Abstract
Within the developing vertebrate retina, particular subtypes of amacrine cells (ACs) tend to arise from progenitors expressing the bHLH transcription factor, Atoh7, which is necessary for the early generation of retinal ganglion cells (RGCs). All ACs require the post-mitotic expression of the bHLH transcription factor Ptf1a, however Ptf1a alone is not sufficient to give subtype identities. Here we use functional and in vivo time-lapse studies in the zebrafish retina to investigate on the developmental programs leading to ACs specification within the subsequent divisions of Atoh7-positive progenitors. We find evidences that the homeobox transcription factor Barhl2 is an AC subtype identity-biasing factor that turns on within Atoh7-positive descendants. In vivo lineage tracing reveals that particular modes of cell division tend to generate Barhl2-positive precursors from sisters of RGCs. Additionally, Atoh7 indirectly impacts these division modes to regulate the right number of barhl2-expressing cells. We finally find that Atoh7 itself influences the subtypes of Barhl2-dependent ACs. Taken together, our study uncovers lineage-related and molecular logic of subtype specification in the vertebrate retina, by showing that specific AC subtypes arise via a particular mode of cell division and a transcriptional network cascade involving the sequential expression of first atoh7 followed by ptf1a and then barhl2.
Keywords: fate determination, cell lineage, Barhl2, subtype specification, retina, zebrafish
INTRODUCTION
A major challenge in vertebrate neurobiology is to understand how the developmental programs of a neural progenitor cell is regulated in vivo in the context of cell lineages and modes of cell division. Within the vertebrate retina, some types of neurons tend to be lineally related or descendants of common progenitor cells (Poggi et al., 2005b; Vitorino et al., 2009; Feng et al., 2010; Brzezinski et al., 2011; Jusuf et al., 2011). The bHLH transcription factor Atoh7 (a.k.a Ath5) is required for RGCs development (Brown et al., 2001; Kay et al., 2001; Vetter and Brown, 2001; Wang et al., 2001; Ghiasvand et al., 2011), and turns on just before mitosis that precedes their birth (Poggi et al., 2005b). One cell from this mitosis differentiates as a RGC. However, many other cell types, including some subtypes of ACs also come from atoh7 expressing progenitors (Poggi et al., 2005b; Feng et al., 2010; Jusuf et al., 2011). The sisters of RGCs must therefore generate these other cell types.
The fates of all retinal neurons that primarily express the inhibitory neurotransmitters GABA or glycine (horizontal cells and ACs) require the expression of the Pancreas transcription factor 1a (Ptf1a) (Fujitani et al., 2006; Dullin et al., 2007; Nakhai et al., 2007; Jusuf et al., 2011). All ACs express Ptf1a, but Ptf1a alone is not sufficient to confer subtype-specificity (Jusuf et al., 2011). However, precursors that express both atoh7 and ptf1a tend to differentiate into specific subtypes of ACs, thus suggesting that other key factors might regulate AC subtypes within this lineage (Jusuf et al., 2011).
Barhl homeobox transcription factors have been implicated in ACs diversity and RGC development downstream of Atoh7 (Poggi et al., 2004; Ding et al., 2009). Targeted disruption of barhl2 alters AC subtype composition and survival of RGCs (Ding et al., 2009). Nothing is known on the lineage-origin of barhl2-expressing cells, the networks in which Barhl2 specifies AC subtypes, or how it works in relation to genes that drive the same (ptf1a) or alternate fates (atoh7). In zebrafish, additional whole genome duplication has generated another barhl paralog (Reig et al., 2007; Schuhmacher et al., 2011). Barhl1.2 is specifically expressed in RGCs, while barhl2 is expressed in ACs (Schuhmacher et al., 2011). This led us to investigate the distinct role of Barhl2 as an AC subtype-biasing factor downstream of Atoh7. We found that barhl2-expressing precursors arise within the Atoh7-lineage. Barhl2 expression, however, does not depend on Atoh7, but on Ptf1a, and is necessary and sufficient for biasing AC subtypes. Additionally, Atoh7 affects the identities of Barhl2-dependent ACs. With timelapse imaging (Poggi et al., 2005b; Poggi et al., 2005a) we traced the origins of Barhl2-positive cells. We found that these cells arise as one of the two post-mitotic daughters of a dividing RGC’s sister, i.e. Barhl2 ACs tend to be nieces of RGCs. Our study provides in vivo evidences that modes of cell division and lineage-restricted cell fate determination programs regulate the correct number of neuronal subtypes within particular progenitor pools.
MATERIALS AND METHODS
Animals and ethics statements
Zebrafish breeding / raising followed standard protocols. Fish were maintained at 26.5°C and embryos raised at 28.5°C or 32°C and staged as described (Kimmel et al., 1995). Fish were housed in three facilities: Fish facility of our German laboratory (built in accordance to Tierschutzgesetz 111, Abs. 1, Nr. 1 and with European Union animal welfare guidelines); fish facility at the University of Cambridge, UK; and FishCore at Monash University, Australia. Each facility is under supervision of and in accordance with local animal welfare agencies. Zebrafish (Danio rerio) embryos of either sex were used exclusively prior to free-feeding stages. Embryos used for whole-mount imaging were treated with 0.0045% 1-Phenyl-2-Thiourea (PTU) (Sigma, Gillingham, Dorset, UK) to delay pigment formation.
Fish lines
Seven transgenic lines expressing GFP, dsRed, gap43-GFP or gap43-RFP under the control of different promoters were used in this study: Tg(barhl2:GFP) line (Kinkhabwala et al., 2011); Tg(ptf1a:GFP) line (Godinho et al., 2005) kindly provided by Steven D. Leach (John Hopkins Medical Institutions, Baltimore, USA); Tg(atoh7:gap43-GFP), Tg(atoh7:GFP), Tg(atoh7:gap43-RFP) and Tg(atoh7:gal4/pUAS:gap43-GFP) lines (Zolessi et al., 2006). For the Tg(ptf1a:dsRed) line (Tg(-5.5ptf1a:DsRed)ia6) we created a plasmid containing 5.5 kb of the 5′ region of the ptf1a gene cloned upstream of DsRed2 in pT2AL200R150G vector (Kawakami, 2004). The plasmid was injected with Tol2 transposase mRNA and F1 progeny of different insertion lines was screened. The ia6 allele faithfully represents the endogenous expression of Ptf1a mRNA and has a comparable expression pattern to that of the previously characterised Tg(ptf1a:GFP) with the dsRed showing only a slight delay in expression (data not shown). Double transgenic lines where generated via outcrossing.
Morpholino injection
Translation blocking morpholino oligonucleotides (MO) obtained from Gene Tools, LLC (Oregon, USA) were reconstituted as 1mM stock solutions in water and injected into the yolk of 1 - 2 cell stage embryos. A MO targeted against a region 44 bp upstream from the translational start site with sequence 5′-TTGCCCAGTAACAACAATCGCCTAC-3′ was used to knockdown ptf1a (10 – 12ng / embryo) (Lin et al., 2004; Jusuf et al., 2011). A MO with sequence 5′TTCATGGCTCTTCAAAAAAGTCTCC-3′was used to knockdown atoh7 (Pittman et al., 2008). A barhl2 translation MO targetting 6 bp upstream from the translational start site with the sequence 5′-AGAAAAGGATGAGCACTCAAGTCGT-3′ was designed and injected at 0.5mM / embryo. Injections of standard control morpholino with sequence 5′-CCTCTTACCTCAGTTACAATTTATA-3′ up to 12 ng had no effect. The 5 bp Barhl2 mismatch MO with sequence 5′-AGAATACGATCAGCACTGAACTCGT-’3′ is also comparable to uninjected (data not shown). Cell-autonomous role of Barhl2 was assessed using transplantation technique. Briefly, 10 – 20 cells were transplanted from blastula stage donors (cells labelled with H2A-GFP or H2B-RFP RNA) into the animal poles of blastula stage host embryos. Integration and survival of transplanted cells was aided by injecting p53 MO with sequence 5′-GCGCCATTGCTTTGCAAGAATTG-3′ into donor embryos. Retinas injected with standard control MO together with p53 MO display normal expression of Barhl2 protein as shown by antibody staining (data not shown).
Overexpression plasmid cloning
The coding sequence of barhl2 was PCR amplified using 5′-ATGGAAGGATCCAGTGGGGCTAGT-3′ and 5′-CCGAGCATGCGGTGTGCC-3′ for which the reverse primer was tagged with the T2A sequence 5′-AGGGCCGGGATTCTCCTCCACGTCACCGCATGTTAGAAGACTTCCTCTGCCCTC-3′ (Kim et al., 2011). H2B-RFP fused coding sequences were PCR amplified using 5′-ATGCCAGAGCCAGCGAAGTCT-3′ and 5′-GATGTACACGGCGCCGGT-3′ primers for which the forward primer was tagged with the T2A sequence 5′-GAGGGCAGAGGAAGTCTTCTAACATGCGGTGACGTGGAGGAGAATCCCGGCCC-3′. A fusion PCR was performed to generate the barhl2-t2a-h2b-rfp product, which was cloned into a pUAS vector containing 16 cassettes of the Upstream Activation Sequence and recognition sequences for I-SceI meganuclease for efficient transgenesis.
Immunohistochemistry
Primary antibodies were diluted in blocking solution: rabbit anti-calretinin (Chemicon AB5054; 1:1000), rabbit anti-Sox2 (Chemicon AB5603, 1:200), mouse anti-parvalbumin (Chemicon MAB1572, 1:1000), rabbit anti-GABA (Sigma A2052, 1:500), rabbit anti-calbindin (Calbiochem PC253L, 1:500), rabbit anti-serotonin (Sigma S5545, 1:50), mouse anti-tyrosine hydroxylase (Millipore MAB 318, 1:100), rabbit anti-Neuropeptide Y (Immunostar 22940, 1:500), rabbit anti-Barhl2 (Santa Cruz Biotechnology sc-68370, 1:50 for immunohistochemistry, 1:1000 for Western blot). Secondary antibodies were goat or donkey anti-mouse, anti-rabbit or anti-goat IgG conjugated to Alexa 488, 546, 594 or 647 fluorophores (1:1,000 - 1:2,000; Molecular Probes, Eugene, OR, USA).
