Abstract
Mounting evidence points to a role for CD4+ T-helper (Th) cell activities in controlling human immunodeficiency virus type 1 (HIV-1) infection. To determine the induction and evolution of Th responses following acute infection, we prospectively analyzed Env- and Gag-specific Th responses longitudinally for 92 patients with acute (n = 28) or early (n = 64) HIV-1 infection (median, 55 days postinfection [DPI]). The probability of detecting HIV-1-specific lymphoproliferative responses was remarkably low, and when present, the responses were more likely to be Gag specific than Env specific (16 versus 5%). Env-specific responses were significantly more common in patients presenting at <30 DPI than in those presenting at 30 to 365 DPI (21 versus 0.5%, P = 0.001). By contrast, Gag-specific responses occurred with similar frequencies among subjects presenting at <30 DPI and 30 to 365 DPI (13 versus 17%, P = 0.6). After treatment, and regardless of the duration of infection before therapy, Gag-specific Th responses predominated. Furthermore, some acutely infected subjects lost detectable Env-specific Th proliferative responses, which failed to reemerge upon treatment. Detailed analysis for one such subject revealed Env-specific lymphoproliferation at 11 DPI but no detectable Env-specific lymphoproliferation or ex vivo gamma interferon (IFN-γ) secretion at multiple subsequent time points. Env-specific CD4+ T-cell clones from 11 DPI recognized six epitopes in both conserved and variable regions within gp120 and gp41, exhibited major histocompatibility complex-restricted cytotoxicity, and secreted high levels of antiviral cytokines. T-cell receptor clonal transcript analyses and autologous virus sequencing revealed that Th cells induced during acute infection were maintained and there were no Th escape mutations. Subsequent analysis for this subject and six of seven others revealed detectable IFN-γ-secreting cells, but only following in vitro gp160 stimulation. In summary, we conclude that Env-specific Th responses are elicited very early in acute infection and may precede Gag-specific responses. The inability to detect Env-specific Th responses over time and despite antiretroviral therapy may reflect low frequencies and impaired proliferative capacity, and viral escape is not necessary for this to occur.
Helper activities of antigen-specific CD4+ T cells mediate control of many viral infections (2, 5, 21), and they appear critical in maintaining human immunodeficiency virus type 1 (HIV-1) suppression. This is best exemplified by the frequent detection of HIV-1 Gag-specific CD4+ T-helper (Th) cells in untreated persons termed long-term nonprogressors (LTNP), who are infected for more than 12 years but do not develop HIV-1 disease (27). By contrast, most untreated persons with recent seroconversion or with chronic HIV-1 infection have low to undetectable levels of HIV-1-specific Th cells, particularly when measured in vitro by antigen-induced lymphoproliferation (19, 24). However, recent studies indicate that Th responses are enhanced in patients who substantially suppress viral replication with use of antiretroviral therapy (ART). This occurs predominantly in patients who initiate treatment soon after acquisition of infection (19, 27) and in those with chronic infection who receive prolonged ART (4, 11). The newly identified HIV-1-specific Th cells following therapy may represent clonotypes that were induced with initial infection but are present at a frequency too low to detect, or they may arise from naïve CD4+ T cells as a result of immune reconstitution following suppression of viremia. One goal of this study was to clarify this issue by monitoring longitudinally patients with primary (acute or early) HIV-1 infection.
The Th responses found to occur in LTNP and treated patients are predominantly directed against Gag epitopes (19, 27). In addition, persons who develop Gag-specific HLA-DR13-restricted responses are more likely to maintain viral suppression (20). However, little is known about the role of Th responses to epitopes expressed by other HIV-1 genes and how these responses evolve in early infection and following treatment. Such findings could support efforts to improve immunity against HIV-1 infection by augmenting the Th responses.
We were particularly interested in investigating Env-specific Th responses for several reasons. Helper epitopes have been identified within the Env protein (3, 14, 17), and CD4+ T-cell lines and clones recognizing Env epitopes can exhibit cytolytic activities (23). Also, we observed that in persons presenting with acute HIV-1 infection prior to seroconversion, Env-specific responses were often detected. By contrast, Env-specific responses were less frequently detected later in infection (26).
In this study, we explored several hypotheses to understand the induction and evolution of the HIV-1 Env-specific helper response following acute infection. First, we sought to determine whether Env-specific responses preferentially emerge early in acute infection and whether delay in treatment leads to greater loss of these responses. Secondly, we explored the possibilities that the autologous viral strains may mutate within the Th epitopic region from immune pressure or that the env sequence of the wild-type strains may be considerably divergent from the env sequence expressed by the proteins used in the assays. It is well recognized that the env gene is highly variable among infected individuals and undergoes marked intraindividual diversification over the course of infection. Longitudinal studies of the evolution of the C2-V5 region of the env gene in HIV-1-infected subjects reveal a divergence rate of about 1% per year from the founder strain (29). Lastly, we considered that the detection of Env-specific Th cells can be limited by the sensitivity of the assay employed, if the memory precursor frequencies are low. Thus, responses were examined by several complementary methods (i.e., lymphoproliferation-assay, gamma interferon (IFN-γ) secretion by enzyme-linked immunospot (ELISPOT) assay, and intracellular cytokine staining (ICS) by flow cytometry).
MATERIALS AND METHODS
Study population.
Subjects with HIV-1 infection were recruited and enrolled at the University of Washington (UW) Primary Infection Clinic (PIC). The UW and Fred Hutchinson Cancer Research Center (FHCRC) human subjects review boards approved the study, and volunteers provided written consent prior to participation. The duration of infection was defined as the time elapsed since the onset of clinical signs and/or symptoms suggestive of acute retroviral syndrome. Determination of the exact time of exposure to HIV-1 and acquisition of infection is frequently not possible, and the time interval from exposure to appearance of symptoms can vary. Thus, the duration of infection was defined as the time since the onset of clinical signs and/or symptoms suggestive of acute retroviral syndrome. If symptoms were absent, the duration of infection was estimated as the midpoint between the last previous documented negative enzyme immunoassay and the recent positive confirmatory test. Some patients at various time points following diagnosis elected to receive combination ART with nucleoside reverse transcriptase inhibitors and either a protease inhibitor or a nonnucleoside reverse transcriptase inhibitor. Patients underwent clinical evaluation and venipuncture for virological studies weekly for the first month, then every 4 weeks for 1 year, and then every 3 months thereafter. Anticoagulated blood for immunological studies was obtained 1 or 2 times prior to treatment and then approximately every 1 to 3 months thereafter.
Virological and T-cell subset analyses.
The amount of HIV-1 RNA in plasma was determined by quantitative branched-chain DNA (bDNA; Chiron, Emeryville, Calif.) and ultrasensitive reverse transcriptase-PCR (RT-PCR) (Roche Molecular Systems, Branchburg, N.J.) assays. Levels were expressed as copies/milliliter, and the lower levels of sensitivity were 500 copies/ml (for the bDNA assay) and 50 copies/ml (for the RT-PCR assay) (7). We report the viral levels measured by the bDNA assay that were >500 copies/ml and those measured by the ultrasensitive RT-PCR assay when the result by bDNA assay was ≤500 copies/ml. Absolute blood CD4+ T-cell count was measured by consensus flow cytometry methodology.
HLA typing.
Class II alleles at the DRB1 and DQB1 loci were identified by PCR using sequence-specific primers (SSP) (Micro SSP DNA typing kit; One Lambda, Inc., Canoga Park, Calif.). The Puget Sound Blood Center Immunogenetics Laboratory, Seattle, Wash., performed these studies according to the manufacturer's instructions.
LPA.
Lymphocyte proliferation assays (LPA) were performed as previously described (19). In brief, fresh peripheral blood mononuclear cells (PBMC) were plated at 105 cells/well in quadruplicate wells containing no antigen, 5 μg of baculovirus-expressed recombinant HIV-1MN gp160 and HIV-1LAI p24 per ml, and 0.15 μg of baculovirus control protein (Protein Sciences, Meriden, Conn.) per ml. On day 6, cells were pulsed with 1 μCi of [3H]thymidine (Perkin-Elmer Life Sciences, Boston, Mass.), and they were washed and harvested 18 h later. The stimulation index (S.I.) was calculated as the mean counts per minute of stimulated cultures divided by mean counts per minute of unstimulated cultures. Responses with S.I. >5.0 were considered positive, based on comparisons in our laboratory with in vitro lymphoproliferation to these proteins by PBMC from HIV-1- uninfected controls. The net counts per min (mean counts per min in stimulated cultures minus mean counts per min in unstimulated cultures) for all positive responses was >1,000 cpm.
