Abstract
Allergic diseases have been increasing in industrialized countries. The environment is thought to have both direct and indirect modulatory effects on disease pathogenesis, including alterating on the allergenicity of pollens. Certain plant proteins known as pathogenesis-related proteins appear to be up-regulated by certain environmental conditions, including pollutants, and some have emerged as important allergens. Thus, the prospect of environmentally regulated expression of plant-derived allergens becomes yet another potential environmental influence on allergic disease. We have identified a novel pathogenesis-related protein allergen, Jun a 3, from mountain cedar (Juniperus ashei) pollen. The serum IgE from patients with hypersensitivity to either mountain cedar or Japanese cedar were shown to bind to native and recombinant Jun a 3 in Western blot analysis and ELISA. Jun a 3 is homologous to members of the thaumatin-like pathogenesis-related (PR-5) plant protein family. The amounts of Jun a 3 extracted from mountain cedar pollen varied up to 5-fold in lots of pollen collected from the same region in different years and between different regions during the same year. Thus, Jun a 3 may contribute not only to the overall allergenicity of mountain cedar pollen, but variable levels of Jun a 3 may alter the allergenic potency of pollens produced under different environmental conditions.
Hypersensitivity to aeroallergens causes allergic diseases (e.g., bronchial asthma, allergic rhinitis, and allergic conjunctivitis) that affect up to 30% of the population (1, 2). Symptoms of rhinitis and asthma due to aeroallergens are major causes of morbidity, lost productivity, and increasing healthcare costs (3). Hypersensitivity to mountain cedar pollen (Juniperus ashei) in the Cupressaceae family causes severe seasonal allergic disease in broad areas of south-central U.S. and northern Mexico. Related species within the cedar family cause similar problems worldwide (4). Allergenic cross-reactivity and protein allergen sequence similarity among these related species suggests that conservation of allergenic proteins occurs across diverse regions and climate conditions. However, the degree of allergenicity of pollens obtained from related species and between specific lots of collected pollens may vary tremendously.
We have recently isolated and cloned the two major extractable proteins of mountain cedar pollen (5). One of them, Jun a 1, was found to be homologous to Cry j 1 and Cha o 1, which are the major allergens of Japanese cedar, Cryptomeria japonica, Taxodiaceae, and Japanese cypress, Chamaecyparis obtusa, Cupressaceae. Here, we describe the cloning and characterization of the second major extractable pollen protein, Jun a 3. Jun a 3 bears no homology to previously described pollen proteins and, thus, defines a new class of pollen allergens within the group 5, pathogenesis-related plant proteins (PR-5)3 (6). We have characterized the IgE-binding capacity of Jun a 3 from patients with mountain cedar hypersensitivity as well as its serum IgE cross-reactivity in patients with hypersensitivity to Japanese cedar. Further, we have investigated the expression of Jun a 3 in different lots of pollen collected from different regions and in different years and suggest that Jun a 3 content may contribute to variable allergenicity in those different lots of pollen. Variability of Jun a 3 expression may be another example of the role of environmental changes in the increasing prevalence and severity of allergic disease.
Materials and Methods
Human sera and polyclonal Abs
Serum samples were obtained from 14 mountain cedar allergic patients in Texas and 35 Japanese cedar allergic patients in Japan and 12 nonatopic donors. Subjects were selected on the basis of clinical history and positive scratch test or capsulated hydrophilic carrier polymer-radioallergosorbent test (Pharmacia, Uppsala, Sweden) results. Total radioimmunosorbent test IgE was measured by (Pharmacia). A polyclonal antiserum to Jun a 3 was produced in New Zealand white rabbits. IgG was purified from this antiserum using protein G Sepharose (Pharmacia) (5). Biotinylated goat anti-human IgE was prepared using an IgG fraction of goat anti-human IgE antiserum (Sigma, St. Louis, MO) and oxysuccinimide-biotin (1:8 molar ratio; Sigma) (5).
ELISA
Sandwich ELISA was performed to detect allergen-specific IgE in patient sera. The IgG fraction of polyclonal anti-Jun a 3 antiserum was used to coat polystyrene plates (Dynex, Chantilly, VA). The wells were washed with saline containing 0.05% Tween 20, and incubated with 60 μg/ml of purified Jun a 3. Sera from pollen-allergic and control patients diluted 1:4 in PBS containing 0.05% Tween 20 (T-PBS) were then incubated in the coated wells for 16 h. IgE was detected with biotinylated goat anti-human IgE in T-PBS followed by avidin-HRP and O-phenylenediamine dihydrochloride/H2O2.
