Abstract
The identification of germline mutations in families with HNPCC is hampered by genetic heterogeneity and clinical variability. In previous studies, MSH2 and MLH1 mutations were found in approximately two-thirds of the Amsterdam-criteria–positive families and in much lower percentages of the Amsterdam-criteria–negative families. Therefore, a considerable proportion of HNPCC seems not to be accounted for by the major mismatch repair (MMR) genes. Does the latter result from a lack of sensitivity of mutation detection techniques, or do additional genes underlie the remaining cases? In this study we address these questions by thoroughly investigating a cohort of clinically selected North American families with HNPCC. We analyzed 59 clinically well-defined U.S. families with HNPCC for MSH2, MLH1, and MSH6 mutations. To maximize mutation detection, different techniques were employed, including denaturing gradient gel electrophoresis, Southern analysis, microsatellite instability, immunohistochemistry, and monoallelic expression analysis. In 45 (92%) of the 49 Amsterdam-criteria–positive families and in 7 (70%) of the 10 Amsterdam-criteria–negative families, a mutation was detected in one of the three analyzed MMR genes. Forty-nine mutations were in MSH2 or MLH1, and only three were in MSH6. A considerable proportion (27%) of the mutations were genomic rearrangements (12 in MSH2 and 2 in MLH1). Notably, a deletion encompassing exons 1–6 of MSH2 was detected in seven apparently unrelated families (12% of the total cohort) and was subsequently proven to be a founder. Screening of a second U.S. cohort with HNPCC from Ohio allowed the identification of two additional kindreds with the identical founder deletion. In the present study, we show that optimal mutation detection in HNPCC is achieved by combining accurate and expert clinical selection with an extensive mutation detection strategy. Notably, we identified a common North American deletion in MSH2, accounting for ∼10% of our cohort. Genealogical, molecular, and haplotype studies showed that this deletion represents a North American founder mutation that could be traced back to the 19th century.
Introduction
Hereditary nonpolyposis colorectal cancer (HNPCC [MIM 114500]) is the most frequent autosomal dominant predisposition to the development of colorectal cancer. It is caused by germline mutations in human homologues of the bacterial mismatch repair (MMR) genes MutL and MutS: MSH2 on chromosome 2p16, MLH1 on 3p21, MSH6 on 2p15, and PMS1 and PMS2 on 7p22 (Fishel et al. 1993; Bronner et al. 1994; Nicolaides et al. 1994; Akiyama et al. 1997; Miyaki et al. 1997). The identification of germline mutations in families with HNPCC and HNPCC-like disease is hampered not only by this locus heterogeneity but also by the clinical variability among families with HNPCC. Apart from a lifetime risk of colorectal cancer of ∼80% (Vasen et al. 1996; Aarnio et al. 1999), individuals with an MMR gene mutation are characterized by an increased risk of tumors of the endometrium, stomach, small intestine, pancreas, hepatobiliary system, urinary tract, ovary, brain, and skin (Lynch 1999). The vast majority of HNPCC-causing mutations have been reported in MSH2 and MLH1 (Peltomaki and Vasen 1997). Accordingly, mutations in these genes give rise to the “classical” HNPCC phenotype (Wijnen et al. 1997; Giardiello et al. 2001), whereas MSH6 mutations have been described in families with more atypical HNPCC (Kolodner et al. 1999; Wijnen et al. 1999; Wu et al. 1999; Wagner et al. 2001). Only a few PMS2 germline mutations have been described so far (Hamilton et al. 1995; Liu et al. 2001).
Identification of HNPCC-causing MMR gene mutations enables at-risk relatives to be informed about their cancer risks and to benefit from intensive surveillance programs that have been proven to reduce their overall mortality by 65% (Jarvinen et al. 2000). To optimize mutation analysis in families with HNPCC and HNPCC-like disease, several clinical criteria have been formulated. Of these, the Amsterdam criteria, established by the International Collaborative Group on HNPCC (Vasen et al. 1991, 1999), are now recognized as the gold standard in HNPCC clinical selection. Previously, MLH1 or MSH2 mutations have been found in only 45%–64% and 0%–47% of the Amsterdam criteria–positive and –negative families, respectively (Giardiello et al. 2001). Since >90% of HNPCC colorectal cancers display microsatellite instability (MSI) (Aaltonen et al. 1994; Fujiwara et al. 1998), in contrast to 12%–18% of the sporadic colorectal cancers (Ionov et al. 1993; Thibodeau et al. 1993), the “Bethesda guidelines” include MSI as an additional selection tool for MMR gene mutation analysis (Rodriguez-Bigas et al. 1997). However, although the application of the Bethesda guidelines does improve the mutation analysis sensitivity, it also results in a remarkable reduction of its specificity compared with the Amsterdam criteria, since a considerable number of families with more atypical phenotypes are included (Syngal et al. 2000). The value of immunohistochemical (IHC) analysis of MMR proteins as a valuable additional selective tool has been shown in several studies (de Leeuw et al. 2000; Lindor et al. 2002; Hendriks et al. 2003), although positive IHC staining is observed in the presence of certain mutation types (Wahlenberg et al. 2002).
The fact that germline MSH2 and MLH1 mutations cannot be identified in a still-considerable proportion of the families with “classical” HNPCC may be explained by the involvement of additional HNPCC genes. MSH6, PMS2, and the more recently described MLH3 and EXO1 genes may represent candidate disease-causing genes for MSH2- and MLH1-negative families (Wu et al. 2001a, 2001b). However, preliminary data indicate that germline mutations in these genes are more likely to either result in atypical HNPCC phenotypes and are rare in families with “classical” HNPCC (MSH6 and PMS2) or represent only mild genetic modifiers (EXO1 and MLH3) (Hamilton et al. 1995; Kolodner et al. 1999; Wijnen et al. 1999; Wu et al. 1999, 2001a, 2001b; Wagner et al. 2001; Jagmohan-Changur et al. 2003). By means of more-sensitive mutation detection protocols (conversion technology or monoallelic mutation analysis [MAMA]), Yan et al. (2000) showed involvement of MSH2 or MLH1 in all the families with “classical” HNPCC analyzed. This suggested that clinically well-defined HNPCC is caused by MSH2 and MLH1 alone and that our efforts should be focused on developing new and/or more-thorough mutation detection approaches. To this end, we analyzed a cohort of families with clinically very well defined HNPCC and HNPCC-like disease from the United States for the presence of MSH2, MLH1, and MSH6 germline mutations, by using different techniques to optimize mutation detection. The results presented here not only provide a comprehensive spectrum of the molecular basis of HNPCC in the United States but also underscore the need for clinical selection for a cost-effective screening of MMR genes.
Patients and Methods
The Henry Lynch HNPCC Cohort
The study included a total of 59 extended families selected for having conditions reminiscent of HNPCC by H. Lynch at the Department of Preventive Medicine and Public Health, Creighton University. The process of genetic counseling employed in the management of these families has been described elsewhere (Lynch et al. 1999). Forty-nine families fulfilled the Amsterdam criteria. Of the 10 Amsterdam criteria–negative families, 9 were characterized by two first-degree relatives with an HNPCC-related tumor (colorectal, endometrial, urinary tract, or small bowel cancer), with one relative diagnosed before the age of 50 years. Only one family did not meet the criteria because of the later (50 years) age at onset of the HNPCC-related tumors. However, one patient from this family presented with multiple HNPCC-related tumors. The majority of the families (80%) encompasses five or more generations, whereas, in the remaining cases, four generations were recorded. More family characteristics are listed in table 1. For each family, the youngest affected relative available was selected for mutation analysis, with only three exceptions: in two families, only a single relative with colorectal carcinoma in situ was available for mutation analysis; in a third one, only a nonpenetrant individual was available. Of the selected probands, 24 had colorectal cancer diagnosed before the age of 50 years (and 17 at the age of 45 years or younger). Eight probands had endometrial cancer (six diagnosed before the age of 50 years), and one had small bowel cancer at age 29 years. Twenty-three probands had multiple HNPCC-related tumors (including sebaceous adenomas).