Analysis of cytokine production by ELISA.
PBMC (2 million/ml) were incubated in 24-well plates in the presence of either HIV-1MN gp160 (5 μg/ml), baculovirus control protein (0.15 μg/ml), or peptides (2 μg/ml). The secretion levels of interleukin-4 (IL-4), IL- 5, and IL-10 in cell culture supernatants harvested after 3 days and those of IFN-γ in cell culture supernatants harvested after 5 days were measured by enzyme-linked immunosorbent assay (ELISA) (Woburn, Boston, Mass.), according to the manufacturer's instructions. The optimal time points for the analysis of individual cytokines were determined in experiments measuring the kinetics of their production and were 5 days for IFN-γ and 3 days for IL-4, IL-5, and IL-10. All samples, standards, and controls were run in duplicate, and the lower limit of detection of the assays was 0.4 pg/ml. A level of cytokine secretion in antigen-stimulated cultures exceeding two times the unstimulated cultures was considered a positive response.
Synthetic HIV-1 peptides.
Synthetic overlapping peptides corresponding to gene products from the HIV-1 subtype B (MN) were obtained through the NIH AIDS Research and Reference Reagent Program (Bethesda, Md.). Most were 20 amino acids in length, with 10-amino-acid overlaps between sequential peptides. The peptides were reconstituted at a concentration of 2 mg/ml in 100% dimethyl sulfoxide (Sigma Chemical Co., St. Louis, Mo.) and were used at a final concentration of 2 μg/ml, unless stated otherwise. The final concentration of dimethyl sulfoxide never exceeded 1%.
CD4+ T-cell cloning, epitope mapping, and CTL activity.
CD4+ T-cell clones were derived as described previously (33) by bulk culture with HIV-1MN gp160 at 5 μg/ml. On day 14, positively selected CD4+ T cells were plated at 0.5 cells/well in 96-well plates with gamma-irradiated allogeneic PBMC and Epstein-Barr virus-transformed B-lymphoblastoid cell lines (B-LCL) as feeders, OKT3 (Ortho Diagnostic System, Raritan, N.J.), and recombinant IL-2. After 2 weeks, cells demonstrating growth and Env-specific proliferation in a [3H]thymidine incorporation assay were further expanded into CD4+ T-cell clones. Epitope mapping and specific HLA restriction analyses were performed by using autologous or HLA- mismatched allogeneic B-LCL pulsed with peptides as stimulator cells in proliferative and ELISPOT assays as previously described (20).
Clones were screened for HIV-1-specific cytotoxic T-lymphocyte (CTL) activity by 51Cr release assay (19) by using autologous B-LCL pulsed with the whole protein or the recognized peptide as targets. Results were expressed as percent specific lysis (23). Spontaneous lysis was less than 20% of maximal lysis.
Antibody blocking of lymphoproliferation.
Monoclonal antibodies (MAbs) recognizing HLA DR (L243), HLA DP (B7/221), and HLA DQ (SPV-L3) framework determinants were used to inhibit major histocompatibility complex class II-restricted lymphoproliferation as previously described (15). The supernatants were generated from hybridoma cell lines secreting L243 and B7/221 (American Type Culture Collection, Rockville, Md.), and SPV-L3 (provided by H. Yssel, DNAX Research Institute, Palo Alto, Calif.) and used at a 1:4 final dilution. These concentrations inhibit proliferation of CD4+ T-cell clones restricted at relevant class II loci by >80% and at irrelevant HLA by <15% (15).
IFN-γ ELISPOT assays.
ELISPOT assays were performed to detect HIV-1-specific IFN-γ-secreting cells as previously described (20). In standard assays, CD8+ T-cell-depleted PBMC were plated (105 cells/well) in 96-well hydrophobic polyvinylidene difluoride-backed plates (Millipore, Bedford, Mass.) precoated with 50 μl of 10 μg of anti-IFN-γ MAb (1-D1K, mouse immunoglobulin G1; Mabtech, Nacka, Sweden) per ml. In stimulated ELISPOT assays, antigen-specific cell lines were first generated from previously cryopreserved PBMC cultured in the presence of 5 μg of HIV-1MN gp160 per ml for 10 days and then were depleted of CD8+ T cells and plated as described above. HIV-1-specific peptides and control peptides (a pool of five irrelevant peptides derived from actin and a highly conserved region of HLA class I α-chain precursor) were added to the wells at a final concentration of 2 μg/ml. Plates were incubated overnight at 37°C in 5% CO2 and then developed. Spot-forming cells (SFC) were counted by using the Immunospot (Cellular Technology Ltd., Cleveland, Ohio) optical reader and are expressed per 105 input cells. The number of Env-specific IFN-γ-secreting cells was calculated by subtracting the SFC in the negative-control wells from those in the antigen-stimulated wells. The positive and negative ELISPOT assay responses were determined by using a combination of standard statistical methods. Specifically, the numbers of SFC per well were assumed to follow a simple Poisson model, and the criterion for a positive assay was based on an exact binomial test statistic. Additionally, the number of SFC was always >20 per 105 cells for a response designated positive. Twelve HIV-1-uninfected donors served as negative controls, and no false positives were observed for either the gp160 protein or the peptide-stimulated ELISPOT assays based on these criteria.
Intracellular immunofluorescent staining for IFN-γ production.
ICS for IFN-γ-producing antigen-specific CD4+ T cells was performed as previously described (6, 31, 32) with modifications. PBMC (2 million/ml) were incubated with HIV-1MN gp160 (final concentration, 5 μg/ml) or peptide (final concentration, 2 μg/ml) in the presence of anti-CD49d (Beckman Coulter, Miami, Fla.) and anti- CD28 (BD Biosciences, San Jose, Calif.) purified MAbs (final concentration, 1.0 μg/ml) at 37°C. At 2 h brefeldin A (Sigma) was added (final concentration, 10 μg/ml), and incubation continued for an additional 4 h. The cells were fixed and permeabilized and then stained with fluorochrome-labeled MAbs (intracellular IFN-γ fluorescein isothiocyanate, CD69 PE, CD4 PerCP-Cy5.5; BD Biosciences) at room temperature for 30 min, washed, and fixed in 2% paraformaldehyde. Analysis was performed within 24 hr with a FACScalibur with Cell Quest software (BD Biosciences).
Class II peptide binding assays.
Peptide-binding assays were performed as described previously (6, 30, 31). Specifically, purified human HLA class II molecules (5 to 500 nM) were incubated with various unlabeled peptide inhibitors and 1 to 10 nM 125I-radiolabeled probe peptide for 48 h in phosphate-buffered saline containing 0.05% Nonidet P-40 in the presence of a protease inhibitor cocktail. Class II peptide complexes were separated from free peptide by gel filtration on TSK200 columns (catalog no. 16215; TosoHaas, Montgomeryville, Penn.), and the fraction of bound peptide was calculated as previously described (30). The titer of each DR preparation was determined in the presence of a fixed amount of the appropriate 125I-radiolabeled peptide to determine the concentration of class II molecules necessary to bind 10 to 20% of the total radioactivity. Inhibitor peptides were typically tested at concentrations ranging from 120 μg/ml to 1.2 ng/ml in two to four completely independent experiments. Under conditions where the concentration of label is less than that of MHC and the 50% inhibitory concentration (IC50) is greater than or equal to the concentration MHC, the measured IC50 values are reasonable approximations of true Kd values. Peptides were classified as binders for each HLA-DR molecule for which the binding capacity was ≤1,000 nM.
TCR Vβ usage by multiplex PCR, TCR sequencing, and longitudinal analysis of clone transcripts.
To assess T-cell receptor (TCR) Vβ usage, total RNA (2 μg) from CD4+ T-cell clones (2 × 106 cells) was reverse transcribed, and five aliquots of the cDNA were amplified for 30 PCR cycles with five multiplex primer sets consisting of a total of 25 5′Vβ-specific sense primers and the 3′Cβ- specific antisense primer (1). The amplified products were electrophoresed on 2% agarose gels. Direct sequencing of the PCR product was performed by using the Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems Inc., Foster City, Calif.) and the specific 5′Vβ and 3′Cβ primers. Once characterized, the clone was tracked in stored or fresh specimens over time using total RNA from 2 million CD4+ T-cell-enriched PBMC. Primers specific for the Vβ in the originally isolated clone and Cβ were utilized in the PCRs. Two elongations of 10 cycles were performed in separate tubes, initiated by either the fluorescent dye-labeled Jβ or the clonotype-specific primer. The two reaction products from each amplified DNA were mixed and loaded in a lane and electrophoresed under denaturing conditions on a 373A DNA sequencer (Applied Biosystems), and the fluorescent profiles were analyzed by GeneScan software (25) through the FHCRC Shared Resources Facilities.
