Abstract
Objective
Linkage of the chromosome 1q21-25 region to type 2 diabetes has been demonstrated in multiple ethnic groups. We performed common variant fine-mapping across a 23Mb interval in a multiethnic sample to search for variants responsible for this linkage signal.
Research Design and Methods
In all, 5,290 SNPs were successfully genotyped in 3,179 T2D cases and controls from eight populations with evidence of 1q linkage. Samples were ascertained using strategies designed to enhance power to detect variants causal for 1q-linkage. Following imputation, we estimate ~80% coverage of common variation across the region (r2>0.8, Europeans). Association signals of interest were evaluated through in silico replication and de novo genotyping in approximately 8,500 cases and 12,400 controls.
Results
Association mapping of the 23Mb region identified two strong signals, both restricted to the subset of European-descent samples. The first mapped to the NOS1AP (CAPON) gene region (lead SNP: rs7538490, OR 1.38 (95% CI, 1.21-1.57), p=1.4×10-6 in 999 cases and 1,190 controls): the second within an extensive region of linkage disequilibrium that includes the ASH1L and PKLR genes (lead SNP: rs11264371, OR 1.48 [1.18-1.76], p=1.0×10-5, under a dominant model). However, there was no evidence for association at either signal on replication, and, across all data (>24,000 subjects), no indication that these variants were causally-related to T2D status.
Conclusion
Detailed fine-mapping of the 23Mb region of replicated linkage has failed to identify common variant signals contributing to the observed signal. Future studies should focus on identification of causal alleles of lower frequency and higher penetrance.
Keywords: chromosome 1q, linkage, association
Genome-wide association (GWA) analysis has provided a powerful stimulus to the discovery of common variants influencing type 2 diabetes (T2D) risk, and, to date, approximately 20 susceptibility loci have been identified with high levels of statistical confidence (1). However, these known variants account for only a small proportion of the inherited component of disease risk (probably <10%) and the molecular basis of the majority of the genetic predisposition to T2D has yet to be established (1).
The success of the GWA approach contrasts with the slow progress which characterised previous efforts to map susceptibility loci through genome-wide linkage (GWL) (2). However, now that many of the common variants of largest effect have been identified (in European-descent populations at least), there are cogent reasons to revisit regions previously-identified through GWL. Firstly, variants within the genomic intervals representing replicated linkage signals can be considered to have raised prior odds for a susceptibility-effect, and this information can be used to prioritise GWA signals (particularly those with only modest evidence of association) for targeted replication. Second, genuine linkage signals are likely to be driven by causal variants -- particularly low frequency SNPs or copy number variants (CNVs) not captured by the commodity GWA arrays -- with effect-sizes larger than those currently detectable by GWA (3). Since alleles with these characteristics will have a more marked impact on individual disease predisposition than the common variants found by GWA, identification of causal variants underpinning replicated linkage signals should accelerate efforts to obtain better predictors of disease (4).
For T2D, there appears to be only limited overlap between the regions identified by GWL and those revealed by GWA (5). Though the discovery of TCF7L2 was prompted by a search for causal variants within a region of replicated T2D-linkage, neither the common variants in TCF7L2, nor those in HHEX and IDE (a second nearby GWA signal) account for that linkage signal (6). Thus, the discovery of TCF7L2 reflects either serendipity or the co-localisation of common and rare causal variants in the same locus, the former driving the association, the latter the linkage. Similarly, whilst common variants in HNF4A have been reported to explain the chromosome 20 linkage signals seen in Finns and Ashkenazim (7,8), these associations have proved difficult to replicate (9).
Chromosome 1q (in particular the 30Mb stretch adjacent to the centromere) ranks alongside the regions on chromosomes 10 and 20 as amongst the strongest in terms of the replicated evidence for GWL to T2D. Linkage has been reported in samples of European (UK, French, Amish, Utah), East Asian (Chinese, Hong Kong) and Native American (Pima) origin (summarised in Supplementary Table 1, and reference 2). The region concerned is gene-rich and contains a disproportionate share of excellent biological candidates (2). The homologous region has also emerged as a diabetes-susceptibility locus from mapping efforts in several well-characterised rodent models (10-13).