Table 1.
Clinical Features of the 59 Families with HNPCC Studied in this Article
| ColorectalCancer | EndometrialCancer | OtherHNPCC-RelatedTumorsa | |
| Mean no. per family | 7.4 | 1.9 | 2.1 |
| Mean age at onset, in years (range) | 45.2 (19–85) | 46.4 (31–77) | 52.8 (27–76) |
Ovary cancer, stomach cancer, urinary tract cancer, and small-bowel cancer.
Eleven families with HNPCC selected for this study and screened only for the founder mutation were diagnosed at the Ohio State University’s James Cancer Hospital, Columbus, Ohio. Among these, five were Amsterdam-positive and, in the remaining six families, there were features highly suggestive of HNPCC, including early onset and/or multiple primary cancers in the proband, one or two first-degree relatives with endometrial cancer, or multiple second-degree relatives with colorectal or endometrial cancer. An additional cohort of 128 patients with colorectal (n=85) or endometrial (n=43) cancer from Ohio was studied. These patients represented all MSI-positive cases (98 MSI-high and 30 MSI-low) from a cohort of 716 unselected, consecutive, newly diagnosed patients with colorectal (n=482) or endometrial (n=234) cancer in the Columbus area, collected in 1998–2000.
Denaturing Gradient Gel Electrophoresis (DGGE)
DNA from at least one affected relative of every kindred was analyzed. Genomic DNA was obtained from peripheral blood lymphocytes. Initial DNA analysis was performed by DGGE and direct sequencing, as described elsewhere (Wijnen et al. 1998b, 1999). In short, MSH2- and MLH1-specific exons were amplified by PCR and were analyzed by GC-clamped DGGE. Exons with altered patterns of migration on DGGE were sequenced, to determine the nucleotide alteration. MSH6 was analyzed by DGGE in families negative for pathogenic mutations in MSH2 or MLH1.
Southern Blot Analysis
Southern analysis of MSH2, MLH1, and MSH6 was performed in the families negative for a pathogenic point mutation after DGGE analysis, according to our previously established protocol (Wijnen et al. 1998a). Analysis of MSH2 was performed with XbaI, HindIII, NsiI, and EcoRI genomic DNA digests, followed by hybridization with three overlapping cDNA probes (encompassing exons 1–7, exons 7–12, and exons 10–16). Analysis of MLH1 was performed with XbaI, ApaI, HindIII, and NsiI digests hybridized with two specific cDNA probes encompassing exons 1–12 and exons 11–19; MSH6 Southern analysis was performed with XbaI and ApaI digests, followed by hybridization with two cDNA probes encompassing exons 1–4 and exons 5–10.
Characterization of the Founder Deletion in MSH2
Molecular Analysis
An apparently identical deletion encompassing exons 1–6 of the MSH2 gene was identified by Southern analysis in seven apparently unrelated families. Primers were designed around the approximate location of breakpoints inferred from the restriction map of the deleted chromosome. The 3′ primer (F3) was located in intron 6 of the MSH2 gene (5′-AAGCATCACAGTTACTGTTG-3′), whereas the 5′ primer (R3) was derived on the basis of genomic sequences ∼2 kb upstream of exon 1 (5′-GCTGAATTAGGTTTTGGAAC-3′). Through use of these primers, a deletion-specific PCR product of ∼1,700 bp was obtained by long range PCR (Roche Molecular Biochemicals). This product was partly sequenced using a third primer (R2) located ∼1 kb upstream of exon 1 (5′-TTTCAATCTGTCGCCCACGC-3′). These primers were also employed to screen two Ohio cohorts.
Genealogical Studies
Genealogical data were obtained from the records of the Mormon Church (FamilySearch Internet Web site) and the Offices of Vital Statistics, as well as from cemeteries’ records and patient recollection.
Haplotype Analysis
To determine the genetic history of the deletion chromosome, chromosome 2 haplotypes were derived from several affected and unaffected individuals by PCR amplification of CA repeat markers proximal and distal to the MSH2 gene. The employed chromosome 2 markers were as follows: tel-D2S119- D2S288-D2S391-MSH2IVS10+12g→a-CA1-D2S123-D2S378-cen. Primers and PCR conditions were as described elsewhere (Gyapay et al. 1994; Wijnen et al. 1994; Hutter et al. 2000). Also, a monochromosomal somatic cell hybrid (see below), containing the founder exon 1–6 deletion, was derived from Ohio family 726NM, to facilitate the study of the haplotype of the mutation-carrying chromosome.
MSI, IHC, and MAMA
MSI and IHC analysis was performed on tumor samples from one index patient negative for MSH2, MLH1, or MSH6 mutation (family 101), as well as from one index patient (family 125) with a missense mutation in MSH2. MSI analysis was performed with the National Cancer Institute–recommended markers (Boland et al. 1998), complemented by markers BAT40, MSH3, and MSH6 (Hendriks et al. 2003). IHC was performed with antibodies against MSH2, MLH1, and MSH6, as described elsewhere (de Leeuw et al. 2000). MSI status in the Ohio cohorts was determined with the NCI-recommended markers. Both MSI-high and -low cases were included.
We have applied MAMA of the alleged mutated allele isolated in a murine background (Papadopoulos et al. 1995; Yan et al. 2000). However, because of the limited availability of appropriate patient material from the 59 families with HNPCC, only few selected cases were analyzed by MAMA, mainly to provide proof of feasibility for this technical approach: the paracentric chromosome 2p inversion (family 140) (Wagner et al. 2002), a deletion of exons 1–13 of MLH1 (family 103), and one family negative for mutations in MSH2, MLH1, or MSH6 (family 143). In brief, somatic cell hybrids containing only one chromosome 2p or 3p allele from the index patient were generated by fusing peripheral blood lymphocytes or lymphoblastoid cell lines within an Msh2-deficient murine epithelial cell line (GMP Conversion Technologies) (Papadopoulos et al. 1995; Yan et al. 2000). Somatic cell hybrids were genotyped by CA repeat markers around the MSH2 and MLH1 genes on chromosomes 2p and 3p, respectively, and were selected for MAMA. Total RNA and protein samples were isolated from the different hybrid cell lines. RT-PCR using random hexamers (Fermentas) was performed, followed by a standard PCR using human-specific primers for MSH2 (forward primer 5′-ATATCATGGAACCAGCA-3′ and reverse primer 5′-TACCTTCATTCCATTACTGG-3′), MLH1 (forward primer 5-GCCATTGTCACAGAGGATAA-3′ and reverse primer 5′-CGAAGGAGTGGTTAAGCAAC-3′) and MSH6 (forward primer 5′-TCGGTAGCGCCTGCTGCCCC-3′ and reverse primer 5′-TAAGTTGTGCCTACCTCCA-3′). In addition, western blot analysis was performed on protein lysates obtained from the somatic cell hybrid lines through use of a rabbit polyclonal antibody raised against amino acids 402–737 of human MSH2 (Smits et al. 2000).
