Abstract
Spliceosome-mediated RNA trans-splicing (SMaRT) has been used previously to reprogram mutant endogenous CFTR and factor VIII mRNAs in human epithelial cell and tissue models and knockout mice, respectively. Those studies used 3′ exon replacement (3′ER); a process in which the distal portion of RNA is reprogrammed. Here, we also show that the 5′ end of mRNA can be completely rewritten by 5′ER. For proof-of-concept, and to test whether 5′ER could generate functional CFTR, we generated a mutant minigene target containing CFTR exons 10–24 (ΔF508) and a mini-intron 10, and a pretrans-splicing molecule (targeted to intron 10) containing CFTR exons 1–10 (+F508), and tested these two constructs in 293T cells for anion efflux transport. Cells cotransfected with target and PTM showed a consistent increase in anion efflux, but there was no response in control cells that received PTM or target alone. Using a LacZ reporter system to accurately quantify trans-splicing efficiency, we tested several unique PTM designs. These studies provided two important findings as follows: (1) efficient trans-splicing can be achieved by binding the PTM to different locations in the target, and (2) relatively few changes in PTM design can have a profound impact on trans-splicing activity. Tethering the PTM close to the target 3′ splice site (as opposed to the donor site) and inserting an intron in the PTM coding resulted in a 65-fold enhancement of LacZ activity. These studies demonstrate that (1) SMaRT can be used to reprogram the 5′ end of mRNA, and (2) efficiency can be improved substantially.
Keywords: CFTR; genetic disease; mRNA repair; SMaRT, trans-splicing
INTRODUCTION
Many recent developments underscore the importance of RNA as a tool and a target in biomedical applications (Sullenger and Gilboa 2002). The application of RNA interference to efficiently and specifically knockdown expression of mammalian gene products has opened a novel avenue for experimental and therapeutic applications designed to reduce levels of an undesirable protein (Shi 2003). From a therapeutic standpoint, it is equally important to recover activities lost in disease states or reprogram mRNA to express a completely different protein. A second RNA technology, mRNA reprogramming by trans-splicing, offers the ability to repair mRNAs, and thus, proteins.
RNA reprogramming has been achieved using group I trans-splicing ribozymes and by spliceosome-mediated RNA trans-splicing (SMaRT). SMaRT is a unique technology with many proven and potential applications (Garcia-Blanco et al. 2000). Recently, we demonstrated the feasibility of using SMaRT to repair endogenous cystic fibrosis (CF) mRNAs encoding the cystic fibrosis transmembrane conductance regulator (CFTR) in human epithelial cells in culture and in a xenograft model (Mansfield et al. 2000; Liu et al. 2002), and factor VIII mRNA in hemophilia A knockout mice (Chao et al. 2003), using 3′ exon replacement (3′ER). During 3′ER, the distal portion of a target mRNA is completely rewritten by trans-splicing sequences between the target pre-mRNA and a repair construct called a pre-trans-splicing molecule (PTM) (Puttaraju et al. 1999, 2001). To effect CFTR 3′ end repair, a PTM was designed to base pair with the 3′ splice site of intron 9 and trans-splice the normal coding sequences of exons 10–24 (in which exon 10 of the target carries the CF ΔF508 deletion) (Mansfield et al. 2000; Liu et al. 2002). 3′ER PTMs are composed of several modular domains that include binding-domain sequences that are complementary to the target pre-mRNA, a spacer sequence, and a 3′ splice site followed by virtually any coding sequence. The 3′ splice site typically includes a branchpoint consensus sequence, polypyrimidine tract, and a splice acceptor AG dinucleotide. We term these latter three elements together with the binding domain and spacer sequence, the trans-splicing domain (TSD). This region constitutes the active site of the PTM, because it contains the sequence elements that are necessary for efficient trans-splicing. All of the PTM domains described above can be changed to alter the efficiency and specificity of SMaRT (Puttaraju et al. 1999, 2001; Mansfield et al. 2000).
The strategy of mRNA correction offers several potential advantages, such as reduced transgene size, elimination of ectopic expression of the repaired product, the acquisition of endogenous regulation, and the ability to include large regulatory elements in vectors with limited packaging capacity. The need to repair mutations throughout an mRNA and the versatility of SMaRT technology led us to perform experiments designed to reprogram the 5′ region of mRNA by 5′ exon replacement (5′ER). The consensus sequence that defines the 5′ donor site, AG/GURAGU, is less extensive than the 3′ splice elements (Moore et al. 1999), and it has remained uncertain whether this site could be efficiently exploited to effect trans-splicing repair. Group I ribozymes have been used to perform trans-splicing reactions in vitro to repair globin mRNA in sickle cell anemia-derived erythroid precursor cells (Lan et al. 1998), and trans-splicing ribozymes have also been used successfully to induce changes in p53 and chloride channels (Watanabe and Sullenger 2000; Rogers et al. 2002) by a 3′ replacement process; to our knowledge, there is no published report demonstrating that ribozymes are capable of repair in a 5′ manner. Additionally, in trypanosomes (Van der Ploeg 1986) and Caenorhabditis elegans (Krause and Hirsh 1987), it has been demonstrated that RNAs are commonly modified by the addition of a splice leader on the 5′ terminus by trans-splicing. This process is different from SMaRT in that the latter does not represent a replacement of sequences between the target and modifying molecule. In the present study, our key questions were as follows: (1) whether the 5′ end of mRNA could be efficiently reprogrammed; (2) how the efficiency of the process could be optimized; and (3) which regions of a pre-mRNA, that is, exon, intron, splice junction, or a combination of these, could be targeted to increase trans-splicing. The ability to reprogram the 5′ region of mRNAs confers a unique advantage to SMaRT that is particularly relevant for diseases in which the mutation(s) are 5′ proximal in the RNA.
