Abstract
The imprinted Igf2 gene is associated with a small upstream region that is differentially methylated on the active paternal allele. We have identified a repressor element within this sequence and shown that repression is probably mediated through a trans- acting factor, GCF2. DNA methylation of this site abrogates both protein binding and repressor activity. Targeting experiments demonstrate that this element plays a role in the repression of the maternal Igf2 gene in vivo.
Keywords: GCF/imprinting/methylation
Introduction
The DNA methylation pattern of the animal genome is established during development through a process of de novo methylation and demethylation which is directed by cis-acting elements (Razin, 1998). In contrast to most of the genome where both alleles are methylated to an equal extent, genomically imprinted domains are characterized by the inclusion of small regions that are differentially methylated on the two alleles (DMRs) (Razin and Cedar, 1994; Constancia et al., 1998). From the developmental point of view, these DMRs fall into two distinct categories. In one type, the differential methylation pattern is established in the gametes and then maintained throughout embryogenesis. These modifications serve to mark the two parental alleles so that they can be identified in somatic cells as being of either maternal or paternal origin. Examples include previously characterized sequences upstream of the H19 gene (Bartolomei et al., 1993) or within the Igf2r intron in the mouse (Stoger et al., 1993). A second class of differential methylation is established post-zygotically (Brandeis et al., 1993; Feil et al., 1994). Since these methyl groups are not put on during gametogenesis, the resulting allele-specific pattern must be generated using structural cues already present on the parental alleles. Although it is generally assumed that these DMRs are involved in the local control of allele-specific gene expression, little is known about their mechanisms of action.
A good model system for studying this phenomenon is the mouse Igf2 domain, which is associated with three distinct post-zygotically established DMRs located at different positions in the vicinity of this gene (Moore et al., 1997). Two of these regions are methylated on the active paternal allele, a pattern that is contrary to the accepted dogma that methylation mediates repression in cis. One model for explaining this anomalous behavior proposes that these regions contain repressor elements that interact with trans-acting factors whose binding can be blocked by DNA methylation (Sasaki et al., 1992; Brandeis et al., 1993; Feil et al., 1995). According to this idea, the maternal unmethylated Igf2 allele is transcriptionally inactive because of these cis-acting repressor elements, while the paternal allele adopts an active expression profile by virtue of DNA modification which prevents repressor function. In keeping with this, it has been shown that both the maternal and paternal copies of the Igf2 gene are transcriptionally inactive when the DMRs are unmethylated on both alleles, as occurs in methylase–/– embryos (Li et al., 1993).
Using gene targeting, it has been demonstrated that a 5 kb region upstream to the Igf2 gene contains repressor sequences (Constancia et al., 2000), but the elements themselves were not mapped and the mechanism of repression not studied. Here, we have used a transfection assay to show that a differentially methylated region (DMR1) within this same stretch of DNA serves as a transcriptional repressor and that its function can be abrogated by DNA methylation. This mechanism is actually mediated by a small element within the DMR, which probably interacts with the previously characterized repressor protein GCF2 (Kageyama and Pastan, 1989; Reed et al., 1998). Using sequence-specific gene targeting in mice, we have demonstrated that this site is required for repressing transcription from the maternal Igf2 allele, indicating that this regulatory mechanism is used in vivo as part of the imprinting process.
Results
The Igf2 upstream region is differentially methylated
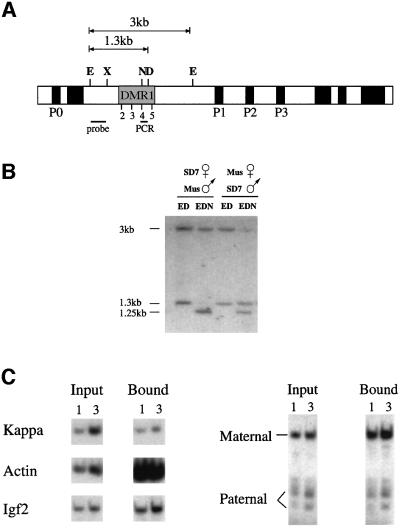
Previous studies have shown that the region upstream of Igf2 in the mouse is differentially methylated (DMR1) in a number of tissues. One of the difficulties in assaying methylation in this region stems from the fact that it contains four closely spaced HpaII sites, which cannot be distinguished easily on Southern blots (Sasaki et al., 1992; Brandeis et al., 1993; Feil et al., 1994). In order to overcome this problem, we utilized the methyl-sensitive enzyme NgoMI, which recognizes HpaII site 4 exclusively (Figure 1A). Mice of a normal Mus m. domesticus genetic background, but with the distal portion of chromosome 7 derived from M.spretus (SD7), were crossed with standard M.m.domesticus mice (Forne et al., 1997) and DNA from the offspring analyzed for DNA methylation. In these animals, it was possible to distinguish between the parental alleles by taking advantage of a spretus-specific polymorphic DraI restriction site located within DMR1. As shown in Figure 1B, regardless of whether the SD7 mouse serves as the mother or the father, the NgoMI site is methylated (∼85%) when derived paternally, but unmethylated when inherited from the mother. These results thus show that there are indeed clearcut differences in methylation between the maternal and paternal alleles in this Igf2 upstream region.