For most antibodies, embryos were fixed in 4% paraformaldehyde (PFA) in 0.1M PB overnight (maximum 2 hours for ChAT immunohistochemistry) at 4°C, rinsed, cryoprotected in 30% sucrose, embedded in OCT and cryosectioned at 14μm thickness. For GABA immunohistochemistry, embryos were fixed in 4% PFA / 0.05% glutaraldehyde, 5mM EGTA, 5mM MgSO4, 0.1% triton-X100 in 0.1M PB for 3 hours at room temperature. All staining steps are performed at room temperature unless stated otherwise. For Sox2 immunohistochemistry antigen retrieval was performed by immersing sections in 0.01M sodium citrate buffer, pH 6.0 at 95 °C for 10 min prior to blocking. All sections were incubated in blocking solution (10% heat-inactivated goat serum (HIGS), 1% bovine serum albumin, 0.2% Triton X-100 in PBS) for 30 minutes (sections) or 60 minutes (wholemounts). For staining with goat anti-ChAT antibody, sections were blocked in 10% donkey serum instead. Sections were incubated in primary antibodies overnight, secondary antibodies for 60 minutes, and nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Sections were mounted with Fluorosave (Calbiochem, Merck Chemicals Ltd., Nottingham, UK) or Mowiol.
Whole-mount single and fluorescent in situ hybridization
In situ mRNA hybridization was performed as described (Lin et al., 2004). The full-length cDNA barhl2 sequence was subcloned from a zebrafish IMAGE clone (IMAGE: 7452725; IRBOp991F0870D, Source BioScience UK Limited, Nottingham, UK) in pME18S-FL3 into a pCS2+ vector to generate digoxigenin- and fluorescein-labelled riboprobes. For antisense probe, we linearized with NotI (Fermentas or New England Biology) and transcribed with Sp6 (mMessage mMachine® Sp6, Ambion). For sense probe, we linearized with BamHI (Fermentas or New England Biology) and transcribed with T7 (mMessage mMachine® T7, Ambion). Atoh7 probes were generated as described (Schuhmacher et al., 2011). Ptf1a probes were generated directly by RTPCR (one step RTPCR kit, Qiagen) using total mRNA extracted from zebrafish embryos of 50 hpf and 5′TTCGAGAGACCACTTGGACA3′ forward primer and T7 tailed 5′-CCAAGCTTCTAATACGACTCACTATAGGGAGAGGCTGAAACACAGATAGTCACAA-3′ reverse primer. Single probe in situ hybridization was done as described (Thisse and Thisse, 2008) with minor modifications. Embryos underwent a stepwise dehydration series into 100% methanol and subsequent rehydration into 0.1% Tween in PBS. Permeabilization was achieved using age-dependent concentrations of proteinase K treatment at room temperature, followed by postfixation in 4% PFA in PBS. After prehybridization, hybridization with digoxigenin-UTP labelled probes (Roche Applied Science) was performed overnight at 65°/68 °C. Signal was detected with nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolyl-phosphate, toluidine salt (NBT/BCIP BM Purple, Roche Products Ltd., Welwyn Garden City, UK).
For double fluorescent whole mount in situ hybridization (FISH) standard digoxigenin- and fluorescein-labelled riboprobes were combined with Tyramide Signal Amplification (TSA), as described in Schumacher et al., 2011. Riboprobes were incubated for 30 minutes (barhl2), 40 minutes (atoh7) or 24 hours (ptf1a). Embryos were kept in the dark for following steps. Embryos were washed with TNT (0.1M Tris pH7.5, 0.15M NaCl, 0.1% Tween20), incubated with 1% H2O2 in TNT for 20 minutes, washed several times and blocked with TNB (2% DIG Block [Roche] in TNT) for 1 hour followed by incubation with anti-digoxigenin-POD Fab fragments (Roche, 1:50 in TNB). Signal was detected using Fluorescein-Tyramide (FITC), Cyanine3-Tyramide (Cy3) or Cyanine5-Tyramide (Cy5) fluorophores (PerkinElmer). Embryos were incubated in DAPI in TNT overnight at 4°C and washed with TNT.
Imaging
NBT/BCIP stained embryos were mounted in 87% glycerol and imaged with Leica DM5000B compound microscope at 10x or 20x. Images were acquired with a Leica CD500 camera using Leica FireCam 1.7.1. FISH embryos were mounted on 100 × 15 mm glass bottom Petri dishes using 1% low melting agarose and z-stacks taken at the Leica SP5 confocal microscope.
Images of fixed and live embryos were acquired on a dissecting stereomicroscope equipped with epi-fluorescence (Leica MZ FLIII). Photomicrography of whole-mount eyes or sections was performed with a laser confocal system (Leica TCS-NT, Leica SpE or leica Sp5 confocal laser scanning microscopes using a Leica 40x, 1.2 NA or Leica 63x, 1.2 NA water immersion objectives) or Nikon fluorescence microscopes, equipped with cooled charge-coupled device (CCD) Hamamatsu Orca cameras and automated z-drive and fluorescence shutters.
At the confocal, excitation was achieved with following laser lines: 405nm (DAPI), 488nm argon (GFP, Alexa 488), 568nm (RFP, DsRed, Alexa 546), 594nm (Alexa 594) and 633nm (Alexa 647). Images were taken through whole-mount fixed and live embryos as described (Poggi et al., 2005b). Sequential image acquisition was performed with emission detected at 500-550nm (FITC), 650-700nm (Cy3), 650-800nm (Cy5) and 400-500nm (DAPI) using individual descanned PMT detectors. Optical sections (40 – 60μm for timelapse and < 100μm for fixed embryos) of 1 μm thickness were taken and Kalmann averaged 2 or 4 times. For time-lapse, images were taken every 5 or 10 minutes for 24 – 42 hours. Motorized XY stage was used to image multiple embryos. Laser power was minimized to avoid bleaching and phototoxicity.
Image processing and cell tracking
Image data was acquired using Leica Application Suite (LAS), Leica TCS NT or Leica LCS software, processed and analysed using Volocity Analysis version 5.3 (Improvision, Coventry, UK). Brightness and contrast were adjusted with Adobe Photoshop CS3 and CS4. Cell tracking was performed using the Volocity classification module (Improvision). Double-labelled Barhl2:GFP/Atoh7:gap43-RFP cells were randomly selected and Atoh7:gap43-RFP positive cells tracked backward in time.
Analysis
Numbers used for each analysis and age (generally 4 or 5 dpf) are indicated in the result section or figure legend. The majority of quantification was performed in the central retina in which the relatively older neurons express the proteins recognised by the antibodies. The region was defined by drawing a straight line through the centre of the lens just central to the ciliary margin as described previously (Holt et al., 1988). The half covering the retina was subdivided into four segments and quantification performed in the central two. For transplanted cells of rare ACs, the whole retina was included. For markers with < 10 cells per section, 5 sections were combined (i.e. serotonin, tyrosine hydroxylase and neuropeptide Y). Subtypes of barhl2-expressing AC subtypes were classified morphologically as described previously (Jusuf and Harris 2009).
Statistical analysis
Statistical tests were performed using Prism software with p < 0.05 used as criterion level. Comparison of morphologically characterised subtypes from barhl2-expressing cells: Fisher’s exact test. Tests used for atoh7 knock down experiment: unpaired t-test with Welch’s correction, Gaussian distribution, 2-tailed (GFP and GABA quantification); Fisher’s exact test, 2-sided and α <0.05 (Serotonin and NY quantification). Tests used for barhl2 over-expression experiment with GABA: Mann-Whitney test, Gaussian approximation, 2-tailed. For barhl2 knockdown transplantation experiments, the binomial test was used, as the number of cells per image was too low to compare means.
RESULTS
Zebrafish barhl2 is expressed differentially in subtypes of inhibitory cells
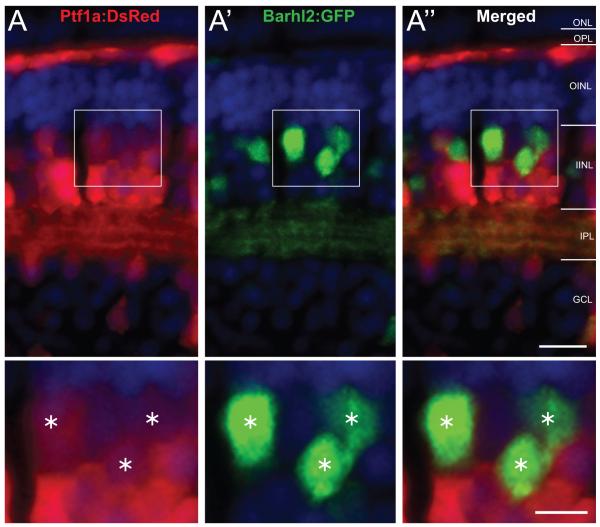
To investigate the possibility that the Barhl2 paralog in the zebrafish plays a specific role as an amacrine subtype specification factor within the Atoh7-lineage, we first assessed its gene expression with respect to the pancreas transcription factor 1a (ptf1a). Ptf1a specifies all inhibitory (defined here as primarily expressing the inhibitory GABA or glycine neurotransmitters) neurons (ACs and HCs) and is excluded from excitatory (defined here as expressing the excitatory glutamate neurotransmitter) cells (Jusuf and Harris, 2009). We used the double transgenic line Tg(ptf1a:dsRed/barhl2:GFP) to assess whether cells expressing barhl2 also express ptf1a. Quantification in 4 days post-fertilization (dpf) Tg(ptf1a:dsRed/barhl2:GFP) transgenic embryos showed that Barhl2:GFP cells co-labelled with Ptf1a:DsRed signal (94.9% ± 0.5% SEM) primarily in ACs (Figure 1). Additionally, the few Barhl2:GFP cells within the ganglion cell layer (GCL) co-expressed Ptf1a:DsRed, suggesting that these are displaced ACs. One striking observation was that, although all Barhl2:GFP cells are Ptf1a-positive, only about 58.5% (± 1.2% SEM) of Ptf1a-expressing ACs are Barhl2:GFP-positive, suggesting that Barhl2 marks a subpopulation of Ptf1a-derived ACs (Figure 1). We found that some Ptf1a:DsRed-positive HCs are also Barhl2:GFP-positive (Mo et al., 2004; Ding et al., 2009). These cells however are mainly located in the retinal periphery, and in these HCs, Barhl2:GFP expression was highly variable (data not shown). Thus, stable barhl2 paralog expression in the zebrafish retina is largely restricted to ACs.
