.14 | 5.23 ± 0.35 | 7.31 ± 2.13 | 17.42 ± 2.17 |
| D-LAK120-AP13 | 1.08 ± 0.62 | 1.22 ± 0.19 | 0.29 ± 0.09 | 0.88 ± 0.06 | 2.36 ± 0.10 | 3.11 ± 0.57 | 3.42 ± 0.16 | 58.61 ± 10.41 |
| D-LAK120-H | 1.54 ± 0.01 | 1.35 ± 0.07 | 0.40 ± 0.01 | 1.26 ± 0.14 | 4.43 ± 0.33 | 4.54 ± 0.30 | 5.21 ± 0.57 | 20.29 ± 2.42 |
| D-LAK120-HP13 | n.d. | n.d. | n.d. | n.d. | 2.88 ± 0.25 | 2.21 ± 0.63 | 2.33 ± 0.30 | 79.61 ± 2.80 |
| D-LAK80-HP13 | n.d. | n.d. | n.d. | n.d. | 0.96 ± 0.07 | 6.50 ± 0.37 | 7.08 ± 1.42 | 8.61 ± 0.35 |
| D-LAK160-HP13 | n.d. | n.d. | n.d. | n.d. | 3.94 ± 0.33 | 7.58 ± 0.42 | 3.95 ± 0.40 | 185.0 ± 18.65 |
Comparison of peptide concentrations (μM) required to inhibit 50% of bacterial growth (MIC50), kill 50% parasites or cancer cells (EC50) or lyse 50% red blood cells (HC50) for 8 designed linear cationic peptides comprising all D amino acids. Results are an average of two or more independently repeated experiments.
Antituberculosis assay
The activities of the D-amino acid peptides against M. tuberculosis H37Ra, an attenuated M. tuberculosis strain commonly used as a standard strain for anti-TB drug testing, were tested in a manner similar to the broth micro-dilution assay described above. Duplicate serial dilutions from 100 to 0.78 μM peptide were prepared in a total volume of 180 μl Middlebrook 7H9 Broth, supplemented with Middlebrook oleic albumin dextrose catalase growth supplement, in 96 well plates. Bacterial suspension (20 μl of a 1 × 106 CFU/ml suspension) was added to each well giving a final volume of 200 μl per well. The plates were sealed and incubated at 37°C with 5% CO2 for between four and five weeks prior to inspection. Negative and 1% growth controls were prepared by preparing wells containing 20 μl of a 1 × 106 CFU/ml or 1 × 104 CFU/ml respectively and 180 μl broth.
Antiplasmodial assay
The activities of the peptides comprising D-amino acids were tested against both the 3D7 and C10 strains of Pl. falciparum using fluorescent nucleic acid binding dye assays modified from (15). Ring stage parasites maintained in AlbuMAX® RPMI 1640 complete medium, were pre-synchronized using magnetic cell sorting and differential lysis with sorbitol. Two-fold serial dilutions of each peptide stock in culture medium of concentrations between 64 μM and 0.064 μM were distributed into either polypropylene 96 well plates (3D7; Greiner Bio-one, Frickhausen, Germany) or polystyrene Nunc MicroWell™ Plates (C10) in a total volume of 50 μl. 150 μl of a thoroughly mixed Pl. falciparum culture at ring stage was added into each well to a final volume of 200 μl, giving a final peptide concentrations ranging from 16 μM to 0.016 μM. The parasitaemia was between 1.5-2%, while the final hematocrit was between 3.5-4%. Conditions were prepared for the SYBR green II assay in triplicate while two further wells were used for slide preparation with Giemsa stain and determination of parasitaemia under light microscope. Wells containing parasite culture (1.5-2% parasitaemia, 3.5-4% final hematocrit) without peptide served as growth control. For time zero controls, 50 μl fresh culture medium and 150 μl malaria culture were added to a separate 96-well plate which was immediately stored at −20°C. The plates were gassed with 5% O2, 5% CO2 and 90% N2 in a sealed gas chamber for 5 min and then incubated at 37°C for a total of 48 hrs. Immediately following incubation, blood smears were prepared from each condition and the plates stored at −20°C. The assay plates, as well as the time zero plate, were thawed at room temperature for the SYBR green assay. After mixing, 100 μl of the contents of each well were transferred to black 96-well plates, 100 μl of SYBR green II/lysis buffer solution (0.2 μl SYBR green II per ml of lysis buffer consisting of 20 mM Tris pH 7.5, 5 mM EDTA, 0.008% wt/vol saponin and 0.08% vol/vol Triton X-100) were added to each well. The plates were incubated in dark for one hour and inspected with a FLUOstar Omega (BMG LABTECH GmbH, Ortenberg, DE) fluorescence plate reader with excitation at 485 nm and emission monitored at 530 nm.
Haemolysis assay
A duplicate dilution series of peptides from 256 to 0.016 μM was prepared in 10 mM Tris, 150 mM NaCl, pH 7.4 buffer. Fresh erythrocytes were washed thoroughly in Tris buffer and diluted to get a final concentration of ~2.5 × 109/ml. 50 μl of this red blood cell (RBC) suspension was added to 450 μl of the prepared peptide dilution series in microcentrifuge tubes giving a final RBC concentration of ~2.5 × 108/ml). The tubes were incubated for 5 min on ice then 30 min at 37°C and a further 5 min on ice before being centrifuged (5 min, 2000 g, 4 °C). 230 μl supernatant from each tube was transferred to a polystyrene Nunc MicroWell™ Plate. To generate a positive control, 230 μl of a 0.1% Triton X-100 solution were added to erythrocyte pellets prepared from tubes challenged with 450 μl Tris buffer only. The resulting suspensions (230 μl) were transferred to the same plate which was read at 540 nm. Percentage haemolysis was calculated relative to the positive control (100%).
MTT assay
Cytotoxicity was evaluated by performing the 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT; Sigma) assay (16). 10,000 adenocarcinomic human alveolar basal epithelial cells (A549) or 10,000 RAW 264.7 Abelson murine leukemia virus transformed macrophages (RAW 264.7) per well were plated in 96-well plates (Costar). The cells were incubated with peptide solutions, prepared in serum free OptiMEM, for four hours at 37°C. Untreated cells were used as a negative control. The cells were then incubated with 200 μl of pre-warmed MTT solution (0.8 mg/ml) per well for a further two hours. The toxicities of the peptides to both mouse monocytic macrophage cell line RAW 264.7 and human lung cancer cell line A549 at different concentrations were evaluated by comparing the absorbance of formazan crystals, dissolved using isopropanol, formed by the MTT reagent at 570 nm.
Circular dichroism
Simultaneous UV absorption and CD spectra were acquired on Chirascan or Chirascan Plus spectrometers (Applied Photophysics, Leatherhead, UK). The instruments were flushed with pure nitrogen gas throughout the measurements. Far-UV spectra were recorded from 260 to 185 nm with a 1 nm spectral bandwidth, 0.5 nm stepsize and a 1 s spectrometer time-per-point. A rectangular 0.5 mm pathlength was employed. Peptides were dissolved in 5 mM Tris-amine buffer at pH 7.2 to a final concentration of around 20 μM. Peptide samples were also prepared in the presence of 50 mM SDS or 50% v/v trifluoroethanol (TFE). Peptide titration with sodium phosphate was carried out in situ (0.5 mm pathlength) using a 1 M sodium phosphate (pH 7.2) containing 20 μM peptide as stock solution. Unless otherwise stated, all spectra were measured at 37°C. During data processing, a spectrum of the peptide-free media or solution was buffer subtracted and Savitsky-Golay smoothing with a convolution width of 5 points applied. CD spectra were normalized for concentration and pathlength and expressed in terms of molar ellipticity per residue. Secondary structure analyses were performed using CDPro (17).