Fig. 1. Igf2 allelic methylation and histone acetylation patterns. (A) Map of the mouse Igf2 gene indicating the promoters (P0–P3), the differentially methylated (DMR1) region including HpaII sites 2–5, restriction sites for E (EcoRI), X (XbaI), N (NgoMI; NgoMIV), D (DraI; only present on the M.spretus allele), the EcoRI–XbaI probe and the position of the PCR fragment used in the immunoprecipitation experiments. (B) Igf2 differential methylation at site 4 was measured by Southern blot analysis of kidney genomic DNA from SD7 hetero zygous mice using the restriction enzymes shown in each lane. After digestion with NgoMI, both the domesticus allele (3 kb) and the spretus allele (1.3 kb) yield a 1.25 kb band. Similar results were obtained using DNA from liver but, in this case, the paternal allele was only 70% methylated. (C) The acetylation state of nucleosomes in DMR1 was determined by immunoprecipitation of mononucleosomes from SD7 heterozygous mice using anti-acetylated H4 histone. DNA from input and bound fractions was extracted and 1 or 3 µl subjected to quantitative radiolabeled PCR with primers specific for sequences from the κ-light chain, β-actin and Igf2 upstream regions (left panel). The Igf2 PCR product was digested with HpaII to identify a polymorphic site on this fragment, present only on the spretus (paternal) allele, analyzed by gel electrophoresis (right panel) and the results quantitated using a phosphoimager. The ratio between the maternal and paternal alleles in the bound fraction was found to be 5.5-fold more than that measured for the input DNA. In a separate experiment, we also determined that the PCR product obtained from total DNA is produced equally from the two alleles (data not shown).
Previous studies have shown that both Igf2 alleles in the nucleus are equally sensitive to DNase I in the promoter region, suggesting that they have similar overall structures (Sasaki et al., 1992; Feil et al., 1995). However, since it is known that DNA methylation can have a profound effect on DNase I sensitivity and chromatin structure in general (Keshet et al., 1986), we asked whether DMR1 may be characterized by local allele-specific differences in structure. It was demonstrated recently, for example, that DNA methylation itself can affect chromatin by mediating the deacetylation of local histone proteins (Eden et al., 1998; P.L.Jones et al., 1998; Nan et al., 1998). In order to determine whether this might be the case for DMR1, we immunoprecipitated nucleosomes rich in acetylated histone H4 from primary fibroblast nuclei of musculus/SD7 F1 mice and then used PCR to assay the representation of several different sequences in this fraction. As shown in Figure 1C, the presence of acetylated histones correlates well with gene activity in control experiments. The constitutive β-actin gene sequence, for example, is highly enriched in this acetylated nucleosome fraction, whereas the unexpressed κ gene sequence is under-represented.
As demonstrated in Figure 1C, DMR1 is relatively rich in acetylated histones. However, when the PCR product was cut with HpaII, which recognizes a polymorphic site that distinguished between the two alleles, one can see immediately that the unmethylated maternal allele is more acetylated than its paternal counterpart (5.5-fold) even though both alleles are equally represented in the starting material before immunoprecipitation. These findings thus suggest that this upstream region may be packaged into a relatively more open chromatin structure when present on the maternal allele, consistent with the idea that this domain is available for interacting with repressor factors, while the paternal allele is relatively closed. Thus, this region appears to carry a chromatin structural imprint.
DMR1 contains a functional repressor element
Since it is the inactive maternal Igf2 allele that is characterized by undermethylation of upstream sequences, it has been suggested that this DMR may harbor a methyl-sensitive repressor element. In confirmation of this prediction, a simple sequence analysis of this region indeed revealed the presence of a well known repressor element that had been shown previously to interact with the protein, GCF (Johnson et al., 1992). Furthermore, this element co-maps precisely with the same HpaII site (site 4) that we have shown to be differentially methylated in vivo (see Figure 1B).
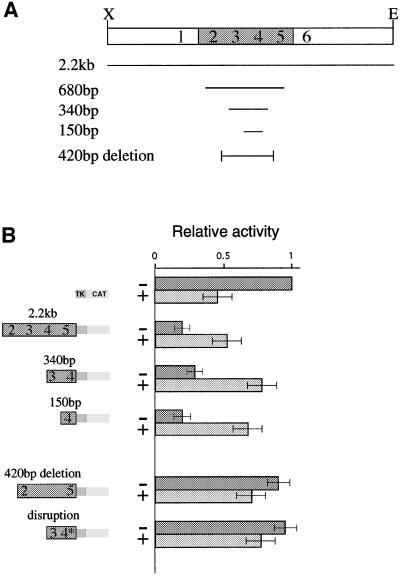
In order to determine directly whether sequences within the DMR1 region actually play a role in the control of Igf2 expression, we studied its effect on transcription by employing a transient transfection assay in human kidney cells (293 cells). To this end, we made a test construct with a CAT reporter gene driven by the herpes thymidine kinase (TK) minimal promoter and examined the influence of DNA inserts from the DMR1 region. The presence of a full 2.2 kb Igf2 upstream region containing all of the differentially methylated HpaII sites (2–5) caused a 4- to 5-fold inhibition of transcription in this system (Figure 2B). Furthermore, this repression was relieved when the plasmid was modified using HpaII methylase. It should be noted that in the absence of repressor sequences, the TK CAT vector itself, which does not contain any HpaII sites in its minimal promoter, is slightly inhibited by this methylation. Despite this background, methylation of the repressor element apparently overcomes inhibition and brings about an increase in transcription.
Fig. 2. DMR1 contains a methylation-sensitive repressor element. Transient transfection into 293 human kidney cells was used to measure repressor activity on a pTK-CAT reporter construct. (A) Map showing the different constructs used in the transient transfection experiments. (B) CAT assays of reporter constructs. The results are shown in graphic form relative to the basic TK-CAT vector. These values were determined by scanning autoradiograms. For each lane, the particular construct used is indicated by a modular diagram made up of fragments from DMR1, the TK promoter and the CAT reporter gene. The last construct (disruption) is the same as the 340 bp fragment but contains a loxP insertion (see text). Constructs were either unmethylated (–) or in vitro methylated by MHpaII (+). The HpaII sites (2–5) located within each fragment are indicated in the diagram. Most of these experiments were repeated 4–5 times and the results shown represent average values (± SD).
These initial experiments indicated that the sequences responsible for repression probably map to the region containing HpaII sites 3 and 4, since insertion of a smaller 340 bp fragment into the test vector brought about a similar degree of repression, which could also be relieved by DNA methylation (Figure 2B). In order to pinpoint further the repressor element itself, we generated a number of reporter constructs carrying the TK CAT gene together with small upstream sequences derived from within a 680 bp fragment (see Figure 2A). These experiments demonstrated conclusively that only fragments containing HpaII site 4 are capable of mediating the repressor effect (data not shown), and a minimal 150 bp fragment was found to be sufficient for both repression and methylation reversal (Figure 2B).