The International 1q consortium represents a coordinated effort by the groups with the strongest evidence for 1q linkage to identify variants causal for that signal. Here, we report efforts to map causal variants using a custom, linkage-disequilibrium mapping approach, predominantly based around common SNP variants, applied to a well-powered set of multi-ethnic samples.
To improve power, ascertainment of T2D cases for this study aimed to enrich for 1q-causal alleles through: (a) a focus on populations and samples that had shown 1q-linkage; (b) selection for positive family history; and (c) for some samples, preferential recruitment on the basis of patterns of IBD-sharing in the 1q region. For each set of cases, we selected a control sample of individuals from the same population. Details of the recruitment have been reported previously (14) and are summarised in the Supplementary material. In all, the case-control part of the study included 2,198 samples (1,000 cases, 1,198 controls) of European descent, 281 (140 cases, 141 controls) of East Asian origin and 285 (144 cases, 141 controls) who were Native American (Pima) (Supplementary Table 2). We also included a small sample of individuals of African-American origin (242 cases, 173 controls), plus an additional 599 Pima individuals (520 affected, 79 nondiabetic after age 45) from 255 families who after combination with the Pima case-control set were used for family-based association analyses.
These samples were submitted to dense-map SNP-typing of the core region of interest (from 147.0 to 169.7 Mb [Build 35]) using a series of 1536-plex BeadArray designs (Golden Gate, Illumina, San Diego). Design of these arrays was contemporaneous with development of dbSNP and HapMap (15). Thus, whilst the first arrays were LD–agnostic and compiled using genomic localisation as the primary consideration for inclusion, subsequent arrays used LD information from the CEU (European ancestry) and CHB/JPT (Asian ancestry) components of HapMap to guide SNP selection and maximise coverage of the region. In all, we designed assays for 6,023 SNPs, of which 5,290 provided reliable data in all populations after passing through our extensive QC (see Supplementary material). We estimate, that following imputation (using the appropriate set of HapMap data as reference), coverage of the region (MAF>0.05; r2>0.8) reaches ~80% in the European and ~72% in East Asian samples. Coverage is harder to estimate (and imputation likely to be less valuable) in Pima and African-American samples since reference data from these populations are not available, though we estimate ~49% coverage in West Africans based on YRI data.
Genotyping quality was generally good, with over 91% SNPs passing QC in each population (see Supplementary Material) and <0.45% of SNPs failing (p<10-4) tests of within-sample HWE. Significant departures from expectation in the distributions of test statistics observed in the Amish and Pima samples (as revealed by QQ-plots – see Supplementary material) likely reflect residual relatedness between subjects from those populations. We adjusted for this (and any population stratification effects) through genomic control methods (16). Association analyses treated each study as a separate stratum and employed standard meta-analysis approaches to deliver estimates of joint effect-size and statistical significance (see Supplementary material). A series of nested meta-analyses were performed including: (a) European-descent samples only (“4-way”); (b) non-African-descent samples only (“7-way”); (c) all samples (“8-way”).
Under an additive model with allele frequency of 0.25, our sample provides ~80% power to detect per-allele odds ratios of >1.36 (“8-way”) or >1.43 (“4-way”) for a=5×10-6. Given that the region covers ~1% of the genome, we consider this a reasonable benchmark for “region-wide” significance (equivalent to consensus genome-wide thresholds of 5×10-8). These power calculations are conservative: given the case ascertainment enrichment strategies employed in this study, we would expect to detect variants with population-level effects in the 1.2-1.3 range. Under reasonable assumptions (three independent alleles contributing to a linkage signal with a locus-specific sibling relative risk of ~1.15), we can expect the effect size of the variants we were seeking to detect (ie those responsible for the 1q-linkage) to be substantially greater than this (e.g. allelic odds ratio 1.6 for a variant with risk allele frequency of 25%). Our study was therefore well-powered to detect putatively-causal alleles within the European and/or combined data sets.
