Abstract
The function of Tic40 during chloroplast protein import was investigated. Tic40 is an inner envelope membrane protein with a large hydrophilic domain located in the stroma. Arabidopsis null mutants of the atTic40 gene were very pale green and grew slowly but were not seedling lethal. Isolated mutant chloroplasts imported precursor proteins at a lower rate than wild-type chloroplasts. Mutant chloroplasts were normal in allowing binding of precursor proteins. However, during subsequent translocation across the inner membrane, fewer precursors were translocated and more precursors were released from the mutant chloroplasts. Cross-linking experiments demonstrated that Tic40 was part of the translocon complex and functioned at the same stage of import as Tic110 and Hsp93, a member of the Hsp100 family of molecular chaperones. Tertiary structure prediction and immunological studies indicated that the C-terminal portion of Tic40 contains a TPR domain followed by a domain with sequence similarity to co-chaperones Sti1p/Hop and Hip. We propose that Tic40 functions as a co-chaperone in the stromal chaperone complex that facilitates protein translocation across the inner membrane.
Keywords: chloroplasts/co-chaperone/protein import/Tic40/TPR
Introduction
Nuclear-encoded chloroplast proteins, translated in the cytosol, must traverse the chloroplast envelope membranes to reach their intended destination. These proteins are synthesized as precursors containing an N-terminal transit peptide. The transit peptide interacts with the import apparatus of the chloroplast envelope to accomplish protein import, and then is removed in the stroma. The import apparatus consists of a complex of proteins located in both the inner and outer envelope membranes, as well as a soluble processing peptidase and molecular chaperones in the stroma. Import components located in the outer membrane are known as Toc (translocon at the outer envelope membrane of chloroplasts) proteins and those in the inner membrane are known as Tic (translocon at the inner envelope membrane of chloroplasts) proteins (Schnell et al., 1997). Among the Toc components identified, Toc159 is cross-linked to precursor proteins during the initial stage of import (Perry and Keegstra, 1994; Ma et al., 1996) and is presumed to be the receptor for transit peptides. Toc159 also exists in a soluble cytosolic form, which suggests that it may be able to switch between a soluble and an integral membrane form and function as the cytosolic receptor for chloroplast precursor proteins (Hiltbrunner et al., 2001; Bauer et al., 2002). Considerable evidence supports the conclusion that Toc75 is the major component of the protein-translocating channel in the outer membrane (Hinnah et al., 1997; Reumann et al., 1999). The function of individual Tic component is much less clear. Among the Tic proteins identified, Tic110 can be co-immunoprecipitated with two stromal chaperones: the Hsp100 family member ClpC (herein called Hsp93) (Akita et al., 1997; Nielsen et al., 1997) and the chloroplast chaperonin cpn60 (Kessler and Blobel, 1996). Tic110 and another Tic component, Tic20, have both been suggested to function as the protein-translocating channel of the inner membrane even though the two proteins have very different secondary structures and membrane topologies (Chen et al., 2002; Heins et al., 2002). Antisense mutant plants of atTic20 show severe chloroplast defects exemplified by pale leaves, reduced accumulation of plastid proteins and significant growth defects. Plastids isolated from the antisense mutant plants are specifically defective in protein translocation across the inner membrane (Chen et al., 2002).
Tic40 has been identified as a putative component of the import apparatus (Wu,C. et al., 1994; Stahl et al., 1999), but has a complicated history in the literature. Originally identified as Com/Cim44, it appeared to be present in both the inner and outer envelope membranes of pea chloroplasts and migrated with an apparent molecular mass of 44 kDa on SDS–PAGE (Wu,C. et al., 1994). Later, it was named Toc36, based on the predicted molecular mass of a Brassica napus clone, although no further evidence was presented to suggest that the protein was in the outer rather than the inner envelope membrane (Ko et al., 1995; Pang et al., 1997). Stahl et al. (1999) then showed that the B.napus clone was truncated at the N-terminus, reassessed the predicted mature size of the protein as 40 kDa in pea and presented evidence that the protein was most likely located in the inner envelope membrane of pea chloroplasts. They showed that the full-length pea clone encoded a transit peptide and named the protein Tic40. Evidence that Tic40 is involved in protein import is limited. It has been shown that Tic40 was associated with an artificially constructed precursor Oee1-Dhfr arrested during import (Wu,C. et al., 1994). It has also been shown that Tic40 could be co-immunoprecipitated with Tic110 from isolated inner membrane vesicles treated with chemical cross-linkers (Stahl et al., 1999). However, because the majority of Tic110 molecules in chloroplasts are not associated with the Tic/Toc translocon complex (Kouranov et al., 1998), it is not clear whether the association between Tic40 and Tic110 alone has any functional significance.
In the work reported here, we investigated the function of Tic40 during chloroplast protein import. We confirmed that Tic40 is localized in the inner envelope membrane and further showed that the large hydrophilic domain is located in the stroma. Two Arabidopsis T-DNA insertion mutants in the atTic40 gene were isolated and exploited to provide evidence of functions of Tic40 in chloroplast protein import. Biochemical analysis and tertiary structure prediction suggest that Tic40 functions as a co-chaperone of the stromal chaperone complex that facilitates translocation of proteins across the inner envelope membrane.
Results
Tic40 is an inner envelope membrane protein with its hydrophilic domain located in the stroma
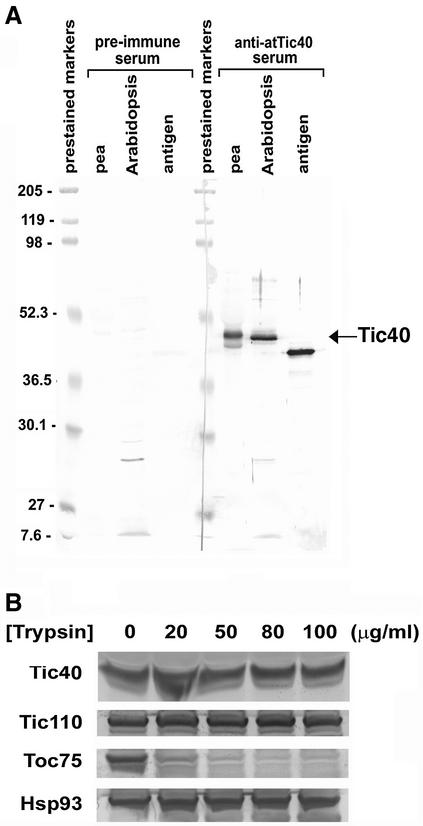
A cDNA clone encoding the Arabidopsis Tic40, atTic40, was isolated and expressed in Escherichia coli, and the resulting proteins were used for antibody production. The antibodies were highly specific for Tic40 in both pea and Arabidopsis (Figure 1A). As has been previously described for pea (Stahl et al., 1999), atTic40 migrates more slowly on SDS–PAGE than predicted based on its molecular mass. The migration rate may be slower because the mature protein contains 11% proline; a high proportion of proline has been reported to cause proteins to appear larger on SDS–PAGE (Postle, 1990). Arabidopsis and pea chloroplast envelopes were separated into outer and inner membranes, and their proteins were analyzed by immunoblotting using antibodies against atTic40. Tic40 was entirely associated with the inner membrane fraction in both pea and Arabidopsis (data not shown).
Fig. 1. The hydrophilic domain of Tic40 is located in the stroma. (A) Characterization of the atTic40 antibodies. Total chloroplast proteins from pea or Arabidopsis were analyzed by immunoblots with antibodies raised against the C-terminal portion of atTic40 or the preimmune serum. Intact chloroplasts containing 5 µg of chlorophyll or antigen containing 0.01 µg of protein were loaded. (B) Treatment of intact pea chloroplasts with various concentrations of trypsin. Digestion was performed at room temperature for 1 h and was terminated by adding a cocktail of protease inhibitors (Tu and Li, 2000). The protease inhibitors were present in all the solutions afterwards. Intact chloroplasts were repurified and analyzed by immunoblots with antibodies against various proteins as labeled on the left.