Protein purification and sequencing
Mountain cedar pollen was obtained from Oklahoma, Arkansas, and Texas, dried under vacuum for 48 h, and stored in amber glass under nitrogen (Bayer, Elkhart, IN). Jun a 3 was purified as described (5). Briefly, mountain cedar pollen was defatted and extracted with 0.125 M ammonium bicarbonate and then ammonium sulfate precipitated (40–80% fraction) and applied to a 214TP510 Vydac HPLC column (Vydac, Hesperia, CA). The elution was performed in acetonitrile (30–50%) in 0.1% trifluoroacetic acid. The relative amounts of Jun a 1 and Jun a 3 in each lot of cedar pollen was determined from the integrated optical densities (215 λ) of the two peaks. The purity of Jun a 1 and Jun a 3 was analyzed by N-terminal amino acid sequencing.
Jun a 3 fragments were generated with trypsin digestion (7). Purified Jun a 3 was reduced, alkylated, and repurified by HPLC. Lyophilized protein was dissolved in 0.1 M Tris-HCl buffer, pH 8, containing 0.01 M CaCl2 and 2 M urea, and N-tosyl-L-phenylalanine chloromethyl ketone-treated trypsin (Promega, Madison, WI) (enzyme/substrate ratio = 1/50) and then incubated for 17 h at 37°C. Tryptic peptides were separated by C18 reverse-phase HPLC (Vydac 218TP52) using a 0–45% gradient of acetonitrile in 0.08% trifluoroacetic acid. N-terminal amino acid sequencing of Jun a 3 and its tryptic peptides were determined using a Perkin-Elmer/Applied Biosystems Precise microsequencer (Norwalk, CT).
Isoelectric point and Western blot analysis
The m.w. of Jun a 3 was determined by SDS-PAGE and the isoelectric point by nonequilibrium pH gradient electrophoresis in slab and tube gels, respectively. After electrophoresis, proteins were either stained with Coomassie blue or colloidal gold (Pierce, Rockford, IL) or transferred to nitrocellulose and incubated with an anti-Cry j 1 (KW-S91; Ref. 8) or biotinylated Con A (Pierce) followed by HRP-streptavidin in 0.3% BSA-TBST and developed with 4-chloro-1-napthol and H2O2.
cDNA cloning
Messenger RNA was purified from frozen pollen with guanidinium thio-cyanate using oligo(dT)-cellulose (Pharmacia). First-strand cDNA was synthesized using oligo(dT) (Life Technologies, Gaithersburg, MD) and reverse transcriptase (Life Technologies). Degenerate PCR sense (5′-AA(C or T)CA(A or G)TG(C or T)GGITA(C or T)ACIGTITGGG-3′) and anti-sense (5′-GTICCIGCIA(G or A)(G or A)TTIACIGTCC-3′) primers were synthesized, based on the sequence of the first 48 N-terminal amino acids of Jun a 3 protein.
Inverse PCR was used to clone unknown 5′ and 3′ sequences (9). Briefly, double stranded DNA was synthesized using Escherichia coli DNA polymerase, DNA ligase, and RNase H and circularized using T4 DNA ligase. Foward (5′-GGAAGCGGCTTGACCAGGGG) and inverse primers (5′-CCTCCGGGCAACCCCGCTG) were synthesized based on the sequence obtained from the PCR fragment described above.
As a second approach to obtain 5′ or 3′ unknown sequences, EcoRI adaptors (Life Technologies) were ligated onto double-stranded cDNAs from a library generated from mountain cedar pollen. These were then inserted into Bluescript II SK-vector (Stratagene, La Jolla, CA), which was used as a template for PCRs in which T3 and T7 primers were paired with the Jun a 3-specific primers (5′-GCTGCACAGTCTCCGGAG, 5′-GCIGG(A or G)CA(A or G)TT(G or A)TCIAC(A or G)TAI). These primers were designed based on sequences obtained from inverse PCR and the amino acid sequence of a tryptic peptide. To identify Jun a 3 sequences, PCR products were separated by agarose gel electrophoresis, transferred to nylon membranes, and hybridized with 32P-labeled osmotin cDNA or digoxigenin-labeled oligonucleotide probes (7, 10).
DNA sequencing and analysis
PCR products were separated by agarose gel electrophoresis, purified by gel extraction (Qiagen, Hilden, Germany), ligated into pCR 2.1 (Invitrogen, San Diego, CA), cloned in E. coli, and sequenced using PCR-based techniques and automated detection (Perkin-Elmer/Applied Biosystems model 373 A DNA sequencer). Sequences were compared against GenBank (Genetics Computer Group, Madison, WI) for homology to known sequences.
Expression of recombinant Jun a 3 (rJun a 3) in E. coli
A full-length cDNA was generated by PCR amplification with engineered NdeI (5′) and EcoRI (3′) sites, inserted into the NdeI and EcoRI sites of pET 30, and transfected into E. coli strain BL21. Synthesis of rJun a 3 was induced in bacterial cultures with 2 mM isopropyl β-D-thiogalactopyranoside. Bacterial cells were harvested by centrifugation, purified, and analyzed by SDS-PAGE and Western blotting (7).