Using CFTR as a model of a genetic disease, and a β-galactosidase repair model to quantify levels of functional restoration, we demonstrate here that PTMs can be engineered to repair mutant mRNA by 5′ER. This work is unique because, to date, alteration of RNA or DNA has focused on 3′ ER. We show that trans-splicing is precise and can replace coding sequences of at least 1.9 kb, and that the efficiency of 5′ER can be increased many fold by targeting different regions of pre-mRNA. This advancement supports broader applications of SMaRT technology and suggests that regardless of position within the transcript, any codon can be corrected or any sequence modified by substitution.
RESULTS
SMaRT modification of mRNA by 5′ exon replacement
Previous work to date has focused primarily on PTMs that utilize a 3′ splice site and have been termed 3′ER PTMs (Puttaraju et al. 1999, 2001; Mansfield et al. 2000). To test the efficiency of repair by 5′ER, we developed a mutant ΔF508 CFTR minigene target and a 5′ER PTM for repairing the target mRNA. This PTM, CFTR-PTM11, contained the normal human CFTR-coding sequences for exons 1–10 (including codon F508 in exon 10), a TSD consisting of a 5′ donor site (AG/GUAAGA), a spacer sequence, and a binding domain of 31 bases complementary to the target. The first binding domain was designed to bind the exon 10-intron 10 junction of the target, with the intention of blocking cis-splicing. The target, CFTR-T11, consisted of mutant exon 10 (ΔF508), a mini-intron 10 containing the 5′ and 3′ ends of intron 10 from the human CFTR gene, and the coding sequence for exons 11 through 24 (Fig. 1A ▶). The repair process for 5′ER is illustrated in Figure 1A ▶.
FIGURE 1.
CFTR model constructs and illustration of trans-splicing by 5′ exon replacement. (A) Detailed structure of a 5′ER PTM (CFTR-PTM11), and a mini-gene target (CFTR-T11). The PTM lacks a poly(A) signal and the target lacks a methionine initiator codon. Modified codons are engineered in exons 9 and 10 of the PTM, and exons 11 and 12 of the target to aid in distinguishing trans-spliced products from cis-spliced or endogenous CFTR products. The position of the three oligonucleotide primers (CF1, CF93, and CF111) used in RT–PCR experiments are indicated by arrows. The diagram shows the PTM binding to the 5′ splice site of intron 10 of the minigene target at the pre-mRNA level. (B) Demonstration of trans-splicing by RT–PCR. Cells were transfected with either PTM plus target, or PTM, target, or vector (pc3.1DNA) alone, or PTM plus target, but without the reverse transcription step (four different negative controls). Cis-spliced (ΔF508) products were detected with primers CF1 and CF111 (right arrow), and trans-spliced products with primers CF93 and CF111 (left arrow). (BD) Binding domain.
CFTR-PTM11 and CFTR-T11 were cotransfected into human embryonic kidney 293T cells (293T), the cells were harvested 48 h after transfection, and total RNA was isolated. Trans- and cis-spliced RNA products were detected by RT–PCR using primers that would specifically amplify cis- (CF1 and CF111) and trans-spliced (CF93 and CF111) products. In those samples that received PTM, target, or pc3.1DNA(-) vector alone, or PTM plus target, but with no reverse transcription, no PCR products were detected. However, in the sample that received PTM and target, a specific product of 265 bp was generated, which is the predicted size for a trans-spliced product between the PTM and target (Fig. 1B ▶). Sequencing of these presumptive cis- and trans-spliced cDNAs confirmed (1) the identity of each product (data not shown); (2) that trans-splicing took place at the intended splice sites; and (3) that the repaired product contained modified codons in exon 10. These three observations demonstrate that the repaired product was generated by trans-splicing between the target and PTM. Semiquantitative RT–PCR assays using primers that were specific for cis- and trans-spliced mRNAs showed that the level of trans-splicing was ~6% of target pre-mRNA cis-splicing (data not shown). These results clearly show that trans-splicing can be used to efficiently reprogram the 5′ end of mRNA by the substitution of target sequences with PTM sequences.