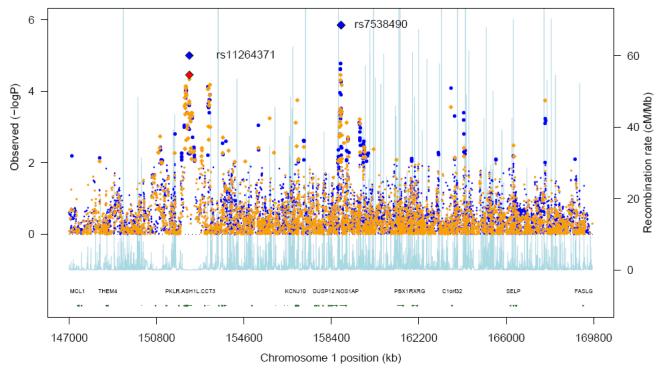
Across these analyses, none of the SNPs showed an association with T2D that withstood genome-wide correction (p<5×10-8) (17). However, two clusters of SNPs showed association signals that approached or exceeded “region-wide” significance thresholds. The first of these, involving rs7538490 and nearby SNPs, mapped to a 51.4kb interval (at ~160.35 Mb) within the first intron of NOS1AP (nitric oxide synthase 1 (neuronal) adaptor protein) with an estimated per-allele OR (in the 4-way, European-only analysis) of 1.38 (95% CI, 1.21-1.57, p=1.4×10-6, additive model, Table 1, Figure 1). Rs7538490 lies ~5.6kb from one of the SNPs (rs10494366) previously shown to influence cardiac repolarisation and QT interval (18): the two SNPs are in modest LD (r2=0.47 in HapMap CEU) and rs10494366 shows some evidence for association with T2D (p=3.1×10-4) in the same 4-way meta-analysis.
Table 1. Association results for rs7538490 in the NOS1AP gene.
| Cohort | N cases | N controls | Risk allele frequency cases |
Risk allele frequency controls |
Additive model OR (95%CI) |
P-value |
|---|---|---|---|---|---|---|
| UK | 443 | 443 | 0.31 | 0.24 | 1.38 (1.13-1.69) | 1.6× 10-3 |
| French | 219 | 239 | 0.34 | 0.29 | 1.21 (0.92-1.60) | 0.16 |
| Utah | 190 | 161 | 0.30 | 0.20 | 1.68 (1.19-2.36) | 2.7 × 10-3 |
| Amish* | 147 | 347 | 0.43 | 0.36 | 1.36 (0.97-1.91) | 0.071 |
|
| ||||||
|
Meta-analysis: 4 European descent
populations |
999 | 1,190 | 1.38 (1.21-1.57) | 1.4 × 10-6 | ||
|
| ||||||
| Shanghai | 77 | 77 | 0.44 | 0.42 | 1.11 (0.71-1.72) | 0.66 |
| Hong Kong | 63 | 64 | 0.57 | 0.55 | 1.10 (0.68-1.78) | 0.70 |
| Pima* | 144 | 141 | 0.46 | 0.44 | 1.09 (0.76-1.57) | 0.62 |
|
| ||||||
| Meta-analysis: 7 non-African populations | 1,283 | 1,472 | 1.31 (1.17-1.46) | 4.3 × 10-6 | ||
|
| ||||||
| African American | 242 | 173 | 0.25 | 0.28 | 0.86 (0.62-1.19) | 0.37 |
|
| ||||||
| Meta-analysis: 8 1qC populations | 1,525 | 1,645 | 1.24 (1.12-1.39) | 5.4 × 10-5 | ||
|
| ||||||
|
| ||||||
| Replication samples | ||||||
|
| ||||||
| Independent WTCCC sample | 1,495 | 2,938 | 0.29 | 0.29 | 0.97 (0.88-1.07) | 0.57 |
|
| ||||||
| W2C vs 58BC | 472 | 1,992 | 0.27 | 0.27 | 1.00 (0.85-1.17) | 0.93 |
|
| ||||||
| UKT2DGC | 3,932 | 4,818 | 0.29 | 0.28 | 1.03 (0.97-1.10) | 0.18 |
|
| ||||||
| DGI | 1,464 | 1,467 | 0.26 | 0.26 | 1.01 (0.87-1.16) | 0.82 |
|
| ||||||
| FUSION | 1,161 | 1,174 | 0.30 | 0.27 | 1.13 (0.97-1.29) | 0.063 |
|
| ||||||
| Meta-analysis: all replication samples | 8,524 | 12,389 | 1.03 (0.98-1.07) | 0.29 | ||
|
| ||||||
|
| ||||||
|
Meta-analysis : all European descent
populations |
9,523 | 13,579 | 1.06 (1.01-1.10) | 0.014 | ||
|
| ||||||
| Meta-analysis: all data | 10,049 | 14,034 | 1.05 (1.01-1.10) | 0.015 | ||
P-value for Amish and Pima was adjusted using estimated lambda for GC
Figure 1. Single-point T2D associations within the 1q region.
