Abstract
A sequence-based prediction method was employed to identify three ligand-binding domains in transferrin-binding protein B (TbpB) of Neisseria meningitidis strain B16B6. Site-directed mutagenesis of residues located in these domains has led to the identification of two domains, amino acids 53 to 57 and 240 to 245, which are involved in binding to human transferrin (htf). These two domains are conserved in an alignment of different TbpB sequences from N. meningitidis and Neisseria gonorrhoeae, indicating a general functional role of the domains. Western blot analysis and BIAcore and isothermal titration calorimetry experiments demonstrated that site-directed mutations in both binding domains led to a decrease or abolition of htf binding. Analysis of mutated proteins by circular dichroism did not provide any evidence for structural alterations due to the amino acid replacements. The TbpB mutant R243N was devoid of any htf-binding activity, and antibodies elicited by the mutant showed strong bactericidal activity against the homologous strain, as well as against several heterologous tbpB isotype I strains.
Through the action of a receptor specific for human transferrin (htf), meningococci are able to acquire iron ions. This transferrin receptor of Neisseria meningitidis is composed of two subunits, TbpA and TbpB (for transferrin-binding proteins A and B) (29), which are associated in a noncovalent manner (1). There is mounting evidence that the receptor consists of one molecule of TbpA associated with two molecules of TbpB (13, 27).
TbpA (100 kDa) is thought to be a porin-like integral membrane protein that is proposed to serve as a channel for the transport of iron across the outer membrane. Based on significant sequence similarities with FepA and FhuA (14), TbpA is proposed to adopt an antiparallel β-barrel structure (28).
TbpB (65 to 90 kDa) is considered to be an outer membrane protein that is anchored to the membrane via the lipidated N-terminal part of the protein (11). There is evidence which indicates that TbpB consists of two globular domains which correspond to the N- and C-terminal halves of the protein (23). It has been shown in vitro that htf binds primarily to the TbpA part of the receptor and with a lower affinity to TbpB (13). The presence of TbpB, however, increases the affinity of TbpA for the iron-loaded form of htf (13), which thus allows the bacterial receptor to compete successfully with the htf receptor.
TbpB has been shown to be a potential candidate for an antimeningococcal vaccine (17). Meningococcal infections represent a major worldwide health problem (2), and there is no vaccine available against serogroup B N. meningitidis. There is no universal vaccine to control and stop this disease. Immunization with TbpB has been shown to elicit a protective immune response in laboratory animals, which has been related to the production of bactericidal antibodies (17, 24). Based on genomic and antigenic features of TbpB, meningococcal strains have been classified into two major families: isotype I, containing TbpB with a mass of ∼68 kDa, and isotype II, harboring TbpB with a mass between 80 and 90 kDa (26).
The study of this protein has largely been hampered by the absence of any three-dimensional structural information about TbpB or related proteins that would allow the generation of a model. According to the present knowledge of the protein, TbpB interacts with two types of ligands. First, it has been demonstrated that TbpA and TbpB form a complex in solution. This association has been shown to be conserved among different human pathogens (9). Recent studies have revealed that both globular domains of TbpB are involved in this interaction (13). Secondly, TbpB interacts with htf. This interaction is restricted to the iron-loaded form of the protein (holo-htf), since the iron-free form (apo-htf) has only a negligible binding affinity (22). Available data on this holo-htf-binding site are not very consistent. It has been demonstrated that htf binding occurs primarily on the N-terminal domain of TbpB (13, 33), but the participation of the C-terminal domain in ligand binding has also been demonstrated (21, 23).
Here, we report the identification of the holo-htf-binding site on TbpB of a representative tbpB isotype I meningococcal strain, N. meningitidis B16B6, using a novel approach based on the prediction of ligand-binding sites from a protein sequence. A recently described prediction method was shown to identify ligand-binding sites from a diverse range of protein database entries with a satisfactory score (10). We have used this method to identify the transferrin-binding site on TbpB. As mentioned above, TbpB binds to two different ligands (TbpA and htf), and the output of the prediction method corresponds to an overall prediction of binding sites for both ligands without being able to differentiate between ligand types. Site-directed mutants were thus prepared in order to verify the implication of the predicted regions in transferrin binding. Immunization experiments were carried out with TbpB mutants devoid of transferrin-binding activity to assess the immunogenicities of these proteins.
MATERIALS AND METHODS
Site-directed mutagenesis, protein expression, and purification of MBP-TbpB fusion proteins.
