Abstract
Although primary mediastinal (thymic) large B-cell lymphoma has been primarily studied, its precise phenotype, molecular characteristics, and histogenesis are still a matter of debate. The International Extranodal Lymphoma Study Group collected 137 such cases for extensive pathological review. Histologically, the lymphomatous growth was predominantly diffuse with fibrosis that induced compartmentalized cell aggregation. It consisted of large cells with varying degrees of nuclear polymorphism and clear to basophilic cytoplasm. On immunohistochemistry, the following phenotype was observed: CD45+, CD20+, CD79a+, PAX5/BSAP+, BOB.1+, Oct-2+, PU.1+, Bcl-2+, CD30+, HLA-DR+, MAL protein+/−, Bcl-6+/−, MUM1/IRF4+/−, CD10−/+, CD21−, CD15−, CD138−, CD68−, and CD3−. Immunoglobulins were negative both at immunohistochemistry and in situ hybridization. Molecular analysis, performed in 45 cases, showed novel findings. More than half of the cases displayed BCL-6 gene mutations, which usually occurred along with functioning somatic IgVH gene mutations and Bcl-6 and/or MUM1/IRF4 expression. The present study supports the concept that a sizable fraction of cases of this lymphoma are from activated germinal center or postgerminal center cells. However, it differs from other aggressive B-cell lymphomas in that it shows defective immunoglobulin production despite the expression of OCT-2, BOB.1, and PU.1 transcription factors and the lack of IgVH gene crippling mutations.
Primary mediastinal (thymic) B-cell lymphoma (PMBL) is a distinct clinicopathological entity, originally described in 1980, 1 officially acknowledged in the REAL Classification in 1994 2 and more recently included in the World Health Organization book on the tumors of the hematopoietic and lymphoid tissues. 3 Phenotypic and molecular studies of this entity have provided controversial results, 4-36 thus challenging a definitive histogenetic interpretation.
In recent times, the field of B-cell lymphoma histogenesis has progressed rapidly because of the increasing availability of well-defined histogenetic markers. Genotypic markers include mutations of immunoglobulin (Ig) genes, which are somatically acquired at the time of B-cell transit throughout the germinal center (GC); mutations of BCL-6 have also been generally regarded as acquired during GC transit. 37-40 Phenotypic markers are represented by expression of the Bcl-6 protein, which is restricted to GC B-cells, and of MUM1/IRF4 (for Interferon Regulatory Factor-4), which denotes the final step of intra-GC B-cell differentiation, as well as subsequent steps of B-cell maturation toward plasma cells. 37-42 Finally, late stages of B-cell maturation are defined by expression of the CD138/syndecan-1 molecule. 41
So far, phenotypic analysis has revealed positivity of PMBL for CD45 and CD20, but negativity for CD3 and a variety of other T-cell markers; CD79a has generally been detected, despite the usual lack of surface and cytoplasmic Ig. 2-5,8,13,16,18,19,21,22,32 In two reported series, CD30 staining was observed in the vast majority of cases, although its was weaker and less homogeneous than in classic Hodgkin’s lymphoma (cHL) and anaplastic large cell lymphoma. 21,28 CD21 and class I and/or II histocompatibility molecules have been claimed to be absent. 4,5,8,14 Bcl-2 protein seems to be generally expressed, 32 while fragmentary data are available concerning the occurrence of some molecules, such as CD10, MUM1/IRF4, PAX5/BSAP (B-cell Specific Activating Protein), Bcl-6, BOB.1, and Oct-2. 31,34
In contrast to diffuse large B-cell lymphoma, 10,31 molecular studies of small series of PMBLs have so far revealed the usual absence of BCL-6 and BCL-2 rearrangements/mutations, 25,30 as well as frequent overexpression of the MAL gene. 29 The latter is located on the long arm of chromosome 2 43 and encodes a protein thought to play a relevant role in membrane trafficking and signaling mediated by specialized, and glycolipid- and cholesterol-enriched, membrane microdomains referred as lipid rafts, 44-46 which might contribute to disease pathogenesis. 29 Mutations of the gene have not been identified and the mechanism of overexpression remains unclear yet. 29 Other reported oncogene abnormalities consist in C-MYC mutations, detection of BCL-2 and REL proto-oncogene amplification and P53 mutations. 17,25,36 Because of the reported lack of BCL-6 mutations, PMBL has been thought to recognize a pre-GC cell derivation. 30-32 This assumption, however, seems to contrast with two recent reports, respectively, showing, in a small series of PMBLs, isotype-switched Ig genes with a high load of somatic hypermutations and variants in the 5′-noncoding region of BCL-6 by single-strand conformation polymorphism analysis. 35,36
The aim of the present report is to focus on the morphological, phenotypic, and molecular characterization of a series of 137 PMBLs collected by the International Extranodal Lymphoma Study Group.
Materials and Methods
Case Composition and Selection
The Centers adhering to International Extranodal Lymphoma Study Group Protocol 9 on PMBL were asked to send to the Hematopathology Unit of Bologna University the pathological material of the 426 cases enrolled in the course of a clinical trial. Histological preparations were obtained in 181 instances. The Hematopathology Units/Surgical Pathology Divisions of the following institutions contributed to the study (listed according to the number of cases submitted): Southampton University, UK; Bologna University, Italy; National Institute of Cancer, Milan, Italy; “La Sapienza” University, Rome, Italy; S. Giovanni Hospital, Bellinzona, Switzerland; Turin University, Turin, Italy; Modena University, Modena, Italy; Udine University, Udine, Italy; S. Maria Nuova Hospital, Reggio Emilia, Italy; Pisa University, Pisa, Italy; S. Giovanni Rotondo Hospital, S. Giovanni Rotondo, Italy; Bern University, Bern, Switzerland; Catanzaro University, Catanzaro, Italy; Coppito Hospital, L’Aquila, Italy; General Hospital, Dolo-Venezia, Italy; S. Maria delle Croci Hospital, Ravenna, Italy; Bianchi-Melacrino-Morelli Hospital, Reggio Calabria, Italy; Perugia University, Perugia, Italy; Bari University, Bari, Italy; S. S. Annunziata Hospital, Taranto, Italy; and General Hospital, Mestre, Italy. Hematoxylin and eosin (H&E)- and Gomori-stained sections were available in all cases, which were integrated by immunohistochemical preparations for CD20/L26 and CD3 or CD45R0/UCHL1 in 11 instances and paraffin blocks in 80 cases. The submitted material corresponded to 36 needle biopsies and 145 surgical samples. All tissue blocks had been fixed in 10% buffered formalin except 10 cases that had undergone Susa fixation. A preliminary screening of the cases was made on H&E to assess the adequacy of both the amount of evaluable material and the preservation of cytological details. Three-μm-thick sections were cut from the paraffin blocks and stained with Giemsa and Gomori.
Immunohistochemistry
Serial sections were cut from the submitted paraffin blocks, coated on naturally charged slides, stored in a warm chamber at 56°C for at least 2 hours, and then rehydrated through repeated washes in Histoclear (National Diagnostics, Atlanta, GA) and graded alcohols. They were used for immunohistochemistry, which was performed by applying the panel of antibodies listed in Table 1 ▶ ; the antibodies, with the exception of the ones raised against PU.1 and MAL protein, were dispensed on a TechMate 500 immunostainer and detected by the alkaline phosphatase-anti alkaline phosphatase (APAAP) technique. 47-49 In particular, all antigens but CD21 and MAL protein were unmasked by treating sections in 1 mmol/L ethylenediaminetetraacetic acid (pH 8.0) in a microwave oven at 900 W for 10 to 15 minutes (two to three cycles of irradiation, 5 minutes each) depending on their cytoplasmic or nuclear location. 50 The sections were then cooled at room temperature for 20 minutes. CD21 was re-exposed by incubating sections in a 0.1% solution of Protease XIV in 0.2 mol/L of Tris-buffered saline (TBS) (pH 7.5) at 37°C for 4 minutes, followed by prolonged washing in cold TBS. The anti-PU.1 antibody was revealed by the EnVision technique. 49 MAL protein expression was evaluated by using a specific monoclonal antibody, 29 after microwave heating in citrate buffer, pH 6.7 (one cycle of 12 minutes at 750 W followed by two cycles of 10 minutes each at 350W): the antibody was revealed by the immunoperoxidase labeled streptavidin biotin (LSAB) technique. 49 In all instances, positive and negative controls were used to assess the reliability of the results, as previously described. 50 The results were graded as follows: +, 75 to 100% of the neoplastic cells positive; +/−, 50 to 75% of the neoplastic cells positive; −/+, 25 to 50% of the neoplastic cells positive; rare, 1 to 25% of the neoplastic cells positive; −, all neoplastic cells negative. The intensity of the staining and the pattern of positivity (nuclear, cytoplasmic diffuse, dot-like, granular, membrane-bound, and so forth) were also recorded.
Table 1.