Tryptophan fluorescence
Fluorescence emission spectra of tryptophan containing peptides were acquired using a Cary Eclipse Fluorescence Spectrophotometer using an excitation wavelength of 280 nm and scanning from 300 nm to 450 nm at 37°C. Peptides were made up to a final concentration of 20 μM using 5 mM Tris buffer at pH 7.0. 900 μl of a peptide solution was titrated with small volumes of phosphate buffer in a 4 mm × 10 mm cuvette. Next, normalized fluorescence intensities were plotted against wavelength and fitted to a log normal distribution, as described (18), in Origin 8 (OriginLab Corporation, Northampton, MA) and the wavelength of the maximum intensities (λmax) were plotted against the concentration of hydrogen phosphate ions.
Size exclusion chromatography
75 μl of peptide solutions prepared in the appropriate buffer at a final concentration of 1 mg/ml were injected onto a Superdex™ 75 10/300 GL column (GE Healthcare, Uppsala, SE). Buffers contained 10 mM Tris-amine, 150 mM NaCl and sodium phosphate at varying concentrations (either 0, 50, 100, 200 or 500 mM) and were adjusted to pH 7.0. Peptides were eluted at a flow rate of 0.5 ml/min on an Agilent 1100 HPLC system with detection at 215 and 280 nm.
Diffusion Ordered SpectroscopY (DOSY)
Samples were prepared in an analogous manner to those used in CD measurements but at a peptide concentration of 100 μM (i) in 5 mM Tris-amine buffer at pH 7.2 and (ii) titrated to 500 mM sodium phosphate at pH 7.2, each buffer containing 5% D2O. NMR spectra were acquired at 298 K on a Bruker Avance 500 MHz spectrometer (Bruker, Coventry, UK) equipped with a cryoprobe. The 1H 90° pulse length was calibrated for each sample and was found to be close to 7.8 μsec for (i) and 15.6 μsec for (ii). Diffusion measurements were made using a double stimulated echo (DSTE) pulse program including bipolar gradient pulses and a longitudinal eddy current delay (LED). The spectral width was set to 12 ppm and the number of scans was set to 32. The relaxation delay was 1 sec and the gradient pulse strength was increased from 5% to 95% of the maximum gradient strength (50 G.cm-1). The diffusion time was set to 120 msec and bipolar gradient pulses of 1.25 msec were applied. Data were processed and diffusion coefficients were extracted by fitting intensities in the manufacturer’s software (TopSpin).
NMR structure determination
The NMR samples consisted of a 1 mM peptide solution also containing 100 mM SDS-d25 with 5 mM Tris(hydroxymethyl-d3)-amino-d2-methane buffer at pH 7. 10% D2O containing trimethylsilyl propanoic acid (TSP) was added for the lock signal and as internal chemical shift reference. The temperature was kept constant at 310 K during the NMR experiments. NMR spectra were acquired on a Bruker Avance 500 MHz spectrometer (Bruker, Coventry, UK) equipped with a cryoprobe. Standard Bruker TOCSY and NOESY pulse sequences were used, with water suppression using a WATERGATE 3-9-19 sequence with gradients (mlevgpph19 and noesygpph19). The 1H 90 degree pulse was calibrated at 37.04 kHz. The TOCSY mixing time was 90 ms, and the mixing time for the NOESY spectra was set to 150 ms. The relaxation delay was 1 s. 2048 data points were recorded in the direct dimension, and either 256 or 512 data points in the indirect dimension. The spectra were processed using Bruker TOPSPIN. The free induction decay was multiplied by a shifted-sine2 window function. After Fourier transformation, the spectra were phase corrected, a baseline correction was applied, and spectra were calibrated to the TSP signal at 0 ppm.
Assignments were carried out with the Sparky software (19) and structure calculations were done with ARIA software (20), using NOE restraints of backbone protons and proline Hδ protons only, and no dihedral angle restraints. We chose to add the Hδ protons of proline to compensate for the fact that this amino acid residue does not have an NH proton on the backbone, and because proline Hδ atoms are easy to assign and do not overlap with other peaks. Restraints involving the sidechain atoms of tryptophan were removed because they violated in every model during and after the structure calculation. The sidechain atoms of other residues were removed because the overlapping peaks from especially lysine and leucine residues made it difficult to unambiguously assign and reliably integrate these peaks. The ARIA software was configured to use manual assignments and not allowed to change them. Proton frequency windows were set to 0.02 and 0.04 for the direct and indirect dimension, respectively. Upper and lower bound corrections were disabled. Structures were calculated using torsion angle MD, with the slow cooling protocol as published (21), followed by refinement in water using default ARIA settings. One hundred structures were calculated in each of eight iterations, and the ten best structures were kept. Network anchoring was enabled during the first three iterations. Results were analyzed with AQUA and PROCHECK_NMR software (22), and python scripts developed in our laboratory. An overview of all backbone NOESY contacts that were used in the structure calculations is available in the supplementary material. Structures of LAK160-P7, LAK160-P10 and LAK160-P12 were deposited in the RCSB Protein Data Bank with accession codes 2l96, 2l99 and 2l9a respectively.
RESULTS
Peptide design
Peptides (Table 1) were designed to adopt amphipathic α-helix conformations in the appropriate environments. The peptides comprise a series of alternating alanine and leucine residues with two lysine residues located close to each of the N- and C-termini. The C-terminus in each peptide is amidated which, together with the four lysines that are distributed elsewhere in the sequences and describe the “charge angle” when the peptide adopts an amphipathic α-helix, confers a nominal charge of +9 to each peptide. We have previously discussed the importance of considering the design of peptides where the angle subtended by the charged residues is varied (23) and recall previous work which describes the activities of cationic α-helical peptides where the hydrophobic moment is either balanced (24) or ignored (25) when altering the charge angle. In the present study we were interested in the ability of proline residues to modulate self-association while considering their effects on the charge angle when adopting α-helical conformations. Consequently, proline-free and proline-containing peptides (Table 1) are designed without attempting to balance the hydrophobic moment.
A number of previous studies (23, 26-28) have noted the enhanced activities of peptides comprising D-amino acids when compared with analogous peptides prepared from L-amino acids, in particular against Pl. falciparum. To pursue this further, we prepared two series of cationic amphipathic peptides comprising all D-amino acids (Supplementary Table 1, Table 1). The first of these (Supp. Tab. 1, D-LAKn) was used to identify which charge angle conferred optimal antibacterial activity. Data obtained for the two E. coli strains, P. aeruginosa and M. tuberculosis indicated that a charge angle of 120° conferred the most potent antibacterial activity and this peptide was used as a template for the second series (Table 1, D-LAK120…). In this series the peptide hydrophobicity was reduced through either altering the ratio of alanine to leucine residues (D-LAK120-A) and/or incorporating histidine residues (D-LAK120-H). Proline residues were included in place of leucine or histidine in three peptides at position 13. The final two peptides in this series reassess the biological effect of varying the charge angle in a proline containing peptide (D-LAKn-HP13).
In light of the conflicting reports regarding the effect of proline incorporation on antibacterial activity, we investigated whether the positioning of proline would affect activity. For reasons of cost, all L-amino acid peptides were used in this part of the study. Three proline-free peptide templates were selected which are defined by the charge angle, 80, 120 or 160 degrees, subtended by the four lysine residues described above. For each template peptide, proline residues were substituted for leucine residues in one of three positions. The proline residues were expected to confer conformational flexibility on the peptides while interrupting the α-helix conformation. A range of proline locations was required in order that the effects of proline location on the conformation adopted in N- and C-terminal segments of differing lengths could be probed.