The construction of a recombinant plasmid corresponding to the expression vector pMAL-c2 (Biolabs) containing a DNA fragment coding for amino acids 2 to 579 of TbpB from N. meningitidis B16B6 (the full-length mature protein devoid of the N-terminal cysteine) in fusion with the maltose-binding protein (MBP) has been described previously (22). Site-directed mutations were introduced into this plasmid using the Quick Change site-directed mutagenesis kit (Stratagene) following the manufacturer's instructions. Lysine (K) or arginine (R) residues were replaced by asparagine (N) or glutamate (E) using pairs of mutagenic oligonucleotides, which are listed in Table 1. Escherichia coli XL-2 Blue (Stratagene) was transformed with the resulting plasmids, and bacterial cultures were grown on Luria broth medium supplemented with 100 μg of ampicillin ml−1. In all cases, the introduction of the desired point mutations was verified by DNA sequencing of the entire tbpB insert.
TABLE 1.
Primers used for site-directed mutagenesis of MBP-TbpB of N. meningitidis strain B16B6
| Mutation | Mutagenic oligonucleotide primersa |
|---|---|
| K51N | 5′GGCTTTGCAGTAAATCTACCTCGCCGG3′ |
| 3′CCGAAACGTCATTTAGATGGAGCGGCC5′ | |
| R54N | 5′GCAGTAAAACTACCTAACCGGAATGCAC3′ |
| 3′CGTCATTTTGATGGATTGGCCTTACGTG5′ | |
| R55N | 5′CTACCTCGCAATAATGCACATTTTAATCC3′ |
| 3′GATGGAGCGTTATTACGTGTAAAATTAGG5′ | |
| R55E | 5′CTACCTCGCGAGAATGCACATTTTAATCC3′ |
| 3′GATGGAGCGCTCTTACGTGTAAAATTAGG5′ | |
| K76N/K77N | 5′GGTTCAATGGATTGGAACAACCTGCAAAGAGG3′ |
| 3′CCAAGTTACCTAACCTTGTTGGACGTTTCTCC5′ | |
| R80N | 5′GAAAAAACTGCAAAACGGAGAACCAAATAG3′ |
| 3′CTTTTTTGACGTTTTGCCTCTTGGTTTATC5′ | |
| R240N | 5′GGGCACACTTTATAATAACAACCGTATTAC3′ |
| 3′CCCGTGTGAAATATTATTGTTGGCATAATG5′ | |
| R243N | 5′GGGCACACTTTATCGTAACAATATTACTC3′ |
| 3′CCCGTGTGAAATAGCATTGTTATAATGAG5′ |
Codons of mutated amino acids are shown in boldface italic.
Protein expression was induced by the addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside, and MBP-TbpB fusion proteins were purified by affinity chromatography as reported previously (21).
Meningococcal strains.
N. meningitidis strains B16B6 (B:2a:P1.2), 2713 (B:NT:P1.2), 2717 (B:NT:P1.2), N106/NK (B:NT:P1.5), and 64/92 (B:2b:NST) were kindly provided by D. Caugant (NIPH, Oslo, Norway) via B. Rokbi (Aventis Pasteur). Each strain was grown in Mueller-Hinton broth medium with the chelator ethylene diamine di-O-hydroxyphenyl-acetic acid (Sigma) for 4 h to allow the expression of the meningococcal transferrin receptor.
Secondary-structure prediction.
The possibility of changes to the secondary structure as a consequence of an amino acid replacement was assessed using the consensus secondary-structure prediction tool of the NPS@ software (3).
Analysis of MBP-TbpB proteins by far-UV CD spectroscopy.
Far-UV circular-dichroism (CD) measurements were made at 25°C with a Jasco (Tokyo, Japan) J-810 spectropolarimeter using cuvettes with a path length of 0.1 mm. The proteins were exhaustively dialyzed against 10 mM Na2HPO4-NaH2PO4 and 150 mM NaCl, pH 7.0. All spectra were corrected using the spectra of the dialysis buffer. The procedure of Taylor and Kaiser (31) was used to determine the α-helix content.
Study of the interaction of MBP-TbpB with holo-htf. (i) Western blot analysis.
The ability of purified MBP-TbpB to bind horseradish peroxidase-conjugated htf (HRP-htf) (Jackson Immuno-Research Laboratories) was evaluated by Western blot analysis as described previously (21). HRP-htf binding was revealed with a colorimetric substrate and quantified by densitometric analysis.
(ii) Surface plasmon resonance (BIAcore) studies.