Repair of CFTR ΔF508 pre-mRNA by 5′ER produces functional ion channels in cells
The next goal of this study was to demonstrate that trans-splicing by 5′ER can produce copies of CFTR that are functional as ion transporters at the cell membrane. CFTR is a transmembrane protein that regulates the flow of chloride ion across the cell membrane in epithelial cells in a cAMP-dependent fashion. For this experiment, we used a CFTR-based PTM (CFTR-PTM30), with a binding domain complementary to exon 10 and its splice sites. Anion efflux was compared in 293T cells transfected with target only (CFTR-T11), PTM only (CFTR-PTM30), or with both. Under baseline conditions, anion efflux was equivalent under all three conditions. Following forskolin and IBMX treatment of 293T monolayers (which is predicted to elevate cAMP levels and stimulate cAMP-dependent ion channel activity), there was a consistent increase in anion efflux transport response in cells cotransfected with target plus PTM. In contrast, there was no response in cells transfected with target alone and only a minor response in cells receiving PTM alone (arrows, Fig. 2 ▶). It is unclear why values for target and PTM alone are not identical, although similar results were obtained for 3′ER CFTR PTMs (S.G. Mansfield, R.H. Clark, M. Puttaraju, L.G. Mitchell, and M.A. Garcia-Blanco, unpubl.). These results suggest that full-length repaired mRNA is generated by trans-splicing and that SMaRT is capable of generating functional CFTR ion channels in cells.
FIGURE 2.
RNA trans-splicing by 5′ exon replacement generates functional ion channels in human cells. HEK 293T cells were transfected with either target alone (CFTR-T11), PTM alone (CFTR-PTM30), or with target and PTM. Cell populations were incubated in medium containing 125Iodine, and then tested for the presence of ion channel efflux activity (in min−1) by stimulation with forskolin and IBMX at intervals of 3 min (time points 7, 10, 13, and 16 min). Forskolin is predicted to elevate cAMP levels and stimulate cAMP-dependant ion channel activity. Only those monolayers that received a PTM and target showed substantial increases in anion efflux at three time-points (arrows). Values represent mean ± standard error of two independent transfections, with the exception of the 4-min time point for PTM plus target, which represents a single data point. (BD) Binding domain.
5′ Exon replacement efficiency can be increased significantly
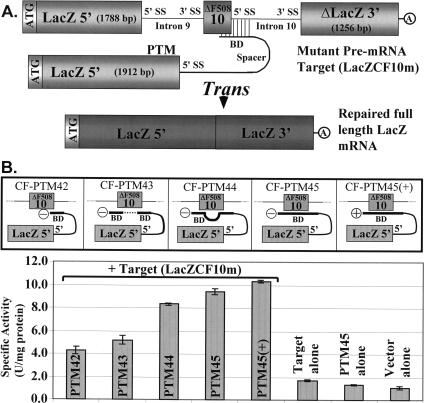
For 3′ER, we had demonstrated previously that increasing the binding domain length from 23 to 153 nucleotides gave much higher trans-splicing efficiency (Puttaraju et al. 2001). To assess how the efficiency of 5′ER can be optimized, we developed a mutant LacZ-CFTR repair model consisting of the LacZ coding unit (minus 124 bases) split into two exons of 1788 and 1256 bp, and separated by CFTR mini-introns 9 and 10, and a mutant exon 10 (ΔF508) (Fig. 3A ▶, LacZCF10m). We engineered PTMs encoding the 5′ portion of LacZ (1912 bp) (Fig. 3A ▶) and a binding domain complementary to different regions of the LacZ-CFTR target (Fig. 3B ▶). Cis-splicing of all exons or just the two LacZ exons (Ibid.) (through exon skipping) does not encode functional β-galactosidase. However, accurate 5′ER trans-splicing between the target and a LacZ PTM is predicted to produce a functional LacZ mRNA (Fig. 3A ▶). To test whether binding different areas of the target could result in increased trans-splicing, we generated five different PTMs with binding domains that would occlude either the 3′ splice site of intron 9, the 5′ splice site of intron 10, exon 10, or a combination of these (Fig. 3B ▶). The binding domain of CF-PTM42 covered the 5′ splice site of intron 10. CF-PTM43 had a binding domain complementary to noncontiguous sequence in the target, such that it would bind to the 3′ and 5′ splice sites of introns 9 and 10, respectively. CF-PTM44 was similar to CF-PTM43, except that we included a 140-bp linker sequence (nonbinding) between the two sequence units that were complementary to the intron 9 and 10 splice sites. The binding domain of CF-PTM45 and CF-PTM45+ was designed to cover the 3′ splice site of intron 9, the entire exon 10, and the 5′ splice site of intron 10. CF-PTM42 to CF-PTM45 were engineered without a bovine growth hormone (bGH) poly(A) signal to reduce the possibility of PTM nucleo-cytoplasmic transport and translation in the absence of trans-splicing. CF-PTM45+ has a poly(A) signal, and was constructed to compare how the presence or absence of this signal affects trans-splicing. Each PTM was transiently cotransfected into 293T cells with target LacZCF10m, and cell lysates were assayed for β-galactosidase activity after 48 h. These experiments showed that occluding the competing exon 10 can significantly enhance trans-splicing efficiency (2.2-fold over the first design; compare CF-PTM42 and CF-PTM45 in Fig. 3B ▶), although an effect due to size of the binding domain cannot be formally excluded. The inclusion of a bGH poly(A) site slightly enhanced the trans-splicing efficiency. Additional experiments testing whether an alpha mosaic virus translational enhancer sequence (Jobling and Gehrke 1987) and a consensus Kozak sequence around the ATG would enhance expression of the mRNA did not show the expected effect, indeed, these modifications caused a slight decrease in activity (data not shown). Finally, in another experiment in which CF-PTM45 was engineered with a cis-acting hammerhead ribozyme (capable of self-cleavage) (Chowrira et al. 1994) with the intention of generating a poly(A) minus PTM with a precise 3′ end, we did not observe any enhancement of trans-splicing (data not shown).