Figure 1.
Barhl2:GFP labels a subpopulation of Ptf1a:DsRed cells. Micrographs of retinal sections from Tg(ptf1a:dsRed/barhl2:GFP). Cell nuclei (in blue) are stained with DAPI. All of the Barhl2-positive cells are positive for varying degrees of Ptf1a. In contrast, only some of the DsRed cells also express GFP, showing that a subpopulation of ptf1a-expressing cells expresses barhl2. Most of these double-labelled cells are found in the amacrine population in the INL. Results quantified from n = 58 eyes, 5358 cells in central retina of 96 hpf embryos. ONL: outer nuclear layer; OPL: outer plexiform layer; OINL: outer half of the inner nuclear layer; IINL: inner half of the inner nuclear layer; IPL: inner plexiform layer; GCL: ganglion cell layer. Asterisks mark cells expressing both GFP and dsRed. Scale bar A” (for A – A”) = 20 μm, scale bar A” inset (for A – A” insets) = 10 μm.
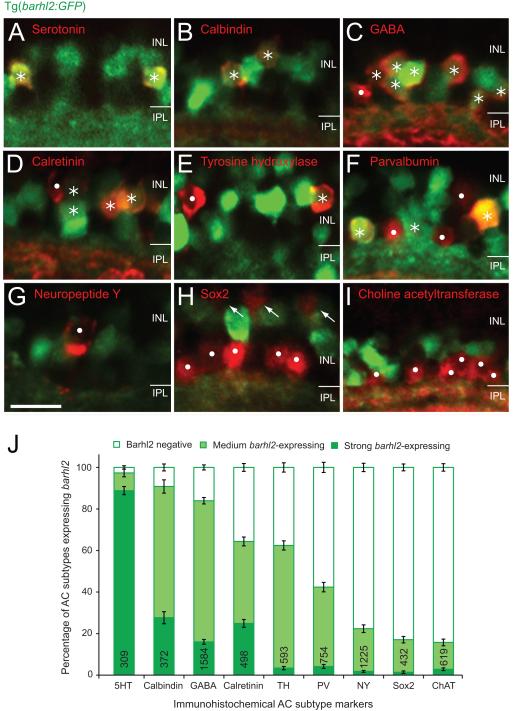
As only 58.5% of Ptf1a-positive ACs turn on Barhl2:GFP, we wondered whether barhl2 is expressed in specific subtypes. Previous studies implicated Barhl2 in biasing specific AC identities (Yazulla and Studholme, 2001; Clemente et al., 2004), but a detailed characterisation of subtypes expressing barhl2 and how this correlates with changes during loss- or gain-of-function studies are still missing. We therefore first performed immunohistochemical staining with nine antibodies in 5 dpf Tg(barhl2:GFP). We specifically chose a range of different markers. Serotonin, tyrosine hydroxylase, neuropeptide Y, and choline acetyletransferase mark non-overlapping individual subtypes based on co-labelling or neurite morphology. Calbindin, parvalbumin and calretinin are calcium-binding markers that may label more than one subtype (e.g. parvalbumin labels 2 subtypes non-overlapping with calretinin labelled cells) (Yeo et al., 2009). Finally, we chose the more general marker GABA, which labels half of the zebrafish amacrine cells (Jusuf and Harris 2009) and likely overlap with some of the subtype specific markers (e.g. serotonin). We found that the vast majority of serotonin, calbindin and GABA-expressing AC types express Barhl2:GFP either strongly (serotonin) or at medium levels (calbindin, GABA; Figure 2A – C, and J), in line with GABAergic subpopulations overlapping with serotonin and/or calbindin labelled subtypes. Calretinin, tyrosine hydroxylase or parvalbumin-expressing amacrine subtypes had more variability with roughly half of the labelled ACs expressing some Barhl2:GFP (Figure 2D – F, and J). Amacrine subtypes labelled by neuropeptide Y, Sox2 or choline acetyltransferase very rarely expressed Barhl2:GFP and never at high levels (Figure 2G – I, and J). These observations are consistent with the hypothesis that Barhl2 may be involved in the differentiation of specific AC subtypes in the zebrafish retina and that differences in expression levels may be important in biasing subtype fates.
Figure 2.
Amacrine markers reveal distinct subtypes expressing barhl2. Micrographs of 120 hpf Tg(barhl2:GFP) embryos immunohistochemically labelled with markers (red). (A – C) Serotonin (5HT, A), calbindin (B) and GABA (C) immunoreactive AC types primarily co-label with Barhl2:GFP. (D – F) Calretinin (D), tyrosine hydroxylase (TH, E) and parvalbumin (PV, F) labelled populations show varying degree and intensities of Barhl2:GFP co-labelling. (G – I) Neuropeptide Y (NY, G), Sox2 (H) and choline acetyltransferase (ChAT, I) label ACs that do not co-label with Barhl2:GFP. Sox2 additionally labels Müller glia cells (arrows, H). Asterisks indicate ACs (red) that co-localise with GFP; dots mark cells that do not express GFP. (J) Quantification of the percentage of different markers co-labelled with Tg(barhl2:GFP), n = 21 – 220 eyes, 309 – 1584 cells. ACs labelled by 5-HT, calbindin and GABA primarily arise from barhl2-expressing cells. ACs labelled by NY, Sox2 and ChAT primarily come from Barhl2-negative or weakly barhl2-expressing cells. ACs labelled with calretinin, TH or PV include both, cells that do and do not arise from barhl2-expressing cells. AC: amacrine cell. Numbers in each bar indicate the number of labelled cells analyzed. Error bars indicate standard error of the mean. INL: inner nuclear layer; IPL: inner plexiform layer. Scale bar G (for A – I) = 20 μm.
Barhl2 functions downstream of Ptf1a to bias amacrine subtypes identities
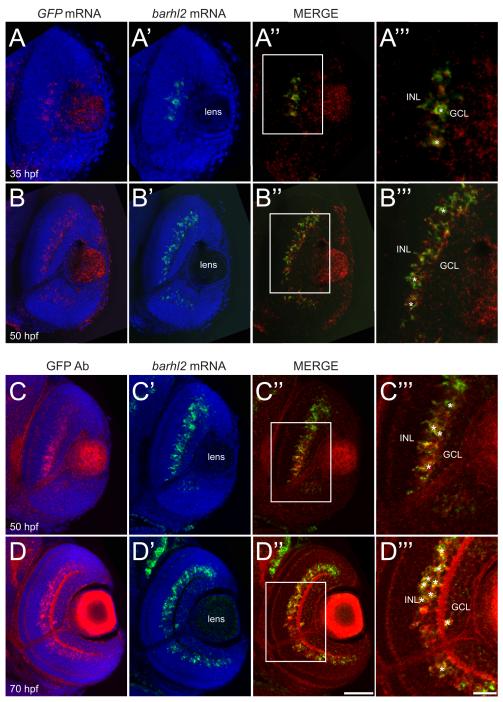
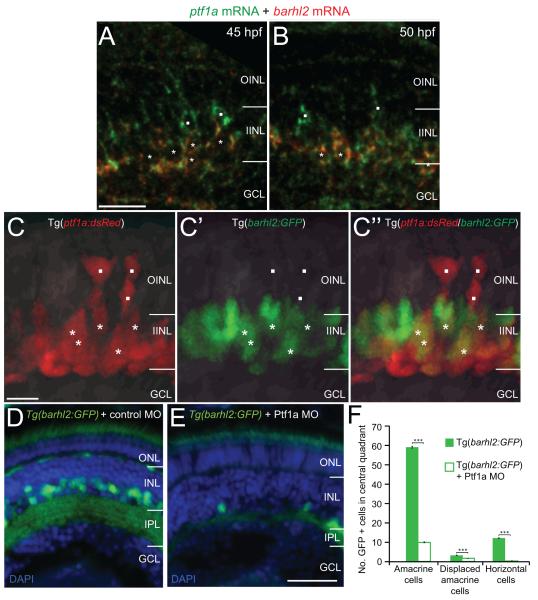
As barhl2 is restricted to some AC subtypes, its expression might be turned on downstream of Ptf1a only in certain AC precursor populations to specify their identities. To first temporally locate barhl2 within the transcriptional cascade that leads to AC specification in vivo, we analysed the dynamics of its expression at the cellular level with respect to ptf1a. The first Barhl2:GFP signal starts at 35 hpf and it faithfully recapitulates the endogenous barhl2 mRNA expression as revealed by double in situ hybridization with GFP mRNA (Figure 3A, B) or GFP protein staining (Figure 3C, D). 3D time-lapse imaging starting from 30-32 hours post-fertilization (hpf) show the first Barhl2:GFP signal at 35 hpf (Movie 1). These Barhl2:GFP positive cells were never seen dividing, always being either in the process of or having finished migrating basally to the inner nuclear layer where ACs reside (Movie 1). Whole mount double fluorescent in situ hybridization shows co-expression of barhl2 and ptf1a mRNAs in individual cells (Figure 4A, B). Cells expressing ptf1a only can be observed apically, suggesting that as cells migrate basally to the future AC layer, they first express ptf1a. Consistently, our 3D in vivo time-lapse imaging shows that within individual developing neurons, Ptf1a:DsRed is turned on apically just after mitosis (Jusuf et al., 2011). In some of these cells Barhl2:GFP is turned on when they have reached the future AC layer (Figure 4C – C”). We tested directly, if Barhl2 functions downstream of Ptf1a, by injecting Ptf1a translational MO or control MO into Tg(barhl2:GFP) embryos. The Ptf1a morphants show a drastic loss in the number of Barhl2:GFP cells (Figure 4D, E) including ACs (59.1 ± 1.2 SEM to 9.02 ± 0.82 SEM; p – value < 0.0001) (Figure 4F). Because Ptf1a morphant cells remain within the retina and are re-specified as excitatory cells (Jusuf et al., 2011), these results demonstrate that barhl2 turns on in inhibitory cell precursors downstream of Ptf1a.