Surface plasmon resonance studies were carried out using the BIAcore (Uppsala, Sweden) 2000 system. The sensor chip CM5 and the amine coupling kit (containing N-hydroxysuccinimide, N-ethyl-N′-[3-diethylamino-propyl]-carbodiimide, and 1 M ethanolamine-hydrochloride, pH 8.5) were also from BIAcore. The buffer used for sample dilution and analysis was 10 mM HEPES- 150 mM NaCl-3.4 mM EDTA-0.005% BIAcore surfactant, pH 7.4 (HBS buffer). Holo-htf was immobilized on activated carboxylated dextran by a covalent linkage involving the amino groups of the protein using the protocol provided by the manufacturer. MBP-TbpB solutions at 1 μM (diluted in HBS buffer) were passed over the sensor chip at a flow rate of 10 μl min−1 for 9 min. The TbpB-htf ligand complex was then washed for 6 min with HBS buffer, and the increase in resonance units with respect to the initial baseline was determined. Data were corrected using the resonance unit change observed for the binding of MBP, and the reported data are the means of three experiments. The sensor chip was regenerated by exposure to 50 mM Tris-HCl-2 M guanidinium hydrochloride-2 M NaCl, pH 8.0. The results of multiple analyses of the same antigen were very similar, indicating that this regeneration procedure does not alter the immobilized ligand.
(iii) ITC.
Isothermal titration calorimetry (ITC) measurements were performed on a VP-Microcalorimeter (MicroCal, Northampton, Mass.). MBP-TbpB samples were exhaustively dialyzed into 10 mM Na2HPO4-NaH2PO4 and 150 mM NaCl, pH 7.0. A solution of 5 mg of holo-htf/ml was made up in dialysis buffer. Heat changes following a series of injections of holo-htf into MBP-TbpB were measured. The ligand concentration was determined using the Micro BCA protein reagent kit (Pierce, Rockford, Ill.).
Preparation of hyperimmune rabbit antisera and IgG.
Hyperimmune sera were raised in rabbits by a first administration of 100 μg of purified MBP-TbpB in Freund's adjuvant and two boosts of 100 μg of purified MBP-TbpB in incomplete Freund's adjuvant, and immunoglobulin G (IgG) was purified from the sera by chromatography on protein A-Sepharose, as described previously (25).
Bactericidal assay.
The bactericidal activities of purified rabbit IgGs specific for the various MBP-TbpB fusions have been determined, as reported earlier (5, 24). The bactericidal titers were expressed as the last dilution of purified IgG in which at least 50% killing compared to the complement control was achieved.
RESULTS
Prediction of binding domains within TbpB from N. meningitidis B16B6.
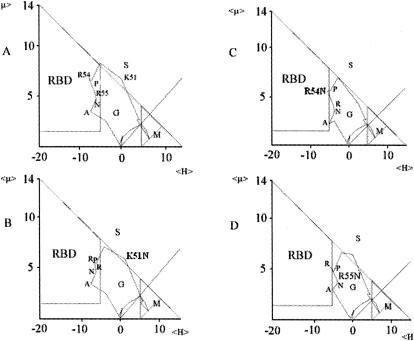
The prediction of ligand-binding sites in the absence of any three-dimensional structural information remains a challenge. However, a recently described method (10) has been shown to identify binding domains with satisfactory reliability. The method is based on the determination of the mean hydrophobicity (<H>) and the mean hydrophobic moment (<μ>) of each residue of the protein by moving a five-residue window along the sequence. In the resulting plot of μ = f(H) (Fig. 1), different regions which provide information on the location of an amino acid in a protein can be identified. This method has been pioneered by Eisenberg et al. (7, 8), who defined three regions, namely, G (globular), S (surface), and M (membrane) (Fig. 1). The scope of this method has been improved by the introduction of a fourth zone called RBD, for receptor binding domain (Fig. 1) (10). The authors demonstrated that amino acid residues present in this zone have an elevated probability of being part of a ligand-binding site. It has also been shown that this technique is a universally applicable method that can be used to define binding sites for a diverse range of ligands. Indeed, this sequence-based method has already been successfully applied in the definition of protein-protein interaction sites for which there is experimental evidence (6, 10, 18). The term RBD was introduced by Gallet et al. (10) and is used here in the sense of a ligand-binding site on a receptor protein, TbpB.
FIG. 1.
Plots of <μH> (10−1 kcal/mol) versus <H> (10−1 kcal/mol) of residues 42SGAAYGFAVKLPRRNAHFNP61 (annotated residues, K51 to A57) of TbpB of N. meningitidis strain B16B6. (A) Residues 53PRRNA57 were found to lie within the RBD zone. (B to D) Evaluation of effects of single-amino-acid replacements on the positions of the residues in the RDB zone, indicating changes in binding potential. K51N has no significant effect on the RBD zone (B); R54N (C) and R55N (D) abolish the binding potential (the RBD zone is almost empty).
