Abstract
The subtelomeric regions of human chromosomes are comprised of sequence homologies shared between distinct subsets of chromosomes. In the course of developing a set of unique human telomere clones, we identified many clones containing such shared homologies, characterized by the presence of cross-hybridization signals on one or more telomeres in a fluorescence in situ hybridization (FISH) assay. We studied the evolutionary origin of seven subtelomeric clones by performing comparative FISH analysis on a primate panel that included great apes and Old World monkeys. All clones tested showed a single hybridization site in Old World monkeys that corresponded to one of the orthologous human sites, thus indicating the ancestral origin. The timing of the duplication events varied among the subtelomeric regions, from ∼5 to ∼25 million years ago. To examine the origin of and mechanism for one of these subtelomeric duplications, we compared the sequence derived from human 2q13—an ancestral fusion site of two great ape telomeric regions—with its paralogous subtelomeric sequences at 9p and 22q. These paralogous regions share large continuous homologies and contain three genes: RABL2B, forkhead box D4, and COBW-like. Our results provide further evidence for subtelomeric-mediated genomic duplication and demonstrate that these segmental duplications are most likely the result of ancestral unbalanced translocations that have been fixed in the genome during recent primate evolution.
Introduction
The end of every human chromosome is capped with 3–20 kb of tandemly repeated TTAGGG sequences (Moyzis et al. 1988). Immediately adjacent to the terminal simple sequence repeat are 100–300 kb of DNA comprised of sequence homologies shared between distinct subsets of chromosomes, referred to as the “subtelomeric region” (Brown et al. 1990). Unique, chromosome-specific DNA is located proximal to this subtelomeric region.
Previous sequence analysis of three human subtelomeric regions suggested a structure comprised of two distinct subtelomeric domains, which are separated by a boundary element comprised of degenerate (TTAGGG)n repeats and putative-origin-of-replication consensus sequences (Flint et al. 1997). The distal subdomain lies adjacent to the (TTAGGG)n sequence and is characterized by a mosaic of short segments of shared sequence homologies from many different chromosomes; these shared homologies are <2 kb in length and suggest the occurrence of frequent exchanges among all telomeres (Flint et al. 1997). The proximal subdomain is comprised of much longer segments (10–40 kb) of shared sequence homologies from fewer chromosomes (Flint et al. 1997), which indicates recent duplications of this domain. These recent duplications could be the result of unbalanced translocations occurring between telomeres during primate evolution.
We recently reported a second-generation set of unique BAC, PAC, and P1 human telomere clones located within 500 kb of the end of each chromosome arm (Knight et al. 2000). In the course of the development and characterization of these unique clones, a number of nonunique genomic clones were also identified from the subtelomeric region, characterized by the presence of cross-hybridization signals on multiple telomeres in a FISH assay. Genomic clones containing such shared sequence homologies can be characterized as being derived from the proximal or distal subtelomeric domains on the basis of their hybridization pattern: clones from the proximal domain generally show strong cross-hybridization on two to three telomeres, whereas clones from the distal domain show weaker cross-hybridization on many additional telomeres.
Further investigations of subtelomeric sequence homologies may shed light on recent gene and genomic duplication events that maintain large segments of interchromosomal sequence homologies. Previous studies have defined three classes of segmental duplication: intrachromosomal (Ji et al. 2000), pericentromeric, and subtelomeric (the last two classes have also been collectively referred to as “transchromosomal” by Eichler [2001]). However, the mechanism behind the generation of subtelomeric duplications remains to be elucidated.
In the present study, we examined the hypothesis that subtelomeric genomic duplications result from ancestral unbalanced translocation events. The evolutionary origin of seven proximal subtelomeric clones was studied using FISH. In addition, to determine the mechanism of subtelomeric duplication, we analyzed the “breakpoint” sequence—defined as the transition between the homologous region and the chromosome-specific sequence—from one of these subtelomeric duplications, which included the human chromosomal region 2q13. The human 2q13 region is the ancestral fusion site of the telomeric regions of two great ape chromosomes (Ijdo et al. 1991). The ancestral subtelomeric sequences at human 2q13 were previously shown to be homologous to 9p (Lese et al. 1999) and 22q (Ning et al. 1996; Wong et al. 1999). It has been proposed that sequence exchanges occur among nonhomologous chromosomes within the subtelomeric regions (Flint et al. 1997; Pryde et al. 1997). The dynamic nature of the subtelomeric region makes it difficult to analyze the origin of subtelomeric sequences. However, the interstitial position of the two ancestral subtelomeric regions at 2q13 makes them unlikely to interact with other subtelomeric sequences. Therefore, this region provides a “molecular fossil” for the study of the evolution of the subtelomeric sequences. Finally, to determine whether these subtelomeric sequence duplications are involved in the creation of gene families, we examined the level of gene homology within these duplicated regions.
Material and Methods
Isolation of Genomic Clones
Human genomic clones were identified by PCR screening of a PAC or BAC library (Incyte Genomics PAC library [RPCI-1]; Incyte Genomics BAC library [Incyte Genomics Release I]). Genomic clones were isolated using the markers shown in table 1. The primer sets for previously unpublished markers are 131N4-T7-F (5′-TGACAGCATTGCTAAGTGCC-3′) and 131N4-T7-R (5′-ATGCTGTGTTTCTGCGAATG-3′).
Table 1.
Proximal Subtelomeric Clones
| CognateTelomere | Subtelomeric Clone(s) (Marker)a | Cross-HybridizationPattern | Human Draft Sequence Equivalentb |
| 2q | cos 2003* (yRM2003) | 8p | RPCI11-91J19, RPCI11-115B2, RPCI11-47F2, and RPCI11-18D5 |
| 4p | BAC 136B17* (AFMA060ZE5) | 1p | RPCI1-296G16 and RPCI11-460I19 |
| 4q | BAC 204M23* (TYAC18 ) | 4p | RPCI11-565A3 |
| 8p | BAC 131N4* (TEL8P069), BAC 57M17 (131N4-T7), and PAC 63M14* (VIJ2205) | 1p | CTA-366D10, RPCI11-139L10, RPCI11-18D5, RPCI11-47F2, RPCI11-1129O4, and CTA-344E3 |
| 22q | cos c202* (RABL2B) | 2q13 | Cosmid n94H12, n1G3 |
| 9p | BAC 305J7* (VIJ2039) | 2q13, 9 pericentromeric | RPCI11-143M1 and RPCI11-174M15 |
| 9q | BAC 120H19 (VIJ2241) | 10p, 16p, 18p | RPCI11-424E7 |
| 10p | BAC 100E1 (TEL10P62) | 9q, 16p, 18p | ND |
| 12p | BAC 8M16 (TYAC14) | 6p, 20q | RPCI11-283I3 and RPCI11-467M14 |
| 17p | BAC 282M15 (TEL17P80) | 11p, 16p | BAC536F19 |
Asterisks (*) indicate clones used for primate panel FISH analysis.