Summary of the Findings Observed at Immunohistochemistry and in Situ Hybridization
| Marker | Cases+ve/casesev |
|---|---|
| CD45 (mAb clone PD7/26+2B11, DAKO, Glastrup, Denmark) | 80 /80 |
| CD20 (mAb clone L26, DAKO) | 91 /91 |
| CD79a (mAb clone JCB117, Prof. D.Y. Mason, Oxford) | 80 /80 |
| PAX5/BSAP (mAb clone 24, Transduction Lab, Lexington, KY) | 80 /80 |
| CD3 (pAb) and/or CD45R0 (mAb clone UCHL1, DAKO) | 0 /91* |
| CD30 (mAb clone Ber-H2, Prof. H. Stein, Berlin, Germany) | 62 /72 |
| HLA-DR (mAb clone DK22, DAKO) | 61 /76 |
| Bcl-2 protein (mAb clone 124, DAKO) | 55 /70 |
| CD10 (mAb clone 56C6, Novocastra, Newcastle, UK) | 15 /71 |
| Bcl-6 protein (mAb clone PG-B6p, Prof. B. Falini, Perugia, Italy) | 31 /66 |
| MUM1/IRF4 (mAb clone Mum-1p, Prof. B. Falini, Perugia) | 46 /61 |
| CD138 (mAb clone MI15, DAKO) | 0 /61 |
| Ig light chains (pAb, DAKO) | 0 /80 |
| Ig light chains m-RNA (PNA probes, DAKO) | 0 /40 |
| BOB.1 (pAb, Santa Cruz, Santa Cruz, CA) | 73 /73 |
| Oct-2 (pAb, Santa Cruz) | 69 /69 |
| PU.1 (mAb clone G148-74, PharMingen, La Jolla, CA)† | 40 /40 |
| CD15 (mAb clone 3CD1, DAKO) | 0 /80 |
| CD68 (mAb clone PG-M1, Prof. B. Falini) | 0 /80 |
| CD21 (mAb clone 1F8, DAKO) | 0 /70 |
| MAL protein (mAb, Dr. M. A. Alonso, Madrid, Spain) | 27 /44 |
Abbreviations: +ve, positive (all cases with a rate of neoplastic cells stained between 10% and 100% are collectively quoted as positive in the present table; deviations from the usual 70 to 100% (+) value are detailed in the result section); ev, evaluable; CD, cluster of differentiation; pAb, polyclonal antibody; mAb, monoclonal antibody.
*The total number includes 11 cases with only basic B- and T-cell markers available.
Footnote: the difference in the number of cases tested with each antibody depends on the amount of sections obtained from the submitted paraffin blocks, which varied from case to case.
†The determination of PU.1, performed in the light of the recently published report of Jundt and colleagues 47 , was performed after the completion of all the other immunostains and was feasible in only 40 cases with left material.
In Situ Hybridization
In 40 cases with abundant material available and optimally preserved tissue as judged by immunostains and morphological details, fluorescein-conjugated peptide nuclei acid (PNA) probes against κ and λ Ig light-chain mRNA (DAKO, Glostrup, Denmark) were applied according to the manufacturer’s instructions. Sections from two cases of multiple myeloma with κ and λ Ig light-chain restriction, respectively, were used as positive controls.
Molecular Biology
Eight-μm-thick sections were cut from 45 paraffin blocks with enough material available for DNA extraction and amplification. DNA purification was performed using a commercial kit (QIAamp DNA mini Kit; Qiagen, Italy) according to the manufacturer’s instructions. 30
IgVH gene rearrangements were amplified with two sets of six VH gene family-specific primers and a JH primer mix in separate reactions for each VH primer, as described. 51,52 The VH primers used in this study hybridize to sequences in the framework region (FR) 1 or FR2 of the respective VH families. Polymerase chain reaction (PCR) was performed for 40 cycles with an annealing temperature of 60°C. PCR products were directly sequenced using a commercially available kit (ThermoSequenase; Amersham Life Sciences) as previouslyreported. 30 Sequences were compared to the V-BASE sequence directory (MRC Centre for Protein Engineering, Cambridge, UK) using MacVector 6.0.1 software (Oxford Molecular Group PLC, Oxford, UK) for comparison of the rearranged IgV genes to the most homologous germline sequences.
Mutational analysis of BCL-6 gene was performed by direct sequencing of three partially overlapping PCR products encompassing nucleotides +404 to +1142 localized in the 5′-noncoding region of the gene. 30 PCR fragments were directly sequenced with appropriate primers, as described above. 30 All mutations detected were reconfirmed on independent PCR products.
Clinical Findings and Statistical Methods
The following clinical information was available in all patients: age, sex, sites of disease, stage, presence of systemic symptoms, International Prognostic Index (IPI), lactate dehydrogenase (LDH) level, bulky disease, outcome, and therapies administered (CHOP/CHOP-like versus MACOP-B/VACOP-B/ProMACE-CytaBOM versus HDS/ABMT). Overall survival was calculated by the Kaplan-Meier method 53 from the date of diagnosis (starting time) until last contact or death for any cause (event). Relapse-free survival was calculated for patients who achieved a complete remission after the first-line therapy from the date of first complete remission to the date of last contact, if alive and nonrelapsed, or to relapse or death (events), whichever came first.
Univariate analysis was performed by the log-rank test 54 or Cox proportional hazard regression model, 55 as appropriate. Multivariate analysis was performed by a Cox model using the stepwise selection method. The chi-square test was used whenever appropriate for comparison of subgroups. Two-sided P values were used throughout.
Results
Case Selection
Forty-four of the 181 submitted cases were excluded because of inadequate sampling or crashing artifacts, which prevented optimal cytological evaluation.
Histological Findings
At Gomori silver impregnation, features of sclerosis were appreciated in 125 of 137 cases. These varied from case to case and from field to field within the same case. In particular, they usually tended to surround groups of lymphomatous elements producing compartmentalization of the tumoral growth (Figures 1, 2, and 3) ▶ ▶ ▶ . In more than half the cases, the latter finding was already evident at H&E because of the presence of thick collagen bands around clusters of neoplastic cells (Figures 1 to 3) ▶ ▶ ▶ . In 14 instances, the sclerotic features were only focal. A vague nodularity was appreciated in 51 instances (Figure 3) ▶ and phenomena of necrosis, at times extensive, in 36.
Figure 1.
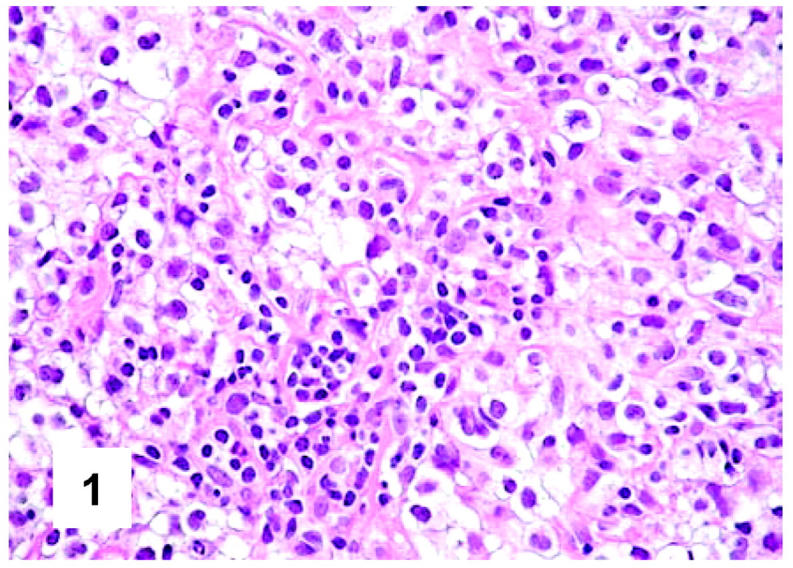
Histopathology of PMBL. The tumoral population homogeneously consists of large cells with clear cytoplasm (H&E, ×400).
Figure 2.
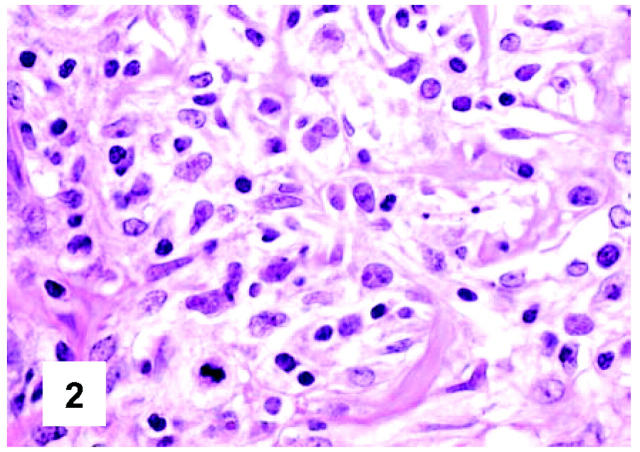
Cytological details of PMBL. Neoplasm cells with clear cytoplasm show some variability both in size and nuclear contours, the latter being at times multilobated (H&E, ×600).
Figure 3.
Fibrotic component of PMBL. Groups of neoplastic cells are surrounded by a delicate meshwork of reticulin fibers (Gomori silver impregnation, ×50).
On cytological grounds, in 103 cases, the tumor uniformly consisted of large cells (measuring >20 μm in diameter) (Figures 1 and 2) ▶ ▶ , whereas in the remaining ones the size of lymphomatous elements ranged from medium to large. The nuclear contours were more often variable within the same case, resulting round, oval, indented, or multilobated (Figure 2) ▶ . The nucleoli comprised within a finely dispersed chromatin were generally medium-sized, at times adjacent to the nuclear membrane (Figures 1 and 2) ▶ ▶ . In all instances, the rim of cytoplasm was relatively wide: at H&E and Giemsa staining, it appeared homogeneously clear or slightly basophilic in 96 and 35 cases, respectively (Figures 1 and 2) ▶ ▶ . A mixture between clear and slightly basophilic cells was appreciated in only six tumors. Mitotic figures were numerous, ranging from 4 to 9/high-power field. Within this context, elements with immunoblastic morphology were occasionally seen, while scattered multinucleated giant cells, at times with Reed-Sternberg-like features, were recorded in 33 instances.