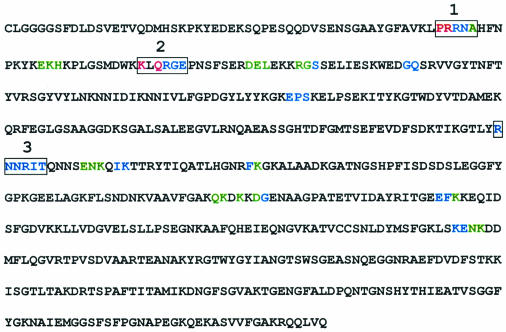
The sequence of TbpB B16B6 has been submitted to this ligand-binding prediction method (10), and amino acids with some degree of binding potential are highlighted in Fig. 2. Based on this initial analysis, three clusters of amino acids with detectable binding potential could be distinguished and were called RBD1, -2, and -3. The identification of RBD1 is illustrated in Fig. 1, which shows the plot of <μH> versus <H> for peptide 42SGAAYGFAVKLPRRNAHFNP61, which contains RBD1 (in boldface). In this plot, it is clear that residues 53 to 57 are found inside the RBD zone, indicating an elevated possibility of forming part of a ligand-binding site, which is referred to here as binding potential. Interestingly, all three RBDs were found to be located in the N-terminal half of TbpB (Fig. 2).
FIG. 2.
Sequence of mature TbpB of N. meningitidis strain B16B6. Amino acids predicted to have a strong (red), moderate (blue), or weak (green) binding potential are highlighted. The three major receptor-binding domains are boxed.
Identification of amino acids within the three RBDs with a major impact on the binding potential.
The potential implication of the three binding domains in htf binding was assessed by site-directed mutagenesis. The choice of amino acid residues to be mutated was based on the prediction of the binding potential (Table 2). The predicted impacts of several amino acid substitutions on the binding potential of RBD1 are shown in Fig. 1. The plot of the unmutated peptide containing RBD1, 42SGAAYGFAVKLPRRNAHFNP61, is shown in Fig. 1A. A similar plot of the same peptide in which K51 (in italics) has been replaced with N is shown in Fig. 1B. It may be noted that the positions of amino acids within the RBD zone have been only slightly altered, and consequently, only a slight decrease in the binding potential of RBD1 can be predicted (Table 2). However, when the two arginine residues located within RBD1 (R54 and R55) are replaced with N, the RBD zone of the resulting plot (Fig. 1C and D) is virtually empty, indicating suppression of the binding potential of this domain (Table 2). Peptides containing RBD2 and -3 were submitted to a similar analysis, and the impacts of amino acid replacements on the receptor binding potential are given in Table 2.
TABLE 2.
Predicted binding potential and qualitative evaluation of binding of point-mutated MBP-TbpB to HRP-htf
| Site(s) of mutation(s) | No. of RBD | Predicted impact of mutation on binding potential of RBD | Relative binding of point-mutated TbpB to htfa |
|---|---|---|---|
| Wild type | 100 | ||
| K51N | 1 | Reduction | 104 |
| R54N | 1 | Suppression | 46 |
| R55N | 1 | Suppression | 0 |
| K76N/K77N | 2 | Reduction | 87 |
| R80N | 2 | Suppression | 125 |
| R240N | 3 | Reduction | 42 |
| R243N | 3 | Suppression | 0 |
| MBP (control) | 0 |
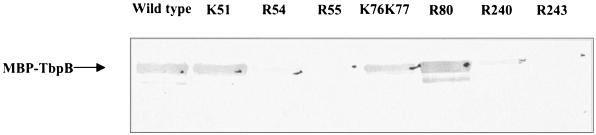
Crude extracts of equivalent amounts of wild-type and mutant MBP-TbpB were analyzed by Western blotting, probed with HRP-htf (Fig. 3), and quantified by densitometric analysis; the htf binding is expressed as a percentage of the wild-type MBP-TbpB, which is considered 100% binding.
Based on these results, a series of TbpB point mutants, which are listed in Table 2, were prepared. The amino acid substitutions were predicted to have no impact on the secondary structure of TbpB, as determined by the NPS@ consensus secondary-structure prediction method (3). This prediction was confirmed by CD spectroscopy (see below).
Binding of wild-type and point-mutated MBP-TbpB to holo-htf.
The binding potentials of wild-type and mutated TbpB to htf have been evaluated using Western blotting, BIAcore, and ITC. A densitometric analysis of the reactivity detected after Western blotting revealed with peroxidase-conjugated htf (Fig. 3) is given in Table 2. It is noted that the R55N (RBD1) and R243N (RBD2) mutants, containing substitutions predicted to suppress htf binding, have lost their affinity for htf. The implication of both amino acids in htf binding is confirmed by a reduction in htf binding seen for the neighboring R54N and R240N mutants. Interestingly, almost no change in htf binding has been noted for point mutants in RBD2 (Table 2).