ND = not determined.
The Primate Panel
Five nonhuman primate cell lines were used in the present study. Lymphoblastoid cell lines were used for three great ape species: chimpanzee (Pan troglodytes), gorilla (Gorilla gorilla), and orangutan (Pongo pygmaeus). Two Old World monkey species were used: a fibroblast cell line (GM03446) was used for macaque (Macaca fascicularis), and a lymphoblastoid cell line was used for baboon (Papio hamadryas). Lymphocytes or lymphoblast cell lines from karyotypically normal humans were used. Metaphase cells were obtained and slides prepared following standard procedures.
FISH
Genomic clones were labeled using a standard nick translation reaction with either directly labeled nucleotides (Spectrum Orange and Spectrum Green; Vysis) or indirectly labeled nucleotides, biotin and digoxigenin, which require detection after hybridization with Avidin–fluorescein isothiocyanate and anti-digoxigenin-rhodamine, respectively (Boehringer Manheim). Labeled genomic probes were hybridized to metaphase spreads from humans and the other primates. At least 10 cells were analyzed using direct microscopic visualization and digital-imaging analysis. Chromosome identification was achieved by inverted 4,6-diamidino-2-phenylindole (DAPI) staining.
Sequence Analysis
Sequence information was obtained from the clones used in the present study, either by end sequencing or by sample sequencing, where DNA from the clones was first digested with EcoRI, then the resulting fragments were subcloned into pBluescript II KS- (Stratagene) and were sequenced with T3 and T7 primers. BLASTN sequence-similarity searches were performed against the nr (nonredundant) and htgs (high-throughput genomic sequences) divisions of GenBank, to look for equivalent clones in the Human Draft Sequence.
BLASTN sequence-similarity searches were also performed to look for paralogous sequences among the 2q13, 9p, and 22q subtelomeric regions. Contigs in unfinished sequences were ordered using the Pipmaker program (Schwartz et al. 2000), on the basis of its paralogous sequences.
When a BAC clone was not completely sequenced, the individual contigs were joined without filling the gap (i.e., force joined). In some cases, the gaps in the sequence were filled by PCR using primers designed on both sides of the gaps. The primers used are 480gap1F (5′-AGCCCAGAAAACCACAGAGG-3′), 480gap1R (5′-GAGAAATCACTGTCCTCTGC-3′), 480C16gap3F (5′-GCACTGATACTCTTTCCTTC-3′), 480C16gap3R (5′-GAGCATGTTTTGAAGAGGTAG-3′), 480C16gap4F (5′-GGCAGACAAAGAGGGTATGG-3′), and 480C16gap4R (5′-CTGCATGCACATGTATGCAT-3′).
Ten nanograms of RPCI11-480C16 BAC DNA were used as PCR template. PCR product was purified using the QIAquick PCR purification kit (Qiagen) and was sequenced on both strands with BigDye Terminator chemistry, following a standard protocol (Applied Biosystems). Sequences from BAC clones covering the same subtelomeric region were assembled into a single sequence through use of Sequencher 4.1 (Gene Code Corporation). Sequence similarities among paralogous regions were analyzed using the PipMaker program (Schwartz et al. 2000). To estimate the overall percentage of similarity between two paralogous regions, repetitive elements were first identified and eliminated using RepeatMasker program (RepeatMasker Web Server). The unique sequences were then aligned using the Blast 2 sequence program (Blast 2 Sequences Web site). The percent similarity was calculated using the formula L(number of matched bases)/L + number of indels × 100 (Horvath et al. 2000).
The Nix program (Nix Application Web site) was used to search for genes and possible open reading frames within the subtelomeric sequences. The ClustalW multiple alignment program (ClustalW Web site) was used to compare the putative gene sequences in the paralogous regions. Primers were designed to verify the divergence of the sequences between two chromosomes. The primers used are 2-22bkF (5′-TCTTCCAAGGCCTAAAAGCC-3′), ACRexon3F (5′-ACCTGGCTTACCTTGTGCCC-3′), 2-22bkR (5′-GACCATGGAGTCAGTAATGAC-3′), 2qbpR (5′-CCCCTACCTGTGTTCTCTTC-3′), 9pbpR (5′-CCCATACAGTGAAGGCATTAC-3′), 2q9pbpF (5′-GGGGCGAGGATTACACCTAC-3′), 2q13UF (5′-TTTGCTACTCTCTCCACACCTG-3′), 2qcenUF (5′-GGTGATTCTTCCACAGGCAC-3′), and 2qdupR (5′-ATGGATTCATCTGTTCCTCC-3′).
DNAs from monochromosomal hybrid cell lines for chromosome 2 (GM11712), chromosome 9 (GM10611), and chromosome 22 (GM13258) were used as templates for PCR. DNAs from monochromosomal hybrid cell lines for chromosome 1 (GM13139), chromosome 3 (GM11713), and chromosome 4 (GM11687) were used as negative controls. All monochromosomal hybrid cell lines are available from the Coriell Cell Repository.
Results
Identification of Proximal Subtelomeric Domain Clones
Table 1 lists the human proximal subtelomeric domain clones that were identified in the course of the present study and the markers used to isolate them. Random short sequences were obtained from these clones, and BLAST searches were performed to identify their corresponding regions in the Human Draft Sequence. All clones show hybridization to the cognate telomere and cross-hybridization to one to three additional telomeres.
FISH Analysis of Proximal Subtelomeric Domain Clones
Proximal subtelomeric clones were used for comparative FISH studies. FISH analysis was performed on metaphase spreads from human and other members of the primate panel, including chimpanzee, gorilla, orangutan, macaque, and baboon.
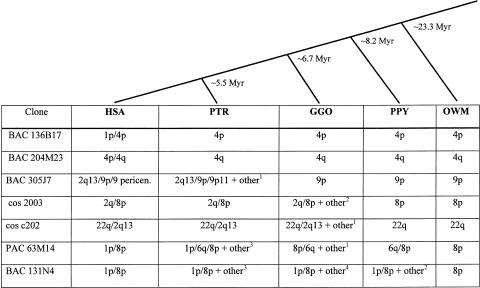
As shown in figure 1, BAC 136B17 from human 4p hybridizes to chromosomes 4p and 1p in human but shows only a single signal in the region orthologous to 4p in chimpanzee and all other species tested. Figure 2 summarizes the results from the analysis of additional subtelomeric duplications. BAC 204M23 shows similar results to those of BAC 136B17: in human, this clone hybridizes to the short- and long-arm telomeres of chromosome 4, but it only shows one signal in all other primates examined. Since these two clones showed duplicated signals only in human and not in the other primates, these duplications most likely occurred after the divergence of humans from the great apes, <3–5 million years ago.