For both pea (Stahl et al., 1999) and Arabidopsis Tic40, a single transmembrane domain is predicted near the N-terminus of the mature protein, which suggests that the predicted large hydrophilic domain of the protein lies on one side of the inner envelope membrane. We next investigated on which side of the inner membrane the Tic40 hydrophilic domain was located. We treated isolated chloroplasts with different concentrations of trypsin, which can penetrate the outer membrane and digest proteins located on the outer surface of the inner membrane, but not proteins in the stroma (Jackson et al., 1998). As shown in Figure 1B, when outer membrane protein Toc75 was degraded by trypsin, Tic40 remained resistant, similar to stromal Hsp93 and Tic110. Therefore, we concluded that Tic40, like Tic110 (Jackson et al., 1998), has the bulk of its polypeptide located in the stroma.
Mutants of atTic40 were defective in chloroplast biogenesis
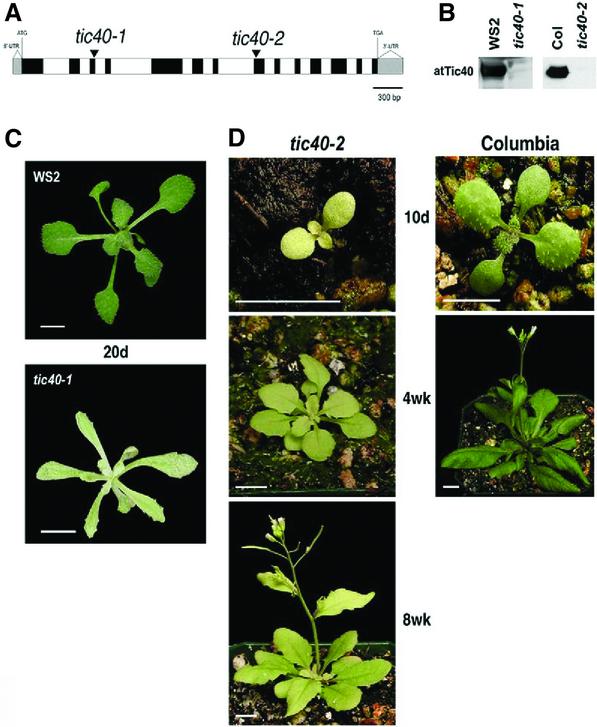
To investigate the role of Tic40, we isolated Arabidopsis mutants lacking functional atTic40. The atTic40 gene is present as a single-copy gene located on chromosome V and contains 14 exons (Jackson-Constan and Keegstra, 2001) (Figure 2A). Two T-DNA insertion mutants were isolated. One of the mutants, tic40-1 (ecotype Wassilewskija WS2), contained a T-DNA insertion in the third exon and the second mutant, tic40-2 (ecotype Columbia), contained a T-DNA insertion in the eighth exon. The mutants had identical phenotypes except that the leaf shape of tic40-1 is long and slender instead of rounded like tic40-2 (Figure 2C and D). Because both mutants are nulls, as indicated by the absence of atTic40 protein (Figure 2B), the difference in leaf shape is most likely due to the difference in ecotypes. The mutant embryos had normal morphology, but pale green coloration at later stages of embryo development (data not shown). The tic40-2 mutant was previously reported to be seedling lethal (Budziszewski et al., 2001). However, we found that the mutant seedlings were still viable, although they were very pale and small and they developed much more slowly than the wild type (Figure 2D). The seedling survival rate was often low if the growth conditions were not optimal. The slower development of the mutant was manifested in a longer life expectancy: tic40 mutant plants survived for 6–8 months, whereas wild-type plants died after 3 months. The chlorophyll content of mutant plants [0.37 mg chlorophyll/g fresh weight (FW)] was less than one-third that of wild-type plants (1.2 mg chlorophyll/g FW) throughout development. By 8 weeks, the tic40 plants had reached the developmental stage of 5-week-old wild-type plants, although they were significantly smaller, with a mass of about one-fifth that of 5-week-old wild-type plants (data not shown).
Fig. 2. Phenotypes of the tic40 mutants. (A) Positions of T-DNA insertion in the two tic40 mutant alleles: solid boxes, exons; open boxes, introns. (B) Both alleles are nulls. Chloroplast proteins from the mutants or the corresponding wild types were analyzed by immunoblotting with antibodies against atTic40: WS2, the Wassilewskija wild-type plant; Col, the Columbia wild-type plant. (C) The tic40-1 mutant and the corresponding WS2 wild-type plants at 20 days (20d). Plants were grown on MS media with 2% sucrose at 22°C under a 16 h day. The bar represents 0.4 cm. (D) The tic40-2 mutant and the corresponding Columbia wild-type plants at 10 days (10d), 4 weeks (4wk) and 8 weeks (8wk). Plants were grown on soil at 22°C under a 16 h day. The bar represents 1 cm.
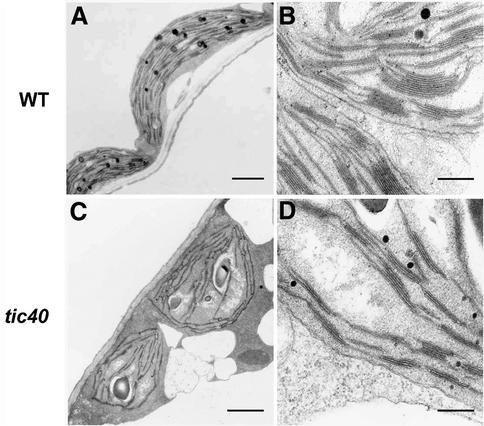
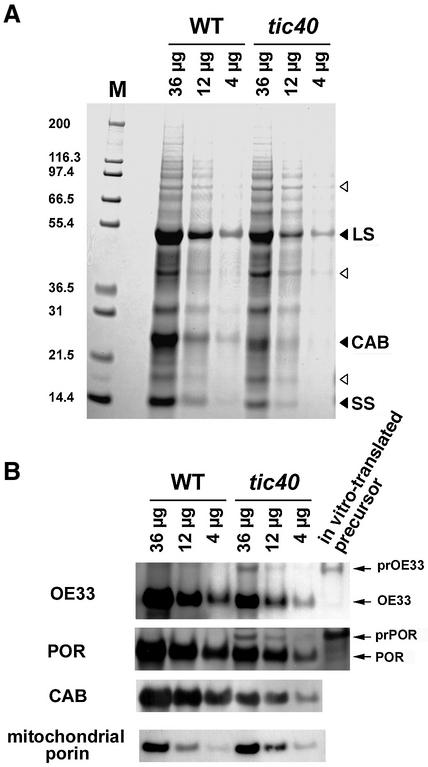
Electron microscopy analysis indicated that the mutant chloroplasts from both alleles were similar in appearance and had fewer granal stacks (Figure 3; data not shown). The quantities of chloroplast proteins, such as the small and large subunits of RuBP carboxylase (SS and LS), chlorophyll a/b binding protein (CAB), the 33 kDa protein of oxygen-evolving complex (OE33) and protochlorophyllide oxidoreductase (POR), were reduced in the tic40 mutant (Figure 4). When equal amounts of total cellular protein were analyzed by SDS–PAGE, proteins in other organelles, e.g. the mitochondrial porin (Figure 4B), appeared to be increased in the mutant because of a reduced amount of chloroplast proteins. Interestingly, the precursors of OE33 (prOE33) and POR (prPOR) were detected in the tic40 mutant but not in the wild type (Figure 4B), suggesting a deficiency in chloroplast protein import. The precursor form of OE33 detected in the mutant also migrated to the same position as the in vitro translated prOE33 on two-dimensional electrophoresis (data not shown), confirming that it was indeed the precursor of OE33 and not an unrelated protein cross-reacting with the antiserum.