FIG. 3.
Western blot analysis of wild-type and point-mutated MBP-TbpB with HRP-htf. Crude extracts with equivalent amounts of expressed wild-type or mutated MBP-TbpB were analyzed for their reactivities to HRP-hTf at 10 μg/ml.
Quantification of ligand binding by Western blotting has to be interpreted with caution, since electrophoretic separation is carried out in the presence of a denaturing agent. Therefore, several TbpB mutants have been analyzed by BIAcore, and the binding of different mutated MBP-TbpB proteins to immobilized holo-htf has been studied. As for the Western blot study, mutation of R55 and R243 with N either strongly reduced or abolished htf binding (Table 3). No affinity was detected for an additionally prepared mutant, R55E, containing an amino acid replacement resulting in an opposite charge. The binding behavior of R80N was similar to that of the wild-type protein.
TABLE 3.
Studies of holo-htf binding to wild-type and point-mutated MBP-TbpB using BIAcore and ITC analyses
| MBP-TbpB | Holo-htf-binding capacity using BIAcorea | Dissociation constant using ITC (nM)b |
|---|---|---|
| Wild type | 1,727 ± 182 | 126 ± 16 |
| R55N | 94 ± 12 | 345 ± 90 |
| R55E | 1 ± 2 | NB |
| R80N | 1,345 ± 162 | 172 ± 74 |
| R243N | 6 ± 5 | NB |
In resonance units.
NB, no binding.
BIAcore analysis involves ligand immobilization, which could potentially lead to a distortion of the binding parameter. To avoid any impact of ligand immobilization on binding behavior, ITC has been used to study the binding affinities to holo-htf (Table 3). In an ITC experiment, both ligands are unmodified and in solution (12). Wild-type MBP-TbpB binds with nanomolar affinity to htf. This binding affinity is reduced for R55N, and no binding is observed for R55E and R243N. R80N binds htf in a fashion similar to that of wild-type TbpB.
In summary, the analyses of point-mutated MBP-TbpBs by three different techniques converge on the conclusion that mutation of R55 and R243 results in a strong reduction of htf binding for TbpB from N. meningitidis B16B6. In contrast, data demonstrate that the R80N mutant has htf-binding characteristics similar to those of the wild-type protein.
CD studies of wild-type and point-mutated MBP-TbpB.
It has been demonstrated in the past that the introduction of single point mutations can result in major structural alterations of the protein, as shown by far-UV CD spectroscopy (20). Under these circumstances, functional changes observed for the mutated protein cannot be attributed only to the amino acid replaced, and the data are generally difficult to interpret. In a previous study (13), it was shown that full-length TbpB has a larger amount of secondary structure than the individual domains.
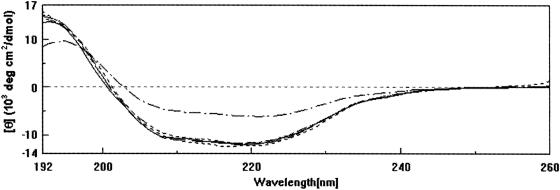
Wild-type and point-mutated MBP-TbpB proteins have been analyzed equally by CD spectroscopy (Fig. 4). The traces are closely superimposable, and minor differences can be attributed to the error associated with the determination of the protein concentration. Using the procedure of Taylor and Kaiser (31), a α-helix content of 29 ± 1% has been determined for all fusion proteins. It can be concluded that all the amino acid substitutions carried out did not result in major structural alterations of the fusion protein. However, we cannot exclude the possibility that mutations gave rise to small conformational rearrangements which did not affect the secondary structure of the protein.
FIG. 4.
Far-UV CD spectra of wild-type and mutated MBP-TbpB proteins. ———, wild-type MBP-TbpB; ———, R55N; - - - - - - - -, R80N; — - - — - - —, R243N. The CD spectrum of mutant R55E, which for clarity is not shown, is superimposable on these spectra. Control, MBP alone (— · — · —).
Determination of the bactericidal titer of antibodies elicited with mutated MBP-TbpB within each RBD region from TbpB B16B6.
TbpB is a potential candidate for inclusion in an antimeningococcal vaccine. In animal experiments, the protein has been shown to induce bactericidal antibodies (17). The influences of amino acid substitutions on the potential of TbpB to induce bactericidal antibodies against the homologous strain B16B6 have been determined (Table 4). Wild-type MBP-TbpB has been shown to induce a bactericidal titer of 1,024, whereas MBP alone did not induce any detectable level of functional antibodies. Point-mutated MBP-TbpB R55N and R80N conserve their capacities to induce bactericidal antibodies, although to a lesser extent. In contrast, point-mutated MBP-TbpB R243N devoid of any htf binding preserves its capacity to induce a strong bactericidal response compared to the wild type.