Finally, three further peptides were designed, comprising all L-amino acids, where additions of hydrophobic phenylalanine residues were made at the N-terminus. The hydrophobic phenylalanine residues were included in order to promote peptide self–association as, in addition to electrostatic effects, aromatic effects are known to contribute to peptide aggregation and fibrillation (29). This allows the effect of proline on self-association driven by hydrophobicity rather than phosphate anion concentration to be tested. Either one or two phenylalanine residues were added to the LAK80 or LAK80-P7 sequences generating either LAK80-F1 or LAK80-F2 and LAK80-F2-P9 respectively.
Biological activities of all D-amino acid AMPs
Based on the initial screen of D-amino acid peptides according to their charge angle, D-LAK120 was selected as the template for a series of peptides (Table 1) where the effects of reducing peptide hydrophobicity and incorporating proline were probed. The activities of these peptides against Gram negative bacteria, M. tuberculosis H37Ra, two strains of Pl. falciparum and two mammalian cancer cell lines were determined as were their hemolytic potential (Table 2, Fig. 1). All of the peptides tested were highly potent against both E. coli and P. aeruginosa and compare very favorably with the activities of naturally occurring AMPs assayed under the same conditions (30). However, no significant differences in activity were observed as a result of any of the modifications. When tested against Pl. falciparum, modest but significant (p < 0.05) improvements in anti-plasmodial activity were sometimes observed in proline containing peptides when compared with the proline free analog. Importantly, the anti-plasmodial activities need to be compared with the HC50 haemolysis data obtained for each peptide as these reveal whether plasmodial toxicity is specific or occurs only as collateral damage associated with infected erythrocyte lysis. HC50 values greater than ten times the EC50 for anti-plasmodial activity for either strain are highlighted in bold (Table 1) and identify three peptides only that are capable of specific anti-plasmodial activity. Light microscopy of erythrocyte cultures of PL. falciparum (Supp. Fig. 2) show the effects of one such specific peptide, D-LAK120-AP13, where a dramatic reduction in parasitaemia is effected without any observable damage to the erythrocyte hosts. Notably, incorporation of proline in the more hydrophilic D-LAK120-A or D-LAK120-H to give D-LAK120-AP13 or D-LAK120-HP13 respectively led to dramatic reductions in haemolytic potential, while anti-plasmodial activity was maintained or improved. This effect was not observed for the most hydrophobic analogues (D-LAK120/D-LAK120-P13). The incorporation of proline in a suitably hydrophilic peptide therefore confers considerable selectivity. Interestingly, altering the charged angle in the D-LAKn-HP13 series also had profound effects on haemolytic potential and hence anti-plasmodial specificity. All of the peptides were toxic to the two mammalian cancer cell lines at low micromolar concentrations. However, modest increases in toxicity were again observed for proline containing peptides when compared with their proline free analogues.
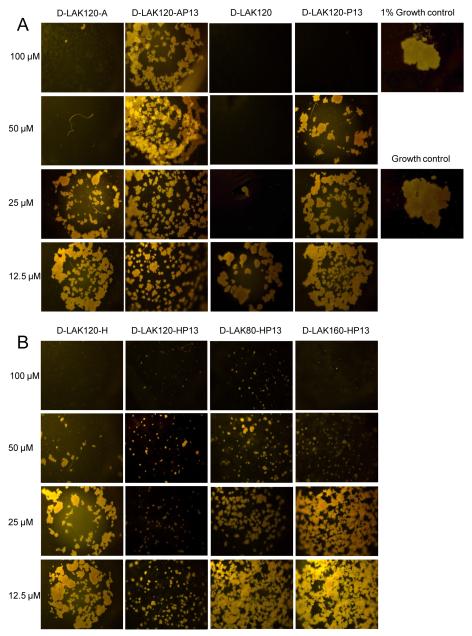
Figure 1.
The ability of eight D-LAK peptides to inhibit the growth of M. tuberculosis H37Ra is shown and compared with a positive growth control (1 × 106 CFU/ml) and a 1% growth control (1 × 104 CFU/ml). When histidine free peptides are compared (A), a reduction of antibacterial activity is observed when proline is incorporated. For histidine containing peptides (B), incorporation of proline increases antibacterial activity. Altering the angle subtended by the lysine residues to either 80 or 160° diminishes peptide potency.
The anti-bacterial activity of the peptides against M. tuberculosis did not follow the pattern observed for the other targets (Fig. 1). Incorporation of proline in D-LAK120-A or D-LAK120 caused a substantial reduction in efficacy (Fig. 1A). This contrasted with the substantial improvement seen for D-LAK120-H when proline was incorporated (Fig. 1B). The resulting peptide, D-LAK120-HP13, had the highest activity against M. tuberculosis of all the peptides tested in the present study. Altering the charged angle in the D-LAKn-HP13 series had a detrimental effect on activity against M. tuberculosis (Fig. 1B).
Ordering and self-association of peptides
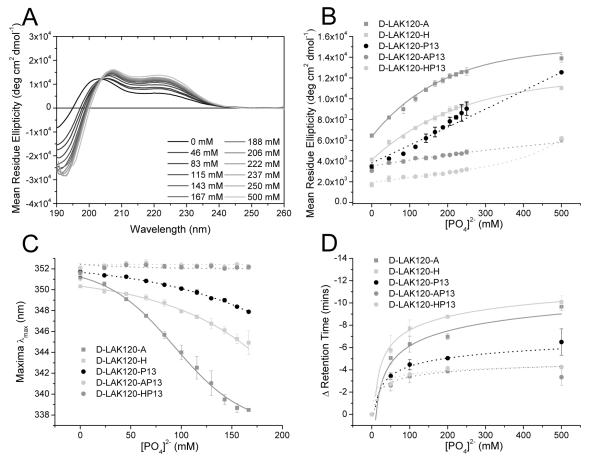
Circular Dichroism (CD) measurements of the peptides in solution or in the presence of membrane mimetic media provide information on the average secondary structure adopted by each peptide in each differing environment. The conformational change in response to the addition of phosphate buffer was observed for both all D- and all L-amino acid peptide families dissolved in aqueous solution (Fig. 2A/B; Supp. Fig. 2-4). Spectra were recorded between 260 and 185 nm with absorbance spectra recorded simultaneously. The phosphate anion absorbs at shorter wavelengths to interfere with the CD signal. Hence with increasing phosphate concentration, data points at shorter wavelengths are removed according to the monitored absorbance. As shown previously for melittin, a membrane lytic α-helical peptide from bee venom, phosphate or other coordinating anions can promote self-association of peptides containing arginine or lysine residues by both minimizing electrostatic repulsion and providing cross-link sites where two lysines may simultaneously hydrogen bond with one orthophosphate “bridging” moiety (13). The comparison of the left handed α-helix inducing effect of phosphate anion on proline-free and proline-containing peptides comprising D-amino acids is made by following the development of the positive band at 220 nm, characteristic of left handed α-helix conformation (Fig. 2A/B). Those peptides containing a proline residue resist the increase in left handed α-helix conformation that is characteristic of the proline free analogues (Fig. 2B). Further, an effect of hydrophobicity can be seen with the most hydrophobic proline containing peptide, D-LAK120-P13, responding with a greater increase in α-helix conformation at increasing phosphate anion concentration when compared with the more hydrophilic analogues. As with melittin, this increase in α-helix content may be expected to be linked to peptide self-association. Support for this comes from measurements of intrinsic tryptophan fluorescence performed in parallel to the CD studies (Fig. 2C) as well as size exclusion chromatography (Fig. 2D) and DOSY NMR (Supp. Fig. 3). Fluorescence emission spectra, resulting from the single tryptophan residue at position 11, were monitored following the addition of increasing amounts of phosphate. Blue shifts and intensity reductions in the fluorescence emission maximum were observed which are consistent with the tryptophan residues in individual peptides moving to a more hydrophobic environment, as would be encountered in an oligomer, and encountering tryptophan residues from