Figure 3.
The Tg(barhl2:GFP) transgenic line faithfully reflects the endogenous barhl2 expression in time and space. (A – B) Whole-mount double fluorescent in situ hybridization against barhl2 mRNA and GFP mRNA on zebrafish retinas counterstained with DAPI (blue) at 35 hpf (A – A”’) and 50 hpf (B – B”’). (C – D) Whole-mount fluorescent in situ hybridization against barhl2 mRNA followed by immunohistochemical labelling against GFP at 50 hpf (C – C”’) and 70 hpf (D – D”’). Co-localization of barhl2 mRNA with barhl2 mRNA or Barhl2:GFP expression (asterisks) occurs in the inner nuclear layer (INL) at all ages and in few cells in the ganglion cell layer (GCL) at 70 hpf. Scale bar D” (for A – D”) = 50 μm, scale bar D”’ (for A”’ – D”’) = 20 μm.
Figure 4.
Ptf1a / barhl2 are sequentially expressed within individual cells, with Ptf1a being necessary for Barhl2:GFP expression. (A, B) Double fluorescent in situ hybridization of barhl2 and ptf1a mRNAs. Ptf1a (green) is expressed in cells apically (squares) and in cells that have migrated basally where they also express barhl2 (red, asterisks) at 45 hpf (A) and 50 hpf (B). (C – C”) Micrographs from in vivo time-lapse of double transgenic Tg(ptf1a:dsRed/barhl2:GFP) embryos at 35 hpf show a similar pattern with apical cells expressing Ptf1a:DsRed alone (squares) and more basal cells co-expressing Barhl2:GFP (asterisks). (D, E) Micrographs of 120 hpf Tg(barhl2:GFP) injected with standard morpholino (D) or Ptf1a morpholino (E). Barhl2:GFP expression is drastically reduced in the Ptf1a morphants. (F) Quantification of GFP labelled cells shows a significant loss of cells in Ptf1a MO in all cell types that usually express Barhl2:GFP, i.e. ACs (inner half of INL), displaced ACs (outermost layer of GCL) and horizontal cells (outermost layer of INL). WT n = 51 eyes, Ptf1a morphant n = 41 eyes. ONL: outer nuclear layer; INL: inner nuclear layer; IPL: inner plexiform layer; GCL: ganglion cell layer; MO: morpholino; ***: p – value < 0.001; error bars indicate standard error of the mean. Scale bar A = 20 μm (for A, B), scale bar C (for C – C”) = 10 μm, scale bar E (for D, E) = 50 μm.
We assessed how the loss of Barhl2 affects the development of ptf1a-expressing neurons. We used a morpholino (MO), which effectively knocked down Barhl2 protein translation as shown by western blot (50 hpf embryos) and antibody staining in hindbrain and retina (72 hpf embryos) (Figure 5). Standard control morpholino injected embryos were comparable to WT (Figure 5F’), as were standard control MO + p53 MO and 5 bp mismatch MO (data not shown). As recent studies implicated Barhl2 in cell survival (Ding et al., 2009; Juraver-Geslin et al., 2011), we firstly assessed activated Caspase-3 immunostaining in Barhl2 morphants, which revealed only a small non-significant increase in retinal apoptosis including in the AC layer (INL, p – value = 0.3; data not shown). Thus, in the absence of Barhl2, the majority of inhibitory Ptf1a:GFP cells remain in the appropriate layers. We next assessed the intrinsic effects of Barhl2 at single-cell level in a normal developing environment. For this we used the technical advantage of the zebrafish model to generate chimeras. Cells from embryos injected with Barhl2 MO or control MO were transplanted into wild-type host embryos (Figure 6A). We first transplanted cells from Tg(ptf1a:GFP) donors injected with H2B-RFP mRNA (to label all donor cells). Overall, the Ptf1a:GFP cells remained as inhibitory cells primarily in the inhibitory amacrine layer (22.96% in WT to 21.2% in morphants, p – value = 0.13, Figure 6B, C). The key question is whether the ACs generated in Barhl2 morphants show changed subtype identity. Using transplantations from WT donors combined with immunohistochemistry, we indeed found that amacrine subtypes that usually express barhl2 were significantly lost in Barhl2 morphant transplants (serotonin 43%, p – value = 0.04, calbindin 38%, p – value < 0.0001, GABA 68%, p – value < 0.0001, Figure 6D – G, J). In contrast, some subtypes that usually do not express barhl2 were strikingly increased (neuropeptide Y 216%, p – value = 0.03), and others mildly increased (ChAT 125%, p – value = 0.14, Figure 6H – J). No evidence was found for a role of Barhl2 in RGC differentiation or survival, as seen in other species (Mo et al., 2004; Poggi et al., 2005b; Ding et al., 2009). These observations demonstrate that the Barhl2 paralog in zebrafish is uniquely dedicated to the AC fates, in which it biases precursors towards generating specific some AC subtypes and away from others. These results also highlight a correlation between the expression level of barhl2 within distinct AC subtypes and its necessity during the development of each subtype.
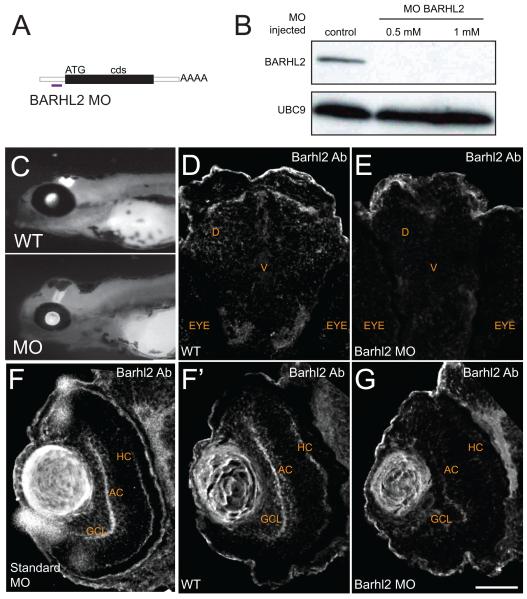
Figure 5.
Zebrafish Barhl2 morpholino causes efficient barhl2 knockdown. (A) A translational blocking morpholino (MO) was designed against 6 base pairs upstream the translational start site of zebrafish barhl2 mRNA. The morpholino efficiency was tested using anti-Barhl2 antibody. (B) Western Blotting of wild type (control) and morphant (0.5 and 1 mM Barhl2 MO) 50 hpf old embryos heads. (C) Micrographs of control and Barhl2 morphant embryos. Morphants look relatively normal, showing only mild general defects in their heads. (D – G) Immunohistochemical labelling of brain (D, E) and retinal (F, F’, G) sections at 50 hpf. Control embryos (D, F) and standard MO injected embryos (F’) show Barhl2 protein in the diencephalon region D around the ventricular zone V of the brain (D) and in the retina (F, F’), in amacrine and horizontal cells (AC, HC) and in cells located in the ganglion cell layer (GCL, F, F’). Barhl2 morphant embryo brain and retina do not stain for Barhl2 protein. Scale bar G (for D – G) = 50 μm.
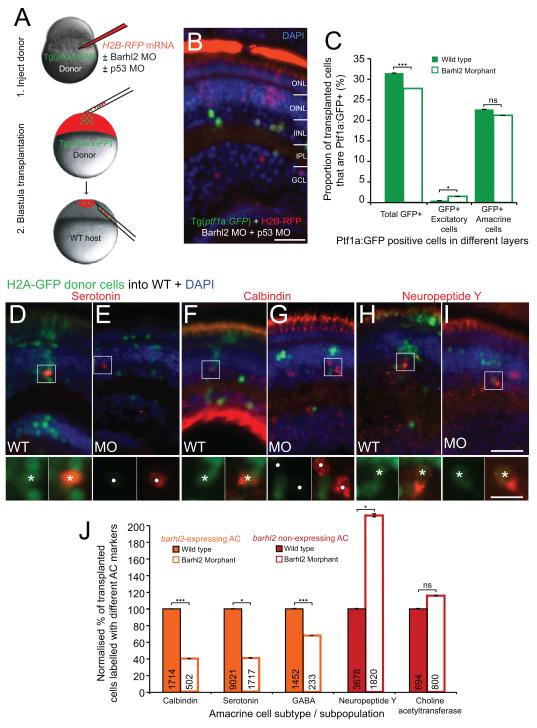
Figure 6.
Barhl2 knockdown causes a subtype fate switch towards alternate inhibitory subtypes. (A) The cell-autonomous effect of Barhl2 loss was assessed with transplantations: Donor Tg(ptf1a:GFP) embryos were injected with H2B-RFP mRNA and p53 MO (to aide cell survival (Robu et al., 2007)), with (knockdown) or without (control) Barhl2 MO. Donor cells were subsequently transplanted into unlabelled WT. (B) Some transplanted H2B-RFP cells express Ptf1a:GFP in both conditions. (C) Quantification reveals a small reduction in the proportion of Ptf1a:GFP labelled donor cells (32% WT to 27.8% morphant, p – value < 0.001) and a few mislocalised Ptf1a:GFP cells in excitatory layers (0.74% WT to 1.49% morphants, p – value = 0.012). Overall, the vast majority of Ptf1a:GFP remain as inhibitory ACs (22.96% WT to 21.2% morphants, p – value = 0.13). (D – I) Immunohistochemically labelled amacrine subtypes (red) in chimeric retinas arise from transplanted H2A-GFP labelled donor (e.g. asterisks for serotonin - D, calbindin - F, neuropeptide Y-H, I) and from unlabelled host cells (circles). (J) Quantification of the proportion of labelled transplanted cells shows varying degrees of loss in subtypes that usually express barhl2 (orange) and increases in AC subtypes that usually do not (red). ONL: outer nuclear layer; OINL: outer half of the inner nuclear layer; IINL: inner half of the inner nuclear layer; IPL: inner plexiform layer; GCL: ganglion cell layer; MO: morphant / morpholino; ns: not significant; *: p – value < 0.05; ***: p – value < 0.001; error bars indicate standard error of the mean. Scale bar B = 20 μm, scale bar I (for D – I) = 25 μm, scale bar I inset (for D – I insets) = 10 μm.