Results
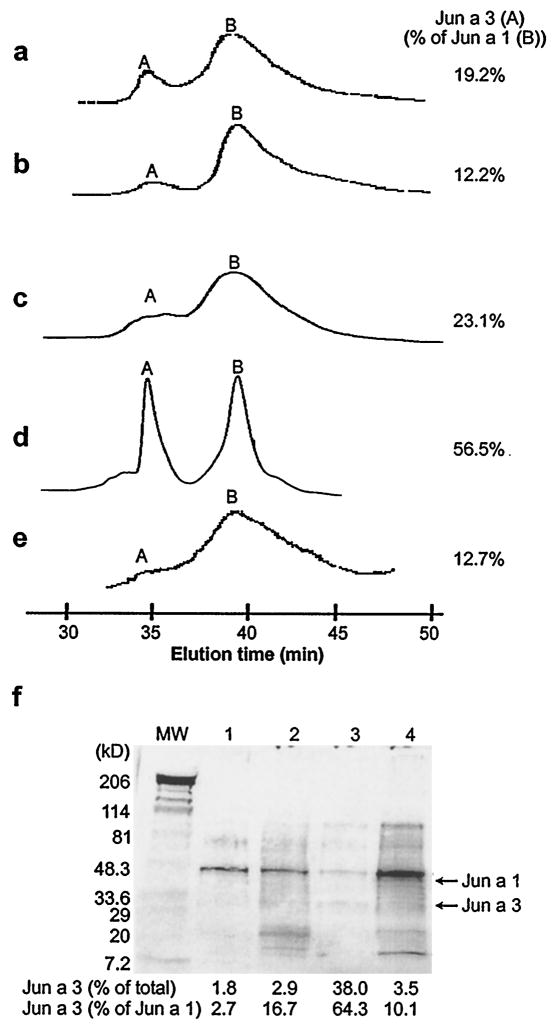
Fig. 1, a–e, shows the HPLC chromatograms of the crude pollen extract of mountain cedar from five different lots of pollen. The area under peak A (Jun a 3) in these lots varied from 12.2 to 56.5% of that of peak B (Jun a 1). Fig. 1f shows the SDS-PAGE stained by colloidal gold (Pierce). In that experiment, the ratio of Jun a 3 and Jun a 1 was analyzed by densitometry. The density of the Jun a 3 band varied from 2.7 to 64.3% of the Jun a 1 band. The 1995 Oklahoma lot of pollen (Fig. 1, d and f, lane 3) expressed a higher proportion of Jun a 3, representing 38% of the total pollen protein. N-terminal amino acid sequence of peak A was analyzed, and a single sequence was obtained, indicating that this peak was not contaminated with other proteins.
FIGURE 1.
HPLC chromatograms of the ammonium sulfate fraction of mountain cedar pollen extracts. The chromatograms of five different lots of mountain cedar pollen are shown. The pollens were harvested in (a) 1992 Arkansas, (b) 1993 Oklahoma, (c) 1993 Arizona, (d) 1995 Oklahoma, and (e) 1998 Texas. The relative percentage of the Jun a 3 peak (A), to the Jun a 1 peak (B) is indicated to the right of each chromatogram. f, Whole pollen was analyzed by SDS-PAGE, transferred to nitrocellulose, and stained with colloidal gold. Lane 1, 1992 Arkansas; lane 2, 1993 Oklahoma; lane 3, 1995 Oklahoma; lane 4, 1998 Texas. The Jun a 3 content as a percent of total protein and relative to Jun a 1, as determined by densitometry, are shown below the each lane.
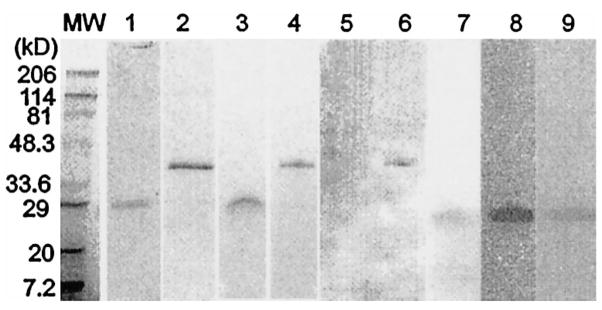
Fig. 2 shows the SDS-PAGE and Western blot analysis of the two peaks (A and B). The 43-kDa protein from peak B reacted with anti-Cry j 1 and was previously identified as Jun a 1 (5). Peak A (Jun a 3) migrated as a 30-kDa protein and weakly bound Con A (data not shown) but not to anti-Cry j 1. Nonequilibrium pH gradient electrophoresis showed that Jun a 3 had isoelectric points of 4.9 and 6.1 (data not shown).
FIGURE 2.