Figure 1.
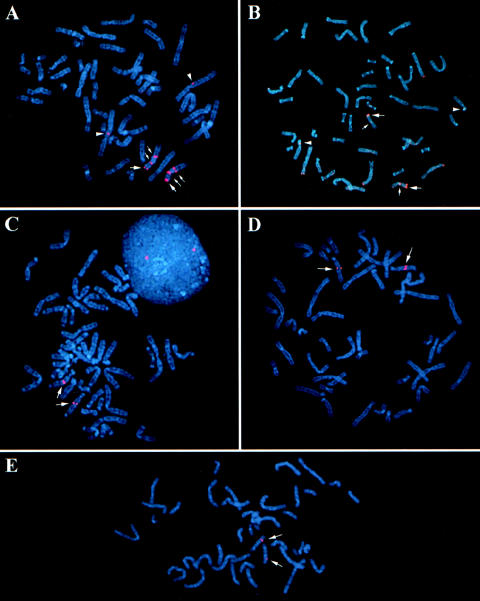
Results of FISH for BAC 136B17 (from human chromosome 4p) on selected members of the primate panel. In human (A), this clone hybridizes to the 4p telomere (large arrows) and to the 1p telomere (small arrows), with the signal intensity being greater on chromosome 4p. Only one hybridization signal (large arrows) is observed for this clone in chimpanzee (B) and macaque (C), both of which correspond to the region orthologous to the human 4p telomere. Similar results were obtained for gorilla, orangutan, and baboon.
Figure 2.
Summary of FISH results from seven human genomic clones tested on the primate panel. The evolutionary lineage between the primates tested is depicted above the table; the molecular time scale used is adopted from Kumar and Hedges (1998) and is indicated in millions of years (Myr). For all clones, the chromosomal localization for FISH hybridization is listed by reference to the human ortholog. The hybridization signal was observed at the telomere region of each chromosome, unless otherwise noted. HSA = human; PTR = chimpanzee; GGO = gorilla; PPY = orangutan; OWM = Old World monkeys (macaque and baboon); pericen. = pericentromeric. Clones listed as having “other” hybridization signals indicate that additional hybridization signals were observed that were characterized by number of signals and chromosomal position, as indicated by the following superscripts: 1 = two telomere regions; 2 = one telomere region; 3 = three telomere regions and one interstitial region; and 4 = three telomere regions.
All other duplications were present in chimpanzee and thus arose >5 million years ago. The results for BAC 305J7 and cosmid c202 are discussed in more detail in the “Ancestral Subtelomeric Regions at Human Chromosome 2q13” subsection. Cosmid 2003, a clone derived from a 2q telomere half-YAC, showed only a single hybridization signal in orangutan and Old World monkeys, but a duplication from 8p to the orthologous region of human 2q occurred in gorilla, chimp, and human. In gorilla, other hybridization signals were observed in addition to the 2q and 8p orthologous sites. PAC 63M14 and BAC 131N4 showed a single hybridization signal only in Old World monkeys. Duplications of sequences orthologous to the human 8p telomere occurred after the divergence of the great apes from the Old World monkeys. In human, both BAC 131N4 and PAC 63M14 hybridize to chromosomes 1p and 8p; however, in most other species examined, these clones showed multiple signals in addition to the two signals observed in human.
As shown in figure 2, for all clones examined, the origin of the duplicated segment could be traced back to a single telomere in Old World monkeys, which corresponded to one of the human orthologous sites. The timing of the duplication event varied among the different subtelomeric regions examined, and some primates were more prone to the occurrence of additional duplication events.
Ancestral Subtelomeric Regions at Human Chromosome 2q13
Human chromosome 2 originated from a fusion between two great ape acrocentric chromosomes (PTR12 and PTR13) (Yunis and Prakash 1982; Ijdo et al. 1991; Wienberg et al. 1994; Kasai et al. 2000). Figure 3 shows the site of the telomere-to-telomere fusion in humans, at 2q13 (Ijdo et al. 1991). The q arm ancestral centromere is localized at 2q21 and was inactivated after the fusion event that resulted in a single stable chromosome (Baldini et al. 1993).
Figure 3.
The genomic region of the ancestral telomere fusion site at human 2q13. An idiogram of human chromosome 2 is shown at the top. An expanded physical map is shown below the idiogram, with distance in kilobases from the telomere fusion site (indicated by the “0” point). The BAC contig that covers the region is shown by blackened rectangles under the chromosome. The physical maps of chromosome 22q (from GenBank accession number NT011526; Dunham et al. 1999) and chromosome 9p are shown below the chromosome 2q13 region, for comparison. On the distal side of the fusion site, the subtelomeric region is homologous to the 68-kb subtelomeric region at 22q, as represented by the rectangular pattern in both chromosomes 2 and 22. Two genes, RABL2A and RABL2B, are within these homologous regions on chromosomes 2 and 22, respectively. The “breakpoint” of this homologous region is within the acrosin (ACR) locus on chromosome 22q. The telomeric repeat (TTAGGG)n is represented by a solid gray box at the end of chromosome 22 and chromosome 9. Chromosomes 2 and 9 share ∼189 kb of homologous subtelomeric sequence (indicated by a checker pattern on both chromosomes). The dashed lines at the telomere of chromosome 9 show a 58-kb region that is not covered by any BAC clones. Two genes, FKHL9 and COBW-like (COBWL), as well as a partial PGM5, are duplicated on chromosomes 2 and 9. Proximal to the ancestral subtelomeric domain on chromosome 2, a 100-kb region (marked by diagonal lines) is duplicated at 2q11.2 (marked by the box with diagonal lines above the clones RPCI11-34G16 and RPCI11-440D17).
We have previously shown by FISH that genomic clones from the 9p and 22q telomeres share sequence homology with the ancestral telomere region at 2q13 in humans (Ning et al. 1996; Lese et al. 1999; Wong et al. 1999). In the present study, to determine the timing of these duplications during primate evolution, FISH analysis was used with human probes from 9p and 22q on metaphase cells from other primates.