While these data show that sequences around HpaII site 4 are sufficient for methyl-sensitive repression in the transfection assay, they do not prove that this element is necessary for the transcriptional repression mediated by DMR1. In order to address this question, we compared the full 2.2 kb DMR1 fragment with a similar sequence that lacks a 420 bp region containing HpaII sites 3 and 4. As shown in Figure 2B, this deletion significantly lowers the level of repression. More importantly, DNA methylation now brings about a slight inhibition of transcription, rather than the enhancement normally seen when the repressor element is present. Similarly, we specifically disrupted site 4 itself by inserting a loxP element in the critical sequence, and this was sufficient to eliminate the repression usually associated with the 340 bp fragment (Figure 2B).
Protein interaction is required for repression
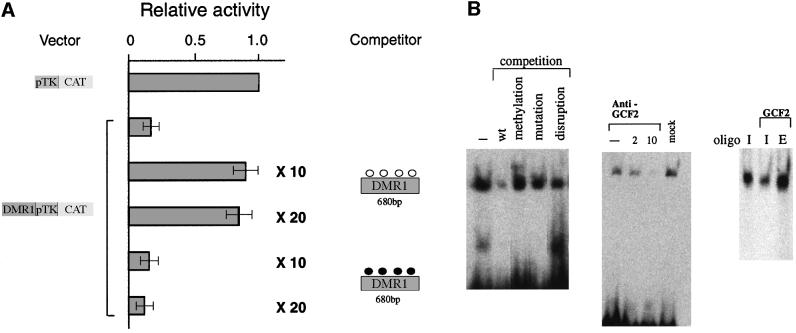
These results suggested that trans-acting repressor factors in the nucleus interact with sequence elements within the Igf2 upstream region, and that DNA methylation prevents this binding. To test this possibility directly, we carried out co-transfection competition experiments using the CAT expression vector together with an excess of a second plasmid containing only 680 bp of DMR1. As seen in Figure 3A, the addition of non-methylated competitor DNA completely reverses the repression profile, while methylated DNA has no effect. This clearly indicates that repressor proteins bind to unmethylated, but not methylated DNA and, since this plasmid was only modified at HpaII sites, it must be these particular sequences that are involved in repression.
Fig. 3. Repression is mediated by protein interactions with DMR1. (A) Co-transfection competition experiments were carried out using a reporter construct containing a 680 bp fragment from DMR1 (see Figure 2A) and competitor plasmid DNA containing only the 680 bp fragment (10- or 20-fold excess in methylated or unmethylated form), and the results (average ± SD) are shown as in Figure 2. The pTk-CAT vector was unaffected by the addition of either unmethylated or methylated competitor DNA (data not shown). (B) Gel shift analysis of a 32P-end-labeled Igf2 upstream oligonucleotide using 293 cell nuclear extract. Competition (left) was carried out using a 45-fold excess of non-radioactive wild-type (wt), methylated or disrupted oligonucleotides (see Materials and methods). Gel shift analysis was also carried out (middle) with the same extract treated with two different concentrations of GCF2 antibody to bring about immunodepletion. In a separate experiment, we removed the antibody complex from the Sepharose beads and confirmed by western analysis that it contained GCF2. A mock experiment in which the extract was passed over protein A–Sepharose beads without antibody is also shown. Extract or pure recombinant GCF2 protein was reacted (right) with either the labeled Igf2 upstream oligonucleotide (I) or an oligonucleotide containing the EGFR recognition sequence (E). A small amount of immunodepleted extract was added to the pure protein in order to stabilize it and enhance binding.
These transfection experiments clearly suggest that HpaII site 4 contains a sequence element that mediates gene repression through interaction with proteins present in the nucleus. To test this hypothesis directly, we carried out gel retardation experiments in vitro (Figure 3B). An end-labeled 20 bp fragment containing HpaII site 4 was clearly shifted when exposed to nuclear extract from human kidney cells (293 cells). This binding activity apparently is sequence specific, since it could be competed out with unlabeled oligonucleotide, but not with identical fragments either mutated or containing a small disruption at the HpaII site. Furthermore, binding was not competed out by a methylated oligonucleotide, consistent with the observation that methylation prevents repression at this site.
Since this binding site contains a GCF motif (Kageyama and Pastan, 1989), we wanted to test the possibility that it is this factor that is responsible for the band shift. GCF, initially characterized as a presumed transcriptional repressor, recently has been shown to be derived from an artifactual cDNA fusion product, which, however, carries the DNA-binding domain of an authentic endogenous protein, GCF2 (Reed et al., 1998; Takimoto et al., 1999). Evidence that GCF2 binds the Igf2 repressor element was obtained by immunodepleting this protein from the nuclear extract using a specific antibody, and this indeed resulted in elimination of the band shift. Furthermore, purified recombinant GCF2 protein itself produces a band shift similar to that seen with the full extract (Figure 3B), and almost identical to a band generated through interaction with a prototypic GCF2 element from the EGFR gene (Reed et al., 1998). These experiments strongly suggest that GCF2 interacts with the Igf2 upstream region and is probably responsible for the repression observed in transfection assays.