Immunohistochemical Findings
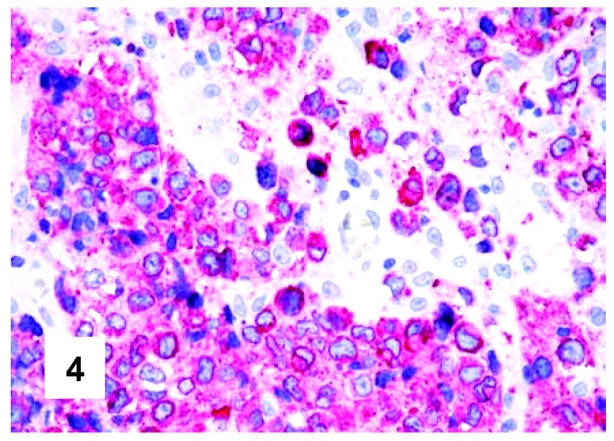
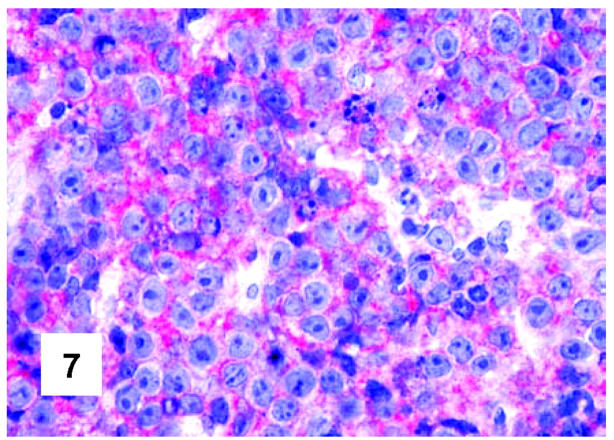
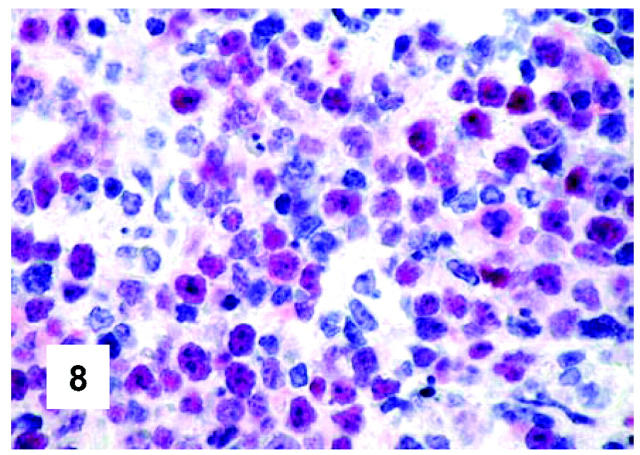
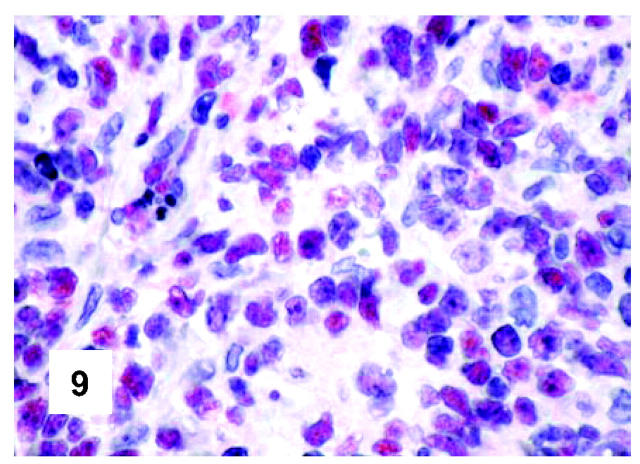
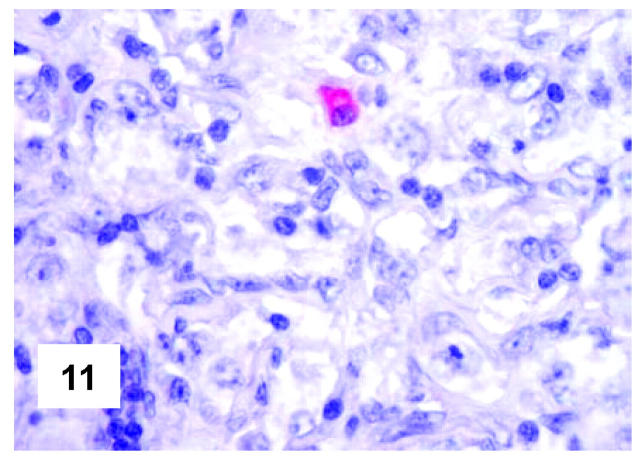
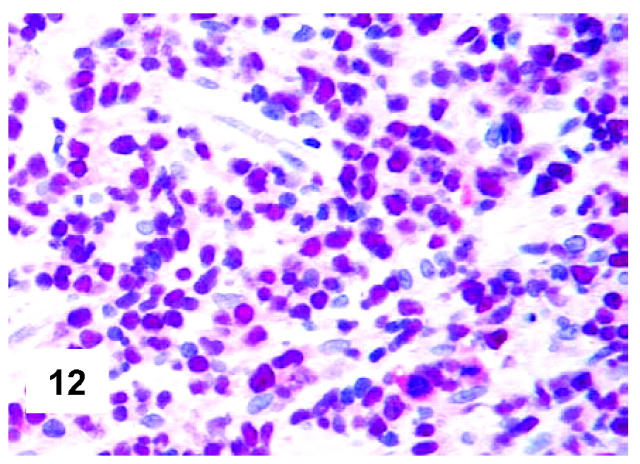
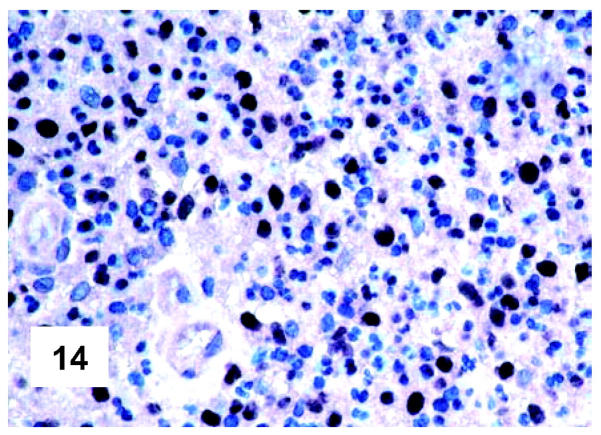
These are summarized in Table 1 ▶ and primarily corresponded to the results obtained in other studies as far as it concerned the expression of the most common markers. 31,32 In particular, B-cell related antigens (CD20, CD79a, and PAX5/BSAP) as well as the leukocyte common antigen (CD45) were positive in all cases (Figure 4) ▶ . However, the staining for CD79a showed some variability both in terms of intensity (which was moderate to weak in 15 cases) and number of positive cells. The latter corresponded to <50% of the neoplastic elements in 19 instances. T-cell markers were limited to reactive T-lymphocytes scattered throughout. CD30 staining was found in 62 of 72 eligible cases: it was more often moderate/weak and limited to 10 to 50% of the lymphomatous cells in 46 of 62 samples (Figure 5) ▶ . HLA-DR was strongly expressed in 61 of 76 cases (Figure 6) ▶ . No single case was found to express CD15, CD21, CD138, and CD68. Bcl-2 product, CD10, Bcl-6 protein, and MUM1/IRF4 were, respectively, detected in 55 of 70, 15 of 71, 31 of 66, and 46 of 61 cases with enough material left. Although some variability of the staining intensity and rate of positive elements was observed with each marker, usually most if not all neoplastic cells were marked by the corresponding antibody (Figures 7, 8, and 9) ▶ ▶ ▶ . Fourteen of the 15 CD10+ cases displayed a reliable Bcl-6 staining. The determination of the MAL protein provided reliable results in 44 of 78 investigated cases, as judged on the basis of the existing internal controls. Twenty-seven of the interpretable cases turned out to express the molecule in question: the content of MAL-positive cells varied from case to case (from 30 to 100% of the neoplastic elements), as did the intensity of the staining (from weak to strong) (Figure 10) ▶ . In eight instances, the positivity was mainly or exclusively confined to the Golgi area. The search for surface and/or cytoplasmic κ and λ Ig light-chains gave negative results in all samples except five which showed an equivocal stain for κ. The latter cases were included among those that underwent in situ hybridization (see below): they all turned out negative at the determination of Ig light chain mRNA (Figure 11) ▶ . The transcription factors BOB.1, Oct-2, and PU.1 were detected in all of the analyzed cases (73, 69, and 40, respectively) (Figures 12, 13, and 14) ▶ ▶ ▶ : the staining was usually nuclear, although simultaneous nuclear and cytoplasmic positivities were recorded in some instances. Finally, the Ki-67 marking (monoclonal antibody Mib-1; DAKO) ranged from 40 to 95% with a median value of 69.5%.