This plot shows the “4-way” (European-descent samples only) meta-analysis using the additive model (Cochran-Armitage trend test). Directly typed SNPs are shown in orange, imputed SNPs in blue. The pale blue track (and secondary y-axis) represent recombination rates (for HapMap CEU) across the region. Blue diamonds represent the strongest association p-value in the two regions taken forward for replication. In the case of the PKLR/ASH1L region, the strongest association was seen for a dominant model (the equivalent additive model result is denoted with the red diamond). Only a small subset of genes within the region is denoted on the gene-track.
The second signal includes ~10 SNPs in a 220kb region of extensive LD at ~152.1 Mb. This region includes the coding sequences of the genes encoding liver pyruvate kinase (PKLR) and ash1 (absent, small, or homeotic)-like (Drosophila) (ASH1L) amongst others. At the lead SNP (rs11264371), the estimated OR for the 4-way analysis was 1.36 (1.18-1.58), p=3.5×10-5, under the additive model. The effect size and significance were marginally greater (1.48 [1.18-1.76], p=1.0×10-5), under a dominant model (Table 2, Figure 1).
Table 2. Association results for rs11264371 in the PKLR/ASH1L region.
| Cohort | N cases | N controls | Risk allele frequency cases |
Risk allele frequency controls |
Additive model OR (95%CI) |
P-value# |
|---|---|---|---|---|---|---|
| UK | 444 | 443 | 0.25 | 0.20 | 1.33 (1.07-1.66) | 0.012 |
| French | 219 | 244 | 0.25 | 0.23 | 1.17 (0.86-1.59) | 0.32 |
| Utah | 190 | 162 | 0.26 | 0.21 | 1.34 (0.94-1.92) | 0.095 |
| Amish* | 147 | 349 | 0.25 | 0.17 | 1.83 (1.19-2.81) | 5.8 × 10-3 |
|
| ||||||
|
Meta-analysis: 4 European descent
populations |
1,000 | 1,198 | 1.36 (1.18-1.58) | 3.5 × 10-5 | ||
|
| ||||||
| Shanghai | 77 | 77 | 0.77 | 0.70 | 1.47 (0.87-2.49) | 0.15 |
| Hong Kong | 63 | 63 | 0.66 | 0.73 | 0.73 (0.42-1.23) | 0.24 |
| Pima* | 144 | 141 | 0.58 | 0.60 | 0.91 (0.64-1.29) | 0.64 |
|
| ||||||
| Meta-analysis: 7 non-African populations | 1,284 | 1,479 | 1.24 (1.10-1.42) | 5.6× 10-4 | ||
|
| ||||||
| African American | 242 | 173 | 0.37 | 0.34 | 1.18 (0.88-1.58) | 0.25 |
|
| ||||||
| Meta-analysis: 8 1qC populations | 1,526 | 1,652 | 1.24 (1.10-1.39) | 2.9 × 10-4 | ||
|
| ||||||
|
| ||||||
| Replication samples | ||||||
|
| ||||||
| Independent WTCCC sample | 1,495 | 2,938 | 0.24 | 0.24 | 1.05 (0.95-1.17) | 0.33 |
|
| ||||||
| W2C vs 58BC | 486 | 1,992 | 0.25 | 0.24 | 1.01 (0.86-1.19) | 0.56 |
|
| ||||||
| UKT2DGC | 3,922 | 4,819 | 0.24 | 0.24 | 1.04 (0.98-1.12) | 0.38 |
|
| ||||||
| DGI | 1,464 | 1,467 | 0.26 | 0.27 | 0.97(0.84-1.12) | 1.00 |
|
| ||||||
| FUSION | 1,161 | 1,174 | 0.25 | 0.27 | 0.90 (0.78-1.03) | 0.60 |
|
| ||||||
| Meta-analysis: all replication samples | 8,528 | 12,390 | 1.02 (0.97-1.06) | 0.54 | ||
|
| ||||||
|
| ||||||
|
Meta-analysis: all European descent
populations |
9,528 | 13,588 | 1.04 (1.00-1.09) | 0.083 | ||
|
| ||||||
| Meta-analysis: all data | 10,054 | 14,042 | 1.04 (1.00-1.09) | 0.069 | ||
P-value for Amish and Pima was adjusted using estimated lambda for GC
Both signals were most prominent in the European samples, and there was no evidence that an equivalent association signal extended to the East Asian, Native American or African-American samples. Though the association p-values for these two signals remained strong in the 8-way meta-analysis of all data (5.4×10-5 for rs7538490 and 2.9×10-4 for rs11264371, Table 1, Table 2), in each case they were driven by the larger European samples. Analyses in the larger Pima family-based association data set also found no evidence of association (rs7538490, p=0.59; rs11264372 [r2 of 1 with rs11264371 in CEU and CHB/JPT HapMap] p=0.79).