The upstream repressor element influences Igf2 expression in vivo
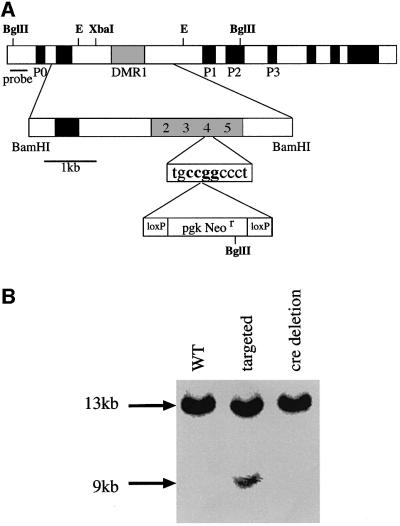
In order to determine whether the repressor element upstream of the Igf2 gene actually plays a role in the control of imprinted expression in vivo, we designed an experiment to disrupt this element by DNA targeting of the endogenous gene in the mouse. To this end, we generated a vector containing 5 kb of sequence upstream of the Igf2 gene and then inserted a loxP-flanked Neor gene into the GCF2 repressor element (HpaII site 4) (see Figure 4A). Embryonic stem (ES) cells were transfected and selected for neomycin resistance, and positive clones were assayed by Southern blot hybridization for targeting events. In eight out of 480 clones we detected an insertion in the genomic Igf2 locus (Figure 4B). The resulting ES clones were injected into blastocysts, and chimeras were used to generate mouse lines with the mutated gene in the germline. These were then crossed with a cre-expressing mouse in order to remove the Neor gene cassette (Lallemand et al., 1998). PCR amplifications of DNA from these clones and subsequent sequencing confirmed that the GCF2 element was indeed disrupted by the presence of a single loxP sequence in one copy of the Igf2 gene (data not shown).
Fig. 4. Genomic targeting of a loxP element into DMR1 HpaII site 4. (A) The targeting construct. A 5 kb BamHI fragment containing DMR1 was subcloned from a genomic library and a loxP-Neor-loxP cassette (bottom line) was then inserted into HpaII site 4, which resides within a GCF2 consensus sequence (see Kageyama and Pastan, 1989; Reed et al., 1998). (B) Southern blot analysis of genomic DNA from wild-type (WT), targeted and cre-deleted ES cells restricted with BglII and probed with the fragments indicated in the map. Only data from the left hand probe are shown in the blot. The targeted allele appears under-represented in this figure because of the presence of DNA from accompanying feeder cells. The nature of the disruption was verified by amplifying and sequencing this region from the cre-deleted mice.
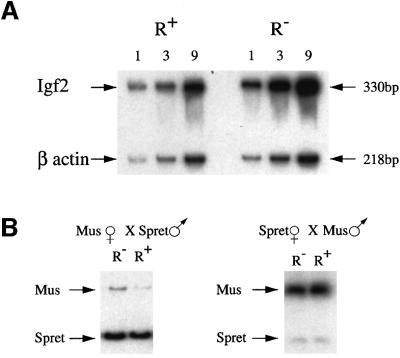
In normal mice, the Igf2 gene is repressed on the maternal allele. It was thus of interest to determine how much of this repression may be due to the presence of the upstream GCF2 element located at HpaII site 4. In order to enable the measurement of transcription exclusively from the maternal allele, we utilized mice in which the paternal allele was deleted. To this end, female mice heterozygous for the repressor mutation (R+/–) were mated to males heterozygous for an Igf2 deletion (Igf2+/–) (DeChiara et al., 1990). Embryos or newborn progeny were then genotyped by PCR analysis and the Igf2–R+/Igf2+R– or Igf2–R+/Igf2+R+ animals then assayed for the level of Igf2 mRNA using quantitative RT–PCR. Initial expression studies on individual whole embryos (12.5–17.5 d.p.c.) showed that the R– mutation had only a small (2.8-fold) but statistically significant (P <0.001) effect (see legend to Figure 5). In order to average out the small variations between embryos, we collected pools of 8–10 embryos for each genotype, and then measured the degree of Igf2 transcription relative to an internal β-actin control. As shown in Figure 5A, these experiments indicated that ∼3-fold more mRNA is made from a gene carrying the repressor element mutation as compared with the wild-type maternal allele. These expression studies were also confirmed by employing an RNase protection assay on the same tissue samples (data not shown). These findings in whole embryos mainly reflect the expression pattern in mesoderm, but similar results were also obtained using RNA derived from placenta or yolk sac and from individual endoderm-derived tissues, such as liver (data not shown). This constitutive pattern, as well as the low magnitude of inhibition, suggests that the GCF2 element is a newly discovered repressor that is independent of the, as yet undefined, sequences responsible for the mesoderm- specific repression previously identified through targeted deletion of a large 5 kb region surrounding DMR1 (Constancia et al., 2000).
Fig. 5. Analysis of Igf2 maternal allele expression in targeted mice. (A) RT–PCR on total RNA extracted from 8–10 whole 12.5 d.p.c. embryos, which were first genotyped by PCR in order to identify and group the R+/Igf2– and R–/Igf2– individuals. PCR was carried out on 1, 3 or 9 µl of cDNA in the presence of [α-32P]dCTP, and the products were run on a 5% acrylamide gel. PCR for β-actin cDNA was carried out in the same tube and served as a control. Igf2 transcription in R– embryos was 2.9-fold higher than in R+ embryos as determined by phosphoimager analysis, after normalization for β-actin. Similar results were also obtained using pooled embryos from an additional inde pendent targeted mouse line. We also carried out expression analysis on individual embryos. In this case, the level of Igf2 mRNA was normalized against β-actin, and the average relative amount of Igf2 in the R+ embryos set at 1 (R+, 1.0 ± 0.2, n = 5; R–, 2.8 ± 0.6, n = 6, P <0.001). (B) RT–PCR was carried out on total RNA from 8–10 whole 12.5 d.p.c. embryos obtained from crosses between M.musculus mice carrying one targeted Igf2 allele (R–) and homozygous SD7 mice (R+). The PCR product was then cut with BsaAI, which recognizes a polymorphic site present only in the M.spretus allele, and was run on a 7% acrylamide gel. In the left panel, the R– allele is derived from the mother while, in the right panel, this allele is paternally derived. The R– maternal allele is 2.8-fold more active than the R+ allele when compared with the spretus paternal allele, as determined by phosphoimager analysis.