Viral diversity at Th epitopes.
The level of amino acid variation within each of the epitopes was analyzed by examining 244 full-length HIV-1 subtype B env sequences obtained from the Los Alamos HIV sequence database. The putative amino acid sequences derived from nucleic acid sequences were aligned and analyzed by using the programs Clustal W (32) and Genetic Data Environment (GDE) (United Kingdom Human Genome Mapping Project Resource Center; http://www.hgmp.mrc.ac.uk/). Amino acid variability was assessed by using the computer program Seqvar, which was kindly provided by Andrew Rambaut (University of Oxford, Oxford, United Kingdom). The program estimates the level of variability at each amino acid position in the sequence alignment by using Shannon entropy. To examine the relative levels of variability over the entire gp160 and to highlight the location of the Th epitopes, a moving average of the estimated variability over three consecutive amino acid positions was plotted. To examine the variability at each amino acid position within the six Env Th epitopes, the site-by-site variability was plotted.
Virus DNA sequencing.
CD14+ monocytes, resting CD4+ T lymphocytes, and activated CD4+ T lymphocytes were purified from PBMC by negative selection, using magnetic bead depletion followed by fluorescent-activated cell sorting. Genomic DNA was isolated from purified cells by using the QIAamp tissue kit (Qiagen, Valencia, Calif.). Viral RNA was isolated from plasma by using the Qiagen viral RNA kit, according to the manufacturer's protocol, and reverse transcribed to cDNA. HIV-1 env gp120 sequences were amplified from cellular DNA and cDNA by using a nested PCR and the following primers: gp120 outer PE0 and PE2 and gp120 inner PE1 and P2 (34). Multiple independent PCR products generated from target sources containing 20 to 200 copies of HIV-1 DNA or cDNA (purified cells or plasma) were cloned and sequenced (34). Six to 17 clone sequences were aligned by using Clustal W (32).
Quantitative PCR detection of TCRBV CDR3 in PBMC and T-cell clones.
Genomic DNA was isolated from PBMC by using the QIAamp DNA blood minikit as recommended by the manufacturer (Qiagen). Clone transcripts in the DNA were quantitated by real-time PCR by using Perkin-Elmer ABI Prism 7700 sequence detector as previously described, with modifications (10, 28). Briefly, 3 μg of genomic DNA was amplified in the presence of 0.5 μM forward (5′-GTTCCCGCTAGGAACCTG-3′) and reverse (5′-GGGTTGGAGTCGGCTGCT-3′) primers and a 50 nM dT-FAM/TAMRA fluorogenic probe (5′-TCCAGGTGAGCCAGGCCATCACTA-3′) (Synthegen, Houston, Tex.). The primers were designed to amplify the TCRBV CDR3 region. After 2 min of incubation at 50°C and 2 min of denaturation at 95°C, 45 cycles (95°C for 20 s and 60°C for 60 s) of amplification were performed. Samples were coamplified with two sets of standards. For the first standard, human placental DNA (Sigma) was used to amplify the β-globin gene in a serial dilution (100,000, 10,000, 1,000, 100, and 10 copies/reaction) to calculate the total number of cells. To make a clone-specific standard, clones were spiked into 1 million HIV-negative unrelated PBMC to create a serial dilution (300,000, 100,000, 30,000, 10,000, 3,000, 1,000, 300, 100, 30, and 10 copies/reaction). The results were analyzed by using Applied Biosystems sequence detection system (version 1.6.3).
Statistical methods.
Longitudinal data were analyzed by using generalized estimating equations to accommodate for possible correlation due to repeated data from the same individual (8). Continuous data were analyzed by using linear regression models, and the probability of responses was determined by using logistic regression models (8). The criterion for a positive ELISPOT assay was based on an exact binomial test statistic, with the numbers of SFC per well assumed to follow a simple Poisson model.
RESULTS
Study population.
Ninety-two HIV-1-infected subjects enrolled in the study. Most were Caucasian males reporting unprotected sex with another male as their predominant risk factor for acquiring infection (Table 1). The subjects were seen at a median of 55 days following the appearance of symptoms; 28 were enrolled within 30 days of symptoms. The majority experienced clinical symptoms during acute infection (Table 1). Twenty-nine subjects remained untreated, and 63 subjects received ART for a median duration of 805 days (range, 168 to 1,659). Therapy was initiated after a median interval of 93 days (range, 9 to 2,608 days) after the appearance of symptoms. Eleven subjects initiated therapy within 30 days, 36 subjects did so between 30 and 365 days, and 16 subjects initiated therapy more than 365 days after the appearance of symptoms (Table 2).
TABLE 1.
Clinical and demographic profile of the 92 study participants with acute and early HIV- 1 infection
| Characteristic | Value |
|---|---|
| Mean age (range) (yr) | 33 (20-58) |
| Gender (no. of subjects) | |
| Male | 91 |
| Female | 1 |
| Race (no. of subjects) | |
| White | 83 |
| Black | 2 |
| Hispanic | 4 |
| Other | 3 |
| Risk behavior associated with HIV-1 acquisition (no. of subjects)a | |
| MSMa | 83 |
| MSM and IDU | 4 |
| Heterosexual | 2 |
| Heterosexual and IDU | 1 |
| Unknown | 2 |
| Median duration of infection at entry (range) (days) | 55 (3-187) |
| No. of subjects (%) with symptomatic acute infection | 75 (82%) |
MSM, men who have sex with men; IDU, injection drug use.
TABLE 2.
Probability of detecting Gag- or Env-specific Th proliferative responses among untreated and treated subjects stratified by duration of infection prior to treatment
| Antigen | No treatment (n = 62) | Probability of detecting response among subjects with:
|
||
|---|---|---|---|---|
| Treatment beginning after duration of infection of (in days):
| ||||
| <30 (n = 11) | 30-365 (n = 36) | >365 (n = 16) | ||
| Gag | 0.16 | 0.49 | 0.44 | 0.42 |
| Env | 0.05 | 0.17 | 0.17 | 0 |
| Gag or Env | 0.19 | 0.49 | 0.52 | 0.42 |
HIV-1-specific LP responses during acute infection and following treatment.
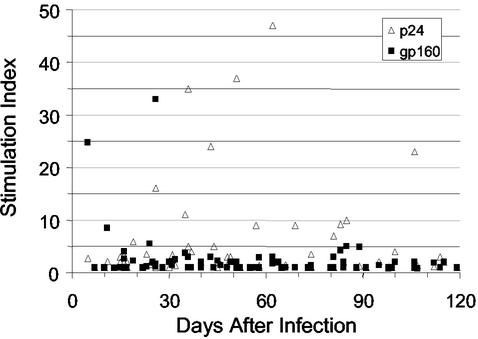
To define the specificities of Th responses induced in HIV-1 infection, we prospectively analyzed Env- and Gag-specific lymphoproliferative (LP) responses among 62 untreated subjects with primary infection, including 33 subjects assessed prior to treatment. We accrued 124 total observations from a mean of two time points and used generalized estimating equations to control for repeated measures for the same subject. Overall, there was a low probability (19%) of detecting HIV-1-specific LP responses in the untreated patients (Table 2). When present, the responses were more likely to be Gag specific than Env specific (16 versus 5%). However, closer inspection revealed distinct differences in the kinetics of Env- and Gag-specific proliferative responses (Fig. 1). When all measurements taken from specimens within the first year of infection were taken into consideration, the probability of identifying Env-specific proliferative responses was significantly higher among subjects with acute infection (<30 DPI) rather than early infection (>30 DPI) (21 versus 0.5%, P = 0.001) (Fig. 1). By contrast, Gag-specific proliferative responses were detected throughout acute and early infection (Fig. 1), and the probability of detecting these among subjects <30 DPI was similar to the probability for those >30 DPI (13 versus 17%, P = 0.64).
FIG. 1.