Barhl2 ACs are nieces of RGCs
The results above demonstrate that Barhl2 acts downstream of Ptf1a to bias specific subtype identities within a subset of post-mitotic AC precursors. How does this cell population and transcriptional cascade relate to the Atoh7-lineage in vivo? Approximately 2/3 of the AC population arises from atoh7-expressing progenitors (Jusuf et al., 2011). Since we found that barhl2-expressing neurons comprise 58.5% of ptf1a-expressing ACs, we wondered whether barhl2-expressing ACs come from atoh7-expressing progenitors. Previous expression analysis on barhl2 and atoh7 did not provide any evidence for mRNA co-localization, e.g. RGCs express atoh7 and not barhl2 and ACs that express barhl2 do not express atoh7 (Schuhmacher et al., 2011). This kind of approach, however, cannot rule out possible lineage-relationships between cells expressing these two factors. In contrast, some of the fluorescent proteins display long perdurance compared to the native mRNAs under whose promoters they are driven. Thus, in the Tg(barhl2:GFP/atoh7:gap43-RFP) double transgenic line in which the RFP is long-lived compared to atoh7 mRNA, we can visualize the respective onset of Barhl2:GFP and Atoh7:gap43-RFP in individual cells using in vivo 3D time-lapse imaging. We started imaging at around 35 hpf, when retinal neurons first express Barhl2:GFP, and asked whether any Barhl2:GFP-positive neurons arise from atoh7-expressing progenitors within the first 6 hours of each time-lapse movie (Figure 7A). Strikingly, we found that almost all of the Barhl2:GFP-expressing cells in this time-window also were Atoh7:gap43-RFP-positives (94.3% ± 2.8% SEM), suggesting that Barhl2-dependent amacrine subtypes indeed arise from the Atoh7-lineage. We compared the distribution of subtypes that express barhl2 with our previously identified subtype biases within the Atoh7-lineage (Jusuf et al., 2011). We thus performed morphological characterisation of individual Barhl2:GFP expressing cells (n = 28, Figure 7B - D) and compared the frequency of subtypes in three categories: We found that Barhl2:GFP subtypes of ACs are underrepresented or overrepresented at similar frequency as are Atoh7+/Ptf1a+ expressing cells when compared to all (Ptf1a+) subtype frequencies. Thus, our Barhl2+ sample was statistically significantly different from WT (p – value = 0.032), but comparable with the Atoh7+/Ptf1a+ population (p – value = 0.55), which our time-lapse analysis showed to be the origin of barhl2-expressing cells.
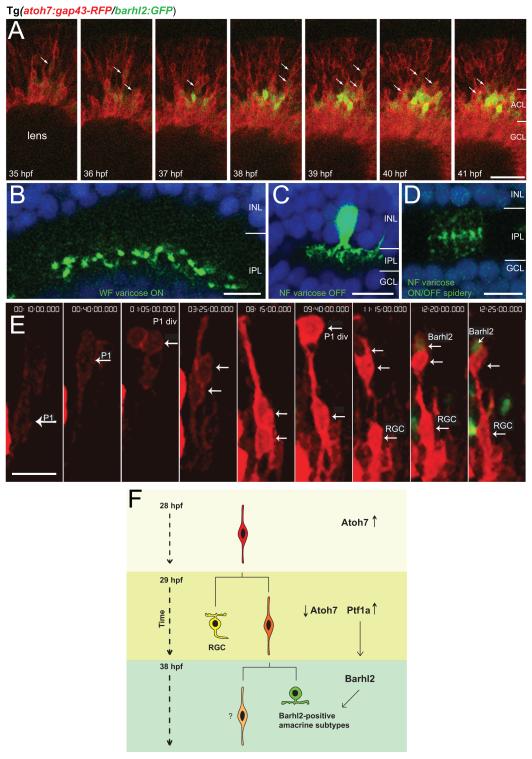
Figure 7.
Barhl2 is expressed in nieces of atoh7-expressing progenitors. (A) Micrographs from time-lapse movie (n = 7 movies) of double Tg(atoh7:gap43-RFP/barhl2:GFP) from 35 hpf. Barhl2:GFP cells almost always express Atoh7:gap43-RFP. Barhl2:GFP expression first occurs in cells that are migrating towards the amacrine layer. Boundaries are indicated for the forming amacrine (ACL) and ganglion cell layer (GCL), which contains the brightly Atoh7:gap43-RFP positive retinal ganglion cells (RGC). Arrows indicate co-labelling. Barhl2:GFP were analysed from the first frame in which they appeared towards the centre of the imaged stack (arrows). (B – D) Examples of individual amacrine cells arising from barhl2 expression. Only one image of a confocal stack is shown, somas for some B and D were located at different depth. (E) Barhl2-positive cells derive from asymmetric divisions of RGC sisters in atoh7-expressing progenitors. 3D reconstruction of confocal stack from time-lapse series (starting at 28 hpf t = 0h:00min) shows the lineage of an Atoh7:gap43-RFP progenitor (P1). After P1 division (white arrow, P1 div, t = 1h:05min), both daughter cells migrate basally (white arrows, t = 3h25min). At t = 8h15 min, one daughter cell migrates apically, dividing at t = 9h40min (white arrow, P2 div), whereas the other daughter cell remains close to the basal surface, differentiating as an RGC. At t = 12h20min one of the daughter cells from the P2 division up-regulates Barhl2:GFP. (F) Scheme summarising the observed lineage tree and sequence of genes expression. Time is indicated on the left. Arrows indicate atoh7 being downregulated in the differentiating RGC and daughter cell while ptf1a is upregulated in the AC precursor (Jusuf and Harris, 2009; Jusuf et al., 2011; Brzezinski et al., 2012). NF: narrow-field; WF: wide-field; P1: progenitor 1; P1 div: cell division of progenitor 1; P2 div: cell division of progenitor 2. Scale bar A = 30 μm, scale bars B – D = 10 μm, scale bar E = 7 μm.
If Barhl2-dependent ACs derive from the Atoh7-lineage, then time-lapse imaging should allow us to understand the lineage relationship between barhl2-expressing amacrine subtypes and RGCs. In our previous 3D time-lapse study we traced individual dividing Atoh7:GFP-expressing progenitors long enough to show that these cells often divided asymmetrically to produce one daughter cell that became a RGC and another daughter which often migrated back toward the apical surface (Poggi et al., 2005b). The time-lapse limitations and lack of appropriate fluorescent reporters did not allow us to follow the fate of the non-RGC daughter cell for more than a few hours, during which time they did not divide again (Poggi et al., 2005b). In the Tg(barhl2:GFP/atoh7:gap43-RFP) double transgenic line, however, it is possible to trace the cellular origin of Barhl2:GFP/Atoh7:gap43-RFP-positive cells. Seven retinas were imaged for a minimum of 20 hours starting from 28 hpf. We traced the lineage of 20 individual Barhl2:GFP/Atoh7:gap43-RFP positive cells. Strikingly, in 19 out of these 20 cases, the Barhl2:GFP positive cell arose from a cell division of an Atoh7:gap43-RFP-positive progenitor, which was identified as the sister of an RGC (Movie 2). In all these 19 cases, the Atoh7:gap43-RFP progenitor generated one Barhl2-positive and one Barhl2-negative daughter cell whose identity remains unknown (Movie 2). In one case out of the 20, we observed that both daughters became Barhl2:GFP-positive. In 14 cases, we were able to reconstruct the lineage of the Barhl2-positive AC from the very onset of Atoh7:gap43-RFP expression. In all of these cases, the lineage started with the asymmetrical division of an Atoh7-positive progenitor, one daughter of which turned up Atoh7:gap43-RFP expression and became a RGC, while the other daughter divided again (see example in Figure 7E). The result of this next division was one Barhl2:GFP-positive cell and one Barhl2:GFP-negative cell (Figure 7E and Movie 2). Taken together, these observations suggest that during RGC genesis in vivo, Barhl2-positive ACs arise as nieces of RGCs, mainly through asymmetric fate outcome of atoh7-expressing progenitors division (Figure 7F).
Barhl2 acts within the Atoh7-lineage to specify subtypes of ACs
Barhl2 is expressed in cells that derive from Atoh7-progenitors, and biases AC identities towards the same subtype identities that arise in the Atoh7-lineage. Therefore, it is important to determine whether Barhl2 by itself is capable in biasing amacrine subtypes in vivo. We investigated this by expressing barhl2 within atoh7-expressing progenitors such that all rather than a subset of these cells would turn on Barhl2. DNA for the H2B-RFP reporter was injected into 1-cell stage embryos in which either pUAS:H2B-RFP (in control retinas) or from pUAS:barhl2-T2A-H2B-RFP (barhl2 misexpressing retinas) was expressed under the control of atoh7 promoter in the Tg(atoh7:gal4/pUAS:gap43-GFP) line. The use of Barhl2-T2A-H2B-RFP in frame fusion allows visualization of barhl2-expressing cells in vivo, and allows simultaneous assessment of their final fate (Kim et al., 2011). In vivo time-lapse analysis confirmed that the expression of barhl2 (H2B-RFP positive cells) indeed occurred prematurely, in dividing Atoh7-positive progenitors (Movie 3). Cell fates of H2B-RFP labelled cells were quantified at 4 dpf (Figure 8). We found no significant changes in the proportion of H2B-RFP positive ACs in the INL (21.02% ± 4.08 SEM control to 26.9% ± 4.15 SEM misexpression, p – value = 0.3255) or HCs (20.26% ± 5.11 SEM control to 19.26% ± 4.0 SEM misexpression, p – value = 0.5157), nor bipolar cells in the outer half of the INL (8.2% ± 5.02 SEM control to 8.9% ± 3.97 SEM misexpression, p – value = 0.1404). However, barhl2 misexpression leads to a significant increase of H2B-RFP positive cells in the GCL (17.68% ± 5.36 SEM control to 34.82% ± 5.8 SEM misexpression, p – value = 0.0017) and a significant loss of H2B-RFP positive cells amongst photoreceptor (the outer nuclear layer (ONL)) cells (32.84% ± 5.76 SEM control to 10.12% ± 5.4 SEM misexpression, p – value = 0.0105). To analyse this effect with respect to AC subtypes, we used antibodies for specific AC populations. Within barhl2-misexpressing H2B-RFP positive cells we found a significant increase in the proportion of GABAergic cells in the INL (15.93% ± 4.86 SEM control to 22.2% ± 4.64 SEM misexpression, p – value = 0.0044) and GCL (7.5% ± 4 SEM control to 28.32% ± 6.05 SEM misexpression, p – value < 0.0001), which was particularly evident in the GCL (Figure 8D). Therefore, the increase in GABA+ cells appears to be at the expense of early made ganglion cells (GABA- in the GCL) and photoreceptors. Notably, neither in the control nor in the misexpression condition were H2B-RFP positive cells ever ChAT-positive (data not shown), a subtype that rarely expresses barhl2. These results suggest that when barhl2 is prematurely expressed in the atoh7-expressing progenitors population, it is itself able to induce within these cells some aspects of Barhl2-dependent AC subtype identities.