The 1993 (lanes 1, 2, and 5–9) and 1995 (lanes 3 and 4) Oklahoma pollen was analyzed further. HPLC peak A (lanes 1, 3, and 5) and peak B (lanes 2, 4, and 6) were examined by SDS-PAGE and Coomassie staining (lanes 1–4), and immunoblotting (anti-Cry j 1; lanes 5 and 6). Both peaks stained with Coomassie blue but only peak B bound anti-Cry j 1 Ab in immunoblotting. Recombinant Jun a 3 (lanes 7–9) stained with Comassie (lane 7), anti-Juna3 (lane 8), and anti-IgE from patients’ sera (lane 9).
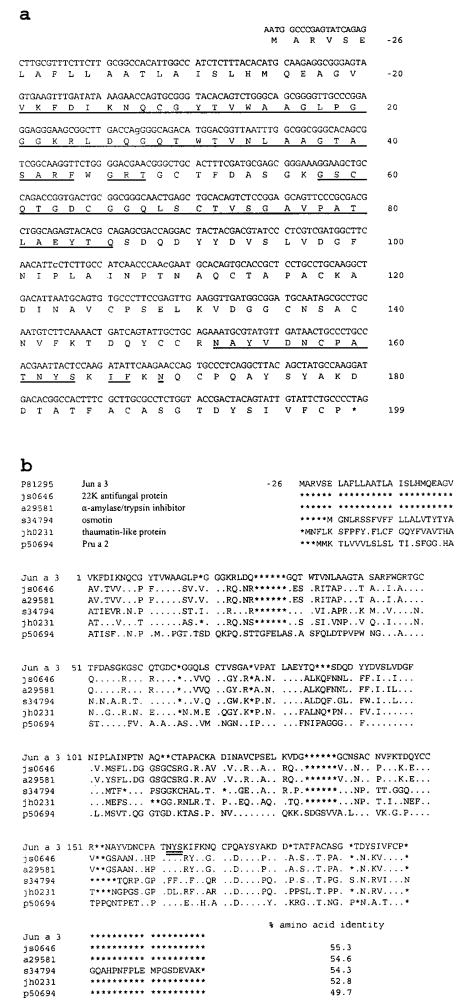
The N-terminal amino acid sequence of the 30-kDa protein (Fig. 3a), Jun a 3, lacked homology with other described pollen allergens (Swiss-prot P81295). Fig. 3b also displays the nucleotide sequence of Jun a 3 obtained by sequencing cDNA clones from three separate mRNA preparations derived from three separate samples of pollen (Fig. 2a; GenBank/EMBL no. AF121776). Much of this sequence was confirmed by N-terminal amino acid sequencing of tryptic peptides. A single potential N-glycosylation site occurred at N162YS. The full-length coding region spans 597 nt. The 26 aa following the first ATG codon were relatively hydrophobic and were not found within the purified protein and most likely represent a signal peptide. A TAG stop codon was identified at position 598, which would result in a mature protein of 199 aa with a calculated molecular mass of 21,174 Da. The amino acid sequence inferred by the nucleotide sequence was identical with that obtained from N-terminal amino acid sequencing of intact Jun a 3 and its tryptic fragments.
FIGURE 3.
Homology analysis. a, Nucleotide and amino acid sequence of Jun a 3 is shown with its N-terminal amino acid sequences of intact protein and tryptic peptides (underlined). b, The amino acid sequence of Jun a 3 is compared with other PR proteins. a29581, α-amylase/trypsin inhibitor (21); js0646, 22K antifungal protein in maize (22); s34794, osmotin in common tobacco (20); jh0231, thaumatin-like protein E2 (19); p50694, Pru a 2 from cherries (24). The potential N-glycosylation site is double underlined.
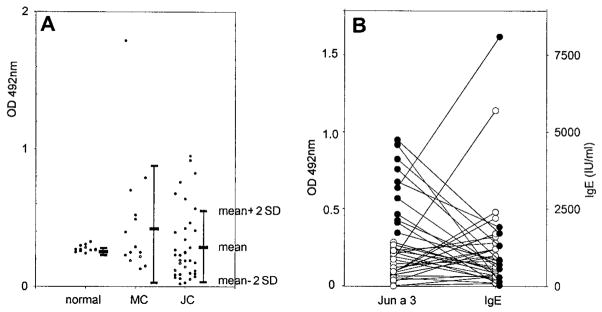
The sera from patients allergic to mountain cedar or Japanese cedar were examined for binding of IgE to Jun a 3 (Fig. 4A). Of 14 mountain cedar sensitive patients, six (42.9%) reacted with Jun a 3. Of 36 Japanese cedar sensitive patients, 12 (33.3%) reacted with Jun a 3. Fig. 4B shows the lack of correlation between the IgE reactivity to Jun a 3 and total IgE levels in the Japanese patients. Further, the total IgE levels in the serum of the Jun a 3-positive patients was not significantly different from that of the Jun a 3-negative patients (1481.73 ± 2277.18 IU/ml vs 1000.19 ± 1154.23 IU/ml). Thus, the binding of IgE to Jun a 3 did not appear to be due to nonspecific binding by high levels of circulating IgE.