BAC CITB-HSP-C 305J7 contains the half-YAC vector-insert junction sequence derived from the chromosome 9p telomere, and in humans it hybridizes to the 9p telomere, the pericentromeric region of chromosome 9, and 2q13, as shown in figure 4A (Lese et al. 1999). Figure 4B shows results in chimpanzee, where BAC 305J7 hybridizes to the telomere that is orthologous to human 9p; to the short-arm side of the centromere of the human 9 ortholog; to the short arm of the chimpanzee chromosome 12 telomere, which is orthologous to the human 2q13 telomere fusion site; and to other telomeric regions. Since only a single signal is present near the centromere of chromosome 9, this sequence was duplicated to the other side of the chromosome 9 centromere after the divergence of chimpanzee from human, which results in the pericentromeric signals observed in human (figs. 4A and 5). For gorilla, orangutan, and Old World monkeys, hybridization signals were observed only at an interstitial site on the human chromosome 9 ortholog (fig. 4C, 4D, and 4E, respectively). These interstitial signals can be explained by the fact that the chromosome 9 ortholog in these species is acrocentric (Yunis and Prakash 1982; Wienberg et al. 1992). As shown in Figure 5A, a pericentric inversion must have occurred after the divergence of the gorilla from the hominoid lineage, to form a metacentric chromosome, as seen in chimpanzee and human. The inversion relocated the interstitial sequence to the telomeric region, followed by a duplication to one side of the centromere. By an unbalanced translocation event, the 9p subtelomeric region was then duplicated to the telomere of the short arm of chimpanzee chromosome 12, which corresponds to the ancestral telomere site at 2q13 in humans (fig. 5B).
Figure 4.
Hybridization of BAC CITB-HSP-305J7 to various primate metaphase cells. In human (A), strong hybridization is visible at the 9p telomere (large arrows) and the pericentromeric region (small arrows) of chromosome 9. A weaker hybridization signal is present at 2q13 (arrow heads). In chimpanzee (B), hybridization signals are present at the sites orthologous to the human 9p telomere (large arrows), 9p11 (small arrows), and 2q13 (arrow heads). Additional hybridization signals were observed in chimpanzee, to two telomeric regions. In gorilla (C), orangutan (D), and macaque (E), only a single hybridization signal (large arrows) is observed, at an interstitial site that is orthologous to the human 9p telomere.
Figure 5.
Proposed sequence of duplication of the 9p subtelomeric sequence. The sequence of changes is indicated by the arrows, proceeding from the top to the bottom of the figure. A, In gorilla (GGO), the human 9p subtelomeric sequence is within an interstitial region of the orthologous chromosome. A pericentric inversion relocated this sequence to the subtelomeric region, as in human (HSA) and chimpanzee (PTR). This 9p subtelomeric region is partially transposed to the centromere of PTR 11 (HSA 9). In human, the copy at the chromosome 9 centromere is further duplicated around the pericentromeric region. B, Duplication of 9p subtelomeric sequence to PTR 12 through a translocation event between the telomeres. Two great ape chromosomes (PTR 12 and PTR 13) fused to form human chromosome 2, which fixed this subtelomeric sequence at an interstitial position.
To examine the timing of the duplication from the telomere of 22q to 2q13, we used a human cosmid FISH probe, ICRFc106C202, which contains the 22q genomic region from the end of the acrosin gene to the proximal subtelomeric domain (Ning et al. 1996; Wong et al. 1997). The results are outlined in figure 2. In human, ICRFc106C202 hybridizes to the 22q telomere and cross-hybridizes to 2q13 (Ning et al. 1996). A similar hybridization signal pattern was observed in chimpanzee; cosmid c202 hybridizes to the orthologous site of the human 22q telomere and to the short arm telomere of chimpanzee chromosome 13, which is orthologous to the human 2q13 telomere fusion site. In gorilla, weak hybridization signals are present, in addition to the orthologous human 22q telomere and 2q13 sites. However, in orangutan and Old World monkeys, the hybridization signal is found only at the telomere of the human 22q ortholog. These results suggest that the 22q sequence is the ancestral copy; it was likely duplicated to the short arm of a great ape chromosome after the divergence of orangutan from the hominoid lineage.
Sequence Analysis of the Ancestral Subtelomeric Regions at 2q13
As shown in figure 3, the 2q13 ancestral telomere fusion site is flanked by two subtelomeric regions. While the proximal region is homologous to the 9p subtelomeric region, the subtelomeric region at the distal side of the fusion site is homologous to the subtelomeric region on chromosome 22q. To compare the size of the homologous region of 2q13 to that of the subtelomeric region of 22q, the RABL2A gene (GenBank accession number AF095351), which is within the ancestral subtelomeric domain of 2q13 and has an almost identical paralog on 22q (RABL2B; Wong et al. 1999), was used to search the GenBank database for a clone from 2q13. One completely sequenced BAC clone (RPCI11-395L14 [GenBank accession number AL078621]) was found to contain the RABL2A locus. This clone spans the end-to-end fusion site of the two great ape chromosomes, as is evident by the degenerative head-to-head arrays of telomeric repeats (TTAGGG)n at nucleotide positions 108305–109115 and flanking telomere adjacent repeat 1 sequences, which are usually found adjacent to the (TTAGGG)n telomeric repeat (Wilkie et al. 1991). As shown in figure 3, the second half of the RP11-395L14 sequence (nucleotide positions 110495–176734) shares strong homology with the 22q subtelomeric sequence (cosmid n94h12 [GenBank accession number AC002056] and cosmid n1G3 [GenBank accession number AC002055]) (Dunham et al. 1999).
Since the end of the 2q13 ancestral subtelomeric sequence in RP11-395L14 is still homologous to the 22q sequence, another GenBank database search was performed to identify sequence overlapping with RPCI11-395L14. Sequence from BAC RPCI11-432G15 (GenBank accession number AC017074) overlaps with the RPCI11-395L14 sequence and contains the “breakpoint” of this homologous subtelomeric region. The “breakpoint” sequences, defined as the transitional sequences between the homologous subtelomeric sequences and sequences unique to different chromosome ends, were determined by aligning the 2q and 22q subtelomeric sequences against each other. The breakpoint is located between exons 3 and 4 of the acrosin gene (ACR) on chromosome 22q, which is 68,681 bp from the 22q telomere (fig. 3).
To confirm that the breakpoint sequence is truly where the divergence of the two subtelomeric sequences occurs—rather than an artifact resulting from misassembly of sequence contigs in the BAC clones covering the homologous region—PCR primers (2-22bkF, ACRexon3F, and 2-22bkR) were designed on both sides of the breakpoint sequence, and DNAs from monochromosomal hybrid cell lines were used as templates for PCR. There are no repetitive elements in the 2q- and 22q-unique sequences; therefore, it is unlikely that the duplication of this ancestral subtelomeric region is due to a repetitive element–mediated translocation.