Another way to distinguish between the maternal and paternal Igf2 gene products is to use mice containing one allele derived from M.musculus and the second from M.spretus. To this end, we carried out reciprocal crosses between M.m.domesticus R–/+ and homozygous SD7 mice, which have two copies of chromosome 7 whose distal portion is derived from M.spretus. Embryos were then assayed for Igf2 mRNA by RT–PCR and allele-specific expression revealed by using a restriction enzyme to detect a polymorphism in the spretus allele. As expected, the paternal allele is expressed at much higher levels than the maternal allele in all mice (Figure 5B). When the repressor mutation is present on the maternal allele, however, expression from this gene copy is elevated ∼3-fold. In contrast, when this mutation is present on the highly active paternal allele, it apparently has little effect on transcription (Figure 5B). It should be noted that the mutation itself did not influence the methylation pattern at other HpaII sites in DMR1 (data not shown). When taken together, these results clearly suggest that the upstream repressor element indeed plays a role in bringing about low expression levels from the maternal allele.
Discussion
Characterization of an Igf2 upstream repressor element
The control of genomic imprinting is obviously a com plicated process, which involves molecular decision making at multiple stages of development. These events leave in their wake clearcut footprints in the form of allele-specific DNA methylation (Razin and Cedar, 1994; Constancia et al., 1998). The methyl moieties not only serve as a means of differentially marking the two parental alleles, but also probably operate directly by influencing chromatin structure and gene expression. Here, we have studied the function of a single DMR located adjacent to the mouse Igf2 gene, and find that this region plays a novel role in the regulation of Igf2 imprinted expression.
On the basis of a transient reporter gene transfection assay, we have identified an element within DMR1 that represses promoter activity 3- to 5-fold, and this sequence was deemed to be both necessary and sufficient for inhibiting transcription in this in vitro system. Furthermore, by employing gene targeting technology to introduce a site-specific mutation in vivo, it was clearly demonstrated that this same element actually also plays a role in the repression of the endogenous maternal Igf2 allele. Biochemical evidence indicates that this repression may be mediated through the action of the ubiquitously expressed (Johnson et al., 1992; GenBank mouse expressed sequence tags) GCF2 protein itself, and transfection experiments using an expression vector have shown that it can also interact with the Igf2 upstream region to bring about functional repression (data not shown).
Role of methylation in imprinting
What is the role of methylation in this process? We have shown that DNA methylation of the Igf2 upstream region can abrogate repression in vitro, probably by preventing interactions with trans-acting factors. In this model, the repressor element would operate mainly on the unmethylated maternal allele while being neutralized by the presence of methylation on the paternal allele. DNA methylation probably protects against repression in two ways. First, the presence of a methyl moiety within the repressor element itself apparently prevents binding of trans-acting factors at this site as shown by in vitro gel shift analysis. In addition, regional DNA methylation, perhaps in concert with other genomic cues, may bring about a change in local histone acetylation, forming a closed nucleosome structure that reduces general accessibility to trans-acting factors. The generation of such a structural imprint may in fact explain why the entire region is differentially methylated even though repression activity is restricted to a single element.
Although not tested directly in this study, a number of previous published experiments also support a role for DNA methylation in vivo. Tucker et al. (1996), for example, demonstrated that DMR1 is unmethylated and the Igf2 gene expressed at very low levels in differentiated methylase–/– ES cells, but this activity was increased when the upstream region became remethylated after the addition of a maintenance methylase cDNA expression vector.
Further evidence for a role for methylation can be derived from targeting experiments in which the H19 3′ enhancer was knocked out. In the absence of this activating element, the Igf2 gene is expressed at very low levels on both alleles, but transcription is clearly lower on the differentially unmethylated maternal copy (Leighton et al., 1995). These data suggest that DNA methylation of the Igf2 upstream region can indeed serve to cancel repressor activity in vivo.
Unlike the situation on the maternal allele, Igf2 is fully expressed on the paternal allele. While this could come about because the presence of methylation within the DMRs prevents repression, it is also possible that the downstream enhancers themselves have an intrinsic ability to overcome repressor effects. Initial experiments using methylase–/– embryos indicated that in the absence of methylation, both Igf2 alleles are repressed, consistent with the idea that methylation may be involved in preventing local repressor activity (Li et al., 1993). However, recent studies suggest that some of this inhibition actually may be due to the effects of undermethylation on imprinting elements located near the H19 gene, and deletion of these sequences causes the Igf2 gene to become fully activated even in methylase–/– embryos (B.K.Jones et al., 1998). Since this induction takes place under conditions where the Igf2 upstream region is probably unmethylated, these data suggest that the downstream enhancer can act in a dominant manner to overcome repression, implying that DNA methylation itself may not be required to ensure full expression on the paternal allele. Considering the fact that these experiments were carried out in very early embryos lacking DNA methylation throughout the genome, it is difficult to derive any meaningful conclusions about the role of local methylation in normal mice, and additional studies are clearly necessary to resolve this issue.
Role of repression in imprinting
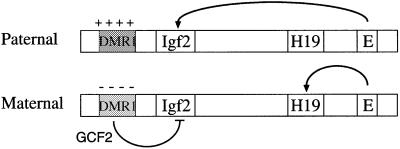
The results presented here provide a new understanding of how imprinting operates in the Igf2/H19 region and also perhaps in other imprinted domains. Theoretically, one can imagine two different strategies for controlling imprinting. One possibility is that the imprinted gene is naturally active in the default state and the major regulatory step is to inhibit transcription from one of the alleles. Alternatively, the default state may be inactive and imprinting would then involve expression enhancement. Our findings suggest a third model, whereby the default state for Igf2 allows a moderate degree of transcription, and imprinting is carried out by a combination of acti vation on one allele and repression on the other (see Figure 6). Activation of the Igf2 gene on the paternal allele undoubtedly is mediated by regional enhancer elements. One of these cis-acting sequences, located 3′ to the H19 gene, serves as an endoderm-specific enhancer that can stimulate either the paternal Igf2 or the maternal H19 gene in a mechanism that involves a methylation-sensitive insulator element (see Reik and Walter, 2001), and it is very likely that this domain harbors additional regional enhancers, which operate in a similar manner for other cell types. The experiments presented herein, however, clearly indicate that the control of enhancer activity alone cannot explain the full extent of differential expression at this locus, and additional regulatory mechanisms are responsible for bringing about further repression on the maternal allele.