Although neither signal was of sufficient effect size to be considered causal for the 1q-linkage signal (the estimated sibling relative risk attributable to these loci in combination is only 1.045) we reasoned that these signals might nevertheless be pointers towards nearby causal variants (of lower frequency but higher penetrance) that were inadequately-tagged by the SNPs we had typed. However, before proceeding to resequencing and fine-mapping, we first sought replication of our findings in independent datasets. Mindful that our case ascertainment strategies may have led to inflated estimates of effect-size compared to those evident in unselected cases, we recognised that very large sample sizes would be required to test the observed associations. Because the signals were clearest in European-descent samples, we focused replication on samples from Northern Europe.
First, we used GWA data from the Wellcome Trust Case Control Consortium (19,20). After removing 429 overlapping cases, we examined 1,495 additional T2D cases and 2,938 controls with Affymetrix 500k data (using imputation to test for association at the lead SNPs in each interval). No evidence of association was evident (rs7538490, p=0.57; rs11264371, p=0.33). Similarly, analysis of GWA data from the Diabetes Genetics Initiative (21) and FUSION (22) studies provided no corroboration of either signal. Furthermore, when analysed jointly (4,549 cases, 5,579 controls), these three studies also failed to reveal any additional common variant signals of interest (p<10-4) across the wider 1q region (23), and no corroboration of any of the lesser signals evident in the 1q consortium analyses.
Finally, we genotyped the two lead SNPs (rs7538490, rs11264371) using fluorogenic 5′-nuclease (Taqman) assays in a further 4,572 cases and 6,941 controls from the UK (the UKT2DGC and W2C/58BC strata in Tables 1 and 2). Once again, there was no evidence of replication. Taking into account all replication samples (~8,500 cases, ~12,400 controls) there was no significant association with T2D (rs7538490, OR 1.03 (0.98-1.07), p=0.29; rs11264371, OR 1.02 (0.97-1.06), p=0.54 [additive], 1.06 (0.99-1.13), p=0.10 [dominant]). Nominal significance was retained when these replication data were combined with the original 1q consortium case-control data (rs7538490, p=0.015; rs11264371, p=0.069), but these associations are unimpressive in either the region-wide or genome-wide context. Even allowing for some heterogeneity of effect-size between the primary and replication data sets (due to ascertainment differences and the “winner’s curse”), there seems no substantive evidence that the association signals observed in NOS1AP and the PKLR/ASH1L region are genuinely associated with T2D.
In summary, we have undertaken a detailed survey of common variants across the region of replicated 1q-linkage, achieving coverage that exceeds that of available GWA data for the region. Despite analysis of multiple ethnic groups in samples sufficiently-powered (in the European-descent component at least) to have detected common variants causal for the linkage, we found no compelling signals.
Should we conclude therefore that the original evidence for 1q linkage was false? Though this possibility cannot be discounted, it is worth considering that recent experience from GWA studies has shown that, for common susceptibility variants at least, effect-sizes are modest, and that none is of magnitude sufficient to generate a linkage signal detectable in achievable sample sizes. Efforts to explain the “missing heritability” for T2D (that is, the disparity between the predisposition attributable to the known loci and independent estimates of overall heritability and familiality) are now shifting towards the search for low-frequency, medium-penetrance alleles. Alleles with these characteristics are likely to have escaped detection through the genome-wide approaches available so far since they would be insufficiently penetrant to be detected with traditional linkage approaches applied to monogenic families, and too infrequent to be reliably identified through GWA studies (4). Yet, low-frequency, medium-penetrance alleles could, particularly if several independent alleles map to the same locus, generate the kinds of linkage signals detectable in family-based studies (as is the case for NOD2/CARD15 and Crohn’s, for example) (24).