Results
Point-Mutation Analysis
A total of 37 germline mutations was detected in MSH2, MLH1, and MSH6 by DGGE analysis, including seven mutations of uncertain pathologic significance (six missense mutations and one in-frame 3-nt deletion) (table 2). In another three families, both a missense and a pathogenic mutation were detected (table 3). Most point mutations (23 of 37) were detected in MLH1. In three independent families, an identical nonsense mutation in MLH1 exon 12 leading to exon skipping was found (Stella et al. 2001).
Table 2.
Summary of Results of the Point Mutation Analysis in Our Cohort[Note]
|
No. of PointMutations ina |
|||
| Gene andMutation Type | AC-PositiveFamilies(N=49) | AC-NegativeFamilies(N=10) | Total (N=59) |
| MLH1: | |||
| Frameshift | 8 | 0 | 8 |
| Nonsense | 2 | 1 | 3 |
| Splice site | 6 | 2 | 8 |
| Missense | 3 |
1 |
4 |
| Subtotal | 19 | 4 | 23 |
| MSH2: | |||
| Frameshift | 4 | 0 | 4 |
| Nonsense | 3 | 1 | 4 |
| Splice site | 1 | 0 | 1 |
| Missense | 1 |
1b |
2 |
| Subtotal | 9 | 2 | 11 |
| MSH6: | |||
| Frameshift | 1 | 0 | 1 |
| In-frame deletion | 0 | 1 | 1 |
| Missense | 1 |
0 |
1 |
| Subtotal | 2 | 1 | 3 |
| Total | 30 | 7 | 37 |
Table 3.
Overview of the Individual Probands and Mutations Detected in the 59 Families[Note]
|
Mutation in |
||||
| Family | ACa | MSH2 | MLH1 | MSH6 |
| 101 | + | |||
| 102 | + | 243delAATG | ||
| 103 | + | Δ exons 1–13 and E578G | ||
| 104 | + | G67R | ||
| 107 | + | IVS5+3a→t | ||
| 108 | + | Δ exons 1–6 | ||
| 109 | + | 733insAACA | ||
| 110 | + | IVS5−1g→c | ||
| 111 | + | 840delA | ||
| 112 | + | A29S | ||
| 113 | + | 466delC | ||
| 114 | + | |||
| 115 | − | A636P | ||
| 116 | + | N583S | IVS7−1g→t | |
| 117 | + | Q298X | ||
| 118 | + | K461X | ||
| 119 | + | 64insA | ||
| 120 | + | Δ exon 6 | ||
| 121 | + | IVS6−2a→g | ||
| 122 | + | Δ exons 1–6b | ||
| 123 | + | Δ exons 1–6b | ||
| 124 | + | Δ exons 1–6b | ||
| 125 | + | T552P | ||
| 126 | + | 129delTC and S473L | ||
| 127 | + | R226L (splice site) | ||
| 128 | + | 52insA | ||
| 129 | + | V185L | ||
| 130 | + | Δ exons 1–6b | ||
| 131 | + | 55delC | ||
| 132 | + | IVS18+1g→a | ||
| 133 | K461X | |||
| 134 | + | IVS6−2a→g | ||
| 135 | + | K461X | ||
| 136 | + | Δ exons 3–8 | ||
| 137 | + | 733insAACA | ||
| 138 | + | Q885X | ||
| 139 | − | IVS13+1g→t | ||
| 140 | + | inv exons 8–16 | ||
| 141 | − | R389X | ||
| 142 | − | IVS9+1g→a | ||
| 143 | − | |||
| 145 | + | 168insA | ||
| 146 | + | |||
| 147 | + | |||
| 148 | + | Δ exons 1–6b | ||
| 149 | + | 806insA | ||
| 150 | + | Δ exon 3 | ||
| 151 | − | |||
| 152 | + | Δ exons 1–6b | ||
| 153 | + | 1087insC | ||
| 154 | − | 1013delCTT | ||
| 155 | + | Δ exons 1–6b | ||
| 157 | + | R680X | ||
| 158 | + | IVS3−3t→g | ||
| 177 | + | Δ 5′ MSH2 | ||
| 178 | − | G244D | ||
| 205 | + | 525delTT | ||
| 207 | − | |||
| 208 | + | M492V | ||
Note.— Δ = deletion.
AC = Amsterdam criteria.
Founder genomic deletion encompassing exons 1–6 of the MSH2 gene.
Detection of Genomic Rearrangements
Southern blot analysis was performed in 29 of the 59 families with HNPCC, where no pathogenic point mutation was found. In total, 14 genomic rearrangements were detected, 12 in MSH2 and 2 in MLH1 (table 4). One of the deletions is a ±13-kb deletion 6 kb upstream of MSH2 (family 177). All families with a rearrangement of MSH2 or MLH1 fulfilled the Amsterdam criteria. In seven families, an identical deletion of exons 1–6 of the MSH2 gene was identified. The further characterization of this deletion is described below. Other deletions and types of rearrangement were also observed in the MSH2 gene: a second but different deletion of at least 20 kb encompassing exons 1–6 (family 108), an ∼2-kb exon 3 deletion (family 150), and a deletion of exons 3–8 of ∼50 kb (family 136). The chromosome 2 paracentric inversion encompassing MSH2 exons 8–16 found in one kindred (family 140) has been reported elsewhere (Wagner et al. 2002). In MLH1, two genomic deletions, encompassing exon 6 (∼2.3 kb; family 120) and exons 1–13 (at least 42 kb; family 103), were identified. No MSH6 genomic rearrangements were found among the 29 families (tables 4 and 5).
Table 4.
Summary of Genomic Rearrangements in the 59 Families with HNPCC
|
Genomic Rearrangement in |
||
| Family | MSH2 | MLH1 |
| 103 | Deletion of exons 1–13 | |
| 108 | Deletion of exons 1–6 | |
| 120 | Deletion of exon 6 | |
| 122 | Deletion of exons 1–6a | |
| 123 | Deletion of exons 1–6a | |
| 124 | Deletion of exons 1–6a | |
| 130 | Deletion of exons 1–6a | |
| 136 | Deletion of exons 3–8 | |
| 140 | Inversion of exons 8–16 | |
| 148 | Deletion of exons 1–6a | |
| 150 | Deletion of exon 3 | |
| 152 | Deletion of exons 1–6a | |
| 155 | Deletion of exons 1–6a | |
| 177 | Upstream deletionb | |
| Total | 12 | 2 |
The founder genomic deletion encompassing exons 1–6 of the MSH2 gene.
An ∼13-kb deletion, the 3′ breakpoint end of which maps 6 kb 5′ of MSH2. The pathogenicity of this deletion could not be investigated.
Table 5.
Overview of the Results of the Complete Mutation Analysis of the MSH2, MLH1, and MSH6 Genes in the 59 Families with HNPCC[Note]
|
No. of Mutations in |
|||
| Gene and Mutation Type | AC-PositiveFamilies(N=49) | AC-NegativeFamilies(N=10) | Total (N=59) |
| MLH1: | |||
| Point mutation | 19 | 4 | 23 |
| Genomic deletion | 2 |
0 |
2 |
| Subtotal | 21 | 4 | 25 (42%) |
| MSH2: | |||
| Point mutation | 10a | 2 | 12 |
| Genomic deletion/inversion | 12 |
0 |
12 |
| Subtotal | 22 | 2 | 24 (41%) |
| MSH6: | |||
| Point mutation | 2 | 0 | 2 |
| In-frame deletion | 0 |
1 |
1 |
| Subtotal | 2 | 1 | 3 (5%) |
| Total | 45 (92%) | 7 (70%) | 52 (88%) |
Note.— In the case of double mutations, only the assumed disease-causing mutation is reported.