HIV-1 Gag and Env-specific Th responses of 62 untreated subjects with acute or early HIV-1 infection. All assays were performed with fresh PBMC. Each symbol represents the measurement for an individual subject at enrollment. The net counts per minute for the positive Env-specific responses ranged from 1,111 to 7,696 (median, 3,479), and for the positive Gag-specific responses, they ranged from 1,128 to 26,956 (median, 3,917).
Next, we determined whether the duration of infection prior to therapy had an impact on the ability to mount a detectable Env- or Gag-specific proliferative response following suppression of viremia with ART (>24 weeks). The 63 treated subjects were stratified into three groups according to the duration of infection prior to treatment initiation: within 30 days, between 30 and 365 days, and more than 365 days (Table 2). As in the untreated group, Gag-specific proliferative responses predominated over Env-specific responses following treatment, and the probabilities of detecting Gag-specific responses were comparable among the three groups (Table 2). Of note, several subjects who initiated therapy even after the first year of infection developed Gag-specific responses, but none of these subjects developed detectable Env-specific responses (Table 2). Moreover, none of the four subjects with Env-specific proliferative responses at the time of enrollment demonstrated these responses again during a mean of 10 (range 3 to 18) subsequent follow-up visits despite the early initiation of ART.
Antiviral activities of Env-specific Th responses.
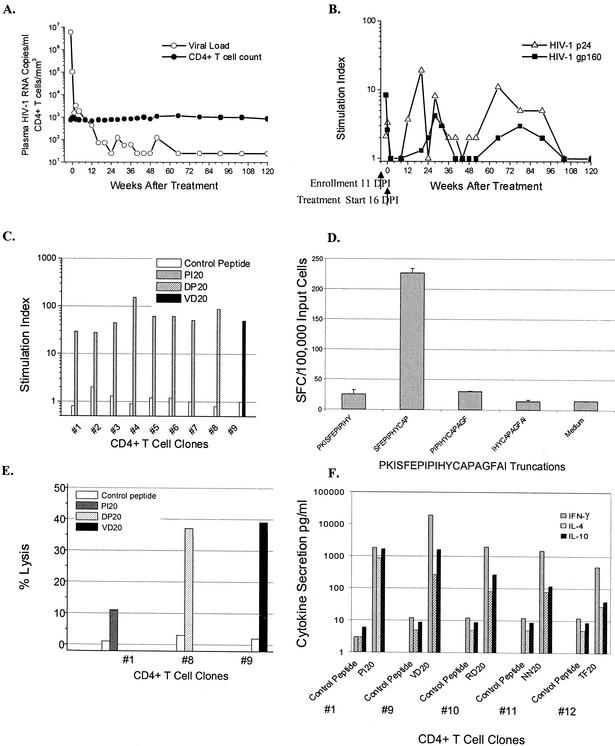
We sought to determine why early Env-specific Th responses are not preserved by patients who initiate early ART. Detailed studies were conducted with stored specimens from subject 1212, who enrolled 11 days after infection and who manifested an early Env-specific Th proliferative response (Fig. 2). On presentation with acute HIV-1 infection, subject 1212 had an initial viral load in plasma of 6 million copies/ml and a CD4+ T-cell count of 755 cells/mm3 (Fig. 2A). Within 5 days (16 DPI) plasma HIV-1 RNA spontaneously declined more than one log to 102,045 copies/ml, and ART was then initiated. Plasma viremia decreased further to <500 copies/ml by 12 weeks and <50 copies/ml by 28 weeks of therapy, and CD4+ T-cell counts remained stable (Fig. 2A). Studies of the Th responses revealed an Env-specific LP response (S.I. of 8) at day 11, which was lost by day 16 and undetectable at all subsequent 18 time points examined over 120 weeks of therapy (Fig. 2B). However, Gag-specific LP responses, absent at two time points (11 and 16 DPI) prior to initiation of therapy, were detected intermittently after therapy (Fig. 2B).
FIG. 2.
Characterization of Env-specific Th responses of subject 1212. (A) Effect of combination ART on plasma viremia (RNA copies/milliliter) and CD4+ T-cell count (no. of cells/cubic millimeter) for subject 1212. The subject presented on day 11, and treatment was initiated on day 16 of infection. (B) Effects of ART on HIV-1-specific Th responses measured by lymphoproliferation and indicated by S.I. All assays were performed with fresh PBMC. (C) Env- specific CD4+ T-cell clones established from peripheral blood taken on day 11 of infection. Clones map to epitopes in gp120 and gp41 (clones 1 to 7, PI20; clone 8, DP20; and clone 9, VD20). (D) Mapping of immunodominant epitope region PI20 by ELISPOT assay. (E) Env- specific clones exhibit cytotoxic activity when incubated with autologous EBV-transformed lymphoblastoid cell lines pulsed with a specific peptide at an E:T ratio of 20 to 1. (F) Secretion of IFN-γ, IL-4, and IL-10 by Env-specific CD4+ T-cell clones after stimulation with the envelope or control peptide (clone 1, PI20; clone 9, VD20; clone 10, RD20; clone 11, NN20; and clone 12, TF20). A level of cytokine secretion in antigen-stimulated cultures exceeding two times that of the unstimulated cultures was considered a positive response.
To understand the antiviral function and fate of the Env-specific responses in subject 1212, we first established Env-specific CD4+ T-cell clones by using cryopreserved PBMC obtained from 11 DPI and then mapped their MHC class II-restricted epitopes by LP assays. Seven (no. 1 to 7) of 12 clones recognized the highly conserved C3 region of gp120 PI20-PKISFEPIPIHYCAPAGFAI (Fig. 2C), and the responses were further mapped to the 12-mer SP12-SFEPIPIHYCAP in an IFN-γ ELISPOT assay (Fig. 2D). In LP assays, clone no. 8 proliferated in response to a peptide in the C1 region of gp120 (DP20-DTEVHNVWATQACVPTDPNP) and clone no. 9 recognized a highly conserved region in gp41 (VD20-VWGIKQLQARVLAVERYLKD) (Fig. 2C). In addition, the Env-specific CD4+ T-cell clones (no. 10 to 12) recognized three additional epitope regions: gp120 RD20-RDKMQKEYALLYKLDIVSID; gp120 NN20-NFTDNAKTIIVHLNESVQIN; and gp41 TF20-TKAKRRVVQREKRAAIGALF (data not shown). Thus, the acute Env-specific Th response was broad, recognizing at least six distinct epitopes throughout the protein.
To determine the class II molecules responsible for presenting the HIV-1 epitopes to the CD4+ T-cell clones, LP assays were performed in the presence and absence of anti-DP, anti-DQ, and anti-DR antibodies. Peptide-specific proliferation of clones no. 1 and no. 9 was inhibited more than 99% by anti-DR antibody, in contrast to <15% inhibition with the anti-DP and anti-DQ antibodies, indicating HLA-DR as the restriction element (data not shown). Further restriction analysis by lymphoproliferation revealed promiscuous binding to multiple HLA-DR molecules (data not shown), and fine mapping to a distinct HLA-DR molecule was not possible for these clones. HLA restriction analyses could not be performed with clones no. 8, 10, 11, and 12 due to limited availability of the clones. The affinity of three of the Env-specific Th epitopes (PI20, DP20, and VD20) to various HLA-DR molecules was determined by using IC50 binding assays (Table 3). Of note, the two epitope regions PI20 and VD20 each bound 7 of the 12 common DRB1, 3, 4, and 5 molecules tested at an IC50 of <1,000 nM, indicating their promiscuous affinity for multiple class II molecules.
TABLE 3.
The binding capacity (IC50) of Th epitope regions for common DRB1, DRB3, DRB4, and DRB5 allelesa
| Env peptide | IC50 of Th epitope region for indicated allele
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
DRB1*
|
DRB3* 0101 | DRB4* 0101 | DRB5* 0101 | |||||||||
| 0101 | 0301 | 0401 | 0404 | 1101 | 1302 | 0701 | 0802 | 0901 | ||||
| DP20 | 686 | − | 16,667 | 3,032 | − | 5,833 | 3,846 | − | − | − | 5,414 | − |
| PI20 | 203 | 7,452 | 182 | 1,485 | − | 758 | 387 | − | 263 | − | 74 | 769 |
| VD20 | 2.3 | 11,371 | 3,462 | 1,069 | 487 | 135 | 193 | 769 | 2,222 | − | 243 | 37 |
Peptides are classified as a binder for a DR molecule if the binding capacity is <1,000 nM (shown in bold); −, IC50 >20,000 nM.