Figure 4.
Immunohistochemical features of PMBL. CD79a staining in a case characterized by almost complete positivity of neoplastic cells (monoclonal antibody JCB117; APAAP technique; Gill’s hematoxylin nuclear counterstaining; ×600).
Figure 5.
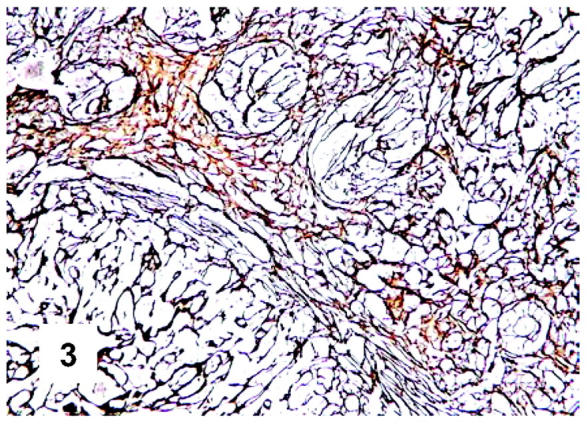
Immunohistochemical features of PMBL. CD30 expression: the staining occurs in the majority of neoplastic cells and is weaker than in classical Hodgkin’s lymphoma and anaplastic large cell lymphoma (monoclonal antibody Ber-H2; APAAP technique; Gill’s hematoxylin nuclear counterstaining; ×400).
Figure 6.
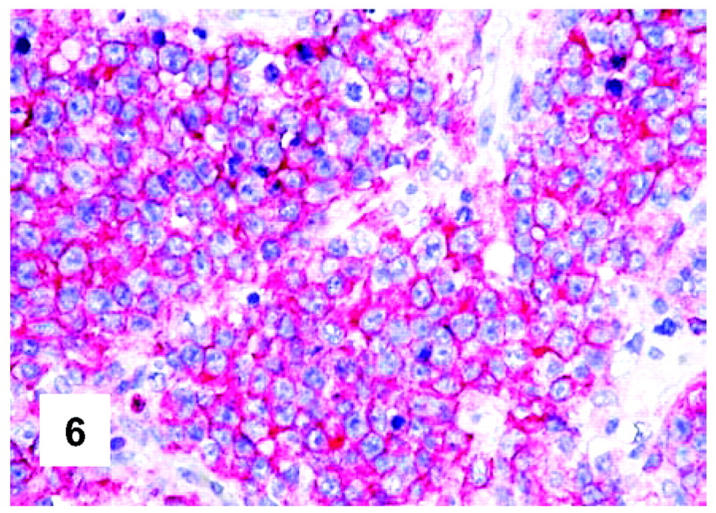
Immunohistochemical features of PMBL. All neoplastic cells carry HLA-DR (monoclonal antibody DK22; APAAP technique; Gill’s hematoxylin nuclear counterstaining; ×500).
Figure 7.
Immunohistochemical features of PMBL. CD10 expression by neoplastic cells (monoclonal antibody 56C6; APAAP technique; Gill’s hematoxylin nuclear counterstaining; ×700).
Figure 8.
Immunohistochemical features of PMBL. Expression of the BCL-6 gene product by neoplastic cells (monoclonal antibody PG-B6p; APAAP technique; Gill’s hematoxylin nuclear counterstaining; ×600).
Figure 9.
Immunohistochemical features of PMBL. The search for the MUM1/IRF4 molecule produces a staining pattern that corresponds to the one observed with the PG-B6p antibody (monoclonal antibody mum-1p; APAAP technique; Gill’s hematoxylin nuclear counterstaining; ×600).
Figure 10.
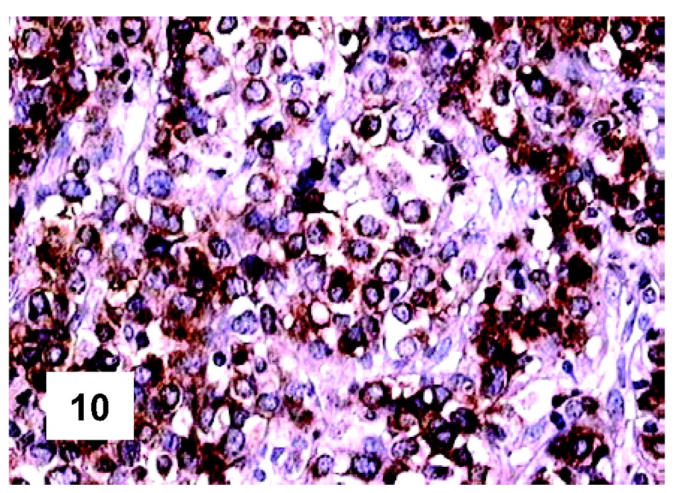
Immunohistochemical features of PMBL. Most neoplastic cells express the MAL protein at the cytoplasmic level: some positivities confined to the Golgi area can also be seen (monoclonal antibody; LSAB technique; hematoxylin nuclear counterstaining; ×500).
Figure 11.
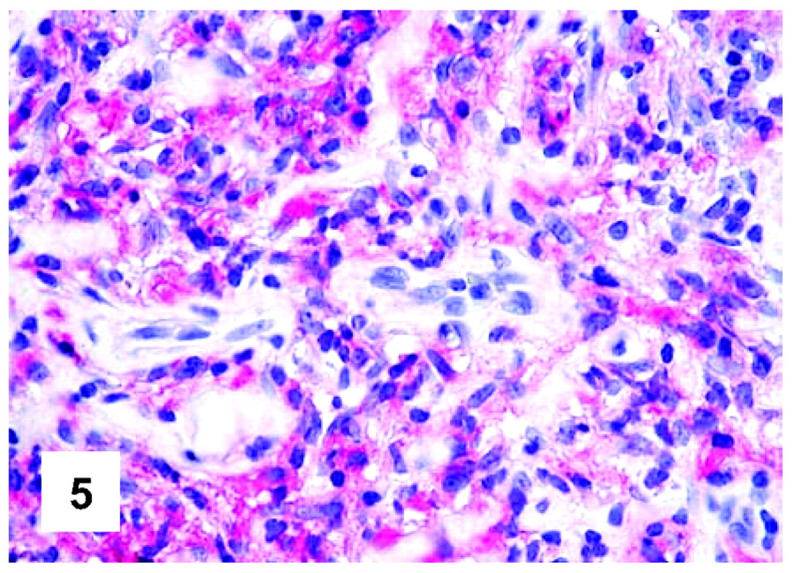
In situ hybridization features of PMBL. One single positive plasma cell is comprised among negative neoplastic elements (in situ hybridization; mRNA κ Ig light chain complementary PNA probe; development in New Fuchsin; Gill’s hematoxylin nuclear counterstaining; ×700).
Figure 12.
Immunohistochemical features of PMBL. All lymphomatous cells react with anti-Oct-2 antibody (polyclonal antibody; APAAP technique; Gill’s hematoxylin nuclear counterstaining; ×500).
Figure 13.
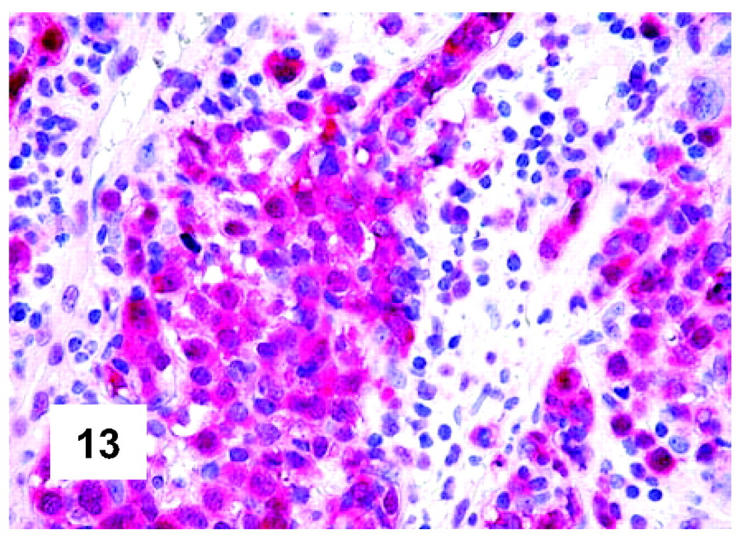
Immunohistochemical features of PMBL. The same pattern is observed at the determination of BOB.1, which is expressed both at the nuclear and cytoplasmic level (polyclonal antibody; APAAP technique; Gill’s hematoxylin nuclear counterstaining; ×400).
Figure 14.
Immunohistochemical features of PMBL. Most neoplastic cells, intermingled with some granulocytes, do express PU.1 (monoclonal antibody; Envision technique; Gill’s hematoxylin nuclear counterstaining; ×500).
In Situ Hybridization
No positivity at the determination of κ and λ Ig light-chain mRNA was appreciated at the neoplastic level in the tested cases, whereas a strong staining was seen in occasional reactive plasma cells comprised within the tumor (Figure 11) ▶ , as well as in the positive controls (Table 1) ▶ .