The 22q subtelomeric and 2q13 ancestral subtelomeric regions share 98.8% overall sequence similarity. To determine whether any change has occurred in these paralogous regions since the ancestral duplication, a dot plot of this 68-kb region was generated by the PipMaker program. Figure 6A demonstrates that these regions share continuous homology from the breakpoint to the end of chromosome.
Figure 6.
A dot plot comparison between the homologous subtelomeric sequences of the 2q13 and 22q regions (A) and the distal subtelomeric domains of 22q and 4p (GenBank accession number Z95704) (B). The subtelomeric 22q sequence contains the acrosin gene (ACR; the last two exons are included), a minisatellite repeat 3′AR, a processed pseudogene U2 snRNP specific polypeptide A (U2snRNP), and the RABL2B gene. All exons are highlighted in yellow, and other sequences are in gray. The degenerative (TTAGGG)n repeat sequence, which marks the boundary of the proximal and distal subtelomeric domains, is highlighted in pink.
To construct a physical map covering the ancestral subtelomeric region that is homologous to the 9p telomere, a GenBank database search was performed to screen for genomic clones overlapping with the first half of RPCI11-395L14 (nucleotide positions 1–110494). The results are summarized in figure 3. Three BAC clones from 2q13 (the first half of RPCI11-395L14, RPCI11-480C16 [GenBank accession number AC016745], and RPCI11-65I12 [GenBank accession number AC016683]) were assembled into a single sequence of 439,061 bp. The first 36 kb contain a distal subtelomeric domain, which shares discontinuous homology with sequences at many other chromosome ends. However, pairwise alignment shows that this distal subtelomeric domain shares no homology with the 22q subtelomeric domain. In addition, degenerative telomeric repeat (TTAGGG)n sequences are not present at the boundary between the distal and proximal domain. The proximal domain of this ancestral subtelomeric region is homologous to that of chromosome 9p. Figure 3 shows a 350-kb BAC contig at the subtelomeric region of 9p. The breakpoint of the homologous subtelomeric region between 2q13 and 9p is at nucleotide position 189490 of 2q13 or nucleotide position 130178 of the 9p sequence contig.
The sequence that is unique to 2q after the breakpoint contains no repetitive elements. However, an Alu sequence was found adjacent to the 9p breakpoint. Since the 2q breakpoint does not contain an Alu element, it is unlikely that the duplication between the 2q13 ancestral subtelomeric and 9p subtelomeric sequence is a result of repetitive element–mediated translocation. Primers (2qbpR, 2q9pbpF, and 9pbpR) were designed from the putative breakpoint sequence, to verify the position of the divergence between the chromosome 2 and 9 sequences. When the primers specific for the 2q breakpoint (2qbpR/2q9pbpF) were used for PCR, the reaction that used chromosome 9 monochromosomal hybrid DNA as a template showed a product. Further sequence analysis showed that this chromosome 2 breakpoint sequence matches chromosome 9 BAC clones located at the centromeric region (RPCI11-403A15 [GenBank accession number AL445925], RPCI11-87H9 [GenBank accession number AL353763], RPCI11-284A21 [GenBank accession number AC025021], and RPCI11-88I18 [GenBank accession number AL161457]), which show discontinuous homology to the 9p subtelomeric sequence.
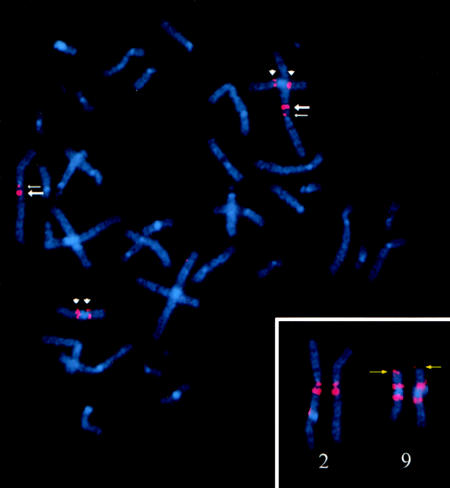
The sequence similarities between the chromosome 2 breakpoint and the chromosome 9 centromeric region were confirmed independently by FISH. Figure 7 shows FISH using a probe for RPCI11-480C16, from the 2q13 region; hybridization signals are present in the chromosome 9 pericentromeric region, at the 9p telomere, and at 2q13 and 2q11.2. This FISH hybridization pattern was examined more closely through use of sequence analysis. A complex sequence-rearrangement pattern was identified at the breakpoint region between the 2q ancestral subtelomeric and 9p subtelomeric sequence. Proximal to the 2q13 ancestral subtelomeric region, at nucleotide positions 216098–317838, the sequence is duplicated to 2q11.2. A 357,785-bp sequence, which is assembled from BACs RPCI11-34G16 (GenBank accession number AC008268) and RPCI11-440D17 (GenBank accession number AC009237), covers the 100-kb duplicated sequence at 2q11.2 (fig. 3). The telomeric breakpoint of the ancestral subtelomeric region is between two L1 repetitive elements, whereas the duplicated region at 2q11.2 is within an L1 and a MER90 element. No repetitive element is found at the centromeric breakpoint for either of the duplicated regions, and no known or predicted genes are present within the duplicated sequence. This intrachromosomal duplication was confirmed independently by PCR through use of primers 2q13UF, 2qcenUF, and 2qdupR.
Figure 7.
Hybridization of BAC RPCI11-480C16 to a human metaphase cell. Two hybridization signals are observed on human 2q: one at 2q13 (large arrows) and one at 2q11.2 (small arrows). Hybridization signals are also observed in the chromosome 9 pericentromeric region (arrow heads). The inset shows enlargements of the chromosome 2 and 9 homologs; the capture time for the Spectrum Orange signal was increased, to show the hybridization signal at the 9p telomere (yellow arrow).
Gene Duplications within the 9p Subtelomeric and 2q13 Ancestral Subtelomeric Regions
Three previously identified genes, phosphoglucomutase 5 (PGM5 [GenBank accession number NM_021965] or PGMRP [GenBank accession number L40933]), forkhead box D4 (FKHL9 [GenBank accession number U13223]) and a protein similar to cobalamin synthesis protein/P47K (COBW-like [GenBank accession number AF257330]) are within the homologous proximal subtelomeric regions. Both paralogous sequences on 2q and 9p contain a degenerate promoter, an incomplete exon 1, and a complete sequence of exon 2 of phosphoglucomutase 5, which spans 39–63 kb from the telomere. Since both paralogous sequences do not contain a functional copy of PGM5, PGM5 is likely located in the pericentromeric region (Edwards et al. 1995).