To determine if the Env-specific CD4+ T cells from subject 1212 were capable of antiviral activities, clones no. 1, 8, and 9, recognizing PI20, DP20, and VD20, respectively, were assessed for lysis of autologous B-LCL pulsed with their specific peptide epitopes. These three clones demonstrated 10 to 39% greater cytotoxicity of targets expressing the Env epitope in contrast to the control peptide at an E:T ratio of 20 to 1 (Fig. 2E). In addition, clones (1, 9, 10, 11, and 12) were examined for their ability to secrete helper and antiviral cytokines in response to stimulation with their putative Env epitope. Culture supernatants were tested for secretion of IL-4 and IL-10 (72-h harvest), and IFN-γ (120-h harvest) to determine the Th profile. The clones 1 and 9, recognizing PI20 and VD20, respectively, secreted high levels of IFN-γ (608- to 1,564-fold over background), IL-4 (53- to 854-fold), and IL-10 (176 to 265-fold) indicating a Th0 profile (Fig. 2F). The clones 10, 11, and 12, recognizing RD20, NN20, and TF20, respectively, secreted predominantly IFN-γ (39- to 160-fold) and lower levels of IL-4 (6- to 16-fold) and IL-10 (4- to 30-fold) (Fig. 2F). Thus, the earliest Env-specific responses in subject 1212 detected at day 11 of infection recognized epitopes in gp120 and gp41, were capable of HIV-1- specific cytolytic activities, and demonstrated potential for antiviral function through secretion of IFN-γ.
Evolutionary changes in Th epitopes in subject 1212.
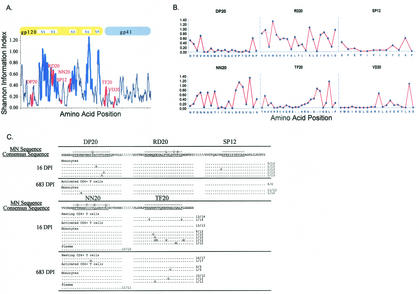
Since the HIV-1 envelope consistently exhibits intrahost and interhost variability, we anticipated that mutations in sequences spanning the Th epitopes, particularly those lying within hypervariable regions, could occur over time. We also considered the possibility that there may be significant divergence between the autologous epitopes and the sequences of the HIV-1MN strain upon which the screening peptides were based. To address these issues, we first mapped the position of the epitopes on the variability plot for the entire gp160 protein. Two of the epitopes (RD20 and NN20) were present within regions of the HIV-1 genome that exhibit a high level of variability, while the others were in regions of lower variability (Fig. 3A). This was also evident in a site-by-site analysis of variability within each of these epitopes (Fig. 3B). A comparison of the MN-derived peptide sequences to the autologous sequences (Fig. 3C) revealed a three- and four-amino-acid difference between the sequences within the RD20 and NN20 epitopes, respectively. A one-amino-acid difference was noted in the DP20 autologous and MN sequences, while the TF20 and SP12 autologous sequences were identical to the MN sequences.
FIG. 3.
Genetic variability in regions encoding Th epitopes and the evolutionary changes in autologous viral sequences. (A) Amino acid variability in Th epitopes. Entropy measurements of amino acid variability mapped across the gp160 coding sequence by using 244 full-length subtype B sequences obtained from the Los Alamos HIV sequence database are shown. The thick blue line indicates the V1-to-V5 variable region, and the thick red line denotes the six Th epitopes recognized by patient 1212. The full gene plot illustrates a moving average of variability at adjoining three-amino-acid windows. Two of the epitopes (RD20 and NN20) mapped to regions of high variability, while the other epitopes mapped to more conserved regions in the protein. (B) Site-by- site analysis of variability. The variability at each amino acid position within the six Th epitopes was examined by plotting the site-by-site variability. (C) Evolutionary changes at the Th gp120 epitopes in autologous viral sequences sampled at 16 and 683 DPI from patient 1212. Identity to amino acid in the consensus sequence is indicated by a dot. Numbers at the ends of the sequences indicate the proportion of clones encoding the given variant. Compartments sampled included monocytes, resting CD4+ T cells, activated CD4+ T cells, and plasma.
Next we examined the possibility that the loss of Th responses following acute infection was due to the evolution of variant sequences leading to a loss of stimulation and Th-cell responsiveness over time. Multiple sequences encoding the five gp120 epitopes (but not the gp41 epitope) were analyzed from samples collected at 16 (mean of 25 clones) and 683 (mean of 27 clones) DPI from subject 1212. We found minimal heterogeneity and change over time in the RD20 and NN20 autologous sequences, even though these epitopes reside in regions of high variability. However, we found several variants in the relatively conserved sequences of DP20 and TF20 from blood monocytes sampled at the first time point but fewer variants in the samples from 683 DPI. Thus, the paucity of envelope-specific immune responses at the later time points in this subject was not explained by viral escape within epitopes recognized during acute infection.
Longitudinal tracking of Env-specific Th responses in subject 1212.
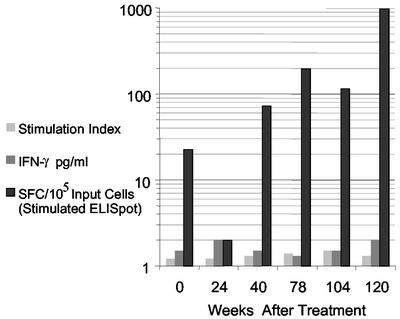
To determine if Env-specific Th cells persisted despite the inability to proliferate to the cognate epitope, we repeated the assays measuring lymphoproliferation and IFN-γ secretion in a batch analysis using PBMC from six time points (weeks 0 [16 DPI], 24, 40, 78, 104, and 120 posttreatment). The Env peptide PI20 was used to stimulate these cells, since the findings above indicated that it was recognized by the majority of clones and remained conserved over the period of study. Neither peptide-specific lymphoproliferation nor IFN-γ secretion (by ELISA) was detected at any of the six time points (Fig. 4). An overnight IFN-γ ELISPOT assay and IFN-γ ICS by flow cytometry (data not shown) were also unable to detect responses, with lower levels of sensitivity in our laboratory of 0.005 and 0.01%, respectively.
FIG. 4.
Longitudinal tracking of Env-specific Th cells from subject 1212. PBMC from six time points between 16 DPI and 27 months posttreatment (weeks 0, 24, 40, 78, 104, and 120 posttreatment) were tested in a batch analysis for lymphoproliferation (S.I. are shown) and IFN-γ secretion by ELISA in response to stimulation with peptide PI20 for 120 h. PBMC from these time points were also stimulated with the peptide for 10 days, and IFN-γ ELISPOT assays were performed with CD8-depleted PBMC poststimulation. Env-specific IFN-γ SFC were detected by stimulated ELISPOT only.
Finally, we hypothesized that the Env-specific Th cells were present at such low frequency that they could not be detected by the conventional assays described above. Thus, PBMC from the six time points were first amplified by in vitro stimulation with the peptide PI20 for 10 days and then assessed qualitatively for IFN-γ secretion in an ELISPOT assay. After stimulation, IFN-γ-secreting cells were detected in the expanded T-cell population at weeks 40, 78, 104, and 120 with levels ranging from 73 to 980 SFC/100,000 input cells (Fig. 4). By contrast, <20 IFN-γ SFC/105 cells were detected in similarly stimulated PBMC from nine HIV-1-seronegative subjects. Subsequent experiments confirmed that the T cells secreting IFN-γ in these assays were within the CD8+ T-cell-depleted PBMC fraction (data not shown).
Longitudinal tracking of Env P120-specific clonal transcripts.