Fig. 3. Electron micrographs of chloroplasts from tic40-1 mutant and WS2 wild-type plants. Samples were taken from cotyledons of 10-day-old seedlings. The bars in (A) and (C) represent 2 µm; those in (B) and (D) represent 200 nm.
Fig. 4. The levels of endogenous chloroplast proteins are reduced in the tic40 mutant. (A) Various amounts of total leaf proteins, as indicated at the top of the lanes, from the tic40-1 mutant and the wild type (WT) were analyzed by SDS–PAGE and stained with Coomassie Blue. Filled triangles indicate the three major chloroplast proteins, which were present in reduced amounts in the mutant. Open triangles indicate three proteins whose amounts are not reduced in the mutants. M, molecular mass markers. (B) Samples were the same as in (A) but were analyzed by immunoblotting with antibodies against OE33, POR, CAB and mitochondrial porin. In addition, in vitro translated OE33 and POR precursor proteins (prOE33 and prPOR) were analyzed alongside the leaf samples and detected using anti-OE33 and anti-POR antibodies.
The tic40 mutant chloroplasts were defective in protein import
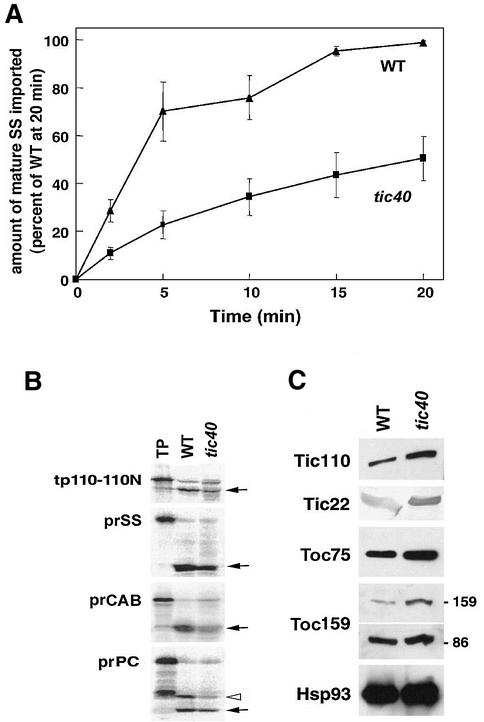
To investigate whether the tic40 mutant plants were defective in the process of chloroplast protein import, we measured the amount of mature SS imported in time-course experiments using chloroplasts isolated from tic40 and wild-type plants. The initial rate of protein import in tic40 chloroplasts was ∼40% of that of wild-type chloroplasts (Figure 5A).
Fig. 5. The tic40 mutant chloroplasts are defective in protein import. (A) Import of [35S]prSS into chloroplasts isolated from Columbia wild-type and tic40-2 plants. Isolated chloroplasts were incubated with [35S]prSS and 4 mM ATP in import buffer under natural light at 22°C. Import was stopped at different times by centrifuging the chloroplasts through 40% Percoll. Chloroplast proteins were subjected to SDS–PAGE. Data points represent the mean (± SE) of three independent experiments. The quantitity of mature SS imported into the wild type at 20 min was set at 100%. (B) Import of various other precursors into isolated chloroplasts. Import was performed at room temperature for 10 min: TP, 10% of the translated precursor proteins that were added to the import assay. The arrows mark the position of the imported mature proteins. The open triangle in the PC samples indicates the intermediate (iPC) in the stroma during PC import (Bauerle et al., 1991). (C) The tic40 mutant chloroplasts contained normal amounts of translocon components. Chloroplast proteins of the tic40-2 mutant and the wild-type plants were analyzed by immunoblotting with antibodies against translocon components as labeled on the left. The 86 kDa degradation product of atToc159 is also shown.
Import was also measured using precursor proteins targeted to other locations within chloroplasts, and in all cases it was reduced in tic40 chloroplasts: the precursor to CAB (prCAB) was reduced to 25%, tp110-110N [a C-terminal truncated Tic110 which is still able to insert into the inner envelope membrane (Lübeck et al., 1997)] to 28% and the precursor to plastocyanin (prPC) to 31% of that of the wild type (Figure 5B). Therefore, atTic40 is important for chloroplast protein import and the reduction of protein import in tic40 mutant is a universal reduction in the general import pathway.
To investigate whether the tic40 mutation altered the amounts of other translocon components through secondary effects, we performed immunoblotting on chloroplasts isolated from wild-type and tic40 plants using antibodies directed against Toc75, Toc159, Tic110, Tic22 and Hsp93. All components were present in tic40 chloroplasts in at least the same amounts as in wild-type chloroplasts (Figure 5C), indicating that the reduced import competence of the tic40 chloroplasts was probably not the result of secondary effects of the mutation reducing the amounts of other import components.
The tic40 mutant chloroplasts were defective in protein translocation across the inner envelope membrane
To dissect the import defect in the tic40 mutant further, the import process was separated into binding and translocation steps by manipulating the ATP concentrations during import reactions. Energy-depleted chloroplasts and [35S]prSS were incubated under a dim green safelight. When 0.1 mM ATP was provided, thereby permitting only binding to occur, the amount of prSS associated with both wild-type and mutant chloroplasts increased similarly (Figure 6A, lanes 2 and 6). After the binding reaction, a portion of the chloroplasts was repurified and incubated with sufficient ATP (5 mM) to permit translocation. The amount of mature SS produced was much lower in the mutant than in the wild-type chloroplasts (Figure 6A, compare lanes 3 and 7). From these results, we concluded that the tic40 mutant chloroplasts were specifically defective in translocation across the inner envelope membrane.