TABLE 4.
Western blot reactivities and bactericidal titers against N. meningitidis strain B16B6 of rabbit TbpB-specific IgG preparations elicited with wild-type or point-mutated MBP-TbpB
| TbpB fusion | Animal no. | TbpB Western blot specificitya | Bactericidal titerb |
|---|---|---|---|
| MBP alone (control) | 1 | − | <4 |
| Wild type | 2 | + | 1,024 |
| 3 | + | 1,024 | |
| R55N | 4 | + | 32 |
| 5 | + | 128 | |
| R80N | 6 | + | 512 |
| 7 | + | 64 | |
| R243N | 8 | + | 4,096 |
| 9 | + | 4,096 |
TbpB-specific antisera raised in rabbits against the different mutated MBP-TbpB fusions were characterized with N. meningitidis B16B6 outer membrane preparations separated after sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransfer to nitrocellulose. +, presence of a band corresponding to TbpB B16B6; −, absence of a band corresponding to TbpB B16B6.
Bactericidal titers were determined on the homologous strain N. meningitidis B16B6 grown under iron-limiting conditions. The bactericidal titer is expressed as the reciprocal of the last purified IgG dilution in the presence of which at least 50% of the initial bacterial load was killed.
Evaluation of the extents of cross-bactericidal activity displayed by antibodies generated with the MBP-TbpB R243N mutant devoid of htf-binding activity.
The TbpB mutant R243N, shown above to induce a high bactericidal antibody titer similar to that of the wild type, was chosen in order to evaluate the cross-bactericidal activity of this immune response to other tbpB isotype I strains. The difficulty of achieving broad cross-bactericidal activity is frequently an obstacle in the development of a protein-based vaccine (24). The bactericidal activities of rabbit IgG raised with the MBP-TbpB R243N mutant toward N. meningitidis strains 2713, 64/92, 106/NK, and 2717 have been determined (Table 5). Strong cross-bactericidal activity against the four heterologous tbpB isotype I strains was obtained with IgG elicited with the TbpB mutant R243N. This high cross-bactericidal immune response is comparable to that observed for the wild-type protein (Table 5).
TABLE 5.
Bactericidal titers of rabbit IgG elicited with wild-type or point-mutated MBP-TbpB R243N toward a range of different tbpB isotype I meningococcal strains
| Antigen | Animal no. | Cross-bactericidal titer on meningococcal tbpB isotype I straina:
|
||||
|---|---|---|---|---|---|---|
| B16B6 | 2713 | 64/92 | 106/NK | 2717 | ||
| Wild-type MBP-TbpB | 1 | 1,024 | 2,048 | ≥4,096 | 128 | 8 |
| 2 | 1,024 | ≥4,096 | >4,096 | 128 | <8 | |
| Mutated MBP-TbpB R243N | 3 | 4,096 | ≥4,096 | >4,096 | 256 | 64 |
| 4 | 4,096 | ≥4,096 | >4,096 | 512 | <8 | |
| MBP alone (control) | 5 | [4] | <8 | <8 | <8 | <8 |
Bactericidal titers were determined on N. meningitidis tbpB isotype I strains B16B6, 2713, 64/92, 106/NK, and 2717 grown under iron-limiting conditions. The bactericidal titer is expressed as the reciprocal of the last IgG dilution in the presence of which at least 50% of the initial bacterial load was killed. In most cases, IgG preparations from two individual rabbit sera were tested and titration was performed twice. The number in brackets indicates incomplete killing.
DISCUSSION
Due to numerous genome-sequencing projects, the amount of available protein sequence information is growing with breathtaking speed. However, the functions associated with a very large number of sequences are unknown, and their discovery is a major challenge of life sciences in this century. Furthermore, the study of a fair number of proteins with known functions is hampered by the absence of any three-dimensional structural information.
In this article, we describe the sequence-based prediction and experimental confirmation of transferrin-binding sites in TbpB. Our data confirm previous successful predictions, such as for the PBP3 protein from E. coli (18). This approach might be a useful tool to obtain information from a sequence about potential ligand-binding sites, which, combined with mutagenesis studies, could lead to the identification or better understanding of protein function.