Molecular Biology
Details of the results are shown in Tables 2 and 3 ▶ ▶ . The IgVH gene region could be amplified in 17 of 45 cases. Presumably, the quality of DNA conservation hampered amplification of IgVH genes in the majority of the remaining cases. Clonal and functional rearrangements were detected in all instances. Nine cases used genes belonging to the VH3 family, four cases used genes belonging to the VH1 family, two cases used genes belonging to the VH4 family, and one case each used genes belonging to the VH2 or VH6 families. Somatic mutations of IgVH genes at a rate ≥2 × 10−2 bp were found in 8 of 17 cases, whereas the remaining cases harbored unmutated IgVH genes.
Table 2.
IgVH Gene Status
| Case number | IgVH gene | % Homology | Frequency of IgVH mutations | Technical approach utilized |
|---|---|---|---|---|
| 560/98 | VH3(DP-50 hv3) | 92.50 | 7.53 × 10−2 bp | FR1/JH |
| 4111/99 | VH3(DP-47) | 95.70 | 4.3 × 10−2 bp | FR1/JH |
| 4365/99 | VH3(DP-50 hv3) | 91.90 | 8.1 × 10−2 bp | FR1/JH |
| 3898/00 | VH1(DP-10 hv1) | 100 | 0 × 10−2 bp | FR1/JH |
| 3906/00 | VH3(VH3-8) | 100 | 0 × 10−2 bp | FR1/JH |
| 28/01 | VH1(DP-10 hv1) | 100 | 0 × 10−2 bp | FR1/JH |
| 29/01 | VH3(DP-50 hv3) | 92.2 | 7.8 × 10−2 bp | FR1/JH |
| 35/01 | VH3(DP-38/9-1) | 100 | 0 × 10−2 bp | FR1/JH |
| 41/01 | VH1(DP-8+) | 91.1 | 8.9 × 10−2 bp | FR1/JH |
| 66/01 | VH3(DP-58 hv3) | 86.10 | 13.9 × 10−2 bp | FR1/JH |
| 72/01 | VH6(DP-74) | 100 | 0 × 10−2 bp | FR1/JH |
| 862/01 | VH2(S12-7) | 99.5 | 0.5 × 10−2 bp | FR1/JH |
| 857/01 | VH1(DP-88 hv1) | 100 | 0 × 10−2 bp | FR2/JH |
| 867/01 | VH4(DP-63) | 100 | 0 × 10−2 bp | FR1/JH |
| 2837/00 | VH3(DP-51+) | 81.8 | 18.2 × 10−2 bp | FR1/JH |
| 2839/00 | VH4(DP-63) | 90.1 | 9.9 × 10−2 bp | FR2/JH |
| 2842/00 | VH3 COS3/BHG | 100 | 0 × 10−2 bp | FR2/JH |
Table 3.
BCL-6 Gene Status
| Case number | BCL-6 mutations | IgVH somatic mutations (≥2 × 10−2 bp) | Bcl-6 protein | MUM1/IRF4 protein |
|---|---|---|---|---|
| 528/95 | WT | Ne | + | + |
| 560/98 | A446G, A468C, T716G, G810C | Yes | − | + |
| 4111/99 | WT | Yes | − | + |
| 4365/99 | WT | Yes | + | + |
| 2700/00 | G799A | Ne | − | − |
| 3898/00 | A581G | GL | Ne | Ne |
| 3900/00 | WT | Ne | Ne | − |
| 3906/00 | WT | GL | + | + |
| 5463/00 | WT | Ne | − | − |
| 28/01 | G590C, G629C | GL | + | − |
| 29/01 | A581C | Yes | − | + |
| 33/01 | C742T | Ne | − | + |
| 35/01 | G759C | GL | − | + |
| 41/01 | WT | Yes | − | + |
| 47/01 | A436C, G478C, G481A, T495A, G518C, G533A | Ne | + | + |
| 64/01 | G518C, C525A, T548A, A551T, C601A, T633C | Ne | − | + |
| 65/01 | G478A, G533C, G540A, A541C, A547T, T554C, G578A, etc | Ne | + | − |
| 66/01 | G790T | Yes | + | − |
| 862/01 | T482C, T554G, C584G, G758C | GL | − | − |
| 838/01 | G708A, G810A | Ne | + | + |
| 857/01 | InsG706, C763T | GL | + | + |
| 866/01 | G485A | Ne | − | + |
| 867/01 | C584T, C765T | GL | − | + |
| 870/01 | T447C, T469A, C479T, G533A, A535C, A551C | Ne | + | + |
| 871/01 | G790A, T814C, T620A | Ne | + | − |
| 874/01 | C444G, G578C, T531C, C572G | Ne | + | + |
| 875/01 | G503A, G641A, C750T, insC770, T838G | Ne | − | + |
| 2837/00 | WT | Yes | − | − |
| 2838/00 | WT | Ne | Ne | Ne |
| 2839/00 | T466C, G804C | Yes | − | + |
| 2840/00 | G766T, C769G, C777T | Ne | + | + |
| 2841/00 | WT | Ne | Ne | Ne |
| 2842/00 | C744A, T739A, G833C | GL | − | + |
| 2843/00 | G488A, A535G, G549A, A551C, T557C, A582G, G621T;* | Ne | + | + |
| 2844/00 | T545G, A561C, C588T, G829A | Ne | − | Ne |
| 3409/00 | WT | Ne | − | Ne |
| 3903/00 | C704T | Ne | Ne | Ne |
Abbreviations: WT, wild type; *, mutation in homozygosis; GL, germline; Yes, presence of IgVH mutations (≥ 2 × 10−2 bp); +, positive result at immunohistochemistry; −, negativity at immunohistochemistry; Ne, not evaluable.
The analysis of BCL-6 gene mutations could be reliably performed in 37 of 45 cases. Twenty-six of these cases (70.2%) carried gene mutations with a mutation rate ranging from 1.13 × 10−3 bp to 18.22 × 10−3 bp (mean value, 4.6 × 10−3 bp). Among the BCL-6 mutated cases, 21 showed positivity for the Bcl-6 protein and/or MUM1/IRF4, 2 were negative for both markers, and 3 did not provide reliable immunohistochemical results. Thirteen of the BCL-6 mutated cases expressed the MAL protein, the remaining ones being either negative (n = 5) or not evaluable (n = 8). Concerning the 11 cases without BCL-6 mutations, 5 were positive at the determination of Bcl-6 and/or MUM1/IRF4, 2 were negative for both, and 4 were not evaluable; only 1 of them expressed the MAL protein, 4 were MAL-negative, and 6 belonged to the group of not interpretable cases.
Clinical Findings
The overall and relapse-free survival of the 137 patients calculated at 144 months corresponded to 85% and 70%, respectively. No significant differences were observed, when these data were stratified according to IPI (0 to 1 versus >2) and type of therapy administered. As to the latter, however, it should be underlined that the cases treated with MACOP-B and so forth or HDS/ABMT definitely predominated over the ones that received CHOP/CHOP-like therapy. A slightly worse outcome was observed in the patients displaying MAL protein expression or BCL-6 gene in germ line configuration: however, the difference between these cases and those with an opposite pattern never achieved statistical significance.
Discussion
The present study provides morphological, phenotypic, and molecular data on a large series of PMBL.
At light microscopy, the vast majority of the tumors showed a diffuse growth pattern and consisted of large cells with polymorphic nuclei and a wide rim of clear cytoplasm. Fibrosis was common with frequent compartmentalization of neoplastic cells. Possible additional findings were focal nodularity, necrosis, vascular invasion, and Reed-Sternberg-like elements. These findings basically correspond to those quoted in the literature: 1-8,12-14,16,19-21,23,26,31,32 in conjunction with immunohistochemical results, they allow a confident diagnosis in most instances, thus definitely disproving the old statement that the recognition of PMBL represents a hard task for the pathologist. 1,6 The only problematic cases in our series were those corresponding to needle biopsies with crushing artifacts and Reed-Sternberg-like elements. Under these circumstances, the differentiation from cHL should be based on the distinctive phenotypic profile of PMBL, which is in turn CD15− and positive for CD45, CD20, CD79a, MAL, BOB.1, Oct-2, and PU.1. 2,3,56
On immunohistochemistry, MAL protein was detected in a percentage of tumors similar to that originally reported and recently confirmed by Copie-Bergman and co-workers. 29,57 As its detection is somewhat fixative-dependent, 29 it might be that the prevalence of positive cases is slightly underestimated. Interestingly, this marker, which has not been so far detected in diffuse large B-cell lymphoma, seems to be characteristic of PMBL. 29 Under physiological conditions it is restricted to some steps of T-cell maturation, being involved in specialized lipid raft-mediated, membrane-trafficking, and signaling pathways. 29,45,46 Thus, in PMBL MAL protein expression might influence both the control of the neoplastic growth and the process of lymphomagenesis. 29
Likewise two previous studies, 21,28 most of the PMBLs of our series showed frequent expression of the CD30 antigen. 58 In contrast, the high frequency of HLA-DR expression in our material is in conflict with previous reports that stated the lack of histocompatibility antigen expression in PMBL. 4,5,8,32,59,60 The differences between our own and previous studies are possibly because of the significant improvement in the antibody affinity and immunohistochemical techniques that occurred during the last few years.