FIGURE 3.
Model system to optimize trans-splicing efficiency. (A) Schematic diagram showing a LacZ PTM binding to a chimeric LacZ-ΔF508-CFTR target at the pre-mRNA level. (B) Trans-splicing efficiency by 5′ exon replacement is increased by binding to more than one splice site. The panels above the histogram shows the location of four different PTM-binding domains (CF-PTM42, CF-PTM43, CF-PTM44, CF-PTM45) targeted to different regions of a defective LacZ-ΔF508 target (LacZCF10m). CF-PTM45 and CF-PTM45(+) are identical, except that the latter has a poly(A) signal and the former does not. The histogram below the five panels compares the repair efficiency of the five PTMs using an in-solution β-galactosidase assay. Transfections, preparation of cell lysates, and assessment of activity was as described previously (Puttaraju et al. 2001). Values are the mean of two independent experiments (± standard error) and are expressed as units of β-galactosidase activity per milligram of total protein. PTMs and the target are cloned in pc3.1DNA(−). The vector-alone sample refers to plasmid pc3.1DNA(−) that does not contain either a PTM or a target. Each transfection was performed with 2 μg of target plasmid and 2 μg of PTM plasmid. All samples contained the same amount of plasmid DNA; PTM-alone and target-alone samples were balanced to 4 μg with vector pc3.1DNA(−). (BD) Binding domain; (SS) splice site; (+), with bGH poly(A) signal; (−), without bGH polyA signal.
To substantiate the results from the in-solution β-galactosidase assays, we also performed in situ staining with several cell populations that had either received PTM and target, PTM alone, or target alone. Cells stained strongly positive for β-galactosidase only in samples that had received a PTM and a target (data not shown). The first series of PTMs described above demonstrate that mRNA can be rewritten at its 5′ end by SMaRT, the process is precise and the efficiency can be altered by binding at different regions in the target, but the process is relatively inefficient using these particular PTMs.
PTMs targeted closer to the target acceptor site produce a significant increase in efficiency
The first set of 5′ER designs (CF-PTM42 to CF-PTM45) for this particular study were made to occlude the 5′ and/or 3′ splice site of CFTR mini-intron 10 and suppress cis-splicing. This theory of blocking the cis-splicing elements had been first tested in 3′ER PTMs, in which binding domains were developed to occlude the branchpoint, polypyrimidine tract, and acceptor site of the target intron. However, more recent work has shown clearly that this design feature is not essential for effective trans-splicing, and that other regions of an intron can be targeted (S.A. Mansfield, P. Du, S. Hiriyanna, R.C. Bartel, and E. Otto, unpubl.). We hypothesized that targeting the 5′ER PTM close to the 3′ end of the target intron would bring the PTM donor site in close proximity to the target acceptor site, but without sterically occluding the 3′ splice site (required for trans-splicing). CF-PTM50 was re-engineered with a binding domain of 125 nucleotides complementary to sequence just upstream (position −62 to −186) of the intron 9 predicted branchpoint. The final construct was termed CF-PTM53 (Fig. 4 ▶). In these constructs, the length of sequence between the donor site and binding domain (referred to as a spacer sequence in 3′ER constructs; Puttaraju et al. 1999) was shortened from 60 to 10 bases, firstly to assess the effect of the spacer on PTM activity, and secondly to ensure that the PTM donor and target acceptor were in close proximity. Both constructs are identical with the exception of the binding domain position and size. Retargeting the PTM in this manner resulted in a 10-fold increase in trans-splicing (Fig. 4 ▶). Thus, although the PTMs differ in their binding-domain size, these data suggest that approximating the PTM near the 3′ splice site enhances the efficiency of trans-splicing quite substantially. At the very least, we can conclude that targeting the PTM to a region near the reactive splice site can lead to high levels of trans-splicing.
FIGURE 4.