other peptides in the oligomer in close proximity leading to self-quenching (31). Peptide bonds from neighboring peptides may also contribute, weakly, to the observed reduction in intensity (32). Qualitatively, the fluorescence data mirrors the CD data with the greatest changes in fluorescence and ellipticity at 220 nm in response to phosphate anion occurring for the same proline free peptides and the least change observed for proline containing peptides suggesting these two parameters are closely linked. To further investigate whether peptide self-association is responsible for the observed conformational and fluorescence emission changes, size exclusion chromatography and DOSY NMR studies were performed in parallel. The change in retention time on a Superdex™ 75 10/300 column is shown as a function of phosphate anion concentration (Fig. 2D). Larger particles elute more rapidly and, with increasing phosphate concentrations, proline free peptides are observed to elute at increasingly shorter time intervals. This trend is more effectively resisted for proline containing peptides and is very similar to the effects observed using either CD or fluorescence emission spectroscopy suggesting that these three techniques are reporting on the same phenomenon. When DOSY NMR was performed for two pairs of proline free/proline containing peptides (Supp. Fig. 3) the results indicated a much better correlation between peptide diffusion coefficients and absolute rather than relative changes in size exclusion retention time. Taken together, this suggests that the changes in secondary structure and shielding of tryptophan from an aqueous environment are closely related to a relative change in particle size. However when set against the diffusion coefficients and considering the absolute retention times, it is apparent that the particles comprising disordered proline containing peptides are larger than those comprising structurally ordered proline free peptides at low phosphate anion concentrations. Whether this corresponds to comprising a larger number of peptides or reflects a less dense packing is unclear at present. Nonetheless, both size exclusion chromatography and NMR diffusion measurements are in agreement that the presence of proline affects the supramolecular arrangement of the peptides in solution.
Figure 2.
Circular dichroism spectra of D-LAK120-A (A) in 5mM Tris buffer solution pH 7.3 titrated with increasing concentrations of phosphate buffer at 37°C. The changes in left handed α-helix content as monitored by the ellipticity at 220 nm is shown as a function of phosphate concentration (B) and compared with corresponding experiments performed with a further four peptides. The shift in the tryptophan emission maximum (C) and the retention times on a size exclusion column (D) were also evaluated as a function of phosphate concentration and are related to the buildup of secondary structure. Lines are to guide the eye.
All L-amino acid containing LAK peptides dissolved in Tris/Cl− buffer adopted mostly disordered conformations while addition of phosphate anions induced either small or more dramatic increases in right handed α-helix conformation content depending on the charge angle and the presence or absence of proline in the primary sequence. For proline-free peptides, LAK80 (Supp. Fig. 4A) and LAK120 (Supp. Fig. 5A), in the absence of phosphate, disordered conformations are adopted that nevertheless indicate equilibrium with an α-helix conformation with notable positive bands at ~190 nm. Conformational equilibria have been demonstrated for other linear peptides (33). Addition of phosphate anion produces a substantial increase in α-helix conformation content with the appearance of strong negative bands at 207 and 220 nm and an increase in the positive band intensity at 190 nm until the effects of phosphate ion absorbance obscure this region. In contrast, the proline-free peptide with a greater charge angle, LAK160 (Supp. Fig. 6A), adopts an α-helix disordered state with much lower α-helix content compared to the two peptides designed to incorporate more acute charge angles. As with the D-amino acid peptides, the incorporation of proline in either the LAK80 or LAK120 sequences further reduces the α-helix content of the disordered state adopted in Tris buffer and enables the peptide to resist the ordering effects induced by the addition of phosphate anions (Supp. Fig 4/5). The effect of phosphate anion on the LAK160 peptides (Supp. Fig. 6) reveals that, within this series, both proline-free and proline-containing peptides resist ordering in solution.
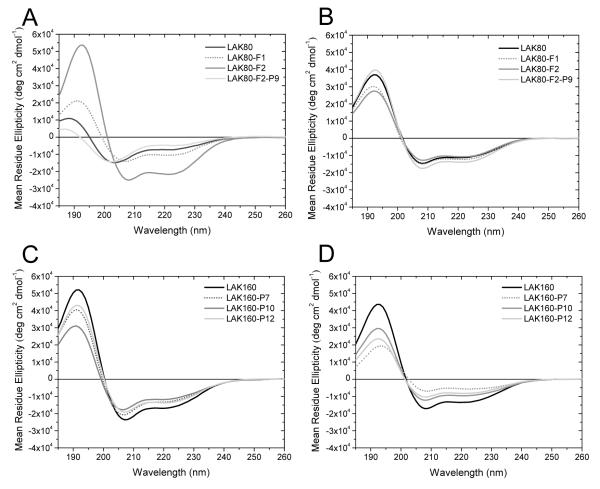
To further probe the relationship between structural ordering and the supramolecular organization of LAK peptides in solution and to investigate the ability of proline residues to interfere with this, further variants of LAK80 and its proline containing analogue LAK80-P7 were studied (Fig. 3A/B). All of these variants, incorporating either a further one or two phenylalanine residues at the C-terminus, adopt conformations with a similarly large α-helix content in the presence of model DMPC/DMPG (75:25) membranes (Fig. 3B). In contrast, in aqueous solution in the absence of phosphate ions, the secondary structure of the four analogues differ considerably (Fig. 3A). LAK80 adopts a mostly disordered conformation while addition of first one and then two hydrophobic phenylalanine residues to the sequence causes respectively a more modest and then substantial increase in α-helix content. The ability of proline to disrupt α-helix conformation induced by hydrophobic interactions is now highlighted by the disordered conformation adopted by LAK80-F2-P9, a peptide incorporating a single proline residue in addition to the two hydrophobic phenylalanine residues at the N-terminus.
Figure 3.
Circular dichroism spectra reveal the effect of peptide hydrophobicity and proline position on secondary structure in aqueous solution and membrane mimicking media. CD spectra are shown for LAK80, LAK80-F1, LAK80-F2 and LAK80-F2-P9 in 5mM Tris buffer solution (A) or DMPC:DMPG (75:25) liposomes (B), and LAK160, LAK160-P7, LAK160-P10 and LAK160-P12 in 50 mM SDS (C) or DMPC:DMPG (75:25) liposomes (D). All spectra were recorded at 37°C.
Antibacterial activities and the positioning of proline residues
The antibiotic activities of the peptides were tested against two separate strains of E. coli and one of P. aeruginosa (Table 3). E. coli (NCTC 9001) is a type strain while TOP10 is a competent strain with a genotype similar to that of DH10B which is deficient in galU, galK and galE (34). Inactivation of galE perturbs the incorporation of glucose into the O-side chain of lipopolysaccharide (LPS) and hence E. coli (TOP10) are expected to have an altered LPS structure. For each peptide, E. coli (TOP10) was more susceptible to AMP challenge than the type strain. All of the designed peptides tested had potent activity against the Gram negative bacteria used in the study with the activities of some peptides entering the nanomolar range even for P. aeruginosa.