Including the splice-site mutation detected by MAMA analysis by Yan et al. (2000).
Characterization of the MSH2 Exon 1–6 Founder Deletion
Molecular Analysis
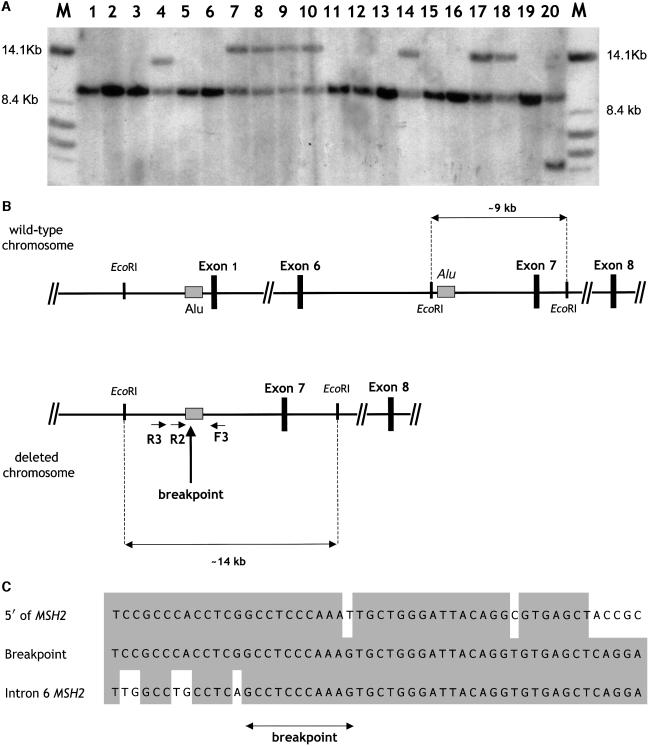
In seven patients, aberrant fragments of ∼14 kb were apparent by Southern analysis after digestion with EcoRI and hybridization with the MSH2 exon 7 probe (fig. 1a). Also, on the basis of decreased intensity of the wild-type fragment and additional hybridizations with exon 1–6 probes (data not shown), the presence of the aberrant EcoRI fragment was indicative of a genomic deletion of exons 1–6 of the MSH2 gene. Digestion by BclI revealed an ∼13-kb aberrant fragment and confirmed the presence of an identical rearrangement in these seven families. To further characterize this recurring North American deletion previously not found among HNPCC samples of European origin (Wijnen et al. 1998a), PCR primers were designed around the approximate location of breakpoints inferred from the restriction map of the deleted chromosome. The 5′ primer (R3) was based on genomic sequences ∼2 kb upstream of exon 1, whereas the 3′ primer (F3) was located in intron 6 of the MSH2 gene. Long range PCR with these primers resulted in a deletion-specific PCR product of ∼1,700 bp, exclusively observed in carriers of the common deletion and not in deletion-negative controls (data not shown). Sequencing of the breakpoint revealed exactly the same nucleotide sequence in all seven families diagnosed with the exon 1–6 deletion by Southern analysis. The breakpoints are positioned within two Alu repeats (fig. 1B and 1C). Thus, the deletion is likely to have arisen through an Alu-mediated recombination. The presence of identical breakpoint sequences in all seven cases is suggestive of a founder mutation, since a frequently recurring recombination event would be likely to result in at least a few single-nucleotide differences.
Figure 1.
A, Southern blot analysis of genomic DNA samples from North American individuals with HNPCC, revealing a common rearrangement. Total genomic DNA was digested with EcoRI and was hybridized with the MSH2 exon 7–specific probe. An aberrant ∼14-kb band was observed in lanes 7, 8, 9, 10, 14, 17, and 18, corresponding to individuals from families 122, 123, 124, 130, 148, 152, and 155, respectively. Aberrant fragments of different sizes are also observed in lanes 4 (family 108, carrying a similar but distinct MSH2 exon 1–6 deletion) and 20 (a previously identified carrier of an MSH2 exon 6 deletion, employed here as positive control). Also note that the presence of additional bands is accompanied by a decreased intensity of the ∼9-kb wild-type fragments encompassing exon 7. The latter was confirmed by hybridizations with exon 1–6 probes and by BclI digestion (data not shown). B, Genomic map of the normal and deleted MSH2 gene. For the sake of clarity, only the EcoRI restriction sites, the Alu repeats involved in the recombination (shaded boxes), and the relevant MSH2 exons (blackened boxes) are depicted. The normal (∼9-kb) and aberrant (∼14-kb) EcoRI restriction fragments also shown in panel A are indicated, as well as the primers (R3, R2, and F3) employed to selectively amplify the deletion breakpoint by PCR. C, Nucleotide sequence of the founder deletion breakpoint. Three distinct sequences are reported from top to bottom: the normal sequence flanking MSH2 at the 5′ side, the deletion breakpoint, and the normal sequence of MSH2 intron 6. The shaded boxes indicate the nucleotide homology between the two Alu repeats present in the 5′ flanking sequence and within intron 6 of MSH2. Because of the extensive homology, it is difficult to pinpoint the exact deletion breakpoint. The arrow indicates its most likely position on the basis of nucleotide homology.
Screening for the Founder Deletion
An additional cohort of 11 clinically selected families with HNPCC from Ohio was investigated for the presence of the common deletion, through use of the PCR primers (R3 and F3) described above. Two additional deletions were detected (families 726NM and CG336). Notably, the same mutation was not found in a consecutive series (n=128) of newly diagnosed MSI-positive patients with colorectal and endometrial cancer derived from a total of 716 Ohio patients studied for MSI.
Haplotype Analysis
To confirm the common genetic origin of the deletion of exons 1–6, haplotype analysis was performed (fig. 2). With the exception of families 124 and CG336, the haplotype of the deleted allele was reconstructed in the seven remaining families, six from the Lynch cohort and one of the two Ohio kindreds (726NM) found to carry the identical MSH2 deletion. A common haplotype is evident in all families, with the one exception of kindred 155, which shows, possibly as the result of mitotic recombination, novel alleles at markers DS288 and D2S119. Additional variation was observed exclusively at more-distant centromeric markers—that is, D2S123 and D2S378 (fig. 2).
Figure 2.
Haplotype analysis of the founder MSH2 deletion chromosome. A total of seven polymorphic markers flanking MSH2 on both the telomeric and centromeric side were employed. The haplotype encompassing the founder MSH2 deletion could be reconstructed in most families, with the exception of family 124 from the Lynch cohort and kindred CG336 from the Ohio cohort, because of a lack of samples from informative relatives. The haplotype of family 726NM was derived from a somatic cell hybrid containing the isolated deletion chromosome in a rodent background. Shaded boxes indicate the alleged founder haplotype. Both alleles are indicated in those cases where the exact phase could not be derived.