Targeting the PTM closer to the intended 3′ splice site results in a substantial increase in trans-splicing efficiency. The panel above the histogram shows a schematic of two different PTMs and the location of their binding domain with respect to the target acceptor site. The binding domain of CF-PTM53 is located ~400 bases downstream of CF-PTM50. Both PTMs have a spacer sequence that is shorter than all previous constructs by 50 bases. Values are mean ± standard error of three separate experiments. (BD) Binding domain.
To dissect whether spacer length was important for efficient trans-splicing, we made two comparisons. Firstly, we compared CF-PTM50 with a long and short spacer (60 bases versus 10 bases) and showed that there was a 7% difference in trans-splicing between the two (n = 6; data not shown), and secondly, we showed that the difference between CF-PTM50 and CF-PTM53 was also very similar whether the spacer was long or short (n = 7, ninefold difference; data not shown). Thus, it appears that in this study, the spacer length (in the range that we tested) did not alter trans-splicing significantly.
The presence of an intron in the PTM increases trans-splicing
For 5′ER to work efficiently, the donor site of the PTM must be efficiently recognized by the splicing machinery. For internal exons, the close proximity of the splice sites that flank an exon are thought to help in exon definition (Berget 1995). Because 5′ER PTMs typically contain a large coding unit followed by a single splice site, we hypothesized that the recognition of this donor site is weak. To test this hypothesis, we introduced two changes to the PTM as follows: (1) we inserted a CFTR mini-intron 9 into the coding unit close to the PTM donor site (to create two exons of 1788 and 124 bp) (Fig. 5 ▶), and (2) we placed an IAS1 sequence element in the trans-splicing domain just downstream of the PTM donor. The latter element (IAS1) is a U-rich sequence that enhances the splicing of the alternative exon IIIb from the fibroblast growth factor receptor 2 gene (Del Gatto-Konczak et al. 2000). The presence of an IAS1 sequence (inserted at position +12 in the trans-splicing domain) produced no significant increase in activity (data not shown). The CFTR mini-intron sequence may serve two functions. Firstly, it may improve exon definition, and secondly, it may serve to increase the level of protein product by mechanisms other than splicing, such as increased mRNA processing, nucleo-cytoplasmic transport, and translation (Lu and Cullen 2003; Nott et al. 2003). In cotransfection experiments, PTMs containing a mini-intron gave a 3.4-fold increase in β-galactosidase activity (Fig. 5 ▶, cf. CF-PTM54 and CF-PTM59). In absolute units of β-galactosidase activity, the 199-mU/mg protein was the highest observed and represented a 65-fold increase over that obtained with the original PTM (CF-PTM42).
FIGURE 5.
Inserting an intron in the PTM coding unit results in a significant improvement in trans-splicing efficiency. The two PTMs shown in the panel above the histogram are identical, except that CF-PTM59 has a 543-bp CFTR mini-intron 9 in the LacZ coding unit. The 3′ LacZ exon in CF-PTM59 is 124 bp long. Values are mean ± standard error of two separate experiments. (BD) Binding domain.
DISCUSSION
5′ Exon replacement
This work is significant because it demonstrates that SMaRT can be used to efficiently reprogram the 5′ end of mRNA. This process, termed 5′ exon replacement (5′ER), has many potential applications, including gene correction, imaging of living cells in vitro or in vivo, or suicide gene therapy. We have shown that (1) trans-splicing by SMaRT can be performed with coding sequences of at least 1.9 kb; (2) trans-splicing is precise; (3) trans-splicing can be achieved by tethering the PTM to the target in several different locations; (4) the efficiency can be increased many fold (65-fold in this study); and (5) functional β-galactosidase and CFTR ion channels can be generated in human cells.
Previous work using SMaRT focused mainly on the application of 3′ER, in which PTMs were engineered to replace the 3′ end of target mRNA (Puttaraju et al. 1999, 2001; Mansfield et al. 2000; Liu et al. 2002). Each form of SMaRT has its own specific design features and potential advantages (Garcia-Blanco et al. 2000). As is true for all forms of mRNA repair, each PTM need only deliver a fraction of the full-length coding sequence used in the conventional approaches for RNA or gene therapy. However, PTMs that perform 3′ER should not possess a methionine initiator codon, which they acquire from the target by trans-splicing. However, 5′ER PTMs do require the methionine codon for expression. Another difference is that 5′ER PTMs can be modified at their 5′ end for the purpose of increasing translation, whereas mRNA modified by 3′ER PTMs will acquire the endogenous target 5′ UTR. Conversely, mRNA modified by 5′ER will contain the endogenous 3′ UTR and poly(A) signal sequence, whereas 3′ER PTMs can benefit by inclusion of a specialized poly(A) signal and 3′ UTR in the construct, which can significantly influence mRNA processing, stability, and transport. However, both modes of repair confer important advantages in the reduction of transgene size, which allows efficient packaging of PTMs into vector delivery systems with limited packaging capacity, for example, AAV, the ability to generate mRNA of normal length, and a much broader choice of regulatory sequences. A second and profound advantage of SMaRT, in general, is the acquisition of endogenous regulation, as the level of the mutant target is controlled by the natural transcriptional machinery. The fact that SMaRT can reprogram mRNA at either the 5′ or 3′ end suggests that any codon within a single mRNA can be modified regardless of position. This significantly broadens the applications of SMaRT and provides a choice of how to repair particular mRNA.