Table 3.
| Peptide |
E. coli (NCTC 9001) |
P. aeruginosa
(PAO1) |
E. coli (TOP10) |
|---|---|---|---|
| LAK80 | 13.05 ± 0.13 | n.d. | n.d. |
| LAK80-P7 | 2.97 ± 0.15 | 2.13 ± 0.77 | 1.26 ± 0.49 |
| LAK80-P10 | 1.62 ± 0.17 | 1.18 ± 0.53 | 0.82 ± 0.03 |
| LAK80-P12 | 1.80 ± 0.27 | 0.86 ± 0.09 | 0.80 ± 0.01 |
| LAK120 | 2.95 ± 0.06 | 4.20 ± 0.92 | 1.47 ± 0.64 |
| LAK120-P7 | 0.80 ± 0.03 | 4.45 ± 0.19 | 0.45 ± 0.10 |
| LAK120-P10 | 0.78 ± 0.01 | 9.16 ± 1.43 | 0.47 ± 0.15 |
| LAK120-P12 | 0.97 ± 0.06 | 2.14 ± 0.53 | 0.68 ± 0.14 |
| LAK160 | 1.11 ± 0.48 | 2.23 ± 0.41 | 0.73 ± 0.40 |
| LAK160-P7 | 0.79 ± 0.41 | 2.98 ± 0.21 | 0.46 ± 0.11 |
| LAK160-P10 | 2.22 ± 0.63 | 4.14 ± 0.23 | 0.45 ± 0.24 |
| LAK160-P12 | 0.48 ± 0.20 | 1.69 ± 0.09 | 0.38 ± 0.08 |
Comparison of minimal inhibitory concentrations (μM) required to inhibit 50% of growth (MIC50) for 12 designed linear cationic peptides. Results are an average of two or more independently repeated experiments.
No significant differences in antibacterial activity were detected between the D-amino acid peptides. Importantly however, small but statistically significant differences in activity could be detected between peptide analogues comprising L-amino acids. Notably, the proline-free peptides with small or intermediate charge angles, LAK80 or LAK120, performed poorly against E. coli (NCTC9001) when compared with the analogue with the larger charge angle, LAK160. Within the LAK80 and LAK120 series, activity against E. coli (NCTC9001) was enhanced by the incorporation of proline (p < 0.05), irrespective of its position in the primary sequence. For the LAK120 series, a similar but less dramatic effect was seen when challenging E. coli (TOP10) but when P. aeruginosa was challenged, the positioning of the proline residue affected activity. In particular, the incorporation of proline in the LAK120 sequence was noted to both increase activity (p < 0.05) when located at position 12 but decrease activity (p < 0.05) when located at position 10.
In contrast with LAK80 and LAK120, LAK160, the proline-free analogue with the largest charge angle, had potent antimicrobial activity against each of the panel of Gram negative organisms. Despite the high potency of the parent peptide, incorporation of proline at position 12 led to a modest but significant increase in activity against E. coli (NCTC9001) and P. aeruginosa (p < 0.05 and p < 0.10 respectively). Substitution of proline for leucine at position 7 had a less notable effect but performing the same substitution at position 10 led to a reduction in activity against the two strains (E. coli (NCTC9001): p < 0.10, P. aeruginosa: p < 0.05). This proline containing analogue, LAK160-P10, was therefore significantly (p < 0.05) less potent than its close relation, LAK160-P12, against both E. coli (NCTC9001) and P. aeruginosa. E. coli (TOP10) was again more sensitive to each of the peptides in this series and no significant differences in activity could be discerned. In summary, both the size of the charge angle and the positioning of the incorporated proline residues affected the activity against Gram negative bacteria.
Since the LAK160 peptides show little tendency to adopt α-helix rich conformations in aqueous solution, the structure adopted in the target membrane environment may contribute to significant differences in observed antibacterial activity. The ability of the LAK160 peptides to adopt such conformations in the presence of membrane mimicking anionic detergent SDS (Fig. 3C) or model DMPC/DMPG (75:25) membranes (Fig. 3D) was confirmed. Micelles composed of the anionic detergent SDS provide a simple model for bacterial membranes, replicating the negative surface charge and hydrophobic/aqueous interface. In the presence of 50 mM SDS, all four LAK160 peptides adopted conformations with a high α-helix content with the positive and negative bands at 190, 207 and 220 nm of lower intensity for the proline containing peptides as would be expected if these residues induced localized disruption of α-helix. However the spectrum and resultant CDPro analysis (Supp. Fig. 7) of one peptide, LAK160-P10, indicated a substantially lower α-helix content in SDS but a higher content in model membranes when compared with the other proline containing analogues. Since this peptide is modestly but significantly less potent against E. coli (NCTC9001) and P. aeruginosa than the other two LAK160 proline containing analogues we investigated the structural details of these three peptides further.
NMR structure determination
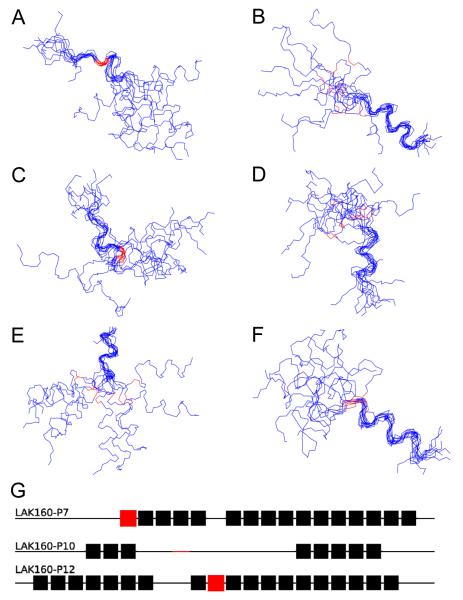
The NMR structures of the three proline containing peptides (Fig. 4) confirm that the peptides adopt mostly α-helix conformation in the presence of 50 mM SDS with conformational flexibility induced through the incorporation of proline. Importantly however, the amount of α-helix conformation, the location and size of the region of flexibility and the apparent role of the proline residues differs from one peptide to another within the LAK160-P series. The backbone atoms for each of the ten lowest energy structures were superimposed and it was found that the structures could be aligned to one of two separate helical regions in each peptide but not over the entire length. This was due to the conformational flexibility induced by the proline. The conformation adopted by the peptides is also shown schematically (Fig. 4G) where the number of structures adopting α-helix conformation at a given residue is used to reveal the predicted regions of high and low secondary structure. Notably, LAK160-P10 has two helical segments which are much smaller than those of either LAK160-P7 or LAK160-P12 which is qualitatively in agreement with the NOESY (Supp. Fig. 8), NOE connectivities (Supp. Fig. 9) and CD spectra (Fig. 2C).
Figure 4.
Superimposition of backbone atoms in each of the ten lowest energy structures for LAK160-P7 (A, B), LAK160-P10 (C, D) and LAK160-P12 (E, F) fitted to either region 1 (A, C, E) or region 2 (B, D, F). Proline residues are shown in red. Also shown is a schematic representation of the ten lowest energy structures where residues that are α-helical in more than eight structures are shown as solid boxes and those that are α-helical in seven or fewer structures are shown as a line (G). Proline residues are again marked in red.