Fig. 6. Model of the Igf2 imprinting mechanism. The diagram shows the maternal and paternal Igf2/H19 alleles with the DMR1 methylated and packaged into a closed chromatin structure containing deacetylated histones on the paternal copy, while the DMR1 on the maternal copy is unmethylated and packaged into a more open structure containing acetylated histones. Enhancer sequences (E) are located 3′ to the H19 gene. On the maternal allele, these sequences activate H19, and Igf2 activity is repressed by elements in DMR1 that bind the GCF2 protein. On the paternal allele, DMR1-mediated repression is abrogated either because enhancer sequences overcome this inhibitory element or because of a closed methylated chromatin structure.
Although the R– genotype causes only a 3-fold increase in Igf2 maternal allele expression, it is very likely that other repressor elements also take part in the control of Igf2 expression. One of these, also located upstream of Igf2, has been shown to be specific for mesoderm-derived tissues (Constancia et al., 2000), and additional elements may be present in this same general region (Hu et al., 1997). Furthermore, DMR2, located in the last exon of the Igf2 gene, contains yet another GCF2 element. Thus, while the repressor sequence in DMR1 serves as a prototype, it is clear that the overall influence of repression on the maternal allele might be greatly amplified through the involvement of multiple repression units acting in concert. It should be noted that GCF2 elements are present in the differentially methylated intronic region of the imprinted mouse Igf2r gene as well as in sequences upstream of human Igf2, and another class of repressor sequences has been identified near H19 (Brenton et al., 1999), suggesting that repression may represent a common strategy for imprinted gene regulation.
Materials and methods
Immunoprecipitation
Nuclei from primary fibroblasts were digested with micrococal nuclease, and mono- and dinucleosomes were then immunoprecipitated using anti-acetylated H4 histone antibody (Sigma) as described (Hebbes et al., 1994). DNA was extracted from input and bound fractions and subjected to quantitative PCR (Eden et al., 1998), using the following primers: 5′-CGCCATGGATGACGATATCG-3′ and 5′-CGAAGCCGGCTTTGCACATG-3′ (β-actin); 5′-CTCCAAGGCAAAGATACAGA-3′ and 5′-GCATTCATTCTCCAGAGAAC-3′ (κ-light chain); and 5′-TCGGGACTCTGTTCCCAGAA-3′ and 5′-GAGGGGTTCTCTACCTTTCC-3′ (Igf2). The Igf2 PCR product from SD7 (Forne et al., 1997) heterozygous mice was digested with HpaII to distinguish between the two alleles.
Transient transfections
A total of 5 × 105 293 human kidney cells were transiently transfected with 1 µg of RSV-gal plasmid (Borras et al., 1988), 0.1 µg of CAT reporter plasmid, and competitor plasmids where indicated, using the calcium phosphate method (Graham and van der Eb, 1973). After 48 h in culture, total protein extract was prepared by three cycles of freezing in liquid nitrogen and thawing at 37°C, and protein concentration was determined by means of the Bradford reagent. CAT activity was adjusted to take into account transfection efficiency by first measuring β-gal (Nielsen et al., 1983), and on this basis using normalized amounts of protein extract for the CAT assay.
The CAT reporter constructs were generated by cloning PCR products from the Igf2 upstream region into pTK-CAT, cut with XbaI–BgIII. PCR primers (1–6 below) used for making each fragment were designed to terminate with XbaI and BgIII sites. pR-TK-CAT was generated by cloning a 2.2 kb EcoRI–XbaI fragment into the upstream XbaI site of pTK-CAT, and a partial 420 bp deletion of DMR1 (pRΔ420-TK-CAT) was generated by digestion of this plasmid with HindIII–AflII, filling-in and self-ligation. For competition experiments, we prepared plasmids containing either the 680 bp PCR fragment (primers 1 and 2) or the 340 bp PCR fragment (primers 3 and 4) inserted into pGEM7 by XbaI–BglII restriction. p4lox-TK-CAT, containing a 380 bp fragment from DMR1 with a loxP element inserted in HpaII site 4, was generated by preparing a PCR fragment (primers 3 and 4) from the cre-deleted targeting vector (see below)
and inserting it into pTK-CAT by XbaI–BgIII digestion. In vitro methylation was carried out using the enzyme MHpaII (New England Biolabs) according to the manufacturer’s protocol.
Targeting
In order to generate a mutation of the Igf2 upstream GCF2 element in vivo, a 5 kb BamHI fragment that includes DMR1 was excised from phage λUp12 isolated from an SV-129 mouse λ genomic library and subcloned into the pIC-20H vector (Marsh et al., 1984). This plasmid (pIC5.0) was digested by NaeI (HpaII site 4) in order to insert a blunt end loxP-flanked Neor cassette that was originally removed from plasmid pLTNL (Mombaerts et al., 1996) by XbaI digestion and end filling. The targeting vector was linearized using SalI and electroporated into GCR ES cells (Constancia et al., 2000). Colonies were screened by isolating DNA and carrying out Southern blot analysis using BglII digestion and hybridization with two probes external to the targeting construct, the 386 bp BamHI–AccI fragment and the 1250 bp BamHI–BglII fragment. Eight out of 480 colonies screened showed homologous recombination in one of the Igf2 alleles. Targeted cells were microinjected into [(C57Black × BALB/C)F1] × [(C57Black × BALB/C)F1] blastocysts and transferred into (C57Black × BALB/C)F1 foster mothers to generate chimeric mice. After germline transmission, the mice were crossed into (C57Black × BALB/C)F1 mice, which express cre early in development (Lallemand et al., 1998), in order to enable deletion of the Neor cassette in the targeted mice. The presence of the targeted mutation was confirmed by PCR analysis of tail DNA using the primers 5′-CCTTCTTGGGGAAAGGTAGAGAAC-3′ and 5′-GGTGATGTTCCTCATTCCAGGGAG-3′, followed by sequencing.