5′ Exon replacement is highly dynamic
This study provided information on design features that may be generally applicable to SMaRT. Two important findings were (1) efficient trans-splicing can be achieved by tethering the PTM at many different locations in a pre-mRNA, and (2) a few changes in PTM design could have a profound effect on the level of trans-splicing. The inclusion of a mini-intron in the PTM-coding region, and retargeting of the PTM close to the target acceptor site, resulted in a 65-fold increase in trans-splicing. The former design feature has also been tested in 3′ER constructs, and resulted in a substantial increase in reporter activity (S.A. Mansfield, R. Hawkins-Clark, M. Puttaraju, L.A. Mitchell, and M.A. Garcia-Blanco, unpubl.). It has been reported previously that including an intron in a cDNA transgene or reporter gene can have a profound impact on protein expression in vitro in cells and in vivo in animal models (Choi et al. 1991; Whitelaw et al. 1991; Zielere and Huynh 2002). The reasons for this are largely unknown and may differ between different cell types and constructs, and the context of the intron insertion (Nott et al. 2003). Certainly, the effect does not seem to be merely a simple requirement for cis-splicing (Whitelaw et al. 1991). Several studies suggest that the process of cis-splicing itself results in the deposition of protein factors on the mRNA, referred to as the exon junction complex (LeHir et al. 2000) that may subsequently affect downstream events such as mRNA transport and translation (for review, see Maniatis and Reed 2002). Some reports also indicate that splicing can substantially impact the downstream process of 3′ end processing and cleavage (Niwa et al. 1990; Lu and Cullen 2003).
Another design feature that significantly affected trans-splicing was retargeting of the PTM close to the target acceptor. In the case of 3′ER PTMs, we have shown recently that targeting a PTM to the 5′ end of a large intron (14 kb) as opposed to the 3′ end resulted in a 10-fold difference (S.A. Mansfield, P. Du, S. Hiriyanna, R. Bartel, and E. Otto, unpubl.). In that case, this difference may be explained by the (temporal) kinetic advantage gained by targeting a PTM to the region of the intron that is first transcribed, that is, the donor site, when the target acceptor site has not been synthesized. However, in the case of trans-splicing by 5′ER, the target acceptor site has to be transcribed before either cis- or trans-splicing can be initiated, and so there is no kinetic advantage gained by having the PTM close to the target acceptor site. The reason here may be fundamentally different. The elongating RNA polymerase II (RNAP II) is thought to interact, via the phosphorylated carboxy-terminal domain of the largest subunit with splicing factors, and these interactions may sequester exons near the polymerase (Morris and Greanleaf 2000; Goldstrohm et al. 2001). In very long introns, which may loop away from the elongating RNAP II, there will be regions adjacent to the exons, but not sequestered by the transcriptional machinery that may be optimal for trans-splicing. In contrast, the intronic regions immediately adjacent to the splice sites may be sterically hindered by the transcriptional machinery.
The demonstration that mRNA can be completely rewritten by 5′ER using SMaRT and that the efficiency of trans-splicing can be increased greatly by subtle changes in PTM structure, demonstrates the dynamic nature of SMaRT. The ability to efficiently perform 5′ER significantly broadens the applications of SMaRT technology.
MATERIALS AND METHODS
Plasmid construction
All PCR products were generated with either REDTaq (Sigma) or cloned Pfu (Stratagene) DNA Polymerase. Target CFTR-T11 (see Fig. 1A ▶) was assembled from two separate PCR fragments, both amplified from an existing plasmid template. PCR primers for amplification contained restriction sites for directed cloning (NheI and ApaI, and ApaI and NotI). PCR products were digested with the appropriate restriction enzymes and cloned into the mammalian expression plasmid pc3.1DNA(−) (Invitrogen) using NheI, ApaI, and NotI sites. Exon 10 was also engineered with two stop codons immediately downstream of a group of three methionines at the 5′ end of exon 10 to ensure that the target could not be translated into a fragment of CFTR. The trans-splicing domains of each PTM were created by annealing different sets of oligonucleotides and ligating them into the appropriate PTM vector using AgeI, HindIII, and EcoRI restriction sites. Oligodeoxynucleotide primers were procured from Sigma Genosys. For the PTMs, the sequence between the putative transcription start site and the 5′ cloning site NheI is TAGAGAACCCACTGCTTACTGGCTTATCGAAATTAAT ACGACTCACTATAGGGAGACCCAAGCTG. For the targets, the sequence up to the XhoI cloning site is TAGAGAACCCACTGCT TACTGGCTTATCGAAATTAATACGACTCACTATAGGGAGAC CCAAGCTGGCTAGCGTTTAAACGGGCCCTCTAGA.