FIGURE 4.
ELISA for cedar allergen-specific IgE and total IgE content of sera from Japanese cedar-hypersensitive patients. A, Allergen-specific IgE levels directed against Jun a 3 in sera from cedar-hypersensitive patients and normal donors were examined by ELISA. MC, Mountain cedar-hypersensitive patients; JC, Japanese cedar-hypersensitive patients. The mean plus 2 SD of the IgE binding to each allergen for a group of 12 normal subjects was regarded as the negative cut off. B, The correlation of IgE reactivity to Jun a 3 with the total IgE levels are shown. The total serum IgE level for the Jun a 3-positive group (●) was 1481.73 ± 2277.18 IU/ml, while that for the Jun a 3-negative group (○) was 1000.10 ± 1154.23 IU/ml.
Recombinant Jun a 3 was expressed in BL21. Lysates of E. coli transformed with plasmid DNA encoding Jun a 3 reacted in Western blot analysis with an anti-Jun a 3 polyclonal Ab and with IgE from cedar-hypersensitive patients (Fig. 1f) (8).
Discussion
The PR proteins represent an important group of human allergens. Several allergenic PR proteins have been biochemically characterized in detail (Table I). The mountain cedar allergen, Jun a 3, characterized in this report was found to be homologous to the PR-5 group of proteins (Fig. 3) (23). No PR-5 group members have been identified previously as pollen allergens. The cherry allergen, Pru a 2, was recently identified as a PR-5 group protein and was suggested to be involved in oral hypersensitivity (24). Bet v 1, the major birch pollen allergen, is a PR protein, but is a member of the PR-10 group. The ubiquitous nature of these families of proteins, their similarity among diverse species, and their ability to be up-regulated under certain environmental conditions makes them an important target of investigation into their role in allergic disease. Further analysis of Jun a 3 and other related PR-5 pollen allergens may reveal important aspects of allergen potency and cross-reactivity.
Table I.
PR proteins
| Family | Source | Properties | Inducible | Reference | |
|---|---|---|---|---|---|
| PR-1 | PR-1 a, b, c proteins | Tobacco leaf | Antifungal | Yes | 12 |
| Tomato leaf | 13 | ||||
| PR-2 | β-1,3-Glucanase | Tobacco leaf | β-1,3-Glucanase | Yes | 14 |
| PR-3 | Chitinase (tobacco), 25kD | Tobacco leaf | Endo-chitinase | 15 | |
| PR-4 | PR-“R” | Tobacco leaf | Antifungal, chitinase | 14 | |
| Hev b 6* | Latex prohevein | 17 | |||
| Bra r 2* | Turnip root | 18 | |||
| Pers a 1* | Avocado | 16 | |||
| PR-5 | Thaumatin | Tobacco | Anti-fungal | Yes | 19 |
| Osmotin | Tobacco leaf | 20 | |||
| α-Amylase/trypsin inhibitor | Maize | 21 | |||
| Anti-fungal protein | Maize | 22 | |||
| Zeamatin | Maize | 23 | |||
| Pru a 2* | Cherry | 24 | |||
| Osmotin-like protein | Paprika | 25 | |||
| PR-6 | Tomato proteinase inhibitor I | Tomato | Proteinase-inhibitor | 26 | |
| PR-7 | Tomato endoproteinase P | Tomato | Endoproteinase | 27 | |
| PR-8 | Cucumber chitinase | Cucumber | Chitinase III | 28 | |
| PR-9 | Tobacco lignin-forming peroxidase | Tobacco | Peroxidase | 29 | |
| PR-10 | Parsley “PR1” | Parsley | Ribonuclease-like | Yes | 30 |
| Bet v 1* | Birch pollen | 31 | |||
| Mal d 1* | Apple | 32 | |||
| Api g 1* | Celery | 33 | |||
| Dau c 1* | Carrot | 34 | |||
| PR-11 | Tobacco class V | Tobacco leaf | Chitinase | 35 |
, Allergens.
In our study, cross-reactivity of Jun a 3 was shown in patients with Japanese cedar hypersensitivity who were never exposed to mountain cedar. This cross-reactivity may be due to the presence of IgE Abs to unidentified Jun a 3-like proteins in Japanese cedar patients. Previous studies suggested a wide degree of cross-reactivity among members of the Cupressaceae (cedar and cypress) family (5). We recently showed that the major pollen protein from mountain cedar, Jun a 1, which is not a PR protein, but has pectate lyase activity, may explain some of that cross-reactivity. Now, the identification of a PR-5 protein as a prominent allergen in pollen from mountain cedar trees may also help to explain not only cross-reactivity, but also variations in the cross-reactivity and potency of pollens among these species.