Interestingly, regardless of the position of the proline residue, all three LAK160 peptides form helices that are flexible near the middle of the sequence although the effect of substituting proline for leucine in these three peptides may differ. Proline does not only act as a helix breaker, but also as a helix stabilising residue. Proline can stabilize an α-helix at its C-terminus by taking part in a helix capping interaction (35), in which the proline side chain caps the hydrophobic α-helix, while proline is the most water soluble of all natural amino acid residues and therefore an ideal candidate for a position at the solvent-exposed α-helix end (35). Both LAK160-P7 and LAK160-P12 have two long α-helix regions separated by a short, one or two residue region which is predominantly unstructured. In contrast, for LAK160-P10, only three turns of α-helix are observed: one turn between Leu5 and Leu7, and two turns from Leu17 to Leu21. Further, although residues Ala8, Lys9, Lys15, Ala16 and Lys22 also have a tendency to form a helical structure, as evidenced by their helical conformation in seven of the ten lowest energy structures, LAK160-P12 exhibits a greater helical region running from Lys2 to Lys22, and is only interrupted at Lys9 and Leu10. Hence even if the mentioned residues are included, LAK160-P10 is only capable of adopting a helical structure from Leu5 to Lys9 and from Lys15 to Lys22, with an unstructured region incorporating Leu10 to Trp14.
The prolines in LAK160-P7 and LAK160-P12 are in the expected position for such interactions to stabilise the N-terminus of the α-helix with the longer helix in LAK160-P12 compared with LAK160-P7 and LAK160-P10 explained by the cooperative nature of α-helix formation. Because the first three residues at the N-terminus can only form one hydrogen bond, a certain minimum number of amino acid residues capable of α-helix formation are needed. LAK160-P12, with proline at position 12 appears to satisfy this condition most effectively. If a well defined α-helix long axis is present then the angle subtended by the charged angles can be measured and the effect of proline substitution determined. The region of conformational flexibility identified in each peptide prevents identification of such an α-helix long axis. However, the structures do indicate that the amphipathic character of each peptide is retained with lysine residues segregated on one side of the structures (Supp. Fig. 10).
DISCUSSION
The prospects for the rational design of peptide antibiotics will be greatly enhanced by an understanding, not only of how such peptides operate, but also by how particular structural features contribute to a given mode of action. At present, a considerable body of work has focused on the interaction of cationic amphipathic α-helical peptides with bacterial membranes (36). While many such peptides have been shown to disrupt bacterial membranes that are rich in anionic lipids, still others have been identified as having alternative modes of action, requiring penetration within the bacteria and inhibition of a number of processes that ultimately lead to bacterial cell death (5). Furthermore, cationic antimicrobial peptides that have been shown to act against bacterial membranes may have multiple modes of action against differing bacteria or indeed multiple effects against the same cell. The lipid composition of bacterial membranes varies considerably from species to species and has been shown to be a good predictor of the activity of some antimicrobial peptides (37) with peptides that are able to effectively cluster anionic lipids in the membranes of Gram negative bacteria, particularly potent against such organisms (38-40). Magainin 2 does not effectively cluster anionic lipids, most likely due to a low density of positive charges, but its membrane disruptive behavior has been shown to depend on membrane composition (41), potentially having the ability to translocate into Gram negative bacteria. Similarly, pleurocidin, a more potent cationic α-helical peptide against Gram negative bacteria than magainin 2 (30), with strong membrane disordering capabilities (42), has been reported to have the capacity to both form membrane pores (43) and inhibit intracellular processes (44). It is likely therefore that alterations in e.g. charge density in a peptide sequence may not only enhance this property at the expense of another but may also promote one mode of action at the expense of another.
Since it remains challenging to isolate the action of a peptide that ultimately leads to bacterial cell death, and since a given peptide may have multiple effects on a bacterial cell, we are presently unable to predict how a given structural change in a peptide will influence antibacterial efficacy and mode of action. The aim of our work therefore is to precisely define the effects of structural alterations in a series of antimicrobial peptides on their biophysical properties and, ultimately, relate both of these to the efficacy and mode of action of the peptide. Here we have investigated the role of conformational flexibility conferred on the peptide through incorporation of the α-helix breaking residue proline. Proline residues in natural antimicrobial peptides and modified sequences define a hinged region which is crucial for antibacterial potency and selectivity (11, 12, 45-52). In buforin II the proline residue is proposed to operate as a translocation factor as the primary mode of antibacterial action for this peptide is considered to be the inhibition of intracellular functions (53). In a recent study we have determined that though buforin has a high affinity for nucleic acids, it adopts a low α-helix content in the presence of Gram negative inner membrane mimicking liposomes and was considerably less potent against such bacteria when compared with magainin and pleurocidin peptides (30). We were interested to see whether peptides that are designed to adopt conformations with high α-helix content but also incorporate proline residues can function as potent antibacterial agents.
In addition to causing a localized disruption of α-helix conformation, proline may have a number of effects on cationic α-helical antimicrobial peptide activity. These include: the prevention of peptide self-association (12) leading to improved access through bacterial lipopolysaccharides to the bacterial inner membrane (54); the perturbation of the charged segment in the helical wheel, a key determinant of activity and selectivity (24); and the ability to translocate across the inner membrane opening up intracellular modes of bactericidal action. Consistent with previous work (12), we have shown that proline has the ability to affect peptide supramolecular organization and that this, in turn, is linked to greater potency against Gram negative bacteria. Related observations have been made for other antimicrobial peptides; the cyclisation of magainin and the substitution of amino acids of varying hydrophobicity on model membrane active peptides led to reductions of antibacterial activity that were attributed to reduced flexibility and greater oligomerisation respectively (55, 56). Importantly however, we have demonstrated that the positioning of the proline residue is crucial to the activity and that the structures adopted by the peptides in the presence of anionic SDS were defined by the location of the proline residue since this affected not only the location of the hinge region but also the conformation adopted in more remote regions of the sequence.
In addition, we have shown that the disruption of peptide structural ordering and its associated effects on supramolecular organization in solution is also linked to the anti-plasmodial selectivity and ability to penetrate erythrocytes of all D-amino acid peptides and influences their cytotoxic activity against mammalian cancer cell lines. Understanding the antibacterial mode of action of AMPs has been the focus of a large body of work. Much less is known about how the same or related peptides act against eukaryotic targets such as Pl. falciparum. The present study shows that the incorporation of proline has a dramatic effect on the haemolytic potential of AMPs but that anti-plasmodial activity is maintained. This indicates that the mechanism of AMP induced haemolysis is distinct from that used to inhibit proliferation of Pl. falciparum. Furthermore, since the infected erythrocyte is not lysed and the parasite effectively inhibited, the inference is that proline containing peptides such as D-LAK120-AP13 or D-LAK120-HP13 are capable of penetrating erythrocytes to attack the intracellular pathogen. Proline free peptides are incapable of this and the observed anti-plasmodial activity is difficult to separate from erythrocyte lysis. While incorporation of proline in AMPs protects human erythrocytes from lysis, its presence enhances toxicity against nucleated cancer cells; again suggesting distinct mechanisms. The absence of cytotoxicity tests against primary human cells precludes any evaluation of the peptides as potential anti-cancer agents but nevertheless suggests this feature may be beneficial. Related observations have shown that the lytic activity of anti-cancer peptides is inhibited by heparin sulphate on the surface of the target cells and that smaller peptides are better able to overcome this barrier (57, 58). Analogously, the incorporation of proline in larger AMPs and the concomitant prevention of self-association may aid their access to tumour cell membranes and enhance cytotoxic activity. Hence, while the extra-cellular barriers may differ from Gram negative bacteria to P. falciparum and mammalian cells, the incorporation of proline may have a common effect in facilitating access to the target membrane.