Detection of low-frequency susceptibility-variants will require new approaches based around next-generation resequencing and large-scale fine-mapping. Genome-wide resequencing of large case-control samples remains economically and logistically unfeasible, but targeted resequencing of selected regions is not, and the future plans of the 1q consortium include deep-resequencing of the 1q region of interest, focusing at least initially on exons and conserved sequence.
Supplementary Material
Acknowledgements
The principal funding was provided as a supplement to NIDDK through R01-DK073490 and as a supplement to U01-DK58026. Other major support has been provided by NIH (T32-AG00219, R01-DK54261, R01-DK54261, K24-DK02673, K07-CA67960, R01-DK39311, U01-DK58026, and intramural funds), the University of Maryland and Arkansas General Clinical Research Centers, the National Center for Research Resources (M01RR14288), the Department of Veteran Affairs and American Diabetes Association (US); “200 Familles pour vaincre le Diabète et l’Obésité” and Association Française des Diabétiques (France); Diabetes UK; The Hong Kong Research Grants Committee (CUHK 4292/99M; 1/04C), Chinese University of Hong Kong Strategic Grant Program (SRP9902) and Hong Kong Innovation and Technology Support Fund (ITS/33/00) (Hong Kong); The National Nature Science Foundation of China (39630150), Shanghai Medical Pioneer Development Project (96-3-004; 996024) and Shanghai Science Technology Development Foundation (01ZB14047) (China). We also recognize the funding support of the UK Medical Research Council (G0601201), the Oxford NIHR Biomedical Research Centre, the General Clinical Research Centers Program, the Baltimore Veterans Administration Geriatric Research and Education Clinical Center and the Wellcome Trust (GR072960). EZ is a Wellcome Trust Research Career Development Fellow. We thank all the subjects participating in this study, and those who contributed to collection of the clinical resources. In particular, we acknowledge members of the Diabetes Genetics Initiative and the Finland-United States Investigation of NIDDM Genetics (FUSION) for sharing data from their studies. For the 1958 Birth Cohort, venous blood collection was funded by the UK Medical Research Council and cell-line production, DNA extraction and processing by the Juvenile Diabetes Research Foundation and the Wellcome Trust.
References
- 1.Prokopenko I, McCarthy MI, Lindgren CM. Type 2 diabetes: new genes, new understanding. Trends Genet. 2008;24:613–621. doi: 10.1016/j.tig.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarthy MI. Growing evidence for diabetes susceptibility genes from genome scan data. Curr Diab Rep. 2003;3:159–167. doi: 10.1007/s11892-003-0040-y. [DOI] [PubMed] [Google Scholar]
- 3.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 4.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 5.McCarthy MI, Zeggini E, Jafar B, Timpson NJ, Al-Hasani N, Frayling TM, Weedon MN, Elliott KS, Lindgren CM, Lango H, Perry JRB, Rayner NW, Freathy RM, Hattersley AT, for the Wellcome Trust Case Control and UKT2D Consortia Analysis of overlap between type 2 diabetes signals identified through genome-wide linkage and association approaches. Diabetes. 2008;57S1:A321. [Google Scholar]
- 6.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 7.Silander K, Mohlke KL, Scott LJ, Peck EC, Hollstein P, Skol AD, Jackson AU, Deloukas P, Hunt S, Stavrides G, Chines PS, Erdos MR, Narisu N, Conneely KN, Li C, Fingerlin TE, Dhanjal SK, Valle TT, Bergman RN, Tuomilehto J, Watanabe RM, Boehnke M, Collins FS. Genetic variation near the hepatocyte nuclear factor-4 alpha gene predicts susceptibility to type 2 diabetes. Diabetes. 2004;53:1141–1149. doi: 10.2337/diabetes.53.4.1141. [DOI] [PubMed] [Google Scholar]