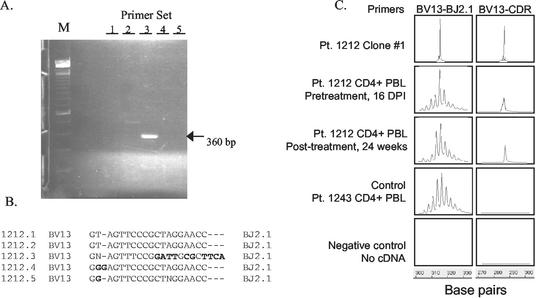
To determine if the Env PI20-specific Th responses elicited following viral suppression with ART represented newly acquired Th cells or the original clonal response detected at day 11, the PBMC from day 16 of infection and week 24 posttreatment were examined for the presence of clone transcripts identical to those found among Env-specific Th clones established at day 11. Using multiplex RT-PCR to amplify the TCRBV family, we found that six of the seven Th clones recognizing Env PI20 at day 11 used BV13 (Fig. 5A). Analysis of the rearranged VDJ region of five of the BV13 clones revealed four unique sequences, with clones 1 and 2 sharing an identical sequence (Fig. 5B). To determine if this common clonotype persisted, cDNA from CD4+ T cells isolated from day 16 and week 24 were amplified by RT-PCR with clonotypic primers based on the dominant sequence. The clonal transcripts were observed at both the early and later time points (Fig. 5C). By contrast, these transcripts were not detected in CD4+ T cells from a different HIV-1-infected subject (no. 1243) (Fig. 5C). Of note, the clonal transcripts were not detected by real-time quantitative PCR performed with DNA, the lower limit of detection of which was 1/10,000 cells. Thus, these studies demonstrate that Env-specific CD4+ T cells elicited during acute infection persist at low frequencies.
FIG. 5.
Longitudinal tracking of Env-specific clonal transcripts from subject 1212. (A) Analysis of TCRBV expression in clone 1212 no. 1 by multiplex RT-PCR and agarose gel electrophoresis. The outside lane (M) contains the molecular weight markers (100-bp ladder). The arrowhead indicates the single distinct band visualized in this gel. The size and location of the band indicate that the clone expressed TCRBV13. Clones 2, 3, 4, 5, and 6 also expressed TCRBV13 (data not shown). (B) Direct DNA sequencing was performed with the amplified products from clones 1, 2, 3, 4, and 5 by using the TCRBC primer. Four unique TCRBV CDR sequences were detected, with clones 1 and 2 sharing identical sequences. Nucleotide mismatches between the CDR sequences for clones 1 and 2 versus clones 3, 4, and 5 are indicated in bold. (C) Detection of clone 1212 no. 1 transcripts before and after treatment. RT-PCR was performed with CD4+-enriched PBMC from weeks 0 and 24 by using primers specific for the Vβ in the clone and Cβ. On each of the amplified DNA samples, two elongations of 10 cycles were performed, initiated by either the fluorescent dye-labeled Jβ or the clonotype primer. The two reaction products were electrophoresed, and the fluorescent profiles were analyzed by GeneScan software. Shown are histograms for spectratyping analysis of CDR3 lengths with the fragment length depicted on the x axis and fluorescent intensity on the y axis. Priming with the Jβ primer depicts a single clonal expansion in clone 1, polyclonal profile in CD4+-enriched PBMC from subject 1212 at weeks 0 (16 DPI) and 24 posttreatment and from subject 1243. Priming with the clonotypic primer depicts the presence of the clone transcripts in clone 1, CD4+-enriched PBMC from subject 1212 at time points week 0 (16 DPI) and week 24 posttreatment, but not in the CD4+-enriched PBMC from subject 1243. The negative control (no cDNA) sample showed no amplification with either Jβ or clonotypic primers.
Envelope-specific IFN-γ secretion in subjects receiving delayed ART.
To test the hypothesis that Env-specific Th cells may be induced and maintained even in subjects who delay initiating ART until after the acute infection stage, we examined the Env-specific Th responses of seven such subjects. Four subjects initiated treatment during the first year, two during the second year, and one during the fourth year of infection (Table 4). The subjects were selected based on the availability of cryopreserved PBMC samples for ELISPOT assays. None of these subjects demonstrated Env-specific proliferative responses prior to initiation of treatment, and the probability of an Env-specific proliferative response after treatment was low (13%), for a mean of 10 posttreatment visits (total, 70 observations). The PBMC (CD8 depleted) from these subjects were tested for the presence of IFN-γ SFC recognizing the whole gp160 protein and Env 20-mer peptide pools after a median duration of 12 months (range, 8 to 24 months) of ART. No responses were detected by standard assays (data not shown). However, Env-specific SFC were detected by stimulated assays in six of the seven subjects (Table 4). Responses to the gp160 protein (median, 64 SFC/105 cells) were identified for four of six subjects, and responses to the peptide pools (82 SFC/105 cells) were found for all six subjects initiating therapy within the first 2 years (Table 4). No IFN-γ SFC were detected for the one subject with advanced disease (CD4+ T-cell count, 61/mm3) who had delayed therapy by approximately 4 years. No positive responses to the Env protein or peptides were identified for 12 HIV-1-uninfected control subjects (Table 4). Thus, despite a delay in therapy of up to 2 years, Env-specific IFN-γ-secreting Th cells were detected for all subjects by stimulated ELISpot assays but not by standard assays.
TABLE 4.
Envelope-specific IFN-γ-secreting Th cells in HIV-1-infected subjects after delayed ARTa
| Subject no. | Data collected at inception of treatment
|
Duration of treatment (days) | No. of IFN-γ SFC/105 input cells
|
|||
|---|---|---|---|---|---|---|
| Duration of infection (days) | Viral load (RNA copies/ml) | CD4+ T-cell count/mm3 | Gp160 protein | Peptide pools | ||
| 1291 | 44 | 1,520 | 530 | 223 | 2 | 72b |
| 1155 | 125 | 131,680 | 559 | 446 | 92 | 138 |
| 1196 | 137 | 49,100 | 378 | 731 | 316 | 104 |
| 1216 | 137 | 38,400 | 257 | 252 | 8 | 52 |
| 1373 | 540 | 29,062 | 528 | 365 | 278 | 99 |
| 1153 | 582 | 28,800 | 742 | 365 | 36 | 21 |
| 1148 | 1,376 | 38,576 | 61 | 392 | 0 | 0 |
| 1885 | Uninfected | 3 | 3 | |||
| 1835 | Uninfected | 9 | 4 | |||
| 1838 | Uninfected | 5 | 0 | |||
| 0941 | Uninfected | 1 | 2 | |||
| ES0000 | Uninfected | 0 | 2 | |||
| 1185 | Uninfected | 4 | 0 | |||
| JM000 | Uninfected | 0 | 0 | |||
| JV000 | Uninfected | 3 | 0 | |||
| 1966 | Uninfected | 0 | NDc | |||
| 1959 | Uninfected | 17 | ND | |||
| 1137 | Uninfected | 2 | ND | |||
| 1242 | Uninfected | 0 | ND | |||
HIV-1-uninfected subjects were not treated, and their viral loads and CD4+ T-cell counts were not measured.
The Env-specific immune response of subject 1291 mapped to Env PI20, the dominant epitope region recognized by subject 1212. Subjects 1212 and 1291 share the DRB1*0701 haplotype.
ND, not done.
DISCUSSION
We have detailed for the first time Th responses to HIV-1, notably Env-specific Th cells, within 5 days of infection. These helper activities, associated with cytotoxicity and antiviral cytokine secretion, represent one of the earliest indicators of acquired immunity in HIV-1 infection. With this study we provide clear evidence that HIV-1-specific Th proliferative responses can occur in primary infection, although the probability of detecting them is low (19%). We demonstrate that Gag-specific LP responses may occur in untreated subjects throughout primary infection (acute, 13%; early, 17%). By contrast, Env-specific Th proliferative responses are more likely to be detected during acute (21%) than during early infection (0.5%) for untreated subjects. Even when using flow-cytometric detection of antigen- induced IFN-γ production, responses to Env determinants are rarely detected in most persons with established HIV-1 infection (26). Thus, some of the earliest Th responses in HIV-1 infection may be directed to epitopes within the HIV-1 envelope, but these become rapidly undetectable by conventional assays measuring lymphoproliferation or IFN-γ secretion within days after infection. Hence, the role of Env-specific Th responses as an important mediator of T-cell help may be underappreciated because of the brief period of detectability and the need to employ sensitive techniques capable of detecting low frequencies of memory precursors.
Within the first year of infection, once ART is initiated, HIV-1 Env- and Gag-specific proliferative responses become more prominent, with threefold increases in the probability of measuring a response by lymphoproliferation. Gag-specific responses climb even if ART is initiated later than 1 year after infection, but this is not the case with Env-specific responses. Some of our findings may help explain why Env-specific proliferative responses do not reemerge under the same conditions.