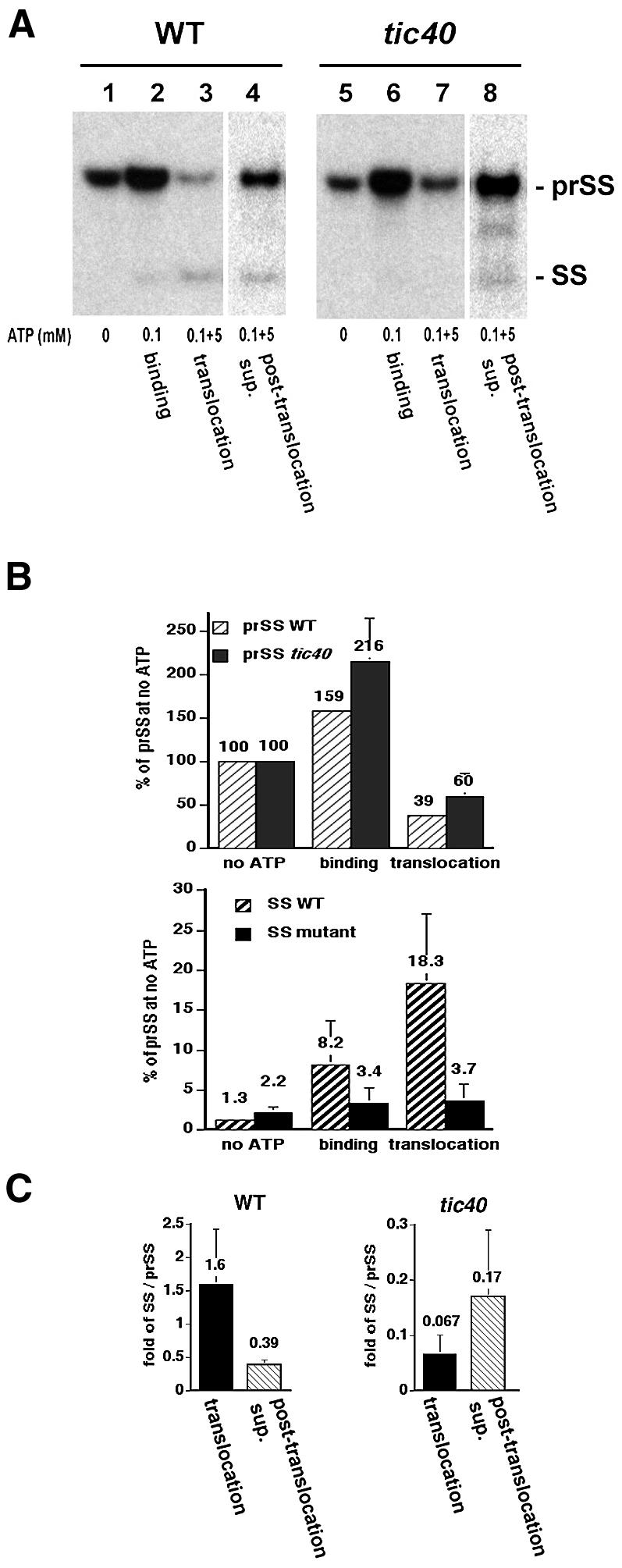
Fig. 6. The tic40 mutant chloroplasts are defective in translocation across the inner envelope membrane. (A) Chloroplasts were isolated from the tic40-1 mutant and kept on ice in the dark for 20 min to deplete internal ATP. Energy-depleted [35S]prSS and the chloroplasts were incubated together under a dim green safelight at room temperature for 5 min in the presence (lanes 2 and 6) or absence (lanes 1 and 5) of 0.1 mM ATP. The chloroplasts were pelleted by centrifugation. Half of the chloroplasts that had been incubated with 0.1 mM ATP were incubated for a further 15 min in import buffer containing 5 mM ATP (0.1 + 5 mM ATP, lanes 3 and 7). The chloroplasts were again pelleted by centrifugation. The recovered chloroplasts were repurified through a 40% Percoll cushion and the contents were analyzed. The supernatant was dried down and dissolved in SDS–PAGE sample buffer. Samples containing equal amounts of protein were loaded in lanes 1–3 and 5–7. Samples containing an equal portion of the total sample were loaded in lanes 4 and 8. The positions of prSS and mature SS are indicated. (B) Quantification of the binding and translocation experiments shown in (A). The amount of prSS when no ATP was added [lanes 1 and 5 in (A)] is set at 100%. Data points represent the mean (± SE) of four independent experiments. The mean value is given at the top of each bar. (C) Quantification of the post-translocation supernatant shown in (A). The amount of SS in lanes 3, 4, 7 and 8 of (A) was compared with the amount of prSS within each sample. Data points represent the mean (± SE) of three independent experiments. The mean value is given at the top of each bar.
We also analyzed the reaction supernatant after the translocation reaction. More precursor proteins were present in the post-translocation supernatant of mutant chloroplasts than of wild-type chloroplasts (Figure 6A, compare lanes 4 and 8), indicating that more precursors were released from the mutant chloroplasts because the precursors could not be translocated. This increase was not due to increased lysis of mutant chloroplasts, because the protein levels in the post-translocation supernatant were similar in both samples (data not shown). Furthermore, a small amount of mature SS was present in the post-translocation supernatant of mutant chloroplasts, even though almost no imported SS could be detected within mutant chloroplasts (Figure 6A, compare lanes 7 and 8, and C). While it is possible that mutant chloroplasts containing processed SS were preferentially lysed, it is more likely that, during translocation, the transit peptide was removed sufficiently early in the transport process that, in the absence of Tic40, translocation could not be completed and the processed SS was released from the chloroplasts.
Tic40 was part of the Tic/Toc translocon
As shown above, tic40 chloroplasts were defective in translocation across the inner envelope membrane. Furthermore, the majority of Tic40 polypeptide was located at the stromal side of the inner membrane. Therefore, it is likely that Tic40 would be associated with translocon components located in the stroma, such as Hsp93 or the hydrophilic domain of Tic110. Indeed, association of Tic40 with Tic110 has been reported in isolated inner membrane vesicles (Stahl et al., 1999). We sought to obtain more evidence that Tic40 was associated with other translocation components. Intact pea chloroplasts were reacted with dithiobis(succinimidylpropionate) (DSP) to cross-link proteins that were in close physical proximity. Total membranes were isolated and their proteins were solubilized with detergent followed by immunoprecipitation with anti-Tic40 antibodies. Tic40 was indeed associated with Tic110 in intact chloroplasts (Figure 7A). In addition, Toc75 and stromal Hsp93 were associated with the Tic40 immunoprecipitate. Reciprocal immunoprecipitation demonstrated that Tic40 was immunopecipitated by anti-Toc75 antibodies (Figure 7B). However, full-length Toc159 or its 86 kDa degradation fragment was not detected. Detection of the 56 kDa degradation fragment of Toc159 was hindered by the presence of IgG heavy chain at the same position of the gel. No association between Tic40 and the chloroplast Hsc70 was observed (Figure 7A). The inner membrane protein IEP21, which is not a Tic component (Kouranov et al., 1998), was not detected, supporting the specificity of the cross-linking (Figure 7A). We were also unable to detect the presence of cpn60 in the Tic40 immunoprecipitate (data not shown). Association with Toc75 and Hsp93 provided further evidence that Tic40 was part of the translocon complex mediating chloroplast protein import.
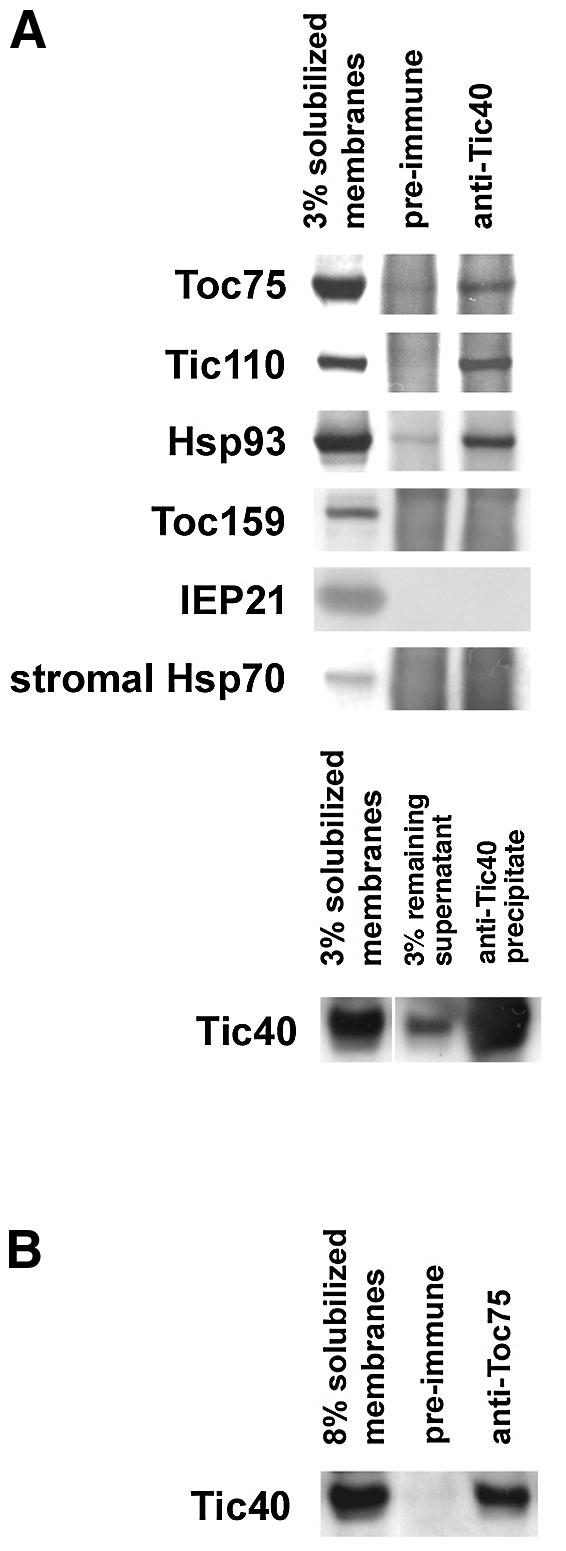
Fig. 7. Tic40 is a component of the translocation apparatus. (A) Intact pea chloroplasts were incubated with DSP for 15 min on ice. After the reaction was stopped, chloroplasts were re-isolated and lysed. Total membranes were isolated and solubilized. Proteins in the solubilized supernatant were immunoprecipitated with antibodies against Tic40. The cross-links in the immunoprecipitates were cleaved by the addition of gel loading buffer containing DTT and analyzed by immunoblotting with antibodies against various proteins as labeled on the left. A minor cross-reactivity of normal rabbit serum with Hsp93 was consistently observed with all the rabbits tested. (B) Reciprocal immunoprecipitation demonstrating association of Tic40 with Toc75. Experiments were the same as in (A), except that immunoprecipitation was performed using anti-Toc75 antibodies.