PR proteins are up-regulated in plants in response to stressors such as drought, freezing temperature, infection by fungi, viruses, or bacteria, ozone, and UV B (36–38). Surplus et al. showed that UV B in Arabidopsis thaliana, Cruciferae (mustard family) induced PR-1, PR-2, and PR-5 group proteins (39). Air pollutants from industry and automobiles are considered cofactors contributing to the recent increase in allergic disease and asthma (40). For example, pollutants have been shown to enhance IgE responses to Japanese cedar pollen (41). The effect of air pollutants on Bet v 1, the major allergen of birch pollen, was examined, and it was found that the distance from an industrial pollutant point sources did not correlate with the expression of Bet v 1 (11). Those authors concluded that other factors including shading, soil properties, and genetics may have stronger influences on the composition of birch pollen allergens.
Our data supports the concept that the levels of PR proteins in allergenic pollens are highly variable, but we did not explore the etiology of that altered expression. Our data does suggest that trees of the same species in different locations and in different years may express variable levels of PR-5 proteins. It is interesting to speculate that alterations in environmental conditions may be the cause of that altered expression of PR proteins, thereby contributing to altered allergenic potency of different pollens.
Acknowledgments
We thank Masanao Watanabe and Tetsuo Oka for providing mAb KW-S91, Rafeul Alam and J. Andrew Grant for providing sera and PBMC from Texas patients, Shin Nouno and Isaac Horiuchi for providing sera from Japanese patients, Ray A. Bressan for providing osmotin cDNA, and J. Wes Padgett and C. Renee Webb for technical assistance. Protein/peptide and DNA sequence analysis were performed in the University of Texas Medical Branch protein chemistry laboratory.
Footnotes
This work was supported by the James W. McLaughlin Fellowship Fund, President’s Cabinet Award, National Institute on Environmental Health Sciences Center for Environmental Science (ES-06676), and the Collaborative Grant Program of the Sealy Center for Structural Biology at the University of Texas Medical Branch.
Abbreviation used in this paper: PR, pathogenesis related.
References
- 1.Nielsen NH, Svendsen UG, Madsen F, Dirksen A. Allergen skin test reactivity in an unselected Danish population. The Glostrup Allergy Study, Denmark. Allergy. 1994;49:86. doi: 10.1111/j.1398-9995.1994.tb00805.x. [DOI] [PubMed] [Google Scholar]
- 2.Midoro-Horiuti T, Nouno S, Seino Y. Skin tests of pollen grains of Taxodiaceae and Cupressaceae in children with bronchial asthma. Acta Paediatr Jpn. 1992;34:501. doi: 10.1111/j.1442-200x.1992.tb00996.x. [DOI] [PubMed] [Google Scholar]
- 3.Malone DC, Lawson KA, Smith DH, Arrighi HM, Battista C. A cost of illness study of allergic rhinitis in the United States. J Allergy Clin Immunol. 1997;99:22. doi: 10.1016/s0091-6749(97)70296-3. [DOI] [PubMed] [Google Scholar]
- 4.Platts-Mills TA, Mueller GA, Wheatley LM. Future directions for allergen immunotherapy. J Allergy Clin Immunol. 1998;102:335. doi: 10.1016/s0091-6749(98)70117-4. [DOI] [PubMed] [Google Scholar]
- 5.Midoro-Horiuti T, Goldblum RM, Kurosky A, Goetz DW, Brooks EG. Isolation and characterization of mountain cedar (Juniperus ashei) pollen major allergen, Jun a 1. J Allergy Clin Immunol. 1999;104:608. doi: 10.1016/s0091-6749(99)70331-3. [DOI] [PubMed] [Google Scholar]
- 6.Van Loon LC, Pierpoint WS, Boller T, Conejero V. Recommendations for naming plant pathogenesis-related proteins. Plant Mol Biol Reporter. 1994;12:245. [Google Scholar]
- 7.Midoro-Horiuti T, Goldblum RM, Kurosky A, Wood TG, Schein CH, Brooks EG. Molecular cloning of mountain cedar (Juniperus ashei) pollen major allergen, Jun a 1. J Allergy Clin Immunol. 1999;104:613. doi: 10.1016/s0091-6749(99)70332-5. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi Y, Nagoya T, Watanabe M, Inouye S, Sakaguchi M, Katagiri S. A new method of counting airborne Japanese cedar (Cryptomeria japonica) pollen allergens by immunoblotting. Allergy. 1993;48:94. doi: 10.1111/j.1398-9995.1993.tb00692.x. [DOI] [PubMed] [Google Scholar]