Two domains, RBD1 and RBD3, are shown to be involved in transferrin binding of TbpB from the meningococcal strain B16B6. In particular R55 and R243, located in these binding domains, are of crucial importance in htf binding. The importance of arginine residues in transferrin binding is also consistent with the fact that this amino acid residue is by far the most frequently found in binding sites (10). Three binding domains were initially predicted, of which only RBD1 and -3 were confirmed to be involved in transferrin binding. Here, we have studied the binding of wild-type and mutant TbpB to htf. Therefore, the possibility that the presence of TbpA in the receptor complex masks or distorts binding sites identified in this study cannot be ruled out. This hypothesis could be verified by studies of htf binding to N. meningitidis producing mutant protein.
Mutation of residues K76, K77, and R80 in RBD2 had only a marginal impact on htf-binding capacity. As mentioned above, the prediction method employed identifies binding domains for both of the TbpB ligands, TbpA and htf. It is therefore likely that RBD2 corresponds to a domain which is involved in an interaction with TbpA. It has been shown (13) that the recombinant N-terminal domain of TbpB binds to TbpA. However, further analysis is needed to verify this hypothesis.
All of the experiments were carried out using TbpB strain B16B6, which raises the question of whether the data obtained are specific for this tbpB isotype I protein or whether a generalization of findings with respect to other neisserial tbpB isotype II strains of N. meningitidis or Neisseria gonorrhoeae is possible.
A sequence alignment of RBD1 and -3 of TbpB derived from tbpB isotype I and II strains of N. meningitidis and N. gonorrhoeae is shown in Table 6. The four major arginine residues within both RBD1 and RBD3, R54, R55, R240, and R243, are fully conserved among the five TbpB sequences from tbpB isotype I strains. This observation clearly indicates that these htf-binding sites are common in these isotype I proteins.
TABLE 6.
Sequence alignment of RBD1 and −3a
| tbpB isotype | Strainb | Amino acidc
|
Reference | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RBD1
|
RBD3
|
||||||||||||
| 53 | 54 | 55 | 56 | 57 | 240 | 241 | 242 | 243 | 244 | 245 | |||
| I | m-B16B6 | P | R | R | N | A | R | N | N | R | I | T | 14 |
| m-64/92 | . | . | . | . | . | . | . | . | . | . | . | 24 | |
| m-2713 | . | . | . | F | W | . | . | . | . | . | . | 16 | |
| m-2717 | . | . | . | F | W | . | . | . | . | . | . | 24 | |
| m-106/NK | . | . | . | F | W | . | . | . | . | . | . | 16 | |
| II | m-BZ83 | K | . | . | . | W | . | . | . | . | . | . | 15 |
| m-90/94 | K | . | . | . | W | . | . | . | . | . | . | 24 | |
| m-SB22 | K | . | . | . | W | . | . | . | . | . | . | 24 | |
| m-32/94 | K | . | . | . | W | . | . | . | . | . | . | 25 | |
| m-NGPB24 | K | . | . | . | W | . | . | . | . | . | . | 16 | |
| m-8680 | K | . | . | . | W | K | . | . | S | V | S | 25 | |
| m-8710 | K | . | . | . | W | . | . | . | A | N | . | 25 | |
| m-901256 | K | . | . | . | R | . | . | . | . | V | . | 24 | |
| m-M982 | K | . | . | . | W | . | . | . | A | S | L | 14 | |
| m-92123 | K | . | . | F | W | . | . | . | . | V | . | 24 | |
| m-BZ163 | K | . | . | . | R | . | . | . | . | V | . | 15 | |
| m-6940 | K | . | . | . | W | . | . | . | . | V | . | 19 | |
| m-M978 | K | . | . | . | W | . | . | . | . | V | . | 19 | |
| m-S3032 | K | . | . | . | W | . | . | . | A | N | Q | 19 | |
| m-MC58 | K | . | . | . | W | . | . | . | A | N | . | 32 | |
| g-FA6642 | K | . | . | . | W | K | . | . | . | V | I | 4 | |
| g-FA19 | K | . | . | . | W | . | . | . | K | V | I | 4 | |
| g-Pgh3-2 | K | . | . | . | R | . | . | . | K | V | I | 4 | |
| g-FA1090 | K | . | . | . | W | K | . | . | M | V | I | 4 | |
For TbpB of N. meningitidis strains belonging to tbpB isotype I and II strains and of N. gonorrhoeae.
Meningococcal sequences are preceded by the letter m; gonococcal sequences are preceded by the letter g.
Dots indicate conservation of the amino acid. The numbers are positions.