At nucleotide positions 103042–103674 of the 2q sequence, an intronless gene is predicted that shares 95% protein similarity to a partial human FKHL9 sequence (Larsson et al. 1995). Since the members of the forkhead gene family share a high degree of protein homology, it is not clear whether this forkhead box gene on 2q would have the same function as its chromosome 9 paralog. The third gene on the homologous proximal subtelomeric domain is a COBW-like protein. Fifteen exons cover nucleotide positions 109130–165069 of the 2q sequence. This 2q gene shares 98% protein similarity with its 9p paralog.
Discussion
The subtelomeric regions of human chromosomes exhibit a dynamic nature. Shared regions of homologies exist between different subsets of chromosomes, allowing the opportunity for mispairing between nonhomologous chromosomes (van Deutekom et al. 1996; Overveld et al. 2000). In the present study, we used FISH and sequence analysis to examine the subtelomeric regions of selected chromosomes in humans and members of a primate panel, to determine the evolutionary origin of subtelomeric shared sequence homologies and the mechanism by which subtelomeric duplications occur.
FISH analysis was used to study the origin and timing of subtelomeric-mediated duplications. All duplicated subtelomeric sequences in human were present at a single orthologous site in Old World monkeys, represented by macaque and baboon. These results demonstrate that these subtelomeric duplications occurred relatively recently, <25 million years ago, after the divergence of the great apes and Old World monkeys. Although all duplicated subtelomeric sequences in human were only present at a single orthologous site in Old World monkeys, the timing of the duplications differed, ranging from ∼5.5 to ∼23.3 million years ago; two duplications occurred after the divergence of Old World monkeys from the great apes, two occurred after the orangutan diverged from the remaining hominoids, one occurred after the gorilla diverged from chimpanzee and human, and two occurred after the divergence of chimpanzee and human. These results suggest that subtelomeric duplications have occurred recently at multiple time points during higher-primate evolution.
Interestingly, some species showed additional duplications (indicated by cross-hybridization) not observed in humans. In particular, chimpanzee and gorilla have gone through multiple cycles of duplication/translocation events, suggesting continued instability after a recent evolutionary rearrangement. Further investigations could examine why these species’ genomes seem more prone to this type of duplication or tolerate the duplications more easily.
Subtelomeric-mediated duplications represent one class of several recently described mechanisms for segmental duplications. By means of FISH (Cheung et al. 2001) and in silico (Bailey et al. 2001) analysis, it has been estimated that 5.4%–10.6% of the human genome shares a high level of sequence homology with other paralogous regions. These segmental duplications can be divided into three classes: intrachromosomal, pericentromeric, and subtelomeric. Intrachromosomal duplications are those that occur within the same chromosome. These duplicons, which share strong sequence homology, flank the critical regions of many microdeletion and microduplication syndromes. The timeline for this class of segmental duplication varies, from >20 million years ago for the Prader-Willi/Angelman syndrome deletion region (Christian et al. 1999) to ∼6–7 million years ago for the CMT1A-REP repeats (Kiyosawa and Chance 1996; Keller et al. 1999; Inoue et al. 2001), which are involved in the microduplication and deletion that cause Charcot-Marie-Tooth neuropathy type 1A and hereditary neuropathy with liability to pressure palsies, respectively (Reiter et al. 1996).
Pericentromeric- and subtelomeric-mediated duplications involve nonhomologous chromosomal regions. Horvath et al. (2000) proposed a two-step model for the dispersal of mosaics of duplicated segments among multiple pericentromeric regions. The first step is a process called “transposition seeding,” where genomic segments from different chromosomal origins were transposed to an ancestral pericentromeric region. This process is estimated to have occurred ∼5–10 million years ago, around the time of the divergence of the human, chimpanzee, and gorilla lineages. In the second step, called “pericentromeric exchange,” large blocks of mosaic duplicated segments were duplicated to the pericentromeric regions of other nonhomologous chromosomes. These duplication events are relatively recent, since they are only seen in human (Horvath et al. 2000).
The mechanism for subtelomeric duplications differs from that observed in the pericentromeric region. Cryptic subtelomeric chromosomal rearrangements are known to occur frequently in humans. It is estimated that subtle subtelomeric rearrangements account for 7.4% of patients with unexplained moderate-to-severe mental retardation (Knight et al. 1999). Therefore, it is possible that, during evolution, small unbalanced translocations occurred at a high enough frequency to contribute to the diversity of the subtelomeric regions. If a cryptic unbalanced translocation did not have a detrimental effect, it could have been fixed in the population during recent primate evolution, providing a mechanism for the creation of these segmental duplications. Our sequencing analysis of the breakpoint sequences in the 2q13 ancestral region supports this hypothesis, since the translocations that occurred to create this region do not appear to use a specific mechanism, such as that used in repetitive element–mediated rearrangements.
Because of a recent duplication, the 2q13 subtelomeric sequence shares strong homology with its ancestral copy at 22q. The continuous strong homology at the distal subtelomeric domain (distal to the degenerative TTAGGG repeat; fig. 6A) is unexpected. Distal subtelomeric domains usually share discontinuous homology with many different chromosomes (fig. 6B) (Flint et al. 1997). Since telomeres are clustered at the nuclear envelope to form a “chromosomal bouquet” during zygotene in meiosis (Scherthan et al. 1996, 2000), it is proposed that the discontinuous homology is a result of exchanges of sequences among nonhomologous chromosomes within the bouquet formation (Flint et al. 1997; Pryde et al. 1997). Since the 2q ancestral subtelomeric sequence is within an interstitial position and, therefore, is unlikely to move to the chromosomal bouquet during meiosis, the sequence exchanges among other nonhomologous chromosomes and 22q should interrupt the continuous homology between these two duplicated subtelomeric sequences. Our data, however, suggest that the 22q distal subtelomeric domain has not been changed since its duplication to the human 2q13 site, which occurred ∼6.7–8.2 million years ago (Kumur and Hedges 1998), after the divergence of the orangutan from the rest of the hominoid lineage. Since the publication by Flint et al. (1997), sequence information from additional subtelomeric regions has become available, and more sequence analyses are needed to determine whether the proximal/distal domain structures are common to every subtelomeric region or to only a subset of telomere regions.