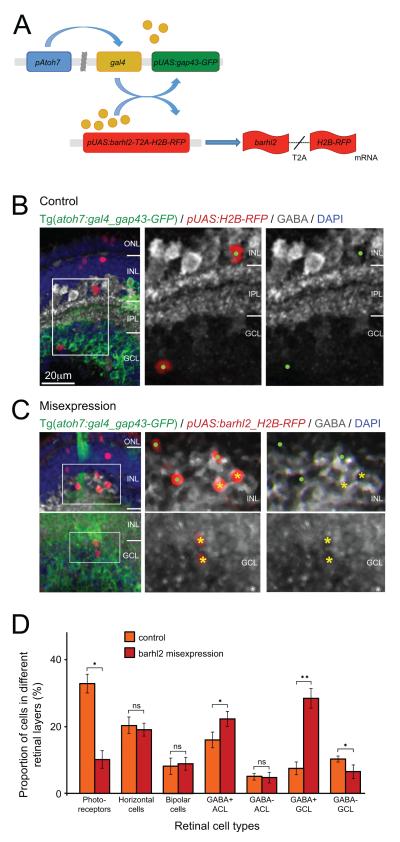
Figure 8.
Misexpression of barhl2 in atoh7-expressing progenitors drives the fate of amacrine subtypes that usually express barhl2. (A) Schematic showing misexpression design: The atoh7 promoter drives expression of Gal4 transcription factor. Gal4 activates the upstream activation sequence (pUAS) promoter to drive expression of gap43-GFP reporter by itself (control) or with Barhl2 and H2B-RFP reporter generated through in frame fusion with T2A peptide. After translation of the barhl2-T2A-H2B-RFP mRNA the T2A sequence will be cleaved and generate separate Barhl2 and H2B-RFP proteins. (B, C) Micrographs of 120 hpf Tg(atoh7:gal4/pUAS:gap43-GFP) retinas with pUAS driving H2B-RFP (control) or barhl2 and H2B-RFP (misexpression), and subsequently labelled for GABA (white). Boxes indicate higher power insets. Cell type and GABA colabeling of H2B-RFP-expressing cells was analysed. Examples of GABA-positive cells (yellow asterisks) and GABA-negative cells (green dots). (D) Quantification of cell fates in control (orange) or barhl2 misexpression (red). There is an increase in GABAergic cells in the INL (15.93% ± 4.86 SEM control to 22.2% ± 4.64 SEM misexpression, p – value = 0.0044) and particularly GCL (7.5% ± 4 SEM control to 28.32% ± 6.05 SEM misexpression, p – value < 0.0001 in GCL) with a concurrent loss in mainly photoreceptors (32.84% ± 5.76 SEM control to 10.12% ± 5.4 SEM misexpression, p – value = 0.01058) and presumed ganglion cells. ONL: outer nuclear layer; INL: inner nuclear layer; IPL: inner plexiform layer; GCL: ganglion cell layer; ns: not significant. Error bars indicate standard error of the mean. * p – value < 0.05, ** p – value < 0.001. Scale bar in B (for B, C) = 20 μm.
Atoh7 affects lineage outcome and number of Barhl2-positive cells
We know that ptf1a expression does not depend on Atoh7 (Jusuf et al., 2011), but as barhl2 is turned on in Atoh7-derived inhibitory precursors, we wondered whether Atoh7 influences the way Barhl2-positive cells arise within the Atoh7-lineage. The Tg(barhl2:GFP) line was outcrossed to atoh7-/- (lakritz) mutants (Kay et al., 2001). Barhl2:GFP expression analysed at 5 dpf is not only retained, but also virtually doubled in the absence of Atoh7 (39.33% ± 0.92 SEM WT to 73.74% ± 2.261 SEM lakritz, p – value < 0.0001; data not shown) with significant increases in the INL (48.33% ± 1.32 SEM to 78.32% ± 2.35 SEM, p – value < 0.0001; data not shown) and GCL (20% ± 1.03 SEM to 63.89% ± 3.81 SEM, p – value < 0.0001; data not shown). How do Barhl2-positive ACs arise in this case? In twelve time-lapse movies of Barhl2:GFP/Atoh7:gap43-RFP cells in lakritz retinas, we consistently observe Atoh7:gap43-RFP-positive cell divisions generating two daughters, only one of which becomes Barhl2:GFP-positive. Interestingly, in eight of these movies, the original Atoh7:gap43-RFP cells first divide once as usual. However, as neither daughter is able to differentiate into a RGC, both daughters divide again, with one cell from each pair starting to express Barhl2:GFP (Movie 4). Thus, when Atoh7 is missing, Barhl2-positive ACs still arise within asymmetric cell divisions from the “Atoh7-lineage”, but in this case there are two rather than only one barhl2-expressing cell generated from each lineage. This different lineage-outcome reveals the mechanism of Barhl2-positive amacrine cell’s increased number in the lakritz mutant.
Atoh7 affects Barhl2-positive amacrine subtypes identity
Are barhl2-expressing subtypes increased equally in the absence of Atoh7? Since GABA labels the majority of Barhl2:GFP cells, we first assessed changes to this cell population. We found that the total proportion of Barhl2:GFP/GABAergic neurons remains unchanged in lakritz mutants (32% WT to 30% lakritz). Because GABAergic neurons consist of multiple subtypes, we next assessed, if specific subtypes are changed within this population. We found that serotonergic amacrine subtypes (usually barhl2-expressing) are significantly increased (+20.6% ± 6 SEM lakritz retina, p – value = 0.0002, Figure 9E – G), thus suggesting that specific barhl2-expressing subtypes are preferentially affected. The most striking change is the large increase in Barhl2:GFP non-GABAergic amacrine cells in the lakritz retina (Figure 9C, D). One possible explanation for this outcome could be that Barhl2-negative subtypes now become Barhl2-positive. To investigate on this possibility, we analysed the population of NY+ amacrine subtypes. We found no significant change in NY+ proportion or GFP expression (+ or − 11% ± 5.5 SEM lakritz retina, p – value = 0.2611). Our data thus show that specific subtypes of Barhl2:GFP+ cells, including 5-HT+ and non-GABAergic populations, are preferentially expanded. Although the increase in amacrine cells can be explained by our lineage analysis, the mechanism by which Atoh7 affects subtype identity remains unclear.
Figure 9.
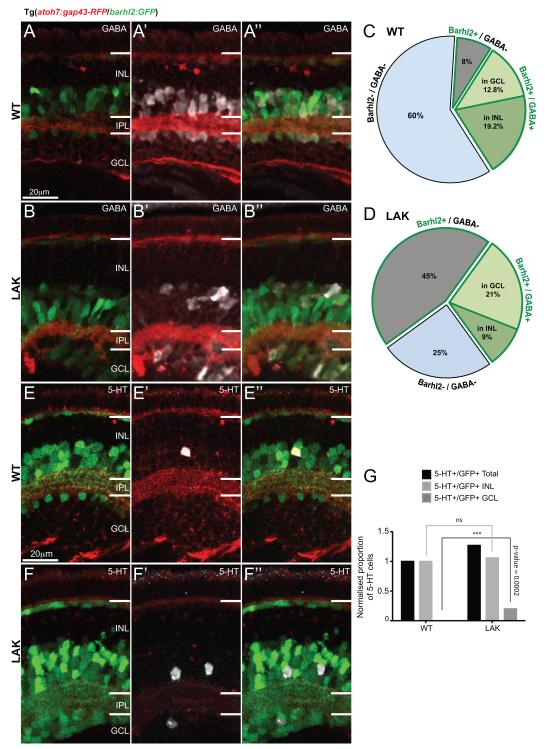
Absence of Atoh7 leads to an increase of barhl2-expressing ACs in the ganglion cell layer. (A, B) Micrographs of 120 hpf Tg(atoh7:gap43-RFP/barhl2:GFP) retinas labelled for GABA in wild type (WT) (A) and lakritz (LAK) (B). (C, D) Quantification of amacrine subpopulations expressed as proportion of total INL and GCL cells. The data indicate that the increase in the number of GFP+ cells in the LAK retina is primarily due to non-GABAergic ACs, as the proportion of GABAergic ACs remains unchanged in total in both GCL and INL in LAK vs. WT (32% WT to 30% lakritz). A redistribution of GABAergic ACs is however observed with an increase of Barhl2+/GABA+ cells in the GCL and a relative decrease of those cells in the INL of the LAK retina in comparison to WT (12.8% WT to 21% LAK, p – value = 0.0008 in GCL and 19.2% WT to 9% LAK, p – value = 0.0014 in INL). Total number of cells is the number of nuclei in the INL and GCL. (E, F) Micrographs of 120 hpf Tg(atoh7:gap43-RFP/barhl2:GFP) retinas labelled for serotonin (5-HT). (G) Quantification of the total proportion of serotonin-positive/Barhl2:GFP-positive (5-HT+/GFP+) neurons shows an increase in 5-HT cells in lakritz specifically in the GCL where they are normally absent (n = 15 – 19 eyes). IPL: inner plexiform layer; INL: inner nuclear layer; GCL: ganglion cell layer; *** p – value < 0.0002. Error bars indicate standard error of the mean. Scale bars = 20 μm.