RT–PCR expression analysis
Total RNA was extracted from cells or tissues using the TriPure Isolation Reagent (Boehringer Mannheim) and 300 ng were treated with 0.1 U of RQ1 DNase I (Promega) for 20 min at 37°C to eliminate any residual DNA contamination. The random reverse transcription reaction was carried out using Promega reagents according to the manufacturer’s protocol. cDNA was diluted 1:5, and 1, 3 or 9 µl were taken for the PCR, which was carried out in the presence of [α-32P]dCTP. The following PCR primers were used: 5′-CGAATTCCAGTGGGGAAGTCGATGTTG-3′ and 5′-TTGGAAGAACTTGCCCACGGGGTATC-3′ (for Igf2 expression on the Igf2– background); 5′-GGCCAAACGTCATCGTCCCCTGAT-3′ and 5′-CTGTCCCTGCTCAAGAGGAGGTCA-3′ (for Igf2 expression in heterozygous SD7 mice); and 5′-CAGCTTCTTTGCAGCTCCTT-3′ and 5′-TCACCCACATAGGAGTCCTT-3′ (for β-actin expression).
Protein binding experiments
Nuclear extract from 293 cell cultures was prepared as described (Lee et al., 1988). Protein binding reactions were carried out for 10 min at room temperature and included 20 ng of DNA probe, 200 ng of poly(dI–dC), 0.1 M Tris–HCl pH 7.4, 2 mM EDTA, 5 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mg/ml bovine serum albumin (BSA), 12.5% glycerol and 20 µg of protein extract in a volume of 10 µl. The Igf2 upstream double-stranded oligonucleotide probe (5′-ACCCCTCTGCCGGCCCTT-3′) and the EGFR probe (5′-GCGACGCGGCCCCCGACGGGC-3′) were end labeled with [α-32P]dCTP using Klenow enzyme. Competitors for Igf2 included the same oligonucleotide synthesized with 5mC at the lone CpG dinucleotide, a mutated oligonucleotide (5′-ACCCCTCTGCCAACCCTT-3′) and an oligonucleotide containing an eight base disruption in the middle of the HpaII site (5′-CTCTGCCTAAATGTATGGC-3′). This latter oligonucleotide was used to mimic the slightly longer loxP in vivo insertion. DNA–protein complexes were run on 4% acrylamide gels in TBE. Purified recombinant GCF2 protein was prepared as previously described (Reed et al., 1998). The affinity-purified GCF2-specific rabbit polyclonal antibody (see Khachigian et al., 1999), but not pre-immune serum, was shown to immunoprecipitate specifically in vitro synthesized GCF2, and western blotting of nuclear extracts from 293 cells revealed a strong band migrating with a molecular mass of ∼160 kDa (data not shown). Immunodepletion was carried out by incubating with anti-GCF2 antibody and then eluting through a protein A–Sepharose column (Dyer and Herzog, 1995).
Acknowledgments
Acknowledgements
We thank M.Mendelsohn and B.Tzuberi for help in the mouse targeting experiments, and G.Kelsey for advice and discussion. We are especially grateful to A.Efstratiadis for the Igf2 knockout mice. This research was supported by grants from the NIH (H.C.), the Israel Cancer Research Fund (H.C.), the Tobacco Research Council (H.C.), the Cancer Research Campaign (W.R.) and the Medical Research Council (W.R.).
References
- Bartolomei M.S., Webber,A.L., Brunkow,M.E. and Tilghman,S.M. (1993) Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev., 7, 1663–1673. [DOI] [PubMed] [Google Scholar]
- Borras T., Peterson,C.A. and Piatigorsky,J. (1988) Evidence for positive and negative regulation in the promoter of the chicken δ1-crystallin gene. Dev. Biol., 127, 209–219. [DOI] [PubMed] [Google Scholar]
- Brandeis M., Kafri,T., Ariel,M., Chaillet,J.R., McCarrey,J., Razin,A. and Cedar,H. (1993) The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J., 12, 3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenton J.D., Drewell,R.A., Viville,S., Hilton,K.J., Barton,S.C., Ainscough,J.F.-X. and Surani,M.A. (1999) A silencer element identified in Drosophila is required for imprinting of H19 reporter transgenes in mice. Proc. Natl Acad. Sci. USA, 96, 9242–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constancia M., Pickard,B., Kelsey,G. and Reik,W. (1998) Imprinting mechanisms. Genome Res., 8, 881–900. [DOI] [PubMed] [Google Scholar]
- Constancia M., Dean,W., Lopes,S., Moore,T., Kelsey,G. and Reik,W. (2000) Deletion of a silencer element in igf2 results in loss of imprinting independent of H19. Nature Genet., 26, 203–206. [DOI] [PubMed] [Google Scholar]
- DeChiara T.M., Efstratiadis,A. and Robertson,E.J. (1990) A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature, 345, 78–80. [DOI] [PubMed] [Google Scholar]
- Dyer R.B. and Herzog,N.K. (1995) Immunodepletion EMSA: a novel method to identify proteins in a protein–DNA complex. Nucleic Acids Res., 23, 3345–3346. [PMC free article] [PubMed] [Google Scholar]
- Eden S., Hashimshony,T., Keshet,I., Cedar,H. and Thorne,A.W. (1998) DNA methylation models histone acetylation. Nature, 394, 842–843. [DOI] [PubMed] [Google Scholar]
- Feil R., Walter,J., Allen,N.D. and Reik,W. (1994) Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development, 120, 2933–2943. [DOI] [PubMed] [Google Scholar]
- Feil R., Handel,M.A., Allen,N.D. and Reik,W. (1995) Chromatin structure and imprinting: developmental control of DNase-I sensitivity in the mouse insulin-like growth factor 2 gene. Dev. Genet., 17, 240–252. [DOI] [PubMed] [Google Scholar]