When we evaluated one patient, subject 1212, within 11 days of infection and 2 years later, we did not observe evidence for viral escape within the Env Th epitopic region, even within variable regions of the envelope protein. However, we acknowledge that additional epitopes within the hypervariable regions of the envelope may be recognized initially, and these were not identified in our study, since responses were screened for recognition of epitopes only within the HIV-1MN laboratory strain. The Th responses targeting epitopes in these hypervariable regions may be the ones that apply immune pressure to viral replication and promote escape mutations. Longitudinal studies to assess the recognition of additional envelope epitopes within the regions where sequence variation has occurred over time are in progress. These regions may be more likely to contain Th epitopes that were recognized in early infection and in which escape mutations have occurred. Additionally, subject 1212 initiated treatment early during acute infection and the rapid control of virus replication may have impeded viral diversification and the emergence of immune escape variants. Besides, minority escape variants replicating at a low rate may not be easily identified in virus strains isolated from genomic DNA. It is also important to consider that the contemporaneous CD8+ cytotoxic-T-cell response during acute infection may exert the predominant immune pressure and that HIV-1 may escape within these class I-restricted epitopes.
Recent studies have demonstrated that HIV-1-specific Th cells are preferentially infected in comparison to other memory CD4+ T cells and that this selection occurs in acute as well as in chronic infection (9). The early evolution of Env-specific Th cells during the phase of high-level viremia may render these cells even more susceptible to either activation-induced apoptosis or death as a consequence of direct viral infection. Hence, here we explored the possibility that the Env-specific Th cells either were deleted from the memory pool or were reduced to such low frequency that they were no longer detectable by conventional assays. CD4+ T cells specific for the immunodominant Env epitope PI20 were not detected posttreatment by lymphoproliferation, ex vivo IFN-γ ELISPOT, or ICS assays. However, PI20-specific IFN-γ-secreting cells were detected following one cycle of in vitro stimulation with gp160. Moreover, TCR clonotypes targeting the immunodominant gp120 epitope region PI20, while not detected by PCR amplification of DNA, were detected by RT-PCR with GeneScan analysis, the latter having severalfold-greater sensitivity than the former. Since no evidence of viral diversification in these epitopic regions was found, these findings indicate that the precursor frequency of Env-specific memory CD4+ T cells may be profoundly low despite control of viremia with ART initiated as early as 16 DPI.
Of note, during the first 24 weeks after initiation of ART, Env-specific Th cells were not detected in the PBMC by conventional assays to measure antigen-specific lymphoproliferation (LPA) or ex vivo IFN-γ secretion (ELISPOT and ICS). Furthermore, antigen-specific IFN-γ SFC were not detected during this time by ELISPOT assays performed with the PBMC after a round of in vitro stimulus with gp160. During these phases of high and declining viremia, the antigen- specific cells are likely of the effector phenotype and die upon in vitro stimulation. Alternately, viremia may have a suppressive effect on the proliferation and function of all Th cells. This effect may be global, as evidenced by impaired memory responses not only to HIV-1 but also to a variety of recall antigens during acute and early infection (24). Inhibition of virus-specific proliferation has also been reported in association with high levels of viremia for subjects who undergo interruptions of ART, with restoration of proliferative responses on resumption of ART and control of virus replication (22).
In six of our seven subjects who had delayed initiation of ART we found demonstrable IFN-γ SFC by stimulated ELISPOT specific for either the Env protein or peptide pools. Our data support the notion that HIV-1-specific Th cells may persist despite long treatment delays, albeit at very low frequencies (<0.005 to 0.01%), and thus are not identified by standard ELISPOT, LP, and ICS assays but require amplification of precursors in vitro for the detection of IFN-γ-secreting cells. Admittedly, this is an unconventional strategy for detecting antigen-specific T cells and provides a qualitative assessment of functional memory cells. Further studies are needed to determine whether there is a critical threshold of functional memory CD4+ T cells that must be maintained to contribute to the control of viral replication by helper activities.
Nevertheless, the mere presence of the Th cells indicates that they may be capable of in vivo boosting with therapeutic HIV-1 vaccination regimens. The emergence of Env-specific Th responses after ART may represent the restoration of the proliferative capacity and amplification of the memory pool, rather than new responses. Several epitopes described here may form important constituents of preventive and therapeutic vaccines against HIV-1. Two of the six epitopes characterized (the immunodominant epitope PI20 and the gp41 epitope VD20) are located in highly conserved domains, and their promiscuous binding properties to common HLA molecules make them particularly attractive candidates for inclusion in vaccines. Of note, the immunodominant epitope in the C3 domain of gp120 (PI20, amino acids [aa] 206 to 225) overlaps with previously defined epitope regions recognized by CD4+ CTL (aa 201 to 225) from an HIV-1LAI gp160 vaccine recipient (12, 13) and CD8+ CTL (aa 202 to 221) from an HIV-1-infected subject (16, 18). The epitope DP20 (aa 62 to 81) in the C1 domain overlaps with an epitope recognized by CD8+ CTL (aa 62 to 80) from an HIV-1-infected subject (16). The gp41 epitope region VD20 was recognized by CD4+ CTL in a recipient of an HIV-1LAI-based gp160 vaccine (14). Thus, the data here suggest that Env-specific Th responses may be boosted even in chronically infected subjects by using immunization strategies.
In conclusion, Env-specific CD4+ Th cells, capable of cytotoxicity and antiviral cytokine secretion, emerge very early in HIV-1 infection, but their proliferative capacity may be lost shortly thereafter. Nevertheless, Env-specific CD4+ memory precursors persist despite long delays in antiviral therapy. The proliferative capacity of these cells is restored after several weeks of virus suppression, although the very low frequencies may preclude their detection by conventional assays measuring lymphoproliferation or cytokine secretion. These findings have important implications for current strategies in the treatment of HIV-1. Notably, boosting Th responses either by supervised treatment interruption or by immunotherapy may be feasible even among those receiving considerably delayed therapy, although the interventions must be attempted only after several months of virus suppression to allow a mature and effective HIV-1- specific Th response. Within our study, we have also defined highly promiscuous epitopes and epitope regions in relatively conserved domains of HIV-1 gp120 and gp41, associated with cytotoxicity and antiviral cytokine secretion, which may be useful in preventive and therapeutic vaccination strategies.
Acknowledgments
This work was supported by National Institutes of Health grants AI01550, AI41535, and AI49109 and University of Washington Center for AIDS Research (CFAR) Subcontract 3332391. U.M. is a recipient of the UW CFAR Clinical Core Award.
We thank C. Stevens for the recruitment of patients, S. Emery and J. Bui for technical assistance, M. Huang for guidance with the design of the primers and probe for quantitative PCR detection of TCRBV CDR3 in clones, T. Smith for data management, L. Wang for assistance with statistical analysis, M. Moerbe and A. Cerna for assistance with preparation of the manuscript, M. Elizaga and H. Horton for critical reviews of the manuscript, Glaxo-Wellcome and Merck Pharmaceuticals for the provision of drugs, and the study participants for their time and effort.
REFERENCES
- 1.Akatsuka, Y., E. G. Martin, A. Madonik, A. A. Barsoukov, and J. A. Hansen. 1999. Rapid screening of T-cell receptor (TCR) variable gene usage by multiplex PCR: application for assessment of clonal composition. Tissue Antigens 53:122-134. [DOI] [PubMed] [Google Scholar]
- 2.Battegay, M., D. Moskophidis, A. Rahemtulla, H. Hengartner, T. W. Mak, and R. M. Zinkernagel. 1994. Enhanced establishment of a virus carrier state in adult CD4+ T- cell-deficient mice. J. Virol. 68:4700-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berzofsky, J. A., A. Bensussan, K. B. Cease, J. F. Bourge, R. Cheynier, Z. Lurhuma, J. J. Salaun, R. C. Gallo, G. M. Shearer, and D. Zagury. 1988. Antigenic peptides recognized by T lymphocytes from AIDS viral envelope-immune humans. Nature 334:706-708. [DOI] [PubMed] [Google Scholar]
- 4.Binley, J. M., D. S. Schiller, G. M. Ortiz, A. Hurley, D. F. Nixon, M. M. Markowitz, and J. P. Moore. 2000. The relationship between T cell proliferative responses and plasma viremia during treatment of human immunodeficiency virus type 1 infection with combination antiretroviral therapy. J. Infect. Dis. 181:1249-1263. [DOI] [PubMed] [Google Scholar]
- 5.Borrow, P., A. Tishon, S. Lee, J. Xu, I. S. Grewal, M. B. Oldstone, and R. A. Flavell. 1996. CD40L-deficient mice show deficits in antiviral immunity and have an impaired memory CD8+ CTL response. J. Exp. Med. 183:2129-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demotz, S., A. Sette, K. Sakaguchi, R. Buchner, E. Appella, and H. M. Grey. 1991. Self peptide requirement for class II major histocompatibility complex allorecognition. Proc. Natl. Acad. Sci. USA 88:8730-8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dewar, R. L., H. C. Highbarger, M. D. Sarmiento, J. A. Todd, M. B. Vasudevachari, R. T. Davey, Jr., J. A. Kovacs, N. P. Salzman, H. C. Lane, and M. S. Urdea. 1994. Application of branched DNA signal amplification to monitor human immunodeficiency virus type 1 burden in human plasma. J. Infect. Dis. 170:1172-1179. [DOI] [PubMed] [Google Scholar]
- 8.Diggle, P. J., K. Y. Liang, and S. L. Zeger. 1995. Analysis of longitudinal data. Oxford University Press, New York, N.Y.