Tic40 functioned in the same stage of the import process as Tic110 and Hsp93
To dissect the function of Tic40 during translocation further, we analyzed the association pattern of Tic40 with importing precursor proteins. An import time-course experiment was performed with [35S]prSS. The reaction was terminated at various time points by diluting with cold import buffer. Chloroplasts were re-isolated and treated with the cross-linker DSP. Total membranes were isolated from these chloroplasts, the proteins were solubilized and the resulting complexes were immunoprecipitated with antibodies directed against various translocon components. Toc75 was associated mostly with prSS (Figure 8), as would be expected from its role early in the import process (Ma et al., 1996). As observed before (Kessler and Blobel, 1996), Tic110 was associated with both prSS and mature SS, but as import progressed mostly mature SS was observed. The association patterns of Tic40 and Hsp93 were similar to that of Tic110 (Figure 8). From these results, we concluded that Tic40 functioned in the same stage of the import process as Tic110 and Hsp93. They were associated with the precursor protein during its translocation across the inner membrane and after its transit peptide had been removed.
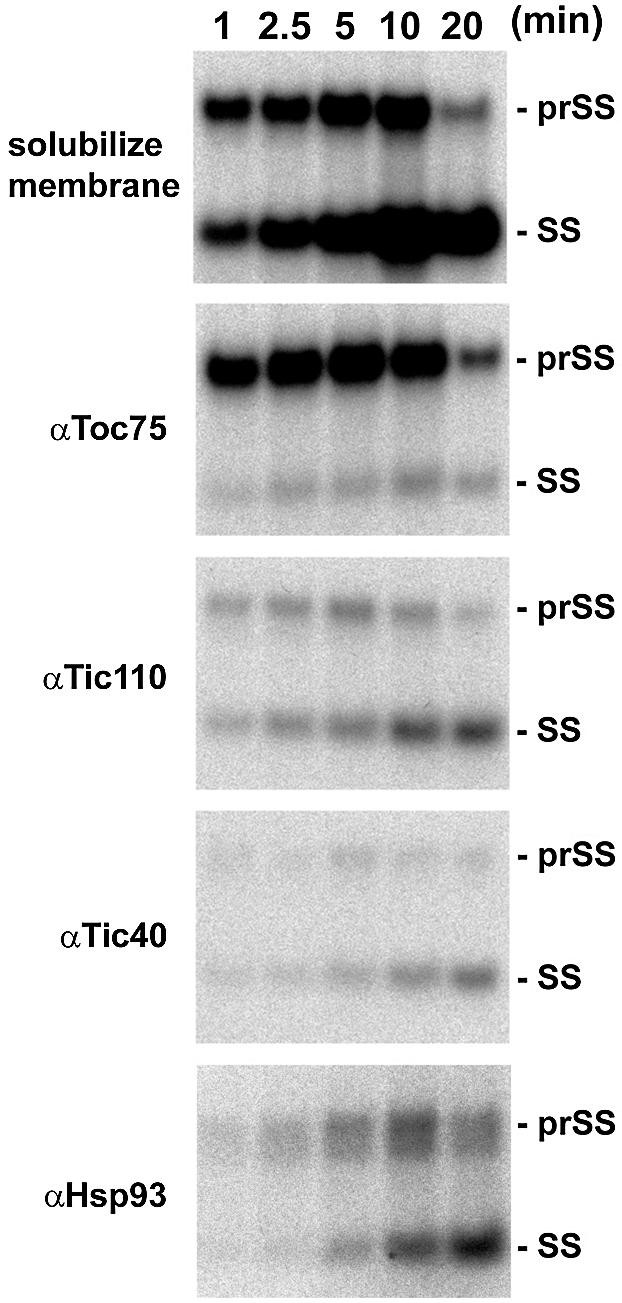
Fig. 8. Tic40 is associated with importing precursor proteins at the same stage as Tic110 and Hsp93. An import time course was performed with [35S]prSS. Reactions were terminated at various time points by diluting with cold import buffer. Chloroplasts were re- isolated, cross-linked with 0.5 mM DSP and lysed hypotonically. Total membranes were isolated; and the proteins were solubilized with Triton X-100 and immunoprecipitated with antibodies against Toc75, Tic110, Tic40 or Hsp93. Immunoprecipitates were analyzed by SDS–PAGE under conditions that cleaved the cross-links. Autoradiographs of the gels showing prSS and SS associated with each translocon component are presented.
The hydrophilic domain of Tic40 resembled the C-terminal half of co-chaperone Hop
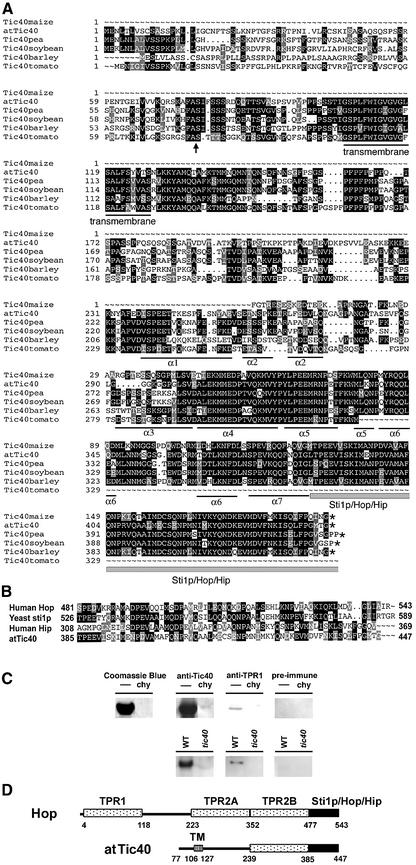
To obtain more structure–function relationship information on Tic40, we identified Tic40 sequences from several plant species from EST databases (Figure 9A). Sequence alignment showed that the best conserved regions are around the transmembrane domain and in the C-terminal half of the hydrophilic domain. The C-terminal end (residues 385–447 of atTic40) has primary sequence homology with the C-terminal globular domain shared by co-chaperones Hop (Hsp70 and Hsp90 organizing protein, also known as p60 or Sti1p) and Hip (Hsp70-interacting protein) (Figure 9B) (Höhfeld et al., 1995; Stahl et al., 1999; Webb et al., 2001). Tertiary structure prediction using the 3D-PSSM program indicated that the Sti1p/Hop/Hip domain in Tic40 is preceded immediately by seven α-helices that form a structure with similarity to the TPR domains of protein phosphatase 5 and Hop (PSSM E values 0.000105 and 0.0992 with 95 and 90% prediction certainty, respectively). This prediction was supported by the result that Tic40 proteins, both the E.coli overexpressed hydrophilic domain and the Arabidopsis chloroplast endogenous protein, were recognized by antibodies against TPR1 (Figure 9C). TPR1 interacts with neurofibromin and Hsc70, and contains a single TPR domain that shares 32% sequence identity with the TPR domain of yeast Sti1p (Murthy et al., 1996; Liu et al., 1999; Wu,S.-H. et al., 2001). Therefore, the structure of Tic40 may resemble that of the C-terminal half of Hop fused to a membrane anchor (Figure 9D).