In agreement with previous reports, 22,31,32 all of the analyzed cases turned out to be negative at the determination of Ig light chains, both at the immunohistochemical and in situ hybridization level, despite the expression of CD79a (ie, the B-cell receptor/Ig-associated molecule). 61 Herein, we provide evidence for the first time that the lack of Ig is not dependent on defective expression of either Oct-2 and BOB.1 or PU.1. Oct-2 and BOB.1 belong to a group of transcription factors, which constitute a novel class of immunohistochemical markers and whose expression correlates with cell lineage and/or the stage of cell differentiation. In particular, it has been reported that the Oct-2, Oct-1, and BOB.1/OCA-B transcription factors are required for GC formation and Ig production: 62,63 their down-regulation in Reed-Sternberg cells of cHL 64,65 has been proposed as a possible explanation for the lack of Ig expression in these cells. 65 PU.1 is a tissue-restricted transcription regulator that is basic for the development of both B-lymphocytes and macrophages. 66-68 Moreover, it binds to a large number of promoters and regulates the expression of genes required for terminal B-cell differentiation, including those encoding for Ig light and heavy chains, RAG-1, CD19, and J-chain. 68,69 Recent studies have shown that PU.1 is not detectable in cultured and primary Reed-Sternberg cells of cHL: along with the absence of Oct-2 and BOB.1, this can contribute to defective Ig gene expression in cHL. 56 Notably, the mechanisms that can explain the lack of Ig production in most cHL cases do not pertain PMBL: in fact, neoplastic cells regularly express Oct-2, BOB.1, and PU.1. The alternative mechanism of defective IgVH gene rearrangements (so-called crippling mutations), 40 which may lead to the lack of Ig synthesis, cannot be applied to our cases either. Thus, the cause of Ig negativity in PMBL remains still obscure and requires further studies.
Finally, the vast majority of our cases expressed Bcl-6 and/or MUM1/IRF4. In keeping with previous observations in diffuse large B-cell lymphoma, 42 these molecules were more often co-expressed by the same cells, an aberrant feature that does not correspond to the mutually exclusive pattern found in normal GCs. 42 In 21% of the cases, a clear-cut CD10 staining was also seen. These findings fit with those recently reported by de Leval and colleagues 34 by studying 19 PMBLs for Bcl-6 and CD10 expression. Although the percentage of Bcl-6+ cases was lower in our series (possibly because of technical reasons), the PMBL phenotype identified in both studies reflects that of GC cells, a concept that indeed contrasts with the assumption more often reported in the literature that the tumor derives from virgin B cells. 32
Because of the histogenetic debate and the results of our phenotypic studies, we decided to focus on the status of IgVH genes and BCL-6 genes. With the exception of a very recent report 36 and on the basis of the results obtained in a small series of cases, it had been stated that the tumor might derive from virgin B cells. 25,30,32 Our results unequivocally suggest a GC-cell derivation for a fraction of PMBLs. In fact, somatic mutations of functioning IgVH gene are detectable in ∼50% of our amplifiable cases. The latter results are in keeping with a recent publication of Laithäuser and colleagues, 35 who reported isotype-switched Ig genes with a high load of somatic hypermutations in 13 PMBLs. Certainly, the efficiency of the technique applied by the latter authors was higher than the one we used, because they analyzed frozen samples instead of routinely processed tissue. Nevertheless, significant analogies do occur between the two studies, because in both IgVH mutations were functioning. As expected, PMBL carrying somatically mutated IgVH genes also harbored BCL-6 mutations in a fraction of cases. Curiously, though, a substantial proportion of PMBL appeared to show BCL-6 mutations in the absence of IgVH mutations. Occurrence of BCL-6 mutations in the absence of IgVH mutations has also been detected in examples of B-cell chronic lymphocytic leukemia, and has been taken to suggest that diverse pathways operate on BCL-6 and IgVH genes, at least in certain biological contexts. 70 On these basis, the sole occurrence of BCL-6 mutations in a given tumor cannot be taken as a marker of GC-derivation in PMBL.
In summary, the present study provides indication that at least a fraction of PMBL is derived from activated GC-related B-cells. This is based on the detection of IgVH gene and BCL-6 somatic mutations, as well as the expression of peculiar molecules, such as CD10, Bcl-6, and MUM1/IRF4. Taking into consideration the results of these molecules, one might hypothesize that the neoplastic cells of GC-related PMBLs correspond to elements that have already undergone the GC selection and are nearby to exit the GC. This might explain the partial loss of CD10 and the variable expression of both Bcl-6 and MUM1/IRF4. The latter antigen in fact starts to be synthesized in the light zone of the GC by cells that have already overcome the GC selection and are going to mature to plasma cells or memory B cells. 42 Remarkably, these transient cells may acquire CD30 antigen expression, as often observed at the periphery of reactive GCs. 71 This plausible hypothesis, however, does not completely solve the histogenetic problem because some tumors lacked both somatic mutations and GC-related protein expression. The latter results might be because of technical artifacts that cannot be completely excluded in our retrospective series, whose material was not treated according to standardized procedures and often remained stored for many years in institutional archives. Alternatively, these germ-line tumors might be derived from virgin B cells, undergoing activation and blastic transformation during the initial phases of the primary immune response. 72 More studies are necessary to clarify this issue.
Acknowledgments
We thank Prof. D.Y. Mason and Prof. H. Stein for kindly providing the monoclonal antibodies JCB117/CD79a and Ber-H2/CD30, respectively; and Dr. Simonetta Poggi, Miss Claudia Valli, Mr. Luigi Chilli, and Miss Federica Sandri for their skilled technical assistance.
Footnotes
Address reprint requests to Professor Stefano A. Pileri, III Cattedra di Anatomia Patologica, Istituto di Ematologia ed Oncologia Medica “L. e A. Seràgnoli,” Università di Bologna, Policlinico S. Orsola, Via Massarenti 9-40138 Bologna, Italy. E-mail: pileri@almadns.unibo.it.
Supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC, Milan; to S. A. P. and B. F.), the Associazione Bolognese per lo Studio dei Tumori Ematologici (ABSTE, Bologna; to S. A. P.), the Fondazione PPG Ferrero (Alba, Italy; to G. G.), and Progetto Strategico Oncologia, CNR-MIUR (Rome; to G. G.).
References
- 1.Lichtenstein A, Levine A, Taylor CR, Boswell W, Rossamn S, Feinstein DI, Lukes RJ: Primary mediastinal lymphoma in adults. Am J Med 1980, 68:509-514 [DOI] [PubMed] [Google Scholar]
- 2.Harris NL, Jaffe ES, Stein H, Banks PM, Chan JK, Cleary ML, Delsol G, De Wolf-Peeters C, Falini B, Gatter KC: A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood 1994, 84:1316-1392 [PubMed] [Google Scholar]
- 3.Jaffe ES, Harris NL, Stein H, Vardiman JW: Tumours of Haematopoietic and Lymphoid Tissues. Jaffe ES Harris NL Stein H Vardiman MJ eds. 2001:pp 175-176 IARC Press, Lyon
- 4.Addis BJ, Isaacson PG: Large cell lymphoma of the mediastinum: a B-cell tumor of probable thymic origin. Histopathology 1986, 10:379-390 [DOI] [PubMed] [Google Scholar]
- 5.Menestrina F, Chilosi M, Bonetti F, Lestani M, Scarpa A, Novelli P, Doglioni C, Todeschini G, Ambrosetti A, Fiore-Donati L: Mediastinal large-cell lymphoma of B-type, with sclerosis: histopathological and immunohistochemical study of eight cases. Histopathology 1986, 10:589-600 [DOI] [PubMed] [Google Scholar]
- 6.Perrone T, Frizzera G, Rosai J: Mediastinal diffuse large cell lymphoma with sclerosis. Am J Surg Pathol 1986, 10:176-191 [DOI] [PubMed] [Google Scholar]