- 8.Love-Gregory LD, Wasson J, Ma J, Jin CH, Glaser B, Suarez BK, Permutt MA. A common polymorphism in the upstream promoter region of the hepatocyte nuclear factor-4 alpha gene on chromosome 20q is associated with type 2 diabetes and appears to contribute to the evidence for linkage in an ashkenazi jewish population. Diabetes. 2004;53:1134–1140. doi: 10.2337/diabetes.53.4.1134. [DOI] [PubMed] [Google Scholar]
- 9.Barroso I, Luan J, Wheeler E, Whittaker P, Wasson J, Zeggini E, Weedon MN, Hunt S, Venkatesh R, Frayling TM, Delgado M, Neuman RJ, Zhao J, Sherva R, Glaser B, Walker M, Hitman G, McCarthy MI, Hattersley AT, Permutt MA, Wareham NJ, Deloukas P. Population-specific risk of type 2 diabetes conferred by HNF4A P2 promoter variants: a lesson for replication studies. Diabetes. 2008;57:3161–3165. doi: 10.2337/db08-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gauguier D, Froguel P, Parent V, Bernard C, Bihoreau MT, Portha B, James MR, Penicaud L, Lathrop M, Ktorza A. Chromosomal mapping of genetic loci associated with non-insulin dependent diabetes in the GK rat. Nature Genetics. 1996;12:38–43. doi: 10.1038/ng0196-38. [DOI] [PubMed] [Google Scholar]
- 11.Galli J, Li LS, Glaser A, Östenson CG, Jiao H, Fakhrai-Rad H, Jacob HJ, Lander ES, Luthman H. Genetic analysis of non-insulin dependent diabetes mellitus in the GK rat. Nature Genetics. 1996;12:31–37. doi: 10.1038/ng0196-31. [DOI] [PubMed] [Google Scholar]
- 12.Masuyama T, Fuse M, Yokoi N, Shinohara M, Tsujii H, Kanazawa M, Kanazawa Y, Komeda K, Taniguchi K. Genetic analysis for diabetes in a new rat model of nonobese type 2 diabetes, Spontaneously Diabetic Torii rat. Biochem Biophys Res Commun. 2003;304:196–206. doi: 10.1016/s0006-291x(03)00548-5. [DOI] [PubMed] [Google Scholar]
- 13.Wallace KJ, Wallis RH, Collins SC, Argoud K, Kaisaki PJ, Ktorza A, Woon PY, Bihoreau MT, Gauguier D. Quantitative trait locus dissection in congenic strains of the Goto-Kakizaki rat identifies a region conserved with diabetes loci in human chromosome 1q. Physiol Genomics. 2004;19:1–10. doi: 10.1152/physiolgenomics.00114.2004. [DOI] [PubMed] [Google Scholar]
- 14.Zeggini E, Damcott CM, Hanson RL, Karim MA, Rayner NW, Groves CJ, Baier LJ, Hale TC, Hattersley AT, Hitman GA, Hunt SE, Knowler WC, Mitchell BD, Ng MC, O’Connell JR, Pollin TI, Vaxillaire M, Walker M, Wang X, Whittaker P, Xiang K, Jia W, Chan JC, Froguel P, Deloukas P, Shuldiner AR, Elbein SC, McCarthy MI. Variation within the gene encoding the upstream stimulatory factor 1 does not influence susceptibility to type 2 diabetes in samples from populations with replicated evidence of linkage to chromosome 1q. Diabetes. 2006;55:2541–2548. doi: 10.2337/db06-0088. [DOI] [PubMed] [Google Scholar]
- 15.The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 17.Dudbridge F, Koeleman BP. Efficient computation of significance levels for multiple associations in large studies of correlated data, including genomewide association studies. Am J Hum Genet. 2004;75:424–435. doi: 10.1086/423738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, West K, Kashuk C, Akyol M, Perz S, Jalilzadeh S, Illig T, Gieger C, Guo CY, Larson MG, Wichmann HE, Marban E, O’Donnell CJ, Hirschhorn JN, Kaab S, Spooner PM, Meitinger T, Chakravarti A. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 19.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS, Wellcome Trust Case Control Consortium (WTCCC) McCarthy MI, Hattersley AT. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 22.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, Ardlie K, Bostrom KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJ, Doney AS, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jorgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CN, Payne F, Perry JR, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjogren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ, Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, McCarthy MI, Boehnke M, Altshuler D. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.