Fig. 9. Sequence analyses of Tic40. (A) Sequence alignment of Tic40 from various plant species. After isolation and sequencing a pea Tic40 cDNA clone, we found several discrepancies between our sequence and the published pea Tic40 cDNA sequence (DDBJ/EMBL/GenBank accession No. AJ243758) (Stahl et al., 1999). Our sequence is shown here and has been submitted to the DDBJ/EMBL/GenBank database under the accession No. AY157668. The partial maize sequence is from the EST clone of accession No. BG550516. The soybean sequence is compiled from EST sequences BF066367+BM522900+BI971146_revers+BE021537+BE211020, the barley sequence from EST sequences BI949343+BJ464357+BI959205+ BJ468493_revers and the partial tomato sequence from EST sequences AW398666+BI934934. The asterisks represent stop codons. The proposed processing site for the transit peptidase is marked by an arrow. The putative transmembrane domain and the seven α-helices of the TPR domain are underlined. The second, fifth and sixth helices in the tertiary structure prediction are composed of non-continuous sequences. The Sti1p/Hop/Hip domain is underlined by a gray box. (B) Sequence alignment of the Sti1p/Hop/Hip domain from yeast Sti1p (accession No. NP014670), human Hop (accession No. NM006819), human Hip (accession No. XP165401) and atTic40. (C) Tic40 was recognized by anti-TPR1 antibodies. In the upper panel, an E.coli overexpressed atTic40 hydrophilic domain was analyzed by SDS–PAGE and revealed by Coomassie Blue staining or immunoblotting with antibodies against Tic40, TPR1 or the preimmune serum of anti-TPR1. In the lanes labeled ‘chy’, the atTic40 proteins had been purposely degraded by chymotrypsin. In the lower panel, total chloroplast proteins from the wild type or the tic40-1 mutant were analyzed by SDS–PAGE and immunoblotting with antibodies against Tic40, TPR1 or the preimmune serum of anti-TPR1. (D) A proposed domain structure of Tic40 compared with that of Hop. The numbers shown are the amino acid residue numbers for human Hop (Scheufler et al., 2000) and atTic40. TM, transmembrane domain.
Discussion
Previous data have shown that anti-Tic40 antibodies could immunoprecipitate a precursor protein arrested at the binding stage of import (Wu,C. et al., 1994) and that Tic40 could be cross-linked to Tic110 in isolated inner membrane vesicles (Stahl et al., 1999). With characterization of the tic40 mutants and further chemical cross-linking and immunoprecipitation analysis in intact chloroplasts, we provide additional evidence that Tic40 is a translocon component important for chloroplast protein import. In addition, we provide evidence that Tic40 functions during translocation across the inner envelope membrane, at a similar stage to that of Tic110 and Hsp93.
The tic40 knockout mutants share several similarities with the tic20 antisense mutants, but also show important differences. Both groups of mutants are much paler and smaller, and they developed more slowly than wild-type plants. Chloroplasts isolated from these mutants were specifically defective in translocation across the inner envelope membrane. However, the most severe tic20 mutants are not viable (Chen et al., 2002), whereas tic40 null mutants are viable. The non-lethal phenotype of tic40 mutants suggests that Tic40 plays an assisting role in protein import. If the primary function of Tic40 is to assist in protein import, it is not clear how a 60% reduction in the rate of precursor translocation leads to reductions in thylakoid biogenesis and developmental defects. We cannot exclude the possibility that Tic40 may have other functions in chloroplast biogenesis. More work is needed to understand how the protein import defects cause the observed phenotypes.
Several lines of evidence indicate that Tic40 functions as part of the stromal chaperone complex that facilitates protein translocation across the inner envelope membrane into the stroma. The hydrophilic domain of the Tic40 polypeptide is located on the stromal side of the inner envelope membrane. It forms a complex with Tic110 and the stromal chaperone Hsp93. Mutant chloroplasts without Tic40 were specifically defective in translocating precursor proteins across the inner membrane. During in vitro import reactions with mutant chloroplasts, significant quantities of the precursor protein, and even some processed mature proteins, were released back into the import reaction when the proteins failed to be translocated. From these observations, we conclude that tic40 mutant chloroplasts have defects in the ability of the import apparatus to interact with precursor proteins and translocate them into the stroma. Tic40 has sequence similarity to co-chaperones Hip and Hop. Hip binds to the ATPase domain of Hsc70 and stabilized Hsc70 in its ADP state. Hop binds to the C-termini of both Hsp70 and Hsp90 and coordinates the functions of these two chaperones (for a review, see Frydman and Höhfeld, 1997). Because Tic40 forms a complex with Tic110 and Hsp93, it may function as a co-chaperone to coordinate the action of Tic110 and Hsp93, and maybe even other as yet unidentified translocon components. Tic40 could also modulate the ATPase activity of Hsp93.
The C-terminal domain of Tic40 that shares sequence similarity with Sti1 and Hip is one of the best conserved regions among Tic40 sequences from various plant species. This region is also conserved from yeast (Sti1p) to human (Hop and Hip) to plant (Tic40), suggesting its importance. Unfortunately, the exact function of this domain is still not clear. Point mutations and deletion of this domain in Hip inhibit maturation of progesterone receptor complexes by blocking the entry of Hsp90 into the complex (Praparanich et al., 1998). In Hip, this domain, plus the preceding region containing the tetrapeptide GGMP repeats, has been shown to bind the ATPase domain of Hsp70 and unfolded protein substrates (Velten et al., 2002).
TPR domains are present in proteins of diverse biological function and are known to mediate protein– protein interaction. Each TPR domain consists of three or more TPR motifs, which are highly degenerated 34 amino acid repeats (Lamb et al., 1995). Each TPR motif forms a pair of antiparallel α-helices that interact with target proteins. Higher conservation in primary sequences is seen only among TPR domains with similar functions. For example, the TPR domain in Hip binds the N-terminal ATPase domain of Hsp70 (Höhfeld et al., 1995) and the TPR domains in Hop and TPR1 bind the C-terminus of Hsp70 or Hsp90 (Scheufler et al., 2000). These two groups of TPR domains have almost no sequence conservation between them. The TPR domain in Tic40 seems to have a similar structure to the TPR domains of Hop and TPR1. However, none of the residues in Hop TPR responsible for direct interaction with the C-terminus of Hsp70 and Hsp90 is present in the predicted TPR domain of Tic40. This suggests that Tic40 binds different target proteins. Tic110 and Hsp93 are the most obvious candidates as the target proteins of Tic40. However, there may be other as yet unidentified translocon components that are regulated by Tic40. It has also been shown that overexpressing a truncated Brassica Tic40 in E.coli could partially complement a temperature-sensitive mutation in secA, which encodes an ATPase component of the bacterial preprotein translocase (Pang et al., 1997). Overexpression of the same construct in wild-type E.coli cells resulted in a reduction in overall SecA levels under normal conditions and a more stable SecA profile when cells were treated with azide (Gordon and Ko, 2002). These results suggest that Tic40 may interact and stabilize SecA. However, in chloroplasts, SecA homolog is shown to function specifically in translocating a subgroup of thylakoid luminal proteins across the thylakoid membrane (Yuan et al., 1994). Further experiments are needed to identify the substrate of Tic40 in chloroplasts and map the interaction among components in the stromal chaperone complex.