Genealogical Studies
By means of genealogical studies, a common ancestry could be traced for five of the nine families found to carry the MSH2 exon 1–6 founder deletion. The alleged ancestor was born around 1814 in Alabama and was presumably of German origin. He married and became a Mormon. From Alabama, he and his family undertook a long journey to Utah, following the Mississippi River through Kentucky and Missouri. Some family members moved to southeastern Iowa, bordering Illinois, and then followed the Missouri River to Nebraska and Wyoming, eventually settling in Utah. However, the ancestor was excommunicated from the Mormon Church and moved, with an unspecified number of children, to California. Two of the families studied here (122 and 152) still carry the same ancestral name and live in California. The other families reside along the trail of their ancestors (A.B., A.W., P.F.F., S.C., Y.K., P.W., R.O., J.F.L., R.F., A.D., and H.T.L., unpublished data).
Alternative Approaches to Unresolved Cases
Even when applying the most thorough mutation detection strategy, unresolved cases—that is, mutation-negative samples or missense mutations of unknown pathogenic relevance—require additional analyses for their elucidation. In our study, we were confronted with a number of such cases. Unfortunately, the limited availability of blood or tumor samples hampered the application of additional approaches to some cases. However, a number of cases provided proof of feasibility for the general strategy (table 6).
Table 6.
Families with Mutations of Unknown Significance and Families without Mutations
|
Test Resultb |
|||||
| Family | ACa | Mutation | MSI | IHC | MAMA |
| 101 | + | High | Decreased expression MSH2 | NP | |
| 104 | + | G67R MLH1 | NP | NP | NP |
| 112 | + | A29S MLH1 | NP | NP | NP |
| 114 | + | NP | NP | NP | |
| 125 | + | T552P MSH2 | High | Normal expression MSH2, MLH1, MSH6 | NP |
| 129 | + | V185L MLH1 | NP | NP | NP |
| 143 | − | NP | NP | Normal expression MSH2, MLH1, MSH6 | |
| 146 | + | NP | NP | NP | |
| 147 | + | NP | NP | NP | |
| 151 | − | NP | NP | NP | |
| 154 | − | 1013delCTT MSH6 | NP | NP | NP |
| 177 | + | D 5′ MSH2 | NP | NP | NP |
| 178 | − | G244D MLH1 | NP | NP | NP |
| 207 | − | NP | NP | NP | |
| 208 | + | M492V MSH6 | NP | NP | NP |
AC = Amsterdam criteria.
NP = not performed.
In one of the families negative for MSH2, MLH1, or MSH6 mutations (family 101), analysis of the corresponding colorectal cancer revealed an MSI-high phenotype and no expression of MSH2 by immunohistochemistry. This kindred was characterized by a classical clinical HNPCC phenotype that is likely the result of a genetic defect in the MSH2 gene that was missed by the DGGE and Southern approaches. In one other individual found to carry an MSH2 missense mutation of unknown pathogenic relevance (family 125), analysis of tumor DNA again revealed an MSI-high phenotype. IHC analysis of the corresponding tumor sections showed normal expression of MSH2, MLH1, and MSH6. Although it is possible that the mutant MSH2 proteins are functionally hampered by the missense mutations, at present we cannot determine the pathogenicity of this mutation (Wahlenberg et al. 2002).
MAMA, or conversion technology, represents a useful and promising approach for the analysis of missense mutations in MSH2 (Papadopoulos et al. 1995; Yan et al. 2000). It has also proven to be a useful tool in the analysis of both splicing errors and genomic alterations (Nakagawa et al. 2002). Somatic cell hybrids containing the alleged mutated chromosome in a murine background can be analyzed by RT-PCR, northern, or western analysis, to ensure expression and stability of the human MSH2 gene. We demonstrated proof of feasibility for this approach in the analysis of the pathogenicity of the chromosome 2p paracentric inversion (Wagner et al. 2002). Moreover, the chromosome 3 allele harboring the MLH1 deletion of exons 1–13 (family 103) did not show any expression of this gene by RT-PCR after conversion (not shown). Conversion analysis of the chromosome 2 and 3 alleles of the index patient negative for mutations in MSH2, MLH1, and MSH6 (family 143) displayed expression of the same genes by RT-PCR and western blot analysis of the somatic cell hybrids.
Discussion
The overall mutation detection rate in our study is remarkably high, compared with most previous studies (Giardiello et al. 2001). In 39 (80%) of 49 families fulfilling the Amsterdam criteria, and in 5 (50%) of the 10 Amsterdam criteria–negative families, a pathogenic mutation was found in one of the three MMR genes analyzed. When the mutations of uncertain significance (table 6) are included, the detection percentages rise to 92% (45 of 49) and 70% (7 of 10) for Amsterdam-positive and -negative families, respectively (table 5). These results are supportive of the extremely high MSH2 and MLH1 mutation detection rate among clinically well-selected HNPCC families. It is safe to assume that extension of the MAMA analysis to all the 59 index patients, improvement of the DGGE strategy, and further functional analysis of the nucleotide variants of as-yet-uncertain pathogenic significance would increase detection rate to close to 95%.
Apart from the more straightforward nonsense and deletion mutations that are expected to be disease-causing by nature, several missense mutations were detected in this study. One of these, the A636P mutation in MSH2, is a founder mutation in the Ashkenazi Jewish population and was proven to be pathogenic (Yuan et al. 1999; Foulkes et al. 2002). Two other missense mutations (G67R and V185L in MLH; families 104 and 129, respectively) were previously described to affect mismatch repair (Shimodaira et al. 1998; Ellison et al. 2001). In one index patient found to carry an MSH2 missense mutation (T552P in family 125), tumor analysis revealed an MSI-high phenotype. As described by Foulkes et al. (2002), one possible approach to the analysis of the pathogenicity of missense mutations is the analysis of large cohorts of affected and healthy individuals. Moreover, cosegregation analysis of the alleged mutation with the disease phenotype within extensive, multigeneration pedigrees can provide strong support for the pathogenicity of the missense substitution. However, definitive proof or disproof of the pathogenicity of such mutations can be obtained only by implementing functional studies. The latter have been primarily performed in yeast (Ellison et al. 2001), although novel protocols based on mammalian expression systems have also been applied successfully (Nakagawa et al. 2002; Trojan et al. 2002; see below).
In a kindred with classical HNPCC that was negative for mutations in MSH2, MLH1, or MSH6 (family 101), analysis of a colorectal cancer showed an MSI-high phenotype and loss of MSH2 protein expression by IHC, indicating that functional impairment of MSH2—for example, by a promotor mutation (Shin et al. 2002) or a splicing error due to a change within an intron—may underlie HNPCC in this family. In another kindred with classical HNPCC (family 158), a mutation was detected, by MAMA, by Yan et al. (2000). By using this technique, these authors found a splice-acceptor mutation in MSH2 that was proven hard to detect by conventional mutation analysis (table 3) (Yan et al. 2000). One other family negative for mutations in MSH2, MLH1, or MSH6 (family 143) was studied by MAMA. This family did not fulfill the Amsterdam criteria but was highly reminiscent of HNPCC, because of the presence of multiple related tumors in an affected individual and colorectal cancer in several individuals. The index patient from this family presented with colorectal cancer at age 56 years. Conversion analysis showed expression of the three MMR genes. Hence, the latter case may represent a bona fide carrier of a genetic defect at a locus other than the major MMR genes. Alternatively, the index patient may represent a phenocopy. This underscores the importance of testing individuals who are most likely, on the basis of clinical features, to carry the familial genetic defect—namely, the presence of HNPCC-related tumors and their age at onset (Wijnen et al. 1998b).