- 7.Isaacson PG, Norton AJ, Addis BJ: The human thymus contains a novel population of B lymphocytes. Lancet 1987, 2:1488-1491 [DOI] [PubMed] [Google Scholar]
- 8.Möller P, Moldenhauer G, Momburg F, Lammler B, Eberlein-Gonska M, Kiesel S, Dorken B: Mediastinal lymphoma of clear cell type is tumor corresponding to terminal steps of B-cell differentiation. Blood 1987, 59:1087-1095 [PubMed] [Google Scholar]
- 9.Scarpa A, Bonetti F, Menestrina F, Chilosi M, Lestani M, Bovolenta C, Zamboni G, Fiore-Donati L: Mediastinal large cell lymphoma with sclerosis: genotypic analysis establishes its B nature. Virchows Arch A Pathol Anat Histopathol 1987, 412:17-21 [DOI] [PubMed] [Google Scholar]
- 10.Aisenberg AC, Wilkes BM, Jacobson JO: The bcl-2 gene is rearranged in many diffuse B-cell lymphomas. Blood 1988, 71:969-972 [PubMed] [Google Scholar]
- 11.Hofmann WJ, Momburg F, Möller P: Thymic medullary cells expressing B lymphocyte antigens. Hum Pathol 1988, 19:1280-1287 [DOI] [PubMed] [Google Scholar]
- 12.Jacobson JO, Aisenberg AC, Lamarre L, Willett CG, Linggod RM, Miketic LM, Harris NL: Mediastinal large cell lymphoma: an uncommon subset of adult lymphoma curable with combined modality therapy. Cancer 1988, 62:1893-1898 [DOI] [PubMed] [Google Scholar]
- 13.Falini B, Pileri S, Martelli MF: Histological and immunohistological analysis of human lymphoma. Crit Rev Oncol Hematol 1989, 9:351-419 [DOI] [PubMed] [Google Scholar]
- 14.Lamarre L, Jacobson JO, Aisenberg AC, Harris NL: Primary large cell lymphoma of the mediastinum: a histologic and immunophenotypic study of 29 cases. Am J Surg Pathol 1989, 13:730-739 [DOI] [PubMed] [Google Scholar]
- 15.Huret JL, Brizard A, Babin P, Schoenwald M, Briault S, Guilhot F, Tanzer J: Translocation (8;9)(q12;p21): a new non-random rearrangement in lymphoid malignancies. Cancer Genet Cytogenet 1990, 45:125-129 [DOI] [PubMed] [Google Scholar]
- 16.Al-Sharabati M, Chittal S, Duga-Neulat I, Laurent G, Mazerolles C, al-Saati T, Brousset P, Delsol G: Primary anterior mediastinal B-cell lymphoma. Cancer 1991, 67:2579-2587 [DOI] [PubMed] [Google Scholar]
- 17.Scarpa A, Borgato L, Chilosi M, Capelli P, Menestrina F, Bonetti F, Zamboni G, Pizzolo G, Hirohashi S, Fiore-Donati L: Evidence of c-myc gene abnormalities in mediastinal large B-cell lymphoma of young adult age. Blood 1991, 78:780-788 [PubMed] [Google Scholar]
- 18.Eichelmann A, Koretz K, Mechtersheimer G, Möller P: Adhesion receptor profile of thymic B-cell lymphoma. Am J Pathol 1992, 141:729-741 [PMC free article] [PubMed] [Google Scholar]
- 19.Lavabre-Bertrand T, Donadio D, Fegueux N, Jessueld D, Taib J, Charlier D, Rousset T, Emberger JM, Baldet P, Navarro M: A study of 15 cases of primary mediastinal lymphoma of B-cell type. Cancer 1992, 50:2561-2566 [DOI] [PubMed] [Google Scholar]
- 20.Lazzarino M, Orlandi E, Paulli M, Boveri E, Morra E, Brusolino E, Kindl S, Rosso R, Astori C, Buonanno MC: Primary mediastinal B-cell lymphoma with sclerosis: an aggressive tumor with distinctive clinical and pathologic features. J Clin Oncol 1993, 11:2306-2313 [DOI] [PubMed] [Google Scholar]
- 21.Falini B, Venturi S, Martelli M, Santucci A, Pileri S, Pescarmona E, Giovannini M, Mazza P, Martelli MF, Pasqualucci L, Ballatori E, Guglielmi C, Amadori S, Poggi S, Sabattini E, Gherlinzoni F, Zinzani PL, Baroni CD, Mandelli F, Tura S: Mediastinal large B-cell lymphoma: clinical and immunohistological findings in 18 patients treated with different third generation regimens. Br J Haematol 1995, 89:780-789 [DOI] [PubMed] [Google Scholar]
- 22.Kanavaros P, Gaulard P, Charlotte F, Martin N, Ducos C, Lebezu M, Mason DY: Discordant expression of immunoglobulin and its associated molecule mb-1/CD79a is frequently found in mediastinal large B cell lymphomas. Am J Pathol 1995, 146:735-741 [PMC free article] [PubMed] [Google Scholar]
- 23.Cazals-Hatem D, Lepage E, Brice P, Ferrant A, d’Agay MF, Baumelou E, Briere J, Blanc M, Gaulard P, Biron P, Schlaifer D, Diebold J, Ardouin J: Primary mediastinal large B-cell lymphoma: a clinicopathologic study of 141 cases compared with 916 nonmediastinal large B-cell lymphomas—A GELA (“Group d’Etude des Lymphomes de l’Adulte”) study. Am J Surg Pathol 1996, 20:877-888 [DOI] [PubMed] [Google Scholar]
- 24.Joos S, Otano-Joos MI, Ziegler S, Bruderlein S, du Manoir S, Bentz M, Möller P, Lichter P: Primary mediastinal (thymic) B-cell lymphoma is characterized by gains of chromosomal material including 9p and amplification of the REL gene. Blood 1996, 87:1571-1578 [PubMed] [Google Scholar]
- 25.Tsang P, Cesarman E, Chadburn A, Liu YF, Knowles DM: Molecular characterization of primary mediastinal B cell lymphoma. Am J Pathol 1996, 148:2017-2025 [PMC free article] [PubMed] [Google Scholar]
- 26.Paulli M, Strater J, Gianelli U, Rousset MT, Gambacorta M, Orlandi E, Klersy C, Lavabre-Bertrand T, Morra E, Manegold C, Lazzarino M, Magrini U, Möller P: Mediastinal B-cell lymphoma: a study of its histomorphologic spectrum based on 109 cases. Hum Pathol 1999, 30:178-187 [DOI] [PubMed] [Google Scholar]
- 27.Abou-Elella AA, Weisenburger DD, Vose JM, Kollath JP, Lynch JC, Bast MA, Bierman PJ, Greiner TC, Chan WC, Armitage JO: Primary mediastinal large B-cell lymphoma: a clinico-pathologic study of 43 patients from the Nebraska lymphoma study group. J Clin Oncol 1999, 17:784-790 [DOI] [PubMed] [Google Scholar]
- 28.Higgins JP, Warnke RA: CD30 expression is common in mediastinal large B-cell lymphoma. Am J Clin Pathol 1999, 112:241-247 [DOI] [PubMed] [Google Scholar]
- 29.Copie-Bergman C, Gaulard P, Mauche-Chretien L, Briere J, Haioun C, Alonso MA, Romeo PH, Leroy K: The MAL gene is expressed in primary mediastinal large B-cell lymphoma. Blood 1999, 94:3567-3575 [PubMed] [Google Scholar]
- 30.Capello D, Vitolo U, Pasqualucci L, Quattrone S, Migliaretti G, Fassone L, Ariatti C, Vivenza D, Gloghini A, Pastore A, Lanza C, Nomdedeu J, Botto B, Freilone R, Bonaiuto D, Zagonel V, Gallo E, Palestro G, Saglio G, Dalla-Favera R, Carbone A, Gaidano G: Distribution and pattern of BCL-6 mutations throughout the spectrum of B-cell neoplasia. Blood 2000, 95:651-659 [PubMed] [Google Scholar]
- 31.Pileri SA, Ascani S, Sabattini E, Fraternali-Orcioni G, Poggi S, Piccioli M, Piccaluga PP, Gamberi B, Zinzani PL, Leoncini L, Falini B: The pathologist’s view point: part II—aggressive lymphomas. Haematologica 2000, 85:1285-1298 [PubMed] [Google Scholar]
- 32.Van Besien K, Kelta M, Bahaguna P: Primary mediastinal B-cell lymphoma: a review of pathology and management. J Clin Oncol 2001, 19:1855-1864 [DOI] [PubMed] [Google Scholar]
- 33.Riguad G, Moore PS, Taruscio D, Scardoni M, Montresor M, Menestrina F, Scarpa A: Alterations of chromosome arm 6p is characteristic of primary mediastinal B-cell lymphoma, as identified by genome-wide allelotyping. Genes Chromosom Cancer 2001, 31:191-195 [DOI] [PubMed] [Google Scholar]
- 34.De Leval L, Ferry JA, Falini B, Shipp M, Harris NL: Expression of bcl-6 and CD10 in primary mediastinal large B-cell lymphoma. Am J Surg Pathol 2001, 25:1277-1282 [DOI] [PubMed] [Google Scholar]
- 35.Laithäuser F, Bäuerle M, Huynh MQ, Möller P: Isotype-switched immunoglobulin genes with a high load of somatic hypermutation and lack of ongoing mutational activity are prevalent in mediastinal B-cell lymphoma. Blood 2001, 98:2762-2770 [DOI] [PubMed] [Google Scholar]
- 36.Palanisamy N, Abou-Elella AA, Chaganti SR, Houldworth J, Offit K, Louie DC, Terayu-Feldstein J, Cigudosa JC, Rao PH, Sanger WG, Weisenburger DD, Chaganti RS: Similar patterns of genetic alterations characterize primary mediastinal large-B-cell lymphoma and diffuse large-B-cell lymphoma. Genes Chromosom Cancer 2002, 33:114-122 [DOI] [PubMed] [Google Scholar]
- 37.Flenghi L, Bigerna B, Fizzotti M, Venturi S, Pasqualucci L, Pileri S, Ye BH, Gambacorta M, Pacini R, Baroni CD, Pescarmona E, Anagnostopoulos, Stein H, Asdrubali G, Martelli MF, Pelicci PG, Dalla-Favera R, Falini B: Monoclonal antibodies PG-B6a and PG-B6p recognize, respectively, a highly conserved and a formol-resistant epitope on the human BCL-6 protein amino-terminal region. Am J Pathol 1996, 148:1543-1553 [PMC free article] [PubMed] [Google Scholar]