- 9.Brooks EG, Filipovich AH, Padgett JW, Mamlock R, Goldblum RM. T-cell receptor analysis in Omenn’s syndrome: evidence for defects in gene rearrangement and assembly. Blood. 1999;93:242. [PubMed] [Google Scholar]
- 10.Nelson DE, Raghothama KG, Singh NK, Hasegawa PM, Bressan RA. Analysis of structure and transcriptional activation of an osmotin gene. Plant Mol Biol. 1992;19:577. doi: 10.1007/BF00026784. [DOI] [PubMed] [Google Scholar]
- 11.Helander ML, Savolainen J, Ahlholm J. Effects of air pollution and other environmental factors on birch pollen allergens. Allergy. 1997;52:1207. doi: 10.1111/j.1398-9995.1997.tb02525.x. [DOI] [PubMed] [Google Scholar]
- 12.Antoniw JF, White RF. The effects of aspirin and polyacrylic acid on soluble leaf proteins and resistance to virus infection in five cultivars of tobacco. Phytophol. 1980;98:331. [Google Scholar]
- 13.Camacho HA, Sanger HL. Gelelectrophoretic analysis of phenol-extractable leaf proteins from different viroid/host combinations. Arch Virol. 1982;74:167. doi: 10.1007/BF01314710. [DOI] [PubMed] [Google Scholar]
- 14.Antoniw JF, Ritter CE, Pierpoint WS, VanLoon LC. comparison of three pathogenesis-related proteins from plants of two cultivars of tobacco infected with TMV. J Gen Virol. 1980;47:79. [Google Scholar]
- 15.VanLoon LC. Regulation of changes in proteins and enzymes associated with active defense against virus infection. In: Wood RKS, editor. Active Defence Mechanisms in Plants. Plenum Press; New York: 1982. p. 247. [Google Scholar]
- 16.Vanek-Krebitz MS, Hsieh LSS. Molecular characterization and purification of conserved pollen and food allergens in avocado (Persea americana) J Allergy Clin Immunol. 1997;99:479. [Google Scholar]
- 17.Beezhold DH, Kostyal DA, Sussman GL. IgE epitope analysis of the hevein preprotein; a major latex allergen. Clin Exp Immunol. 1997;108:114. doi: 10.1046/j.1365-2249.1997.d01-983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanninen AR, Mikkola JH, Kalkkinen N, Turjanmaa K, Ylitalo L, Reunala T, Palosuo T. Increased allergen production in turnip (Brassica rapa) by treatments activating defense mechanisms. J Allergy Clin Immunol. 1999;104:194. doi: 10.1016/s0091-6749(99)70135-1. [DOI] [PubMed] [Google Scholar]
- 19.Edens L, Heslinga L, Klok R, Ledeboer AM, Maat J, Toonen MY, Visser C, Verrips CT. Cloning of cDNA encoding the sweet-tasting plant protein thaumatin and its expression in. Escherichia coli Gene. 1982;18:1. doi: 10.1016/0378-1119(82)90050-6. [DOI] [PubMed] [Google Scholar]
- 20.Singh NK, LaRosa PC, Handa AK, Hasegawa PM, Bressan RA. Hormonal regulation of protein synthesis associated with salt tolerance in plant cells. Proc Natl Acad Sci USA. 1987;84:739. doi: 10.1073/pnas.84.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malehorn DE, Borgmeyer JR, Smith CE, Shah DM. Characterization and expression of an antifungal zeamatin-like protein (Zlp) gene from. Zea mays Plant Physiol. 1994;106:1471. doi: 10.1104/pp.106.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vigers AJ, Roberts WK, Selitrennikoff CP. A new family of plant antifungal proteins. Mol Plant-Microbe Interactions. 1991;4:315. doi: 10.1094/mpmi-4-315. [DOI] [PubMed] [Google Scholar]
- 23.Richardson MV. A possible function for thaumatin and a TMV-induced protein suggested by homology to a maize inhibitor. Nature. 1987;327:432. [Google Scholar]
- 24.Inschlag C, Hoffmann-Sommergruber K, O’Riordain G, Ahorn H, Ebner C, Scheiner O, Breiteneder H. Biochemical characterization of Pru a 2, a 23-kD thaumatin-like protein representing a potential major allergen in cherry (Prunus avium) Int Arch Allergy Immunol. 1998;116:22. doi: 10.1159/000023920. [DOI] [PubMed] [Google Scholar]
- 25.Leitner A, Jensen-Jarolim E, Grimm R, Wüthrich B, Ebner H, Scheiner O, Kraft D, Ebner C. Allergens in pepper and paprika: immunologic investigation of the celery-birch-mugwort-spice syndrome. Allergy. 1998;53:36. doi: 10.1111/j.1398-9995.1998.tb03771.x. [DOI] [PubMed] [Google Scholar]