Lakritz mutants retain some cells in the ganglion cell layer of the retina, which have been attributed to displaced amacrine cells (Kay et al., 2001). Does this simply reflect the tendency of ACs to occupy the now RGC-free most basal positions in the retina (Kay et al., 2001), or rather it indicates a selective increase in specific subclasses of amacrine cells (Feng et al., 2010), e.g. the Barhl2:GFP cells deriving from the Atoh7-lineage? To gain more insights into this question we analysed GABA as marking a large proportion of the usually Barhl2+ population, as well as serotonin and neuropeptide Y as subtype specific markers that were usually Barhl2+ or Barhl2-respectively. We found GABAergic Barhl2:GFP increased in the GCL (12.8% WT to 21% lakritz, p-value = 0.0008) and decreased in the INL (19.2% WT to 9% lakritz, p-value = 0.0014) (Figure 9A - D). Similarly, serotonergic Barhl2:GFP cells, which are normally only found in the INL (Figure 9E – G) are now increased due to their appearance in the GCL (+20.2% increase, p – value = 0.0002, Figure 9E - G). Finally, even though the proportion of NY+ remained unchanged in the lakritz retina, some of these cells were redistributed to the GCL (11% increase in the lakritz retina compared to the WT, p – value = 0.0009, data not shown). These observations together suggest that amacrine subtypes get redistributed to the ganglion cell layer in the lakritz mutants, regardless of whether they are re-specified or increased in number.
DISCUSSION
Although neural cell fate determination factors are being studied in various model organisms, the occurrence of particular patterns and interactions within individual cell lineages can only be assessed in vivo in the zebrafish model system. In this study we use a combination of functional and in vivo time-lapse analyses to uncover novel aspects of lineage-related transcriptional networks and cell-fate outcomes in the developing vertebrate retina. In particular, we here demonstrate that the zebrafish Barhl2 paralog biases particular ACs in vivo, and is turned on in particular Atoh7-lineages.
In the zebrafish retina, 71% of inhibitory neurons (all horizontal and about 60% of amacrine cells) derive from Atoh7 progenitors. These are distinct in subtype composition from the remaining third of amacrine subtypes that derive from non-Atoh7 progenitors (Jusuf et al., 2011). Most amacrine subtypes can arise from either progenitor, but the probability of assuming a particular subtype fate correlates with whether the AC arose from an Atoh7-positive or -negative lineage. Experimentally induced co-expression of ptf1a in all Atoh7-positive progenitors leads to a selective increase in the ACs that normally derive from the Atoh7-lineage (Jusuf et al., 2011). Yet Atoh7 is not essential for the specification of any of the amacrine subtypes, and ptf1a is expressed in all ACs, suggesting that other factors must be key to biasing subtype specification. Here we find that 58.5% of amacrine cells express barhl2, and that most Barhl2 cells arise from atoh7-expressing progenitors, thus providing evidence that Barhl2 may be this “other” factor. Consistent with this hypothesis, we found that barhl2-expressing subtypes were similarly underrepresented and overrepresented when compared to our previously characterised Atoh7+/Ptf1a+ lineage. Furthermore, knocking down or increasing barhl2 expression within Atoh7-progenitors is sufficient to bias the differentiation of amacrine cells in predictable directions. Taken together these observations provide an explanation as to why the ptf1a-expressing progenitors in the Atoh7-lineage, but not those from the non-Atoh7 lineage, bias differentiation toward specific amacrine subtypes and not others (Jusuf et al., 2011).
Barhl2 has been implicated in the development of ACs, and specification and survival of RGCs downstream of atoh7 in the mouse and Xenopus retina (Mo et al., 2004; Poggi et al., 2004; Wang and Harris, 2005; Ding et al., 2009). Here we find that the zebrafish barhl2 paralog is turned on exclusively in post-mitotic, inhibitory Ptf1a-positive ACs and horizontal cells. We also find that barhl2 is preferentially expressed at different levels in specific amacrine subtypes (such as GABA, 5-HT and calbindin expressing), whereas it is almost completely absent in others (NY and ChAT). In accordance with data from mouse, the absence of Barhl2 function leads to an altered amacrine subtype composition (Ding et al., 2009). Specifically, subtypes that are normally Barhl2-positive (GABA, serotonin and calbindin) decrease at the expense of the ones that normally do not express barhl2 (ChAT and NY). Decreases in GABAergic population with increases in cholinergic AC are consistent with the previous mouse data (Ding et al., 2009) and these may indeed represent homologous subtypes. However, other subtypes may present preferential alternate fates, as the proportional increase in NY cells far outweighed the modest and statistically not significant increase in ChAT+ cells. Although some of the markers may label overlapping amacrine subtype populations, our results show specific losses in subtypes that express barhl2 most strongly. We therefore show for the first time a strong direct correlation between expression levels of barhl2 and cell-autonomous necessity for Barhl2 in each of these specific subtypes, rather than just a general role in relative subtype composition.
Unlike in other vertebrates (Poggi et al., 2004; Ding et al., 2009), the zebrafish barhl2 is not likely implicated in RGC differentiation or survival. Several observations support this conclusion: (1) barhl2 expression is restricted to ptf1a-expressing inhibitory cell precursors and is turned on only after the division of sister cell of an RGC; (2) Overexpression of barhl2 is sufficient for amacrine subtypes, but not RGCs, even if overexpressed prematurely under the control of the atoh7 promoter; (3) knocking down barhl2 does not show significant changes in RGC genesis; (4) barhl2 expression depends on the expression of ptf1a and not on the expression of atoh7. All of these observations are in agreement with Barhl2 acting exclusively in inhibitory cell precursors. The function of barhl genes in RGCs genesis or maintenance might instead be retained by the duplicated barhl1.2 paralog; which is expressed in RGCs downstream of Atoh7 (Schuhmacher et al., 2011).
What is then the relationship between barhl2 and atoh7? Previous expression analysis suggested that a negative feedback might occur between the two genes, possibly as a result of the conservation of an ancestral feature existing in the invertebrate Drosophila melanogaster between the homologues atonal and barH genes (Lim and Choi, 2003; Schuhmacher et al., 2011). Our in vivo timelapse study highlighted particular lineage-relationships between Atoh7-dependent RGCs and barhl2-expressing amacrine subtypes. Our results clearly demonstrate that the expression of the two genes is mutually exclusive at cellular level; they also show that Barhl2-positive ACs arise as nieces of RGCs, which depend on Atoh7. This observation, combined with results from loss- and gain-of-function assigns the timing of Barhl2 action in post-mitotic precursors in which atoh7 has been already down-regulated (Skowronska-Krawczyk et al., 2005; Le et al., 2006; Brzezinski et al., 2012) and the switch from RGC to inhibitory cell fates has already occurred via the expression of ptf1a. Thus, Barhl2 in vivo is not well positioned to act as a feedback repressor of atoh7. The intermediate ptf1a expression however, might in fact repress atoh7 within this lineage consistent with data in chick (Lelievre et al., 2011) (Figure 7F).
Conversely, we found that barhl2 expression does not need functional Atoh7. Atoh7 might therefore exert an indirect repressive function on barhl2. A recent study in the mouse shows that in Math5/atoh7-null retina there is a precocious expression of AC fate determinants, thus suggesting that Atoh7 prevents ACs from being generated prematurely (Feng et al., 2010). Yet our in vivo time-lapse analysis of barhl2:GFP/atoh7:gap43-RFP progenitors through two rounds of cell division shows that the onset of Barhl2:GFP within individual cell lineages is unvaried in the absence of Atoh7. Consistently, in both wild type and lakritz retina, Barhl2-dependent ACs tend to arise from divisions of “Atoh7:gap43-RFP” cells in which one daughter turns on Barhl2:GFP and the other does not. However, we also find that Atoh7 affects the lineage-outcome of retinal progenitor cells. Therefore, the increase in the number of Barhl2-positive cells is rather due to the fact that without Atoh7, both daughters of one Atoh7:gap43-RFP progenitor divide again to generate an additional Barhl2-positive daughter. This is coherent with a co-temporaneous study by He et al. (2012 in press), in which randomly selected lineages in the zebrafish retina were traced in time-lapse. In these lineages, approximately 80% of all RGCs arise from asymmetric divisions in which one daughter differentiates as an RGC and the other daughter divides again. Results from He et al. also show that in the absence of Atoh7, there is a marked decrease in such divisions.
Although the onset of barhl2 is independent of Atoh7 action, our results clearly show that Atoh7 does affect barhl2-expressing amacrine subtypes identity. While GABAergic/Barhl2-positive amacrines remain unchanged in the lakritz retina, there is a striking increase in the non-GABAergic/Barhl2-positive cells and in the normally rare serotonin/Barhl2-positive ACs. Thus, Atoh7 might be permissive for some Barhl2-dependent amacrine subtypes (such as GABA) while it might repress others (e.g. serotonin). In the atoh7-null retinas of the mouse, a change in amacrine subtype composition has also been noted, with certain amacrines increasing at the expense of others (Feng et al., 2010). Also in agreement with these studies, our analysis of the lakritz retina shows an overall increase in displaced ACs (Kay et al., 2001; Feng et al., 2010). This general increase was happening regardless of whether ACs where particular Barhl2-positive or Barhl2-negative (such as for NY) subtypes.
It remains a major challenge to identify cell fate determination networks and how they operate within the context of lineage in the developing vertebrate CNS. Recent studies suggest that there are stochastic mechanisms at work in determining fate choice within clones (Gomes et al., 2011; He et al., 2012 in press), and perhaps that is why factors like Barhl2 bias rather than strictly determine fate choice, but our results showing the rather strict expression of Barhl2 within theAtoh7-lineage argue that the expression of at least some cellular determinants in the retina are strongly influenced lineage. We have shown that the zebrafish retina is an excellent model system to address this challenge as this system allows us to follow in vivo modes of cell divisions within individual lineages, concurrently with the expression of specific cell fate determinants. Here, we highlighted the lineage of individual atoh7-expressing progenitors through two rounds of cell divisions, and our functional results suggest that interactions involving the division mode, and the transcriptional network of Atoh7, Ptf1a and Barhl2 promote the orderly generation of RGCs and amacrine subtypes within this lineage.