Segmental duplications at subtelomeric regions increase gene copy number. Recently, transcription was reported in an olfactory-receptor gene (OR-A) that is localized in many subtelomeric regions (Linardopoulou et al. 2001). The paralogous sequence we studied at the 2q13 ancestral subtelomeric region also contains functional genes. The duplication of the 22q subtelomeric sequence created a functional paralog of RABL2B (RABL2A) at 2q13. Two acrosin-gene exons are also duplicated in the ancestral subtelomeric region. The shuffling of exons by chromosome rearrangements may create new genes with novel functions (de Souza et al. 1996; Viale et al. 1998; Courseaux and Nahon 2001; Inoue et al. 2001). However, since no exon is predicted within 28 kb of the chromosome 2 unique sequence proximal to the subtelomeric sequence translocation breakpoint, it is unlikely that the translocation of the acrosin-gene exons generated a novel gene.
In the present study, we found that the region that is homologous between the 9p subtelomeric region and the 2q ancestral subtelomeric region contains two known genes, FKHL9 and COBW-like. From our FISH results with BAC 305J7 and those of Larsson et al. (1995), we conclude that it is possible that more copies of these two genes exist within the pericentromeric region of chromosome 9, since additional FISH hybridization signals were observed in this region.
The ultimate goal of the Human Genome Project is to fully sequence 23 pairs of human chromosomes, from one telomeric end to the other. Recently, a link was made between the telomeres of each chromosome arm and the draft human genome sequence (Riethman et al. 2001). These data provide invaluable resources for physical mapping at human telomere regions. In the present study, we have characterized homologous regions between different telomeres and have demonstrated that ancestral unbalanced translocations are one mechanism for the creation of subtelomeric duplications. Additional targeted efforts are necessary to examine other subtelomeric regions, to gain a complete understanding of the dynamic nature of the telomeric regions of human chromosomes.
Note added in proof.—Since submission of this manuscript, the timeline of the duplication at 22q/2q13 subtelomeric region has been reported (Bailey et al. 2002). The results match those reported in the present article.
Acknowledgments
We thank Drs. Christine O'Keefe and Evan E. Eichler, for providing us with the Papio hamadryas lymphoblastoid cell line, and Dr. Harold Riethman, for useful discussions regarding telomere structure and evolution. This work was supported, in part, by National Institutes of Health grant 5 R01 HD36715-03 (to D.H.L.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Blast 2 Sequences, http://www.ncbi.nlm.nih.gov/gorf/bl2.html
- ClustalW Mutiple Sequence Alignment, http://searchlauncher.bcm.tmc.edu/multi-align/Options/clustalw.html
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for RABL2A [accession number AF095351], RPCI11-395L14 [accession number AL078621], cosmid n94h12 [accession number AC002056], cosmid n1G3 [accession number AC002055], RPCI11-432G15 [accession number AC017074], RPCI11-480C16 [accession number AC016745], RPCI11-65I12 [accession number AC016683], RPCI11-34G16 [accession number AC008268], RPCI11-440D17 [accession number AC009237], RPCI11-403A15 [accession number AL445925], RPCI11-87H9 [accession number AL353763], RPCI11-284A21 [accession number AC025021], RPCI11-88I18 [accession number AL161457], PGM5 [accession number NM_021965], PGMRP [accession number L40933]), FKHL9 [accession number U13223]), COBW-like [accession number AF257330], the physical map of chromosome 22q [accession number NT011526]), and the subtelomeric domain of 4p [accession number Z95704])
- Nix Application, http://www.hgmp.mrc.ac.uk/Registered/Webapp/nix/
- RepeatMasker Web Server, http://ftp.genome.washington.edu/cgi-bin/RepeatMasker
References
- Bailey J, Yavor A, Massa H, Trask B, Eichler E (2001) Segmental duplications: organization and impact within the current human genome project assembly. Genome Res 11:1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Viggiano L, Misceo D, Horvath JE, Archidiacono N, Schwartz S, Rocchi M, Eichler EE (2002) Human-specific duplication and mosaic transcripts: the recent paralogous structure of chromosome 22. Am J Hum Genet 70:83–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini A, Ried T, Shridhar V, Ogura K, D. Aiuto L, Rocchi M, Ward D (1993) An alphoid DNA sequence conserved in all human and great ape chromosomes: evidence for ancient centromeric sequences at human chromosomal regions 2q21 and 9q13. Hum Genet 90:577–583 [DOI] [PubMed] [Google Scholar]
- Brown WR, MacKinnon PJ, Villasante A, Spurr N, Buckle VJ, Dobson MJ (1990) Structure and polymorphism of human telomere-associated DNA. Cell 63:119–132 [DOI] [PubMed] [Google Scholar]
- Cheung VG, Nowak N, Jang W, Kirsch IR, Zhao S, Chen XN, Furey TS, et al (2001) Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature 409:953–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH (1999) Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13). Hum Mol Genet 8:1025–1037 [DOI] [PubMed] [Google Scholar]
- Courseaux A, Nahon JL (2001) Birth of two chimeric genes in the Hominidae lineage. Science 291:1293–1297 [DOI] [PubMed] [Google Scholar]
- de Souza SJ, Long M, Gilbert W (1996) Introns and gene evolution. Genes Cells 1:493–505 [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe B, Chissoe S, Hunt A, Collins J, Bruskiewich R, et al (1999) The DNA sequence of human chromosome 22. Nature 402:489–495 [DOI] [PubMed] [Google Scholar]
- Edwards Y, Putt W, Fox M, Ives J (1995) A novel human phosphoglucomutase (PGM5) maps to the centromeric region of chromosome 9. Genomics 30:350–353 [DOI] [PubMed] [Google Scholar]
- Eichler EE (2001) Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet 17:661–669 [DOI] [PubMed] [Google Scholar]
- Flint J, Bates GP, Clark K, Dorman A, Willingham D, Roe BA, Micklem G, Higgs DR, Louis EJ (1997) Sequence comparison of human and yeast telomeres identifies structurally distinct subtelomeric domains. Hum Mol Genet 6:1305–1314 [DOI] [PubMed] [Google Scholar]
- Horvath J, Viggiano L, Loftus B, Adams M, Archidiacono N, Rocchi M, Eichler E (2000) Molecular structure and evolution of an alpha satellite/non-alpha satellite junction at 16p11. Hum Mol Genet 9:113–123 [DOI] [PubMed] [Google Scholar]