In conclusion, our findings suggest that the environmental dependent structure adopted by antimicrobial peptides remains a determinant of activity against, and selectivity towards, a range of targets including Gram negative bacteria, Pl. falciparum and M. tuberculosis and that the main contribution of proline hinge regions is to modify the secondary structure and supramolecular assembly of the peptides in solution. This general feature may facilitate access to target membranes and affect the mode of interaction on arrival. The three dimensional structures of three proline containing peptides show that significant changes in structure and activity are determined by the positioning of proline residues and that careful consideration of these effects is required to optimize potency against each target pathogen.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the Medical Research Council (NIRG G0801072/87482 to AJM), the Wellcome Trust (VIP Award to AJM and Capital Award for the KCL Centre for Biomolecular Spectroscopy), the University of London Central Research Fund (AR/CRF/B), Applied Photophysics Ltd, the Research Fund for Control of Infectious Diseases (RFCID 11100682; JKWL and AJM) and the Hong Kong SAR University of Hong Kong Seed Funding Programme (JKWL). We are grateful to Drs K.D. Bruce and T. Spasenovski for their assistance in setting up antibacterial testing in the Molecular Microbiology Research Laboratory (IPS), the Tuberculosis Laboratory, Grantham Hospital, Hong Kong for hosting experiments and RC Hider for valuable discussions.
REFERENCES
- 1.Hancock REW, Sahl H-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nature Biotech. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 2.Reddy KVR, Yedery RD, Aranha C. Antimicrobial peptides: premises and promises. Int. J. Antimicrob. Agents. 2004;24:536–547. doi: 10.1016/j.ijantimicag.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Marr AK, Gooderham WJ, Hancock REW. Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr. Opin. Pharmacol. 2006;6:468–472. doi: 10.1016/j.coph.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Gordon YJ, Romanowski EG, McDermott AM. A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr. Eye Res. 2005;30:505–515. doi: 10.1080/02713680590968637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 6.Dathe M, Wieprecht T. Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells. Biochim. Biophys. Acta. 1999;1462:71–87. doi: 10.1016/s0005-2736(99)00201-1. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Benz R, Hancock REW. Influence of proline residues on the antibacterial and synergistic activities of α-helical peptides. N-Terminal analogs of cecropin A: synthesis, antibacterial activity, and conformational properties. Biochemistry. 1999;38:8102–8111. doi: 10.1021/bi9904104. [DOI] [PubMed] [Google Scholar]
- 8.Andreu D, Merrifield RB, Steiner H, Boman HG. N-Terminal analogs of cecropin A: synthesis, antibacterial activity, and conformational properties. Biochemistry. 1985;24:1683–1688. doi: 10.1021/bi00328a017. [DOI] [PubMed] [Google Scholar]
- 9.Gazit E, Boman A, Boman HG, Shai Y. Interaction of the mammalian antibacterial peptide cecropin P1 with phospholipid vesicles. Biochemistry. 1995;34:1683–1688. doi: 10.1021/bi00036a021. [DOI] [PubMed] [Google Scholar]
- 10.Chen HC, Brown JH, Morell JL, Huang CM. Synthetic magainin analogues with improved antimicrobial activity. FEBS Lett. 1988;236:462–466. doi: 10.1016/0014-5793(88)80077-2. [DOI] [PubMed] [Google Scholar]
- 11.Song YM, Yang S-T, Lim SS, Kim Y, Hahm K-S, Kim JI, Shin SY. Effects of L- or D-Pro incorporation into hydrophobic or hydrophilic face of amphipathic α-helical model peptide on structure and cell selectivity. Biochem. Biophys. Res. Commun. 2004;314:615–621. doi: 10.1016/j.bbrc.2003.12.142. [DOI] [PubMed] [Google Scholar]
- 12.Yang S-T, Lee JY, Kim H-J, Eu Y-J, Shin SY, Hahm K-S,, Kim JI. Contribution of a central proline in model amphipathic α-helical peptides to self-association, interaction with phospholipids, and antimicrobial mode of action. FEBS J. 2006;273:4040–4054. doi: 10.1111/j.1742-4658.2006.05407.x. [DOI] [PubMed] [Google Scholar]
- 13.Tatham AS, Hider RC, Drake AF. The effect of counterions on melittin aggregation. Biochem. J. 1983;211:683–686. doi: 10.1042/bj2110683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nature Protocols. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 15.Bacon DJ, Latour C, Lucas C, Colina O, Ringwald P, Picot S. Comparison of a SYBR Green I-based assay with a Histidine-Rich Protein II enzyme-linked immunosorbent assay for in vitro antimalarial drug efficacy testing and application to clinical isolates. Antimicrob. Agents Chemother. 2007;51:1172–1178. doi: 10.1128/AAC.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alley MC, Scudiero DA, Monks A, Hursey ML, Czerwinski MJ, Fine DL,, Abbott BJ, Mayo JG, Shoemaker RH, Boyd MR. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 1988;48:589–601. [PubMed] [Google Scholar]
- 17.Sreerama N, Woody RW. Estimation of protein secondary structure from CD spectra: Comparison of CONTIN, SELCON and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 18.Ladokhin AS, Jayasinghe S, White SH. How to Measure and Analyze Tryptophan Fluorescence in Membranes Properly, and Why Bother? Anal. Bioch. 2000;285:235–245. doi: 10.1006/abio.2000.4773. [DOI] [PubMed] [Google Scholar]
- 19.Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: [Google Scholar]
- 20.Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, Nilges M. ARIA2: Automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 21.Fossi M, Oschkinat H, Nilges M, Ball LJ. Quantitative study of the effects of chemical shift tolerances and rates of SA cooling on structure calculation from automatically assigned NOE data. J. Magn. Reson. 2005;175:92–102. doi: 10.1016/j.jmr.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 22.Laskowski RA, Rullmann JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 23.Mason AJ, et al. Structural determinants of antimicrobial and antiplasmodial activity and selectivity in histidine-rich amphipathic cationic peptides. J. Biol. Chem. 2009;284:119–133. doi: 10.1074/jbc.M806201200. [DOI] [PubMed] [Google Scholar]
- 24.Wieprecht T, Dathe M, Epand RM, Beyermann M, Krause E, Maloy WL, MacDonald DL, Bienert M. Influence of the angle subtended by the positively charged helix face on the membrane activity of amphipathic, antibacterial peptides. Biochemistry. 1997;36:12869–12880. doi: 10.1021/bi971398n. [DOI] [PubMed] [Google Scholar]
- 25.Jin Y, Hammer J, Pate M, Zhang Y, Zhu F, Zmuda E, Blazyk J. Antimicrobial activities and structures of two linear cationic peptide families with various amphipathic β-sheet and α-helical potentials. Antimicrob. Agents Chemother. 2005;49:4957–4964. doi: 10.1128/AAC.49.12.4957-4964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishikawa M, Ogawa K. Occurrence of D-histidine residues in antimicrobial poly(arginyl–histidine), conferring resistance to enzymatic hydrolysis. FEMS Microbiol. Lett. 2004;239:255–259. doi: 10.1016/j.femsle.2004.08.043. [DOI] [PubMed] [Google Scholar]