- Forne T., Oswald,J., Dean,W., Saam,J.R., Bailleul,B., Dandolo,L., Tilghman,S.M., Walter,J. and Reik,W. (1997) Loss of the maternal H19 gene induces changes in Igf2 methylation in both cis and trans. Proc. Natl Acad. Sci. USA, 94, 10243–10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F.L. and van der Eb,A.J. (1973) A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology, 52, 456–467. [DOI] [PubMed] [Google Scholar]
- Hebbes T.R., Clayton,A.L., Thorne,A.W. and Crane-Robinson,C. (1994) Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J., 13, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J.-F., Vu,T.H. and Hoffman,A.R. (1997) Genomic deletion of an imprint maintenance element abolishes imprinting of both insulin-like growth factor II and H19. J. Biol. Chem., 272, 20715–20720. [DOI] [PubMed] [Google Scholar]
- Johnson A.C., Kageyama,R., Popescu,N.C. and Pastan,I. (1992) Expression and chromosomal localization of the gene for the human transcriptional repressor GCF. J. Biol. Chem., 267, 1689–1694. [PubMed] [Google Scholar]
- Jones B.K., Levorse,J.M. and Tilghman,S.M. (1998) Igf2 imprinting does not require its own DNA methylation or H19 RNA. Genes Dev., 12, 2200–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.L., Veenstra,G.J.C., Wade,P.A., Vermaak,D., Kass,S.U., Landsberg,N., Strouboulis,J. and Wolffe,A.P. (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet., 19, 187–191. [DOI] [PubMed] [Google Scholar]
- Kageyama R. and Pastan,I. (1989) Molecular cloning and characterization of a human DNA binding factor that represses transcription. Cell, 59, 815–825. [DOI] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz,J. and Cedar,H. (1986) DNA methylation affects the formation of active chromatin. Cell, 44, 535–543. [DOI] [PubMed] [Google Scholar]
- Khachigian L.M., Santiago,F.S., Rafty,L.A., Chan,O.L., Delbridge,G.J., Bobik,A., Collins,T. and Johnson,A.C. (1999) GC factor 2 represses platelet-derived growth factor A-chain gene transcription and is itself induced by arterial injury. Circ. Res., 84, 1258–1267. [DOI] [PubMed] [Google Scholar]
- Lallemand Y., Luria,V., Haffner-Krausz,R. and Lonai,P. (1998) Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site specific recombinase. Transgenic Res., 7, 105–112. [DOI] [PubMed] [Google Scholar]
- Lee K.A.W., Binderif,A. and Green,M.R. (1988) A small scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal. Tech., 5, 22–31. [DOI] [PubMed] [Google Scholar]
- Leighton P.A., Saam,J.R., Ingram,R.S., Stewart,C.L. and Tilghman,S.M. (1995) An enhancer deletion affects both H19 and Igf2 expression. Genes Dev., 9, 2079–2089. [DOI] [PubMed] [Google Scholar]
- Li E., Beard,C. and Jaenisch,R. (1993) Role for DNA methylation in genomic imprinting. Nature, 366, 362–365. [DOI] [PubMed] [Google Scholar]
- Marsh J.L., Erfle,M. and Wykes,E.J. (1984) The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene, 32, 481–485. [DOI] [PubMed] [Google Scholar]
- Mombaerts P., Wang,F., Dulac,C., Chao,S.K., Nemes,A., Mendelsohn,M., Edmondson,J. and Axel,R. (1996) Visualizing an olfactory sensory map. Cell, 87, 675–686. [DOI] [PubMed] [Google Scholar]
- Moore T., Constancia,M., Zubair,M., Bailleul,B., Feil,R., Sasaki,H. and Reik,W. (1997) Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc. Natl Acad. Sci. USA, 94, 12509–12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X., Ng,H.-H., Johnson,C.A., Laherty,C.D., Turner,B.M., Eisenman,R.N. and Bird,A. (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature, 393, 386–389. [DOI] [PubMed] [Google Scholar]
- Nielsen D.A., Chou,J., MacKrell,A.J., Casadaban,M.J. and Steiner,D.F. (1983) Expression of a preproinsulin–β-galactosidase gene fusion in mammalian cells. Proc. Natl Acad. Sci. USA, 80, 5198–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A. (1998) CpG methylation, chromatin structure and gene silencing—a three-way connection. EMBO J., 17, 4905–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A. and Cedar,H. (1994) DNA methylation and genomic imprinting. Cell, 77, 473–476. [DOI] [PubMed] [Google Scholar]
- Reed A.L., Yamazaki,H., Kaufman,J.D., Rubinstein,Y., Murphy,B. and Johnson,A.C. (1998) Molecular cloning and characterization of a transcription regulator with homology to GC-binding factor. J. Biol. Chem., 273, 21594–21602. [DOI] [PubMed] [Google Scholar]
- Reik W. and Walter,J. (2001) Genomic imprinting: parental influence on the genome. Nature Rev. Genet., 2, 21–32. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Jones,P.A., Chaillet,J.R., Ferguson-Smith,A.C., Barton,S.C., Reik,W. and Surani,M.A. (1992) Parental imprinting: potentially active chromatin of the repressed maternal allele of the mouse insulin-like growth factor II (Igf2) gene. Genes Dev., 6, 1843–1851. [DOI] [PubMed] [Google Scholar]
- Stoger R., Kubicka,P., Liu,C.-G., Kafri,T., Razin,A., Cedar,H. and Barlow,D.P. (1993) Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell, 73, 61–71. [DOI] [PubMed] [Google Scholar]
- Takimoto M., Mao,P., Wei,G., Yamazaki,H., Miura,T., Johnson,A.C. and Kuzumaki,N. (1999) Molecular analysis of the GCF gene identifies revisions to the cDNA and amino acid sequences. Biochim. Biophys. Acta, 1447, 125–131. [DOI] [PubMed] [Google Scholar]
- Tucker K.L., Beard,C., Dausman,J., Jackson-Grusby,L., Laird,P.W, Lei,H., Li,E. and Jaenisch,R. (1996) Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not non imprinted genes. Genes Dev., 10, 1008–1020. [DOI] [PubMed] [Google Scholar]