In total, only two mutations were found in MSH6. One is a frameshift mutation, and the other is an in-frame deletion of 3 nt of unknown significance (in families 153 and 154, respectively). Both families are characterized by patients with endometrial cancer and by a relatively late onset of colorectal cancer, in agreement with the atypical phenotypic features associated with MSH6 mutations (Kolodner et al. 1999; Wijnen et al. 1999; Wu et al. 1999; Wagner et al. 2001). The relatively low incidence of MSH6 mutations is interesting in view of the percentages (8% and 30% of the Amsterdam-positive and -negative kindreds, respectively) of the families with HNPCC in which no mutation could be detected. It is likely that genes like MSH6 that cause an overlapping though distinct phenotype are responsible for a small subset of the families with HNPCC and for a larger proportion of the atypical kindreds.
The high detection rate in our study is partially due to the screening for and identification of larger genomic rearrangements. The latter enabled us to identify one common ∼20-kb deletion, encompassing exons 1–6 of the MSH2 gene, that was proven to represent a founder mutation in the North American population. The origin of this mutation could be traced back to the beginning of the 19th century. Nucleotide sequence analysis of the nine deletion breakpoints did not reveal any base pair difference, as would be expected from a recurring recombination event. Also, haplotype analysis of the seven of the nine families with HNPCC carrying the common deletion by CA repeat markers flanking the MSH2 gene was in agreement with the founder hypothesis. In the majority of the cases, a region of ∼8 cM is conserved. This is in agreement with a relatively young age of the founder deletion. Accordingly, although the alleged ancestor was of German origin, we were unable to find the same deletion in an extensive analysis of European patients with HNPCC (Wijnen et al. 1998a) and in a cohort of 89 German families with HNPCC (authors' unpublished data). As confirmed by the PCR screening of the small but clinically selected Ohio cohort, this mutation represents ⩾10% of the disease-causing mutations among the Midwestern white families with HNPCC. Further analyses are needed to determine the frequency of this founder in other U.S. populations. Notably, screening of the MSI-positive Ohio cohort (n=128, 98 of whom were MSI-high and 30 of whom were MSI-low) for the presence of the founder MSH2 deletion failed to detect any additional carrier. The latter finding does not affect the alleged frequency of the founder mutation, since it is known that a large fraction of the MSI-high cases represent sporadic tumors due to hypermethylation of the MLH1 promoter. Moreover, a large fraction of sporadic CRC is known to be MSI-low (Halford et al. 2002). Therefore, this finding emphasizes the importance of clinical selection.
Seven additional genomic rearrangements were detected in our cohort. Five of the genomic rearrangements were in MSH2. Notably, in the case of family 177, the 13-kb deletion maps 6 kb 5′ of MSH2. Presumably, this deletion encompasses upstream regulatory sequences, although no evidence for its true pathogenicity could be demonstrated in this study. Also, in view of the high frequency of genomic rearrangements detected in the MSH2 gene in our (data not shown) (Wijnen et al. 1998a) and other laboratories (Charbonnier et al. 2002), this locus may contain sequences prone to recombination. Indeed, genomic sequences immediately upstream of MSH2 and some of its larger introns (introns 6, 7, and 8; 13.3 kb, 15.6 kb, and 17.4 kb, respectively) do contain many repetitive sequences, like Alu and other repeats (from our own analysis by RepeatMasker [RepeatMasker Web Server]) (Charbonnier et al. 2002). These repeats are known to mediate recombination events between partially homologous sequences resulting in genomic deletions, insertions, inversions, or other more-complex rearrangements (Wagner et al. 2002). On the basis of our findings, mutation analysis must include screening for genomic rearrangements, since these represent a frequent cause of disease at the major MMR genes (in this study, 24% of the total HNPCC burden and 50% of the cases due to MSH2). The characterization of the breakpoints of the most common genomic rearrangements will allow the development of PCR-based screening protocols (present study) to circumvent more cumbersome and time-consuming methods such as Southern blotting.
Overall, accurate and expert clinical selection of the studied families greatly improves mutation detection, especially when combined with a thorough and extensive methodological approach. Thirty-four (69%) of the Amsterdam criteria–positive families in our cohort had two additional relatives with an HNPCC-related tumor, exceeding the Amsterdam criteria requirements. In all these families, an MSH2 or MLH1 mutation was detected. Also, a pathogenic mutation was detected in all families fulfilling the revised Amsterdam criteria, only one of which was in MSH6. This indicates that the fulfillment of the Amsterdam criteria by three patients with colorectal cancer is not necessarily an indication of the presence of an MSH2 or MLH1 mutation. This is probably because of the high frequency of colorectal cancer in the general population, often with relatively early age at onset, and because of other forms of familial clustering of colorectal cancer cases. The presence of additional relatives with HNPCC-related tumors considerably increases the chance of MSH2 or MLH1 mutation detection. Patients with multiple HNPCC-related tumors or with sebaceous tumors reminiscent of Muir-Torre syndrome were observed in mutation-positive families that did not fulfill the Amsterdam criteria. Conversely, we were unable to detect the disease-causing mutation in one family with “classical” HNPCC (family 101). A colorectal tumor sample of the index patient from this family displayed an MSI-high phenotype and loss of MSH2 expression.
In conclusion, improved clinical selection forms the basis of optimal mutation detection in HNPCC. Mutation analysis in “classical” HNPCC should focus on the two major MMR genes—that is, MSH2 and MLH1. We underscore that, apart from point mutations, genomic rearrangements strongly contribute to the HNPCC mutation spectrum. MSI and IHC analyses will direct further mutation detection strategies in the mutation-negative families with classical HNPCC and in the more atypical HNPCC cases. This approach has also led to the identification of a common North American deletion in MSH2 accounting for ∼10% of our cohort. Genealogical, molecular, and haplotype studies showed that this common deletion represents a North American founder mutation that could be traced back to the 19th century.
Acknowledgments
This study was supported by the Dutch Cancer Foundation and by National Institutes of Health grants CA67941 and CA169058.