Supplementary Material
Acknowledgements
We thank J.L. Matéo Cerdán and D.S. Mosti for advice with statistical analyses; S. Schultz for help with Western Blot experiments; B. Wittbrodt, E. Leist, A. Saraceno, M. Majewski, B. Seiferling, T. Kellner, L. Schertel, C. Mueller, Julian Cocks for fish maintenance and technical assistance. We are grateful to M. Carl, J. Wittbrodt, L. Centanin, G. Lupo and M. Zigman for valuable discussion and comments on the manuscript. This work was supported by DFG Research grant (PO 1440/1-1) to LP, Wellcome Trust Programme Grant (ID RG49253) to WAH and Australian National Health and Medical Research Council CJ Martin training fellowship (ID 454798) to PRJ. SA and AP are funded by LGFG (Funding program of the State of Baden-Württemberg). We also would like to thank J. Wittbrodt for generous support.
REFERENCES
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JAt, Prasov L, Glaser T. Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev Biol. 2012 doi: 10.1016/j.ydbio.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JAt, Kim EJ, Johnson JE, Reh TA. Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina. Development. 2011;138:3519–3531. doi: 10.1242/dev.064006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente D, Porteros A, Weruaga E, Alonso JR, Arenzana FJ, Aijon J, Arevalo R. Cholinergic elements in the zebrafish central nervous system: Histochemical and immunohistochemical analysis. J Comp Neurol. 2004;474:75–107. doi: 10.1002/cne.20111. [DOI] [PubMed] [Google Scholar]
- Ding Q, Chen H, Xie X, Libby RT, Tian N, Gan L. BARHL2 differentially regulates the development of retinal amacrine and ganglion neurons. J Neurosci. 2009;29:3992–4003. doi: 10.1523/JNEUROSCI.5237-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dullin JP, Locker M, Robach M, Henningfeld KA, Parain K, Afelik S, Pieler T, Perron M. Ptf1a triggers GABAergic neuronal cell fates in the retina. BMC developmental biology. 2007;7:110. doi: 10.1186/1471-213X-7-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Xie ZH, Ding Q, Xie X, Libby RT, Gan L. MATH5 controls the acquisition of multiple retinal cell fates. Mol Brain. 2010;3:36. doi: 10.1186/1756-6606-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Luo H, Qiu F, Burlison J, Long Q, Kawaguchi Y, Edlund H, MacDonald RJ, Furukawa T, Fujikado T, Magnuson MA, Xiang M, Wright CV. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006;133:4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- Ghiasvand NM, Rudolph DD, Mashayekhi M, Brzezinski JAt, Goldman D, Glaser T. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat Neurosci. 2011;14:578–586. doi: 10.1038/nn.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho L, Mumm JS, Williams PR, Schroeter EH, Koerber A, Park SW, Leach SD, Wong RO. Targeting of amacrine cell neurites to appropriate synaptic laminae in the developing zebrafish retina. Development. 2005;132:5069–5079. doi: 10.1242/dev.02075. [DOI] [PubMed] [Google Scholar]
- He JC, Zhang G, Almeida AD, Cayouette M, Simons BD, Harris WA. How variable clones build an invariant retina. Neuron. 2012 doi: 10.1016/j.neuron.2012.06.033. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Bertsch TW, Ellis HM, Harris WA. Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron. 1988;1:15–26. doi: 10.1016/0896-6273(88)90205-x. [DOI] [PubMed] [Google Scholar]
- Juraver-Geslin HA, Ausseil JJ, Wassef M, Durand BC. Barhl2 limits growth of the diencephalic primordium through Caspase3 inhibition of beta-catenin activation. Proc Natl Acad Sci U S A. 2011;108:2288–2293. doi: 10.1073/pnas.1014017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusuf PR, Harris WA. Ptf1a is expressed transiently in all types of amacrine cells in the embryonic zebrafish retina. Neural Dev. 2009;4:34. doi: 10.1186/1749-8104-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusuf PR, Almeida AD, Randlett O, Joubin K, Poggi L, Harris WA. Origin and determination of inhibitory cell lineages in the vertebrate retina. J Neurosci. 2011;31:2549–2562. doi: 10.1523/JNEUROSCI.4713-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 2004;77:201–222. doi: 10.1016/s0091-679x(04)77011-9. [DOI] [PubMed] [Google Scholar]
- Kay JN, Finger-Baier KC, Roeser T, Staub W, Baier H. Retinal ganglion cell genesis requires lakritz, a Zebrafish atonal Homolog. Neuron. 2001;30:725–736. doi: 10.1016/s0896-6273(01)00312-9. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, Choi SY. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kinkhabwala A, Riley M, Koyama M, Monen J, Satou C, Kimura Y, Higashijima S, Fetcho J. A structural and functional ground plan for neurons in the hindbrain of zebrafish. Proc Natl Acad Sci U S A. 2011;108:1164–1169. doi: 10.1073/pnas.1012185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Wroblewski E, Patel S, Riesenberg AN, Brown NL. Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev Biol. 2006;295:764–778. doi: 10.1016/j.ydbio.2006.03.055. [DOI] [PubMed] [Google Scholar]
- Lelievre EC, Lek M, Boije H, Houille-Vernes L, Brajeul V, Slembrouck A, Roger JE, Sahel JA, Matter JM, Sennlaub F, Hallbook F, Goureau O, Guillonneau X. Ptf1a/Rbpj complex inhibits ganglion cell fate and drives the specification of all horizontal cell subtypes in the chick retina. Dev Biol. 2011;358:296–308. doi: 10.1016/j.ydbio.2011.07.033. [DOI] [PubMed] [Google Scholar]
- Lim J, Choi KW. Bar homeodomain proteins are anti-proneural in the Drosophila eye: transcriptional repression of atonal by Bar prevents ectopic retinal neurogenesis. Development. 2003;130:5965–5974. doi: 10.1242/dev.00818. [DOI] [PubMed] [Google Scholar]
- Lin JW, Biankin AV, Horb ME, Ghosh B, Prasad NB, Yee NS, Pack MA, Leach SD. Differential requirement for ptf1a in endocrine and exocrine lineages of developing zebrafish pancreas. Dev Biol. 2004;274:491–503. doi: 10.1016/j.ydbio.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Mo Z, Li S, Yang X, Xiang M. Role of the Barhl2 homeobox gene in the specification of glycinergic amacrine cells. Development. 2004;131:1607–1618. doi: 10.1242/dev.01071. [DOI] [PubMed] [Google Scholar]
- Nakhai H, Sel S, Favor J, Mendoza-Torres L, Paulsen F, Duncker GI, Schmid RM. Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development. 2007;134:1151–1160. doi: 10.1242/dev.02781. [DOI] [PubMed] [Google Scholar]
- Pittman AJ, Law MY, Chien CB. Pathfinding in a large vertebrate axon tract: isotypic interactions guide retinotectal axons at multiple choice points. Development. 2008;135:2865–2871. doi: 10.1242/dev.025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggi L, Zolessi FR, Harris WA. Time-lapse analysis of retinal differentiation. Current opinion in cell biology. 2005a;17:676–681. doi: 10.1016/j.ceb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Poggi L, Vitorino M, Masai I, Harris WA. Influences on neural lineage and mode of division in the zebrafish retina in vivo. J Cell Biol. 2005b;171:991–999. doi: 10.1083/jcb.200509098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggi L, Vottari T, Barsacchi G, Wittbrodt J, Vignali R. The homeobox gene Xbh1 cooperates with proneural genes to specify ganglion cell fate within the Xenopus neural retina. Development. 2004;131:2305–2315. doi: 10.1242/dev.01099. [DOI] [PubMed] [Google Scholar]
- Reig G, Cabrejos ME, Concha ML. Functions of BarH transcription factors during embryonic development. Dev Biol. 2007;302:367–375. doi: 10.1016/j.ydbio.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher LN, Albadri S, Ramialison M, Poggi L. Evolutionary relationships and diversification of barhl genes within retinal cell lineages. BMC evolutionary biology. 2011;11:340. doi: 10.1186/1471-2148-11-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronska-Krawczyk D, Matter-Sadzinski L, Ballivet M, Matter JM. The basic domain of ATH5 mediates neuron-specific promoter activity during retina development. Molecular and cellular biology. 2005;25:10029–10039. doi: 10.1128/MCB.25.22.10029-10039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Vetter ML, Brown NL. The role of basic helix-loop-helix genes in vertebrate retinogenesis. Seminars in cell & developmental biology. 2001;12:491–498. doi: 10.1006/scdb.2001.0273. [DOI] [PubMed] [Google Scholar]
- Vitorino M, Jusuf PR, Maurus D, Kimura Y, Higashijima S, Harris WA. Vsx2 in the zebrafish retina: restricted lineages through derepression. Neural Dev. 2009;4:14. doi: 10.1186/1749-8104-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Harris WA. The role of combinational coding by homeodomain and bHLH transcription factors in retinal cell fate specification. Dev Biol. 2005;285:101–115. doi: 10.1016/j.ydbio.2005.05.041. [DOI] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazulla S, Studholme KM. Neurochemical anatomy of the zebrafish retina as determined by immunocytochemistry. J Neurocytol. 2001;30:551–592. doi: 10.1023/a:1016512617484. [DOI] [PubMed] [Google Scholar]
- Yeo JY, Lee ES, Jeon CJ. Parvalbumin-immunoreactive neurons in the inner nuclear layer of zebrafish retina. Exp Eye Res. 2009;88:553–560. doi: 10.1016/j.exer.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Zolessi FR, Poggi L, Wilkinson CJ, Chien CB, Harris WA. Polarization and orientation of retinal ganglion cells in vivo. Neural Dev. 2006;1:2. doi: 10.1186/1749-8104-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.