Materials and methods
Materials
Percoll silica gel, trypsin (from bovine pancreas), thermolysin (type X from Bacillus thermoproteolyticus rokko) and Mg-ATP were obtained from Sigma (St Louis, MO). Nylon mesh was obtained from Sefar America Inc. (Kansas City, MO).
Growth of plants and isolation of chloroplasts
Pea (Pisum sativum cv. Little Marvel) plants were grown for 8–12 days in a greenhouse at 20°C under natural light or in growth chambers under a 12 h light/12 h dark cycle at 19°C. Chloroplasts were isolated as described (Fitzpatrick and Keegstra, 2001). For Arabidopsis, plants were grown on 2% sucrose, MS-supplemented agar for 4 weeks and chloroplasts were isolated using the protoplast procedure as described (Fitzpatrick and Keegstra, 2001). In all cases, Arabidopsis seeds were vernalized for 24–48 h at 4°C after sowing.
Production of antibodies
For antibodies against atTic40, a truncated version of the atTic40 cDNA, encoding the region from the methionine residue 138 to the C-terminal end, was cloned into the pET21a vector (Novagen, Madison, WI) with a C-terminal His6 tag. The truncated Tic40 protein was expressed in E.coli and purified from the soluble phase on a nickel column. Another batch of antibodies was produced with full-length atTic40 precursor fused at the C-terminus of glutathione transferase (GST) using the pGEX-5X-1 vector (Amersham Pharmacia). The overexpressed fusion proteins were purified from the soluble phase of E.coli cell lysates by glutathione-conjugated Sepharose (Amersham Pharmacia).
Isolation of tic40 mutants
The tic40-1 mutant was obtained from the Arabidopsis Biological Resource Center in the Feldmann Collection (stock number cs2774). When grown on media containing kanamycin, F2 progeny from a cross between cs2774 and WS2 showed a segregation ratio of ∼1:2:1 wild-type sensitive/wild-type resistant/mutant resistant, indicating a single T-DNA insertion linked to the mutant phenotype. The tic40-2 mutant was isolated by screening T2 seeds from a T-DNA insertion population for seedling-lethal phenotypes (Budziszewski et al., 2001; line 2490). T3 seeds from single T2 resistant plants were collected and two ‘subfamilies’ were identified with a single T-DNA insertion locus (resistant-to-sensitive ratios of 1.9 and 3.0). All of the 40 T3 resistant plants segregated the mutant phenotype in the T4 generation. The T-DNA insertion position was identified by TAIL-PCR [tic40-1 (Liu et al., 1995)] or plasmid rescue [tic40-2 (Budziszewski et al., 2001)]. In tic40-1, the T-DNA left-border sequence starts behind base 707 (amino acid residue 122), if the translation initiation site is counted as base 1. In tic40-2, the T-DNA left-border sequence starts behind base 2395 (amino acid residue 316).
Translation of precursor proteins
The plasmid containing the cDNA clone for prSS was transcribed in vitro and translated using labeled [35S]methionine as described previously (Perry et al., 1991). Other precursor substrates (for Tic40, Toc75, tp110-110N, CAB, PC) were transcribed and translated in a coupled system containing reticulocyte lysate (Promega Corp., Madison, WI). The system contained T7 (Tic40, tp110-110N), T3 (Toc75) or SP6 (PC, CAB) RNA polymerases and [35S]methionine.
Protein import and thermolysin treatments
For import experiments, wild-type and tic40 plants were grown on agar plates (Fitzpatrick and Keegstra, 2001). Under these conditions, because the wild-type plants grew no more than 1.5 cm in diameter, there was no dramatic difference in size between wild type and tic40. Import assays were standardized to contain the same amount of chlorophyll for the wild-type and tic40 chloroplasts. The number of chloroplasts per milligram of chlorophyll was not significantly different between wild-type and tic40 chloroplast preparations. Chloroplasts at a final concentration of 0.16 mg chlorophyll/ml were incubated with 4–10% (v/v) precursor in 100–300 µl of import buffer supplemented with up to 5 mM ATP. Reactions were incubated at room temperature and stopped by sedimenting the intact chloroplasts through a 40% Percoll cushion at 4°C. Pellets were washed once with import buffer. Thermolysin treatment of chloroplasts was carried out as described (Fitzpatrick and Keegstra, 2001).
Cross-linking and immunoprecipitation
Cross-linking was carried out in pea chloroplasts as described (Akita et al., 1997). Essentially, isolated pea chloroplasts were incubated with 0.5 mM DSP in import buffer for 15 min and quenched with 50 mM glycine for 15 min on ice. Chloroplasts were re-isolated and lysed by resuspending in a hypotonic buffer (25 mH HEPES–KOH pH 8.0, 4 mM MgCl2). Total membranes were collected by pelleting the lysate at 125 000 g for 45 min. Membranes were solubilized by 1% Triton in immunoprecipitation buffer (Akita et al., 1997) and the lysate was clarified by centrifuging at 100 000 g for 5 min. The supernatant was diluted with an equal volume of immunoprecipitation buffer to reduce the Triton concentration to 0.5%. Solubilized membranes were immunoprecipitated with anti-Tic40 antibodies and protein A–conjugated agarose. For the experiment shown in Figure 7B, anti-Toc75 antibodies were first cross-linked to protein A–agarose using the Seize X Protein A Immunoprecipitation Kit (Pierce, Rockford, IL). This is necessary because Tic40 often co-migrates with the IgG heavy chain. The immunoprecipitates were washed and cross-links were cleaved with β-mercaptoethanol or dithiothreitol (DTT) in the SDS–PAGE sample buffer. The antibodies against Hsp70 were raised against the C-terminal 10 kDa fragment of an Arabidopsis cytosolic Hsp70 (Wu,S.-H. et al., 1994) and they cross-reacted with at least two Hsp70 proteins in chloroplasts (data not shown), one in the total membrane fraction and at least one in the stroma (Marshall et al., 1990). The antibodies against TPR1 were raised against the full-length human TPR1.
Gel quantification and immunoblotting
Gels containing radiolabeled proteins were subjected to fluorography and exposed to X-ray film (Eastman Kodak) or quantified using the PhosphorImager (Molecular Dynamics SP, Sunnyvale, CA) or the Fuji FLA-5000 Imaging System (Fujifilm, Tokyo). For immunoblots, secondary antibodies were linked to alkaline phosphatase or horseradish peroxidase.
Acknowledgments
Acknowledgements
We thank the ABRC for the tic40-1 mutant seeds, Dr David Meinke (Oklahoma State University) for the co-segregation analysis on tic40-2, Dr Hwa Dai for the monoclonal antibodies against maize mitochondrial porin, Dr Chung Wang for antibodies against TPR1 and Hsp70, Dr Neil Hoffman for antibodies against CAB and OE33, and Dr Farhad Forouhar for assistance in tertiary structure prediction of Tic40.