- 27.Hong SY, Oh JE, Lee K-H. Effect of D-amino acid substitution on the stability, the secondary structure, and the activity of membrane-active peptide. Biochem. Pharmacol. 1999;58:1775–1780. doi: 10.1016/s0006-2952(99)00259-2. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Mant CT, Farmer SW, Hancock REW, Vasil ML, Hodges RS. Rational design of α-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J. Biol. Chem. 2005;280:12316–12329. doi: 10.1074/jbc.M413406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellesia G, Shea J-E. What determines the structure and stability of KFFE monomers, dimers, and protofibrils? Biophys. J. 2009;96:875–886. doi: 10.1016/j.bpj.2008.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lan Y, Ye Y, Kozlowska J, Lam JKW, Drake AF, Mason AJ. Structural contributions to the intracellular targeting strategies of antimicrobial peptides. Biochim. Biophys. Acta. 2010;1798:1934–1943. doi: 10.1016/j.bbamem.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lakowicz JT. Principles of Fluorescence Spectroscopy. Plenum Press; New York: 1983. [Google Scholar]
- 32.Chen Y, Liu B, Yu H-T, Barkley MD. The peptide bond quenches indole fluorescence. J. Am. Chem. Soc. 1996;118:9271–9278. [Google Scholar]
- 33.Drake AF, Siligardi G, Gibbons WA. Reassessment of the electronic circular dichroism criteria for random coil conformations of poly(L-lysine) and the implications for protein folding and denaturation studies. Biophys. Chem. 1988;31:143–146. doi: 10.1016/0301-4622(88)80019-x. [DOI] [PubMed] [Google Scholar]
- 34.Durfee T, et al. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J. Bacteriol. 2008;190:2597–2606. doi: 10.1128/JB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aurora R, Rose GD. Helix capping. Protein Sci. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 37.Epand RM, Rotem S, Mor A, Berno B, Epand RF. Bacterial Membranes as Predictors of Antimicrobial Potency. J. Am. Chem. Soc. 2008;130:14346–14352. doi: 10.1021/ja8062327. [DOI] [PubMed] [Google Scholar]
- 38.Epand RM, Epand RF. Bacterial membrane lipids in the action of antimicrobial agents. J. Pept. Sci. 2010;17:298–305. doi: 10.1002/psc.1319. [DOI] [PubMed] [Google Scholar]
- 39.Epand RF, Maloy L, Ramamoorthy A, Epand RM. Amphipathic helical cationic antimicrobial peptides promote rapid formation of crystalline states in the presence of phosphatidylglycerol: lipid clustering in anionic membranes. Biophys. J. 2010;98:2564–2573. doi: 10.1016/j.bpj.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Epand RF, Maloy WL, Ramamoorthy A, Epand RM. Probing the “charge cluster mechanism” in amphipathic helical cationic antimicrobial peptides. Biochemistry. 2010;49:4076–4084. doi: 10.1021/bi100378m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregory SM, Pokorny A, Almeida PFF. Magainin 2 revisited: a test of the Quantitative Model for the all-or-none permeabilization of phospholipid vesicles. Biophys. J. 2009;96:116–131. doi: 10.1016/j.bpj.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mason AJ, Chotimah INH, Bertani P, Bechinger B. A spectroscopic study of the membrane interaction of the antimicrobial peptide Pleurocidin. Mol. Membr. Biol. 2006;23:185–194. doi: 10.1080/09687860500485303. [DOI] [PubMed] [Google Scholar]
- 43.Saint N, Cadiou H, Bessin Y, Molle G. Antibacterial peptide pleurocidin forms ion channels in planar lipid bilayers. Biochim. Biophys. Acta. 2002;1564:359–364. doi: 10.1016/s0005-2736(02)00470-4. [DOI] [PubMed] [Google Scholar]
- 44.Patrzykat A, Friedrich CL, Zhang L, Mendoza V, Hancock REW. Sublethal concentrations of Pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob. Agents Chemother. 2002;46:605–614. doi: 10.1128/AAC.46.03.605-614.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobayashi S, Takeshima K, Park CB, Kim SC, Matsuzaki K. Interactions of the novel antimicrobial peptide Buforin 2 with lipid bilayers: proline as a translocation promoting factor. Biochemistry. 2000;39:8648–8654. doi: 10.1021/bi0004549. [DOI] [PubMed] [Google Scholar]
- 46.Kobayashi S, Chikushi A, Tougu S, Imura Y, Nishida M, Yano Y, Matsuzaki K. Membrane translocation mechanism of the antimicrobial peptide buforin 2. Biochemistry. 2004;43:15610–15616. doi: 10.1021/bi048206q. [DOI] [PubMed] [Google Scholar]
- 47.Park CB, Yi K-S, Matsuzaki K, Kim MS, Kim SC. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: the proline hinge is responsible for the cell penetrating ability of buforin II. Proc. Natl. Acad. Sci. USA. 2000;97:8245–8250. doi: 10.1073/pnas.150518097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao Y, Herrera AI, Bommineni YR, Soulages JL, Prakash O, Zhang G. The central kink region of fowlicidin-2, an α-helical host defense peptide, is critically involved in bacterial killing and endotoxin neutralization. J. Innate Immun. 2009;1:268–280. doi: 10.1159/000174822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J-K, Lee S-A, Shin S, Lee J-Y, Jeong K-W, Nan YH, Park YS, Shin SY, Kim Y. Structural flexibility and the positive charges are the key factors in bacterial cell selectivity and membrane penetration of peptoid-substituted analog of Piscidin 1. Biochim. Biophys. Acta. 2010;1798:1913–1925. doi: 10.1016/j.bbamem.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 50.Lee SA, Kim YK, Lim SS, Zhu LW, Ko H, Shin SY, Hahm KS, Kim Y. Solution structure and cell selectivity of piscidin 1 and its analogues. Biochemistry. 2007;46:3653–3663. doi: 10.1021/bi062233u. [DOI] [PubMed] [Google Scholar]
- 51.Pukala TL, Brinkworth CS, Carver JA, Bowie JH. Investigating the importance of the flexible hinge in caerin 1.1: solution structures and activity of two synthetically modified caerin peptides. Biochemistry. 2004;43:937–944. doi: 10.1021/bi035760b. [DOI] [PubMed] [Google Scholar]
- 52.Porcelli F, Buck B, Lee D-K,, Hallock KJ, Ramamoorthy A, Veglia G. Structure and orientation of pardaxin determined by NMR experiments in model membranes. J. Biol. Chem. 2004;279:45815–45823. doi: 10.1074/jbc.M405454200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park CB, Kim HS, Kim SC. Mechanism of action of the antimicrobial peptide buforin II: Buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Comm. 1998;244:253–257. doi: 10.1006/bbrc.1998.8159. [DOI] [PubMed] [Google Scholar]
- 54.Papo N, Shai Y. Can we predict biological activity of antimicrobial peptides from their interactions with model phospholipid membranes? Peptides. 2003;24:1693–1703. doi: 10.1016/j.peptides.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 55.Unger T, Oren Z, Shai Y. The effect of cyclization of magainin 2 and melittin analogues on structure, function, and model membrane interactions: implication to their mode of action. Biochemistry. 2001;40:6388–97. doi: 10.1021/bi0026066. [DOI] [PubMed] [Google Scholar]
- 56.Avrahami D, Oren Z, Shai Y. Effect of multiple aliphatic amino acids substitutions on the structure, function, and mode of action of diastereomeric membrane active peptides. Biochemistry. 2001;40:12591–603. doi: 10.1021/bi0105330. [DOI] [PubMed] [Google Scholar]
- 57.Fadnes B, Rekdal Ø, Uhlin-Hansen L. The anticancer activity of lytic peptides in inhibited by heparan sulphate on the surface of the tumour cells. BMC Cancer. 2009;9:183. doi: 10.1186/1471-2407-9-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fadnes B, Uhlin-Hansen L, Lindin I, Rekdal Ø. Small lytic peptides escape the inhibitory effect of heparan sulfate on the surface of cancer cells. BMC Cancer. 2011;11:116. doi: 10.1186/1471-2407-11-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.