DNA sequencing
DNA templates were sequenced at the UNC-CH Automated DNA Sequencing Facility on a model 373A DNA Sequencer (Applied Biosystems) using a Taq Dideoxy Terminator Cycle Sequencing Kit (Applied Biosystems) and 10 pmole of primer per reaction.
Cell culture and transfections
Constructs were cotransfected in human embryonic kidney (HEK) 293T or 293 cells (1.5 × 106 cells per 60-mm poly-d-lysine-coated dish) using LipofectaminePlus (Invitrogen), and the cells were harvested 48 h after the start of transfection. HEK 293T cells were grown in Dulbecco’s Modified Eagle’s Medium (Invitrogen) supplemented with 10% v/v fetal bovine serum (Hyclone). All cells were kept in a humidified incubator at 37°C and 5% CO2.
RT–PCR
Total RNA was isolated using a MasterPure RNA isolation kit (Epicenter Technologies) and used in RT–PCR (by use of an EZ-RT-PCR kit; Applied Biosystems) as described previously (Puttaraju et al. 1999). Each reaction contained 250 ng of total RNA and 120 ng of a 5′ and 3′ specific primer in a 60-μL reaction volume. RT–PCR products were electrophoresed on 2% SeaKem agarose gels. The oligonucleotide primers used to generate cis- and trans-spliced products are 5′-GACCTCTGCAGACTTCACTTC TAATGATGATTATGG-3′ (CF1), 5′-CGCTGGAAAAACGAGC TTGTTG-3′ (CF93), and 5′-ACTCAGTGTGATTCCACCTTCTC 3′ (CF111)
β-galactosidase in-solution assay and in situ staining
Total cellular protein from cells transfected with expression plasmids was isolated by freeze thaw method and assayed for β-galactosidase activity using a β-gal assay kit (Invitrogen). Protein concentration was measured by the dye-binding assay using Bio-Rad protein assay reagents (BIO-RAD). Cells were monitored for the expression of functional β-galactosidase using a β-gal staining kit (Invitrogen).
Iodide efflux assays
Anion efflux was measured by loading cell monolayers with 125Iodine as described previously (Venglarik et al. 1990; Strong et al. 1993). The baseline efflux rate was determined during two collection periods (2 min each); cells were then stimulated with 10 M forskolin plus 5 mM IBMX, and the cAMP-stimulated efflux rate was measured during three additional collection periods (3 min each).
Acknowledgments
This work was supported by grants from the National Institutes of Health (I R43 DK56526-03) and the Cystic Fibrosis Foundation (MANSFI00GO). We thank Carl Baker (NCI) for his critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
BD, binding domain
CFTR, cystic fibrosis transmembrane conductance regulator
5′ER, 5′ exon replacement
3′ER, 3′ exon replacement
TSD, trans-splicing domain
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5101903.
REFERENCES
- Berget, S.M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270: 2411–2414. [DOI] [PubMed] [Google Scholar]
- Chao, H., Mansfield, S.G., Bartel, R.C., Hiriyanna, S., Mitchell, L.G., Garcia-Blanco, M.A., and Walsh, C.E. 2003. Phenotype correction of hemophilia A mice by spliceosome-mediated RNA trans-splicing. Nat. Med. 9: 1015–1019. [DOI] [PubMed] [Google Scholar]
- Choi, T., Huang, M., Gorman, C., and Jaenisch, R. 1991. A generic intron increases gene expression in transgenic mice. Mol. Cell. Biol. 11: 3070–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowrira, B.M., Pavco, P.A., and McSwiggen, J.A. 1994. In vitro and in vivo comparison of hammerhead, hairpin, and hepatitis δ virus self-processing ribozyme cassettes. J. Biol. Chem. 269: 25856– 25864. [PubMed] [Google Scholar]
- Del Gatto-Konczak, F., Bourgeois, C.F., Le Guiner, C., Kister, L., Gesnel, M.C., Stevenin, J., and Breathnach, R. 2000. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 20: 6287–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Blanco, M.A., Puttaraju, M., Mansfield, S.G., and Mitchell, L.G. 2000. Spliceosome-mediated RNA trans-splicing in gene therapy and genomics. Gene Ther. Reg. 1: 141–163. [DOI] [PubMed] [Google Scholar]
- Goldstrohm, A.C., Greenleaf, A.L., and Garcia-Blanco, M.A. 2001. Co-transcriptional splicing of pre-messenger RNAs: Considerations for the mechanism of alternative splicing. Gene 277: 31–47. [DOI] [PubMed] [Google Scholar]
- Jobling, S.A. and Gehrke, L. 1987. Enhanced translation of chimeric messenger RNAs containing a plant viral untranslated leader sequence. Nature 325: 622–625. [DOI] [PubMed] [Google Scholar]