Electronic-Database Information
The URLs for data presented herein are as follows:
- FamilySearch Internet, http://www.familysearch.org/Eng/Search/frameset_search.asp
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HNPCC) [PubMed]
- RepeatMasker Web Server, http://ftp.genome.Washington.edu/cgi-bin/RepeatMasker
References
- Aaltonen LA, Peltomaki P, Mecklin JP, Jarvinen H, Jass JR, Green S, Lynch HT, Watson P, Tallqvist G, Juhola M (1994) Replication errors in benign and malignant tumors from hereditary nonpolyposis colorectal cancer patients. Cancer Res 54:1645–1648 [PubMed] [Google Scholar]
- Aarnio M, Sankila R, Pukkala E, Salovaara R, Aaltonen LA, de la Chapelle A, Peltomaki P, Mecklin JP, Jarvinen HJ (1999) Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int J Cancer 81:214–218 [DOI] [PubMed] [Google Scholar]
- Akiyama Y, Sato H, Yamada T, Nagasaki H, Tsuchiya A, Abe R, Yuasa Y (1997) Germ-line mutation of the hMSH6/GTBP gene in an atypical hereditary nonpolyposis colorectal cancer kindred. Cancer Res 57:3920–3923 [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S (1998) A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58:5248–5257 [PubMed] [Google Scholar]
- Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A, Tannergård P, Bollag R, Godwin A, Ward DC, Nordenskjöld M, Fishel R, Kolodner R, Liskay M (1994) Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature 368:258–261 [DOI] [PubMed] [Google Scholar]
- Charbonnier F, Olschwang S, Wang Q, Boisson C, Martin C, Buisine MP, Puisieux A, Frebourg T (2002) MSH2 in contrast to MLH1 and MSH6 is frequently inactivated by exonic and promoter rearrangements in hereditary nonpolyposis colorectal cancer. Cancer Res 62:848–853 [PubMed] [Google Scholar]
- de Leeuw WJ, Dierssen J, Vasen HF, Wijnen JT, Kenter GG, Meijers-Heijboer H, Brocker-Vriends A, Stormorken A, Moller P, Menko F, Cornelisse CJ, Morreau H (2000) Prediction of a mismatch repair gene defect by microsatellite instability and immunohistochemical analysis in endometrial tumours from HNPCC patients. J Pathol 192:328–335 [DOI] [PubMed] [Google Scholar]
- Ellison AR, Lofing J, Bitter GA (2001) Functional analysis of human MLH1 and MSH2 missense variants and hybrid human-yeast MLH1 proteins in Saccharomyces cerevisiae. Hum Mol Genet 10:1889–1900 [DOI] [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R (1993) The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75:1027–1038 [DOI] [PubMed] [Google Scholar]
- Foulkes WD, Thiffault I, Gruber SB, Horwitz M, Hamel N, Lee C, Shia J, et al (2002) The founder mutation MSH2*1906G→C is an important cause of hereditary nonpolyposis colorectal cancer in the Ashkenazi Jewish population. Am J Hum Genet 71:1395–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Stolker JM, Watanabe T, Rashid A, Longo P, Eshleman JR, Booker S, Lynch HT, Jass JR, Green JS, Kim H, Jen J, Vogelstein B, Hamilton SR (1998) Accumulated clonal genetic alterations in familial and sporadic colorectal carcinomas with widespread instability in microsatellite sequences. Am J Pathol 153:1063–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardiello FM, Brensinger JD Petersen GM (2001) AGA technical review on hereditary colorectal cancer and genetic testing. Gastroenterology 121:198–213 [DOI] [PubMed] [Google Scholar]
- Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, Bernardi G, Lathrop M, Weissenbach J (1994) The 1993–94 Genethon human genetic linkage map. Nat Genet 7:246–339 [DOI] [PubMed] [Google Scholar]
- Halford S, Sasieni P, Rowan A, Wasan H, Bodmer W, Talbot I, Hawkins N, Ward R, Tomlinson I (2002) Low-level microsatellite instability occurs in most colorectal cancers and is a nonrandomly distributed quantative trait. Cancer Res 62:53–57 [PubMed] [Google Scholar]
- Hamilton SR, Liu B, Parsons RE, Papadopoulos N, Jen J, Powell SM, Krush AJ, Berk T, Cohen Z, Tetu B, Burger PC, Wood PA, Taqi F, Booker SV, Peterson GM, Offerhaus GJA, Tersmette AC, Giardiello FM, Vogelstein B, Kinzler KW (1995) The molecular basis of Turcot’s syndrome. N Engl J Med 332:839–847 [DOI] [PubMed] [Google Scholar]
- Hendriks Y, Franken P, Dierssen J, de Leeuw W, Wijnen J, Tops C, Breuning MH, Brocker-Vriends A, Vasen H, Fodde R, Morreau H (2003) Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumours. Am J Pathol 162:469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter P, Couturier A, Rey-Berthod C (2000) Two common forms of the human MLH1 gene may be associated with functional differences. J Med Genet 37:776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M (1993) Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 363:558–561 [DOI] [PubMed] [Google Scholar]
- Jagmohan-Changur S, Poikonen T, Virpi L, Launonen V, Wikman F, Orntoft TF, Moller P, Vasen H, Tops C, Kolodner RD, Mecklin JP, Jarvinen H, Bevan S, Houlston RS, Aaltonen LA, Fodde R, Wijnen JT, Karhu A (2003) Exo1 variants occur commonly in normal population: evidence against a role in hereditary nonpolyposis colorectal cancer. Cancer Res 63:154–158 [PubMed] [Google Scholar]
- Jarvinen HJ, Aarnio M, Mustonen H, Aktan-Collan K, Aaltonen LA, Peltomaki P, De La Chapelle A, Mecklin JP (2000) Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology 118:829–834 [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Tytell JD, Schmeits JL, Kane MF, Gupta RD, Weger J, Wahlberg S, Fox EA, Peel D, Ziogas A, Garber JE, Syngal S, Anton-Culver H, Li FP (1999) Germ-line msh6 mutations in colorectal cancer families. Cancer Res 59:5068–5074 [PubMed] [Google Scholar]
- Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, Walsh-Vockley C, Petersen GM, Walsh MD, Leggett BA, Young JP, Barker MA, Jass JR, Hopper J, Gallinger S, Bapat B, Redston M, Thibodeau SN (2002) Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol 20:1043–1048 [DOI] [PubMed] [Google Scholar]
- Liu T, Yan H, Kuismanen S, Percesepe A, Bisgaard ML, Pedroni M, Benatti P, Kinzler KW, Vogelstein B, Ponz de Leon M, Peltomaki P, Lindblom A (2001) The role of hPMS1 and hPMS2 in predisposing to colorectal cancer. Cancer Res 61:7798–7802 [PubMed] [Google Scholar]
- Lynch HT (1999) Hereditary nonpolyposis colorectal cancer (HNPCC). Cytogenet Cell Genet 86:130–135 [DOI] [PubMed] [Google Scholar]
- Lynch HT, Watson P, Shaw TG, Lynch JF, Harty AE, Franklin BA, Kapler CR, Tinley ST, Liu B, Lerman C (1999) Clinical impact of molecular genetic diagnosis, genetic counseling, and management of hereditary cancer, part II: hereditary nonpolyposis colorectal carcinoma as a model. Cancer 86 Suppl 11:1637–1643 [DOI] [PubMed] [Google Scholar]
- Miyaki M, Konishi M, Tanaka K, Kikuchi-Yanoshita R, Muraoka M, Yasuno M, Igari T, Koike M, Chiba M, Mori T (1997) Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet 17:271–272 [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Yan H, Lockman J, Hampel H, Kinzler KW, Vogelstein B, De La Chapelle A (2002) Allele separation facilitates interpretation of potential splicing alterations and genomic rearrangements. Cancer Res 62:4579–4582 [PubMed] [Google Scholar]
- Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD, Venter JC, Dunlop MG, Hamilton SR, Petersen GM, de le Chapelle A, Vogelstein B, Kinzler KW (1994) Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 371:75–80 [DOI] [PubMed] [Google Scholar]