- Ijdo JW, Baldini A, Ward DC, Reeders ST, Wells RA (1991) Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc Natl Acad Sci USA 88:9051–9055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Dewar K, Katsanis N, Reiter L, Lander E, Devon K, Wyman D, Lupski J, Birren B (2001) The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes. Genome Res 11:1018–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Eichler E, Schwartz S, Nicholls R (2000) Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res 10:597–610 [DOI] [PubMed] [Google Scholar]
- Kasai F, Takahashi E, Koyama K, Terao K, Suto Y, Tokunaga K, Nakamura Y, Hirai M (2000) Comparative FISH mapping of the ancestral fusion point of human chromosome 2. Chromosome Res 8:727–735 [DOI] [PubMed] [Google Scholar]
- Keller M, Seifried B, Chance P (1999) Molecular evolution of the CMT1A-REP region: a human- and chimpanzee-specific repeat. Mol Biol Evol 16:1019–1026 [DOI] [PubMed] [Google Scholar]
- Kiyosawa H, Chance PF (1996) Primate origin of the CMT1A-REP repeat and analysis of a putative transposon-associated recombinational hotspot. Hum Mol Genet 5:745–753 [DOI] [PubMed] [Google Scholar]
- Knight SJ, Lese CM, Precht KS, Kuc J, Ning Y, Lucas S, Regan R, Brenan M, Nicod A, Lawrie NM, Cardy DL, Nguyen H, Hudson TJ, Riethman HC, Ledbetter DH, Flint J (2000) An optimized set of human telomere clones for studying telomere integrity and architecture. Am J Hum Genet 67:320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SJ, Regan R, Nicod A, Horsley SW, Kearney L, Homfray T, Winter RM, Bolton P, Flint J (1999) Subtle chromosomal rearrangements in children with unexplained mental retardation. Lancet 354:1676–1681 [DOI] [PubMed] [Google Scholar]
- Kumur S, Hedges S (1998) A molecular timescale for vertebrate evolution. Nature 392:917–920 [DOI] [PubMed] [Google Scholar]
- Larsson C, Hellqvist M, Pierrou S, White I, Enerback S, Carlsson P (1995) Chromosomal localization of six human forkhead genes, freac-1 (FKHL5), -3 (FKHL7), -4 (FKHL8), -5 (FKHL9), -6 (FKHL10), and -8 (FKHL12). Genomics 30:464–469 [DOI] [PubMed] [Google Scholar]
- Lese CM, Fantes JA, Riethman HC, Ledbetter DH (1999) Characterization of physical gap sizes at human telomeres. Genome Res 9:888–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou E, Mefford HC, Nguyen O, Friedman C, van Den Engh G, Farwell DG, Coltrera M, Trask BJ (2001) Transcriptional activity of multiple copies of a subtelomerically located olfactory receptor gene that is polymorphic in number and location. Hum Mol Genet 10:2373–2383 [DOI] [PubMed] [Google Scholar]
- Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR (1988) A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA 85:6622–6626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y, Rosenberg M, Biesecker LG, Ledbetter DH (1996) Isolation of the human chromosome 22q telomere and its application to detection of cryptic chromosomal abnormalities. Hum Genet 97:765–769 [DOI] [PubMed] [Google Scholar]
- Overveld P, Lemmers R, Deidda G, Sandkuijl L, Padberg G, Frants R, van Der Maarel S (2000) Interchromosomal repeat array interactions between chromosomes 4 and 10: a model for subtelomeric plasticity. Hum Mol Genet 9:2879–2884 [DOI] [PubMed] [Google Scholar]
- Pryde FE, Gorham HC, Louis EJ (1997) Chromosome ends: all the same under their caps. Curr Opin Genet Dev 7:822–828 [DOI] [PubMed] [Google Scholar]
- Reiter L, Murakami T, Koeuth T, Pentao L, Muzny D, Gibbs R, Lupski J (1996) A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet 12:288–297 [DOI] [PubMed] [Google Scholar]
- Riethman HC, Xiang Z, Paul S, Morse E, Hu XL, Flint J, Chi HC, Grady DL, Moyzis RK (2001) Integration of telomere sequences with the draft human genome sequence. Nature 409:948–951 [DOI] [PubMed] [Google Scholar]
- Scherthan H, Jerratsch M, Li B, Smith S, Hulten M, Lock T, de Lange T (2000) Mammalian meiotic telomeres: protein composition and redistribution in relation to nuclear pores. Mol Biol Cell 11:4189–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H, Weich S, Schwegler H, Heyting C, Harle M, Cremer T (1996) Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol 134:1109–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer K, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W (2000) PipMaker: a web server for aligning two genomic DNA sequences. Genome Res 10:577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Bakker E, Lemmers RJ, van der Wielen MJ, Bik E, Hofker MH, Padberg GW, Frants RR (1996) Evidences for subtelomeric exchanges of 3.3 kb tandemly repeated units between chromosome 4q35 and 10q26: implications for genetic counseling and etiology of FSHD1. Hum Mol Genet 5:1997–2003 [DOI] [PubMed] [Google Scholar]
- Viale A, Ortola C, Richard F, Vernier P, Presse F, Schilling S, Dutrillaux B, Nahon JL (1998) Emergence of a brain-expressed variant melanin-concentrating hormone gene during higher primate evolution: a gene “in search of a function”. Mol Biol Evol 15:196–214 [DOI] [PubMed] [Google Scholar]
- Wienberg J, Jauch A, Ludecke H, Senger G, Horsthemke B, Claussen U, Cremer T, Arnold N, Lengauer C (1994) The origin of human chromosome 2 analyzed by comparative chromosome mapping with a DNA microlibrary. Chromosome Res 2:405–410 [DOI] [PubMed] [Google Scholar]
- Wienberg J, Stanyon R, Jauch A, Cremer T (1992) Homologies in human and Macaca fuscata chromosomes revealed by in situ suppression hybridization with human chromosome specific DNA libraries. Chromosoma 101:265–270 [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Higgs DR, Rack KA, Buckle VJ, Spurr NK, Fischel-Ghodsian N, Ceccherini I, Brown WR, Harris PC (1991) Stable length polymorphism of up to 260 kb at the tip of the short arm of human chromosome 16. Cell 64:595–606 [DOI] [PubMed] [Google Scholar]
- Wong ACC, Ning Y, Flint J, Clark K, Dumanski JP, Ledbetter DH, McDermid HE (1997) Molecular characterization of a 130-kb terminal microdeletion at 22q in a child with mild mental retardation. Am J Hum Genet 60:113–120 [PMC free article] [PubMed] [Google Scholar]
- Wong ACC, Shkolny D, Dorman A, Willingham D, Roe BA, McDermid HE (1999) Two novel human RAB genes with near identical sequence each map to a telomere-associated region: the subtelomeric region of 22q13.3 and the ancestral telomere band 2q13. Genomics 59:326–334 [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Prakash O (1982) The origin of man: a chromosomal pictoral legacy. Science 215:1525–1529 [DOI] [PubMed] [Google Scholar]