- Krause, M. and Hirsh, D.A. 1987. trans-spliced leader sequence on actin mRNA in C. elegans. Cell 49: 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, N., Howrey, R.P., Lee, S.W., Smith, C.A., and Sullenger, B.A. 1998. Ribozyme-mediated repair of sickle β-globin mRNAs in erythrocyte precursors. Science 280: 1593–1596. [DOI] [PubMed] [Google Scholar]
- Le Hir, H., Moore, M.J., and Maquat, L.E. 2000. Pre-mRNA splicing alters mRNP composition: Evidence for stable association of proteins at exon-exon junctions. Genes & Dev. 14: 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Jiang, Q., Mansfield, S.G., Puttaraju, M., Zhang, Y., Zhou, W., Cohn, J.A., Garcia-Blanco, M.A., Mitchell, L.G., and Engelhardt, J.F. 2002. Partial correction of endogenous ΔF508 CFTR in human cystic fibrosis airway epithelia by spliceosome-mediated RNA trans-splicing. Nat. Biotech. 20: 47–52. [DOI] [PubMed] [Google Scholar]
- Lu, S. and Cullen, B.R. 2003. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9: 618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T. and Reed, R. 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506. [DOI] [PubMed] [Google Scholar]
- Mansfield, S.G., Kole, J., Puttaraju, M., Yang, C.C., Garcia-Blanco, M.A., Cohn, J.A., and Mitchell, L.G. 2000. Repair of CFTR mRNA by spliceosome-mediated RNA trans-splicing. Gene Ther. 7: 1885–1895. [DOI] [PubMed] [Google Scholar]
- Moore, M.J., Query, C.C., and Sharp, P.A. 1999. Splicing of precursors to mRNA by the spliceosome. In The RNA world (eds. R.F. Gesteland and J.F. Atkins), pp. 303–357. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Morris, D.P. and Greenleaf, A.L. 2000. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 22: 39935–39943. [DOI] [PubMed] [Google Scholar]
- Niwa, M., Rose, S.D., and Berget, S.M. 1990. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes & Dev. 4: 1552–1559. [DOI] [PubMed] [Google Scholar]
- Nott, A., Meislein, S.H., and Moore, M.J. 2003. A quantitative analysis of intron effects on mammalian gene expression. RNA 9: 607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttaraju, M., Jamison, S.F., Mansfield, S.G., Garcia-Blanco, M.A., and Mitchell, L.G. 1999. Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat. Biotech. 17: 246–252. [DOI] [PubMed] [Google Scholar]
- Puttaraju, M., DiPasquale, J., Baker, C.C., Mitchell, L.G., and Garcia-Blanco, M.A. 2001. Messenger RNA repair and restoration of protein function by spliceosome-mediated RNA trans-splicing. Mol. Ther. 4: 105–114. [DOI] [PubMed] [Google Scholar]
- Rogers, C.S., Vanoye, C.G., Sullenger, B.A., and George, A.L. 2002. Functional repair of a mutant chloride channel using a trans-splicing ribozyme. J. Clin. Invest. 110: 1783–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. 2003. Mammalian RNAi for the masses. Trends Genet. 19: 9–12. [DOI] [PubMed] [Google Scholar]
- Strong, D.J., Wilkinson, D.J., Mansoura, M.K., Henze, K., Yang, Y., Wilson, J.M., Cohn, J.A., Dawson, D.C., Frizzell, R.A., and Collins, F.S. 1993. Expression of an abundant alternatively spliced form of the cystic fibrosis transmembrane conductance regulator (CFTR) gene is not associated with a cAMP-activated chloride conductance. Hum. Mol. Genet. 2: 225–230. [DOI] [PubMed] [Google Scholar]
- Sullenger, B.A. and Gilboa, E. 2002. Emerging clinical applications of RNA. Nature 418: 252–258. [DOI] [PubMed] [Google Scholar]
- Van der Ploeg, L.H. 1986. Discontinuous transcription and splicing in trypanosomes. Cell 47: 479–480. [DOI] [PubMed] [Google Scholar]
- Venglarik, C.J., Bridges, R.J., and Frizzell, R.A. 1990. A simple assay for agonist-regulated Cl and K conductances in salt-secreting epithelial cells. Am. J. Physiol. 259: C358–C364. [DOI] [PubMed] [Google Scholar]
- Watanabe, T. and Sullenger, B.A. 2000. Induction of wild-type p53 activity in human cancer cells by ribozymes that repair mutant p53 transcripts. Proc. Natl. Acad. Sci. 97: 8490–8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw, C.B., Archibald, A.L., Harris, S., McClenaghan, M., Simons, J.P., and Clark, A.J. 1991. Targeting expression to the mammary gland: Intronic sequences can enhance the efficiency of gene expression in transgenic mice. Transgenic Res. 1: 3–13. [DOI] [PubMed] [Google Scholar]
- Zieler, H. and Huynh, C.Q. 2002. Intron-dependent stimulation of marker gene expression in cultured insect cells. Insect Mol. Biol. 11: 87–95. [DOI] [PubMed] [Google Scholar]