References
- Akita M., Nielsen,E. and Keegstra,K. (1997) Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol., 136, 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J., Hiltbrunner,A., Weibel,P., Vidi,P.-A., Alvarez-Huerta,M., Smith,M.D., Schnell,D.J. and Kessler,F. (2002) Essential role of the G-domain in targeting of the protein import receptor atToc159 to the chloroplast outer membrane. J. Cell Biol., 159, 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerle C., Dorl,J. and Keegstra,K. (1991) Kinetic analysis of the transport of thylakoid lumenal proteins in experiments using intact chloroplasts. J. Biol. Chem., 266, 5884–5890. [PubMed] [Google Scholar]
- Budziszewski G.J. et al. (2001) Arabidopsis genes essential for seedling viability: isolation of insertional mutants and molecular cloning. Genetics, 159, 1765–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Smith,M.D., Fitzpatrick,L. and Schnell,D.J. (2002) In vivo analysis of the role of atTic20 in protein import into chloroplasts. Plant Cell, 14, 641–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick L.M. and Keegstra,K. (2001) A method for isolating a high yield of Arabidopsis chloroplasts capable of efficient import of precursor proteins. Plant J., 26, 59–66. [DOI] [PubMed] [Google Scholar]
- Frydman J. and Höhfeld,J. (1997) Chaperones get in touch: the Hip–Hop connection. Trends Biochem. Sci., 22, 87–92. [DOI] [PubMed] [Google Scholar]
- Gordon B. and Ko,K. (2002) The plastid translocon component TOC36 exhibits an affinity for the bacterial protein translocation process. Arch. Biochem. Biophys., 404, 147–157. [DOI] [PubMed] [Google Scholar]
- Heins L., Mehrle,A., Hemmler,R., Wagner,R., Kuchler,M., Hormann,F., Sveshnikov,D. and Soll,J. (2002) The preprotein conducting channel at the inner envelope membrane of plastids. EMBO J., 21, 2616–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltbrunner A., Bauer,J., Vidi,P.-A., Infanger,S., Weibel,P., Hohwy,M. and Kessler,F. (2001) Targeting of an abundant cytosolic form of the protein import receptor atToc159 to the outer chloroplast membrane. J. Cell Biol., 154, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnah S.C., Hill,K., Wagner,R., Schlicher,T. and Soll,J. (1997) Reconstitution of a chloroplast protein import channel. EMBO J., 16, 7351–7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhfeld J., Minami,Y. and Hartl,F.U. (1995) Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell, 83, 589–598. [DOI] [PubMed] [Google Scholar]
- Jackson D.T., Froehlich,J.E. and Keegstra,K. (1998) The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J. Biol. Chem., 273, 16583–16588. [DOI] [PubMed] [Google Scholar]
- Jackson-Constan D. and Keegstra,K. (2001) Arabidopsis genes encoding components of the chloroplast protein import apparatus. Plant Physiol., 125, 1567–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F. and Blobel,G. (1996) Interaction of the protein import and folding machineries in the chloroplast. Proc. Natl Acad. Sci. USA, 93, 7684–7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko K., Budd,D., Wu,C., Seibert,F. and Kourtz,L. (1995) Isolation and characterization of a cDNA clone encoding a member of the Com44/Cim44 envelope components of the chloroplast protein import apparatus. J. Biol. Chem., 270, 28601–28608. [DOI] [PubMed] [Google Scholar]
- Kouranov A., Chen,X., Fuks,B. and Schnell,D.J. (1998) Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol., 143, 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J., Tugendreich,S. and Hieter,P. (1995) Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem. Sci., 20, 257–259. [DOI] [PubMed] [Google Scholar]
- Liu F., Wu,S., Hu,S., Hsiao,C. and Wang,C. (1999) Specific interaction of the 70 kDa heat shock cognate protein with the tetratricopetide repeats. J. Biol. Chem., 274, 34425–34432. [DOI] [PubMed] [Google Scholar]
- Liu Y.-G., Mitsukawa,N., Oosumi,T. and Whittier,R.F. (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J., 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Lübeck J., Heins,L. and Soll,J. (1997) A nuclear-coded chloroplastic inner envelope membrane protein uses a soluble sorting intermediate upon import into the organelle. J. Cell Biol., 137, 1279–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Kouranov,A., LaSala,S.E. and Schnell,D.J. (1996) Two components of the chloroplast protein import apparatus IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. J. Cell Biol., 134, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J., DeRocher,A., Keegstra,K. and Vierling,E. (1990) Identification of heat shock protein hsp70 homologues in chloroplasts. Proc. Natl Acad. Sci. USA, 87, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy A., Bernards,A., Church,D., Wasmuth,J. and Gusella,J. (1996) Identification and characterization of two novel tetratricopeptide repeat-containing genes. DNA Cell Biol., 15, 727–735. [DOI] [PubMed] [Google Scholar]
- Nielsen E., Akita,M., Davila-Aponte,J. and Keegstra,K. (1997) Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J., 16, 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang P., Meathrel,K. and Ko,K. (1997) A component of the chloroplast protein import apparatus functions in bacteria. J. Biol. Chem., 272, 25623–25627. [DOI] [PubMed] [Google Scholar]
- Perry S.E. and Keegstra,K. (1994) Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell, 6, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S.E., Li,H.-m. and Keegstra,K. (1991) In vitro reconstitution of protein transport into chloroplasts. Methods Cell Biol., 34, 327–344. [DOI] [PubMed] [Google Scholar]
- Postle K. (1990) TonB and the Gram-negative dilemma. Mol. Microbiol., 4, 2019–2025. [DOI] [PubMed] [Google Scholar]
- Praparanich V., Chen,S. and Smith,D. (1998) Mutation of Hip’s carboxy-terminal region inhibits a transitional stage of progesterone receptor assembly. Mol. Cell. Biol., 18, 944–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Davila-Aponte,J. and Keegstra,K. (1999) The evolutionary origin of the protein-translocating channel of chloroplastic envelope membranes: identification of a cyanobacterial homolog. Proc. Natl Acad. Sci. USA, 96, 784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheufler C., Brinker,A., Bourenkov,G., Pegpraro,S., Morder,L., Bartunik,H., Hartl,F. and Moarefi,I. (2000) Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell, 101, 199–210. [DOI] [PubMed] [Google Scholar]
- Schnell D.J., Blobel,G., Keegstra,K., Kessler,F., Ko,K. and Soll,J. (1997) A consensus nomenclature for the protein-import components of the chloroplast envelope. Trends Cell Biol., 7, 303–304. [DOI] [PubMed] [Google Scholar]
- Stahl T., Glockmann,C., Soll,J. and Heins,L. (1999) Tic40, a new ‘old’ subunit of the chloroplast protein import translocon. J. Biol. Chem., 274, 37467–37472. [DOI] [PubMed] [Google Scholar]
- Tu S.-L. and Li,H.-M. (2000) Insertion of OEP14 into the outer envelope membrane is mediated by proteinaceous components of chloroplasts. Plant Cell, 12, 1951–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velten M., Gomez-Vrielynck,N., Chaffotte,A. and Ladjimi,M.M. (2002) Domain structure of the HSC70 cochaperone, HIP. J. Biol. Chem., 277, 259–266. [DOI] [PubMed] [Google Scholar]
- Webb M.A., Cavaletto,J.M., Klanrit,P. and Thompson,G.A. (2001) Orthologs in Arabidopsis thaliana of the Hsp70 interacting protein Hip. Cell Stress Chaperones, 6, 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Seibert,F.S. and Ko,K. (1994) Identification of chloroplast envelope proteins in close physical proximity to a partially translocated chimeric precursor protein. J. Biol. Chem., 269, 32264–32271. [PubMed] [Google Scholar]
- Wu S.-H., Wang,C., Chen,J. and Lin,B.-L. (1994) Isolation of a cDNA encoding a 70 kDa heat-shock cognate protein expressed in vegetative tissues of Arabidopsis thaliana. Plant Mol. Biol., 25, 577–583. [DOI] [PubMed] [Google Scholar]
- Yuan E., Henry,R., McCaffery,M. and Cline,K. (1994) SecA homolog in protein transport within chloroplasts: evidence for endosymbiont-derived sorting. Science, 266, 796–798. [DOI] [PubMed] [Google Scholar]