- 38.Pasqualucci L, Migliazza A, Fracchiolla N, William C, Neri A, Baldini L, Chaganti RS, Klein U, Kuppers R, Rajewsky K, Dalla-Favera R: BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci USA 1998, 95:11816-11821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen HM, Peters A, Baron B, Zhu X, Storb U: Mutations of BCL-6 in normal B cells by the process of somatic hypermutation of Ig genes. Science 1998, 280:1750-1752 [DOI] [PubMed] [Google Scholar]
- 40.Kuppers R, Klein U, Hansmann ML, Rajewsky K: Cellular origin of human B-cell lymphomas. N Engl J Med 1999, 341:1520-1529 [DOI] [PubMed] [Google Scholar]
- 41.Gaidano G, Carbone A: MUM-1: a step ahead toward the understanding of lymphoma histogenesis. Leukemia 2000, 4:1209-1215 [DOI] [PubMed] [Google Scholar]
- 42.Falini B, Fizzotti M, Pucciarini A, Bigerna B, Marafioti T, Gambacorta M, Pacini R, Alunni C, Natali-Tanci L, Ugolini B, Sebastiani C, Cattoretti G, Pileri S, Dalla-Favera R, Stein H: A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF-4 protein in a subset of germinal center B cells, plasma cells and activated T-cells. Blood 2000, 95:2084-2092 [PubMed] [Google Scholar]
- 43.Alonso MA, Barton DE, Francke U: Assignment of the T cell differentiation gene MAL to human chromosome 2, region cen-q13. Immunogenetics 1988, 27:91-95 [DOI] [PubMed] [Google Scholar]
- 44.Millán J, Alonso MA: MAL, a novel integral membrane protein of human T lymphocytes, associates with glycosylphosphatidylinositol-anchored proteins and Src-like tyrosine kinases. Eur J Immunol 1998, 28:3675-3684 [DOI] [PubMed] [Google Scholar]
- 45.Alonso MA, Millán J: The role of lipid rafts in signaling and membrane trafficking in T lymphocytes. J Cell Sci 2001, 114:3957-3965 [DOI] [PubMed] [Google Scholar]
- 46.Millán J, Montoya MC, Sancho D, Sánchez-Madrid F, Alonso MA: Lipid rafts mediate biosynthetic transport to the T cell uropod subdomain and are necessary for uropod integrity and function. Blood 2002, 99:978-984 [DOI] [PubMed] [Google Scholar]
- 47.Pileri S, Falini B, Sabattini E, Bigerna B, Gherlinzoni F, Tazzari PL: Immunohistochemistry of malignant lymphomas. Advantages and limitations of the new monoclonal antibodies working in paraffin sections. Haematologica 1991, 76:226-234 [PubMed] [Google Scholar]
- 48.Pileri SA, Sabattini E: A rational approach to immunohistochemical analysis of malignant lymphomas on paraffin wax sections. J Clin Pathol 1997, 50:2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sabattini E, Bisgaard K, Ascani S, Poggi S, Piccioli M, Ceccarelli C, Pieri F, Fraternali-Orcioni G, Pileri SA: Envision Plus: a new immunohistochemical method of choice for diagnostics and research. Critical comparison with the APAAP, ChemMate CSA, LABC, and SABC techniques. J Clin Pathol 1998, 51:506-511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pileri SA, Roncador G, Ceccarelli C, Piccioli M, Briskomatis A, Sabattini E, Ascani S, Santini D, Piccaluga PP, Leone O, Damiani S, Ercolessi C, Sandri F, Pieri F, Leoncini L, Falini B: Antigen retrieval techniques in immunohistochemistry: comparison of different methods. J Pathol 1997, 183:116-123 [DOI] [PubMed] [Google Scholar]
- 51.Kuppers R, Rajewsky K, Hansmann M-L: Diffuse large cell lymphomas are derived from mature B cells carrying V region genes with a high load of somatic mutation and evidence of selection for antibody expression. Eur J Immunol 1997, 27:1398-1405 [DOI] [PubMed] [Google Scholar]
- 52.Fais F, Gaidano G, Capello D, Gloghini A, Ghiotto F, Roncella S, Carbone A, Chiorazzi N, Ferrarini M: Immunoglobulin V region gene use and structure suggest antigen selection in AIDS-related primary effusion lymphoma. Leukemia 1999, 13:1093-1099 [DOI] [PubMed] [Google Scholar]
- 53.Kaplan EL, Meier P: Non-parametric estimation from incomplete observation. J Am Stat Assoc 1958, 53:457-481 [Google Scholar]
- 54.Peto R, Pike MC, Armitage P, Breslow NE, Cox DR, Howard SW, Mantel N, McPherson K, Peto J, Smith PG: Design and analysis of randomized clinical trials requiring prolonged observation of each patient. II. Analysis and examples. Br J Cancer 1977, 35:1-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cox DR: Regression models and life-tables (with discussion). J Royal Statist Soc 1972, 34:187-220 [Google Scholar]
- 56.Jundt F, Kley K, Anagnostopoulos I, Schulze Pröbsting K, Greiner A, Mathas S, Scheidereit C, Wirth T, Stein H, Dörken B: Loss of PU.1 expression is associated with defective immunoglobulin transcription in Hodgkin and Reed-Sternberg cells of classical Hodgkin disease. Blood 2002, 99:3060-3062 [DOI] [PubMed] [Google Scholar]
- 57.Copie-Bergman C, Plonquet A, Alonso M, Boulland ML, Marquet J, Divine M, Möller P, Leroy K, Gaulard P: MAL expression in lymphoid cells: further evidence for MAL as a distinct molecular marker of primary mediastinal large B-cell lymphomas. J Clin Pathol 2002, 55(Suppl 1):A16. [DOI] [PubMed] [Google Scholar]
- 58.Falini B, Pileri S, Pizzolo G, Dürkop H, Flenghi L, Stirpe F, Martelli MF, Stein H: CD30 (Ki-1) molecule: a new cytokine receptor of the tumor necrosis factor receptor superfamily as a tool for diagnosis and immunotherapy. Blood 1995, 85:1-14 [PubMed] [Google Scholar]
- 59.Lamarre L, Jacobson JO, Aisenberg AC, Harris NL: Primary large cell lymphoma of the mediastinum: a histologic and immunophenotypic study of 29 cases. Am J Surg Pathol 1989, 13:730-739 [DOI] [PubMed] [Google Scholar]
- 60.Rodriguez J, Pugh WC, Romaguera JE, Cabanillas F: Primary mediastinal large cell lymphoma. Hematol Oncol 1994, 94:175-192 [DOI] [PubMed] [Google Scholar]
- 61.Mason DY, Cordell JL, Brown MH, Borst J, Jones M, Pulford K, Jaffe E, Ralfkiaer E, Dallenbach F, Stein H, Pileri S, Gatter KC: CD79a: a novel marker for B-cell neoplasms in routinely processed tissue samples. Blood 1995, 86:1453-1459 [PubMed] [Google Scholar]
- 62.Schubart DB, Rolink A, Kosco-Vilbois MH, Botteri F, Matthias P: B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature 1996, 383:538-542 [DOI] [PubMed] [Google Scholar]
- 63.Qin XF, Reichlin A, Luo Y, Roeder RG, Nussenzweig MC: OCA-B integrates B cell antigen receptor-, CD40L- and IL 4-mediated signals for the germinal center pathway of B cell development. EMBO J 1998, 17:5066-5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Re D, Muschen M, Ahmadi T, Wickenhauser C, Staratschek-Jox A, Holtick U, Diehl V, Wolf J: Oct-2 and Bob-1 deficiency in Hodgkin and Reed Sternberg cells. Cancer Res 2001, 61:2080-2084 [PubMed] [Google Scholar]
- 65.Stein H, Marafioti T, Foss HD, Laumen H, Hummel M, Anagnostopoulos I, Wirth T, Demel G, Falini B: Down-regulation of BOB. 1/OBF. 1 and Oct2 in classical Hodgkin disease but not in lymphocyte predominant Hodgkin disease correlates with immunoglobulin transcription. Blood 2001, 97:496-501 [DOI] [PubMed] [Google Scholar]
- 66.Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA: The macrophage and B cell-specific transcription factor PU.1 is related to ets oncogene. Cell 1990, 61:113-124 [DOI] [PubMed] [Google Scholar]
- 67.Henderson A, Calame K: Transcriptional regulation during B cell development. Annu Rev Immunol 1998, 16:163-200 [DOI] [PubMed] [Google Scholar]
- 68.DeKoter RP, Singh H: Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 2000, 288:1439-1441 [DOI] [PubMed] [Google Scholar]
- 69.Schwarzenbach H, Newell JW, Matthias P: Involvement of Ets family factor PU.1 in the activation of immunoglobulin promoters. J Biol Chem 1993, 27:898-907 [DOI] [PubMed] [Google Scholar]
- 70.Sahota SS, Davs Z, Hamblin TJ, Stevenson FK: Somatic mutation of bcl-6 genes can occur in the absence of VH mutations in chronic lymphocytic leukemia. Blood 2000, 95:3534-3540 [PubMed] [Google Scholar]
- 71.Pileri S, Rivano MT, Raise E, Gualandi G, Gobbi M, Martuzzi M, Gritti FM, Gerdes J, Stein H: The value of lymph node biopsy in patients with the acquired immunodeficiency syndrome (AIDS) and the AIDS-related complex (ARC): a morphological and immunohistochemical study of 90 cases. Histopathology 1986, 10:1107-1129 [DOI] [PubMed] [Google Scholar]
- 72.Knowles D: Neoplastic Hematopathology. Knowles DM eds. 2001:pp 263-298 Williams & Wilkins, Philadelphia