- 9.Douek, D., J. Brenchley, M. Betts, D. Ambrozak, B. Hill, Y. Okamoto, J. Casazza, J. Kuruppu, K. Kunstman, S. Wolinsky, Z. Grossman, M. Dybul, A. Oxenius, D. Price, M. Connors, and R. Koup. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95-98. [DOI] [PubMed] [Google Scholar]
- 10.Gibson, U. E., C. A. Heid, and P. M. Williams. 1996. A novel method for real time quantitative RT-PCR. Genome Res. 6:995-1001. [DOI] [PubMed] [Google Scholar]
- 11.Haslett, P. A., D. F. Nixon, Z. Shen, M. Larsson, W. I. Cox, R. Manandhar, S. M. Donahoe, and G. Kaplan. 2000. Strong human immunodeficiency virus (HIV)-specific CD4+ T cell responses in a cohort of chronically infected patients are associated with interruptions in anti-HIV chemotherapy. J. Infect. Dis. 181:1264-1272. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, R. P., S. A. Hammond, A. Trocha, R. F. Siliciano, and B. D. Walker. 1994. Epitope specificity of MHC restricted cytotoxic T lymphocytes induced by candidate HIV-1 vaccine. AIDS Res. Hum. Retrovir. 10:S73-S75. [PubMed] [Google Scholar]
- 13.Johnson, R. P., S. A. Hammond, A. Trocha, R. F. Siliciano, and B. D. Walker. 1994. Induction of a major histocompatibility complex class I-restricted cytotoxic T- lymphocyte response to a highly conserved region of human immunodeficiency virus type 1 (HIV-1) gp120 in seronegative humans immunized with a candidate HIV-1 vaccine. J. Virol. 68:3145-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kent, S. J., P. D. Greenberg, M. C. Hoffman, R. E. Akridge, and M. J. McElrath. 1997. Antagonism of vaccine-induced HIV-1-specific CD4+ T cells by primary HIV-1 infection: potential mechanism of vaccine failure. J. Immunol. 158:807-815. [PubMed] [Google Scholar]
- 15.Koelle, D. M., L. Corey, R. L. Burke, R. J. Eisenberg, G. H. Cohen, R. Pichyangkura, and S. J. Triezenberg. 1994. Antigenic specificities of human CD4+ T- cell clones recovered from recurrent genital herpes simplex virus type 2 lesions. J. Virol. 68:2803-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieberman, J., J. A. Fabry, D. M. Fong, and G. R. Parkerson III. 1997. Recognition of a small number of diverse epitopes dominates the cytotoxic T lymphocyte response to HIV type 1 in an infected individual. AIDS Res. Hum. Retrovir. 13:383-392. [DOI] [PubMed] [Google Scholar]
- 17.Lieberman, J., J. A. Fabry, M. C. Kuo, P. Earl, B. Moss, and P. R. Skolnik. 1992. Cytotoxic T lymphocytes from HIV-1 seropositive individuals recognize immunodominant epitopes in Gp160 and reverse transcriptase. J. Immunol. 148:2738-2747. [PubMed] [Google Scholar]
- 18.Lieberman, J., P. R. Skolnik, G. R. Parkerson III, J. A. Fabry, B. Landry, J. Bethel, and J. Kagan. 1997. Safety of autologous, ex vivo-expanded human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte infusion in HIV-infected patients. Blood 90:2196-2206. [PubMed] [Google Scholar]
- 19.Malhotra, U., M. M. Berrey, Y. Huang, J. Markee, D. J. Brown, S. Ap, L. Musey, T. Schacker, L. Corey, and M. J. McElrath. 2000. Effect of combination antiretroviral therapy on T-cell immunity in acute human immunodeficiency virus type 1 infection. J. Infect. Dis. 181:121-131. [DOI] [PubMed] [Google Scholar]
- 20.Malhotra, U., S. Holte, S. Dutta, M. M. Berrey, E. Delpit, D. M. Koelle, A. Sette, L. Corey, and M. J. McElrath. 2001. Role for HLA class II molecules in HIV-1 suppression and cellular immunity following antiretroviral treatment. J. Clin. Investig. 107:505-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matloubian, M., R. J. Concepcion, and R. Ahmed. 1994. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J. Virol. 68:8056-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNeil, A. C., W. L. Shupert, C. A., Iyasere, C. W. Hallahan, J. A. Mican, R. T. Davey, Jr., and M. Connors. 2001. High-level HIV-1 viremia suppresses viral antigen- specific CD4+ T cell proliferation. Proc. Natl. Acad. Sci. USA 98:13878-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musey, L., Y. Hu, L. Eckert, M. Christensen, T. Karchmer, and M. J. McElrath. 1997. HIV-1 induces cytotoxic T lymphocytes in the cervix of infected women. J. Exp. Med. 185:293-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musey, L. K., J. N. Krieger, J. P. Hughes, T. W. Schacker, L. Corey, and M. J. McElrath. 1999. Early and persistent human immunodeficiency virus type 1 (HIV-1)- specific T helper dysfunction in blood and lymph nodes following acute HIV-1 infection. J. Infect. Dis. 180:278-284. [DOI] [PubMed] [Google Scholar]
- 25.Pannetier, C., J. Levraud, A. Lim, J. Even, and P. Kourilsky. 1997. The immunoscope approach for the analysis of T cell repertoires, p. 287-325. In J. R. Oksenberg (ed.), The antigen T cell receptor: selected protocols and applications. Landes Bioscience, Chapman & Hall, Georgetown, Tex.
- 26.Pitcher, C. J., C. Quittner, D. M. Peterson, M. Connors, R. A. Koup, V. C. Maino, and L. J. Picker. 1999. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat. Med. 5:518-525. [DOI] [PubMed] [Google Scholar]
- 27.Rosenberg, E. S., J. M. Billingsley, A. M. Caliendo, S. L. Boswell, P. E. Sax, S. A. Kalams, and B. D. Walker. 1997. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science 278:1447-1450. [DOI] [PubMed] [Google Scholar]
- 28.Ryncarz, A. J., J. Goddard, A. Wald, M. L. Huang, B. Roizman, and L. Corey. 1999. Development of a high-throughput quantitative assay for detecting herpes simplex virus DNA in clinical samples. J. Clin. Microbiol. 37:1941-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sidney, J., S. Southwood, C. Oseroff, M. d. Guercio, A. Sette, and H. Grey. 1998. The measurement of MHC/peptide interactions by gel infiltration. Curr. Protocols Immunol. 18:3.1-3.19.
- 31.Southwood, S., J. Sidney, A. Kondo, M. F. del Guercio, E. Appella, S. Hoffman, R. T. Kubo, R. W. Chesnut, H. M. Grey, and A. Sette. 1998. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 160:3363-3373. [PubMed] [Google Scholar]
- 32.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walter, E. A., P. D. Greenberg, M. J. Gilbert, R. J. Finch, K. S. Watanabe, E. D. Thomas, and S. R. Riddell. 1995. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N. Engl. J. Med. 333:1038-1044. [DOI] [PubMed] [Google Scholar]
- 34.Zhu, T., D. Muthui, S. Holte, D. Nickle, F. Feng, S. Brodie, Y. Hwangbo, J. I. Mullins, and L. Corey. 2002. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14+ monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J. Virol. 76:707-716. [DOI] [PMC free article] [PubMed] [Google Scholar]