- Papadopoulos N, Leach FS, Kinzler KW, Vogelstein B (1995) Monoallelic mutation analysis (MAMA) for identifying germline mutations. Nat Genet 11:99–102 [DOI] [PubMed] [Google Scholar]
- Peltomaki P, Vasen HF (1997) Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology 113:1146–1158 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Bigas MA, Boland CR, Hamilton SR, Henson DE, Jass JR, Khan PM, Lynch H, Perucho M, Smyrk T, Sobin L, Srivastava S (1997) A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: meeting highlights and Bethesda guidelines. J Natl Cancer Inst 89:1758–1762 [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Filosi N, Shibata H, Suzuki T, Radice P, Kanamaru R, Friend SH, Kolodner RD, Ishioka C (1998) Functional analysis of human MLH1 mutations in Saccharomyces cerevisiae. Nat Genet 19:384–389 [DOI] [PubMed] [Google Scholar]
- Shin KH, Shin JH, Kim JH, Park JG (2002) Mutational analysis of promoters of mismatch repair genes hMSH2 and hMLH1 in hereditary nonpolyposis colorectal cancer and early onset colorectal cancer patients: identification of three novel germ-line mutations in promoter of the hMSH2 gene. Cancer Res 62:38–42 [PubMed] [Google Scholar]
- Smits R, Hofland N, Edelmann W, Geugien M, Jagmohan-Changur S, Albuquerque C, Breukel C, Kucherlapati R, Kielman MF, Fodde R (2000) Somatic Apc mutations are selected upon their capacity to inactivate the beta-catenin downregulating activity. Genes Chromosomes Cancer 29:229–239 [DOI] [PubMed] [Google Scholar]
- Stella A, Wagner A, Shito K, Lipkin SM, Watson P, Guanti G, Lynch HT, Fodde R, Liu B (2001) A nonsense mutation in MLH1 causes exon skipping in three unrelated HNPCC families. Cancer Res 61:7020–7024 [PubMed] [Google Scholar]
- Syngal S, Fox EA, Eng C, Kolodner RD, Garber JE (2000) Sensitivity and specificity of clinical criteria for hereditary non-polyposis colorectal cancer associated mutations in MSH2 and MLH1. J Med Genet 37:641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau SN, Bren G, Schaid D (1993) Microsatellite instability in cancer of the proximal colon. Science 260:816–819 [DOI] [PubMed] [Google Scholar]
- Trojan J, Zeuzem S, Randolph A, Hemmerle C, Brieger A, Raedle J, Plotz G, Jiricny J, Marra G (2002) Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology 122:211–219 [DOI] [PubMed] [Google Scholar]
- Vasen HF, Mecklin JP, Khan PM, Lynch HT (1991) The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC). Dis Colon Rectum 34:424–425 [DOI] [PubMed] [Google Scholar]
- Vasen HF, Watson P, Mecklin JP, Lynch HT (1999) New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 116:1453–1456 [DOI] [PubMed] [Google Scholar]
- Vasen HF, Wijnen JT, Menko FH, Kleibeuker JH, Taal BG, Griffioen G, Nagengast FM, Meijers-Heijboer EH, Bertario L, Varesco L, Bisgaard ML, Mohr J, Fodde R, Khan PM (1996) Cancer risk in families with hereditary nonpolyposis colorectal cancer diagnosed by mutation analysis. Gastroenterology 110:1020–1027 [DOI] [PubMed] [Google Scholar]
- Wagner A, Hendriks Y, Meijers-Heijboer EJ, de Leeuw WJ, Morreau H, Hofstra R, Tops C, Bik E, Brocker-Vriends AH, van der Meer C, Lindhout D, Vasen HF, Breuning MH, Cornelisse CJ, van Krimpen C, Niermeijer MF, Zwinderman AH, Wijnen J, Fodde R (2001) Atypical HNPCC owing to MSH6 germline mutations: analysis of a large Dutch pedigree. J Med Genet 38:318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, van der Klift H, Franken P, Wijnen J, Breukel C, Bezrookove V, Smits R, Kinarsky Y, Barrows A, Franklin B, Lynch J, Lynch H, Fodde R (2002) A 10-Mb paracentric inversion of chromosome arm 2p inactivates MSH2 and is responsible for hereditary nonpolyposis colorectal cancer in a North-American kindred. Genes Chromosomes Cancer 35:49–57 [DOI] [PubMed] [Google Scholar]
- Wahlenberg SS, Schmeits J, Thomas G, Loda M, Garber J, Syngal S, Kolodner RD, Fox EA (2002) Evaluation of microsatellite instability and immunohistochemistry for the prediction of germ-line MSH2 and MLH1 mutations in hereditary nonpolyposis colon cancer families. Cancer Res 62:3485–3492 [PubMed] [Google Scholar]
- Wijnen J, de Leeuw W, Vasen H, van der Klift H, Moller P, Stormorken A, Meijers-Heijboer H, Lindhout D, Menko F, Vossen S, Moslein G, Tops C, Brocker-Vriends A, Wu Y, Hofstra R, Sijmons R, Cornelisse C, Morreau H, Fodde R (1999) Familial endometrial cancer in female carriers of MSH6 germline mutations. Nat Genet 23:142–144 [DOI] [PubMed] [Google Scholar]
- Wijnen J, Fodde R, Khan PM (1994) DGGE polymorphism in intron 10 of MSH2, the HNPCC gene. Hum Mol Genet 3:2268 [DOI] [PubMed] [Google Scholar]
- Wijnen J, Khan PM, Vasen H, van der Klift H, Mulder A, van Leeuwen-Cornelisse I, Bakker B, Losekoot M, Moller P, Fodde R (1997) Hereditary nonpolyposis colorectal cancer families not complying with the Amsterdam criteria show extremely low frequency of mismatch-repair-gene mutations. Am J Hum Genet 61:329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen J, van der Klift H, Vasen H, Khan PM, Menko F, Tops C, Meijers Heijboer H, Lindhout D, Moller P, Fodde R (1998a) MSH2 genomic deletions are a frequent cause of HNPCC. Nat Genet 20:326–328 [DOI] [PubMed] [Google Scholar]
- Wijnen JT, Vasen HF, Khan PM, Zwinderman AH, van der Klift H, Mulder A, Tops C, Moller P, Fodde R (1998b) Clinical findings with implications for genetic testing in families with clustering of colorectal cancer. N Engl J Med 339:511–518 [DOI] [PubMed] [Google Scholar]
- Wu Y, Berends MJ, Mensink RG, Kempinga C, Sijmons RH, van der Zee AG, Hollema H, Kleibeuker JH, Buys CH, Hofstra RM (1999) Association of hereditary nonpolyposis colorectal cancer-related tumors displaying low microsatellite instability with MSH6 germline mutations. Am J Hum Genet 65:1291–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Berends MJ, Post JG, Mensink RG, Verlind E, van der Sluis T, Kempinga C, Sijmons RH, van der Zee AG, Hollema H, Kleibeuker JH, Buys CH, Hofstra RM (2001a) Germline mutations of EXO1 gene in patients with hereditary nonpolyposis colorectal cancer (HNPCC) and atypical HNPCC forms. Gastroenterology 120:1580–1587 [DOI] [PubMed] [Google Scholar]
- Wu Y, Berends MJ, Sijmons RH, Mensink RG, Verlind E, Kooi KA, van der Sluis T, Kempinga C, van der Zee AG, Hollema H, Buys CH, Kleibeuker JH, Hofstra RM (2001b) A role for MLH3 in hereditary nonpolyposis colorectal cancer. Nat Genet 29:137–138 [DOI] [PubMed] [Google Scholar]
- Yan H, Papadopoulos N, Marra G, Perrera C, Jiricny J, Boland CR, Lynch HT, Chadwick RB, de la Chapelle A, Berg K, Eshleman JR, Yuan W, Markowitz S, Laken SJ, Lengauer C, Kinzler KW, Vogelstein B (2000) Conversion of diploidy to haploidy. Nature 403:723–724 [DOI] [PubMed] [Google Scholar]
- Yuan ZQ, Wong N, Foulkes WD, Alpert L, Manganaro F, Andreutti-Zaugg C, Iggo R, Anthony K, Hsieh E, Redston M, Pinsky L, Trifiro M, Gordon PH, Lasko D (1999) A missense mutation in both hMSH2 and APC in an Ashkenazi Jewish HNPCC kindred: implications for clinical screening. J Med Genet 36:790–793 [DOI] [PMC free article] [PubMed] [Google Scholar]