- 26.Bryant J, Green TR, Gurusaddaiah T, Ryan CA. Proteinase inhibitor II from potatoes: isolation and characterization of its protomer components. Biochemistry. 1976;15:3418. doi: 10.1021/bi00661a004. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigo I, Vera P, Tornero P, Hernandez-Yago J, Conejero cDNA cloning of viroid-induced tomato pathogenesis-related protein P23: characterization as a vacuolar antifungal factor. Plant Physiol. 1993;102:939. doi: 10.1104/pp.102.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metraux JP, Streit L, Staub T. A pathogenesis-related protein in cucumber is a chitinase. Physiol Mol Plant Pathol. 1988;33:1. [Google Scholar]
- 29.Diaz-De-Leon F, Klotz KL, Lagrimini LM. Nucleotide sequence of the tobacco (Nicotiana tabacum) anionic peroxidase gene. Plant Physiol. 1993;101:1117. doi: 10.1104/pp.101.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Somssich IE, Schmelzer E, Kawalleck P, Hahlbrock K. Gene structure and in situ transcript localization of pathogenesis-related protein 1 in parsley. Mol Gen Genet. 1988;213:93. doi: 10.1007/BF00333403. [DOI] [PubMed] [Google Scholar]
- 31.Breiteneder H, Pettenburger K, Bito A, Valenta R, Kraft D, Rumpold H, Scheiner O, Breitenbach M. The gene coding for the major birch pollen allergen Bet v 1 is highly homologous to a pea disease resistance response gene. EMBO J. 1989;8:1935. doi: 10.1002/j.1460-2075.1989.tb03597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanek-Krebitz M, Hoffmann-Sommergruber K, Laimer dCM, Susani M, Ebner C, Kraft D, Scheiner O, Breiteneder H. Cloning and sequencing of Mal d 1, the major allergen from apple (Malus domestica), and its immunological relationship to Bet v 1, the major birch pollen allergen. Biochem Biophys Res Commun. 1995;214:538. doi: 10.1006/bbrc.1995.2320. [DOI] [PubMed] [Google Scholar]
- 33.Hoffmann-Sommergruber K, Vanek-Krebitz M, Ferris R, O’Riordain G, Susani M, Hirschwehr R, Ebner C, Ahorn H, Kraft D, Scheiner O, Breiteneder H. Isolation and cloning of Bet v 1-homologous food allergens from celeriac (Api g 1) and apple (Mal d 1) Adv Exp Med Biol. 1996;409:219. doi: 10.1007/978-1-4615-5855-2_30. [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann-Sommergruber K, O’Riordain G, Ebner C, Kraft D, Scheiner O, Breiteneder H. Characterization of Dau c 1, the Bet v 1 homologous protein from carrot, and its crossreactivity with Bet v 1 and Api g 1. J Allergy Clin Immunol. 1997;99:140. doi: 10.1046/j.1365-2222.1999.00529.x. [DOI] [PubMed] [Google Scholar]
- 35.Melchers LS, Apotheker-de GM, Avan der Knaap J, Ponstein AS, Sela-Buurlage MB, Bol JF, Cornelissen BJ, van den Elzen PJ, Linthorst HJ. A new class of tobacco chitinases homologous to bacterial exo-chitinases displays antifungal activity. Plant J. 1994;5:469. doi: 10.1046/j.1365-313x.1994.5040469.x. [DOI] [PubMed] [Google Scholar]
- 36.Van Loon LC, Gerritsen YAM. Protease activity and pathogenesis-related proteins in virus-infected Samsun NN tobacco leaves. Plant Sci. 1989;63:141. doi: 10.1007/BF00020536. [DOI] [PubMed] [Google Scholar]
- 37.Linthorst HJM. Pathogenesis-related proteins of plants. Crit Rev Plant Sci. 1981;10:123. [Google Scholar]
- 38.Ernst D, Schraudner M, Langebartels C, Sandermann H., Jr Ozone-induced changes of mRNA levels of β-1,3-glucanase, chitinase and ‘pathogenesis-related’ protein 1b in tobacco plants. Plant Mol Biol. 1992;20:673. doi: 10.1007/BF00046452. [DOI] [PubMed] [Google Scholar]
- 39.Surplus SL, Jordan BR, Murphy AM, Carr JP, Thomas B, Mackerness SAH. Ultraviolet-B-induced responses in Arabidopsis thaliana: role of salicylic acid and reactive oxygen species in the regulation of transcripts encoding photosynthetic acidic pathogenesis-related proteins. Plant Cell Environ. 1998;21:685. [Google Scholar]
- 40.Anto JM, Sunyer J. A point-source asthma outbreak. Lancet. 1986;1:900. doi: 10.1016/s0140-6736(86)90999-2. [DOI] [PubMed] [Google Scholar]
- 41.Maejima KT. Comparison of the effect of various fine particles on IgE antibody production in mice inhaling Japanese cedar pollen allergens. J Toxicol Environ Health. 1997;52:231. doi: 10.1080/00984109708984062. [DOI] [PubMed] [Google Scholar]