The TbpB sequences of N. meningitidis tbpB isotype II strains and of N. gonorrhoeae strains show 37 to 45% sequence identity with the B16B6 sequence. However, both RBDs are generally conserved in the sequence alignment. R54 and R55 are entirely conserved, whereas R240 and R243 are partially conserved. Interestingly, RBD3 is fully conserved for the five strains BZ83, 90/94, SB22, 32/94, and NGPB24, which form part of the previously identified BZ83-like subisotype (24). For the amino acid residues in RBD3s from other tbpB isotype II strains, conservation of the positive charge is frequently observed (K instead of R).
An analogous prediction of RBDs using the tbpB isotype II strain M982 has identified amino acids with ligand-binding potential in both N- and C-terminal domains of TbpB (data not shown). This preliminary result was in agreement with previous observations indicating that the C-terminal domain could also be involved in binding to either TbpA or htf (4, 21, 23).
The two htf-binding sites of B16B6 TbpB are located within the N-terminal domain of TbpB, described previously as the minimal htf-binding domain (13, 33). Recent studies of the TbpB protein of Moraxella catarrhalis (30) have led to the identification of six different regions in the N-terminal half which are implicated in htf binding. It has been proposed that these six regions interact with the six previously identified (23) sequence fragments present on the C-terminal lobe of htf. Both M. catarrhalis and N. meningitidis TbpBs use htf as a ligand. The TbpB studied here is isotype I, whereas the Moraxella protein resembles isotype II proteins, which are characterized by larger size. A sequence alignment of both proteins shows that the N-terminal domain of the Neisseria protein is ∼90 amino acids shorter than its Moraxella counterpart (data not shown). However, this alignment also shows that RBD1 and RBD3 described in this article correspond to two of the binding regions reported for Moraxella TbpB, htf-binding regions 1 and 5, respectively (30). We have predicted additional amino acid sequences with moderate or weak binding potential (Fig. 2); however, they have not been further studied by mutagenesis. Several of these additional sites, namely, 155EPS157, 250ENKQIK255, and 270FK271, were found to align with binding regions identified for Moraxella TbpB. Only two of the six binding regions in the Moraxella TbpB, namely, regions 3 and 4 (30), were shown to have Neisseria counterparts, which were not predicted in our study to be involved in htf binding. Furthermore, the six htf-binding regions identified for Moraxella TbpB did not align with RBD2. This is consistent with our data, which show that RBD2 is not involved in htf binding. A high binding potential has been predicted for this domain (Fig. 2). We propose the implication of this domain in binding to TbpA, which remains to be studied in detail. Furthermore, our data are consistent with a report by Gorringe et al. (A. R. Gorringe, L. I. Irons, P. Aisen, P. Zak, and A. Robinson, Proc. Ninth Int. Pathog. Neisseria Conf., Winchester, England, p. 140-141, 1994) who demonstrated that antibodies generated by immunization with the peptide corresponding to amino acids 43 to 56 (comprising RBD1) of the tbpB isotype II strain M982 inhibit the binding of holo-htf to TbpB. Taken together, these data point to a general role for RBD1 and RBD3 in htf binding for a diverse range of different TbpB proteins.
The overall aim of this study, apart from its contribution to the general knowledge of the protein, is to explore the possibility of TbpB as a vaccine candidate against meningococcal disease. However, vaccination with TbpB implies that the antigen may, immediately after injection, form a complex with holo-htf present in human body fluids. This may mask certain epitopes and may cause conformational alterations to the antigen. To avoid a postinjection modification of TbpB, attempts are made to alter the transferrin-binding site in a manner that prevents htf from binding without perturbing the overall structure or reducing its capacity to induce bactericidal antibodies. This goal has clearly been achieved here. Western blot analysis and BIAcore and ITC studies (Tables 2 and 3) provide evidence that TbpB R243N is devoid of any htf-binding activity, and CD spectroscopy (Fig. 4) shows that this mutation did not perturb the overall structure of TbpB. Rabbit sera derived from immunization experiments with this TbpB mutant had similarly strong cross-bactericidal activities compared to that of the wild-type against other tbpB isotype I strains. The TbpB mutant R243N described in this study can thus be regarded as a vaccine candidate.
Acknowledgments
We thank Luc Aujame for critical reading of the manuscript. We are grateful to Sophie Fraysse, Sophie Naville, Marie-Claire Nicolai, and Frédéric Gréco for technical assistance. We also thank Michel Chevalier, Mireille Latour, and Marie-José Quentin-Millet for helpful discussions.
REFERENCES
- 1.Boulton, I. C., A. R. Gorringe, R. J. Carr, B. Gorinsky, C. L. Joannou, and R. W. Evans. 1997. Characterisation of the meningococcal transferrin-binding protein complex by photon correlation spectroscopy. FEBS Lett. 414:409-413. [DOI] [PubMed] [Google Scholar]
- 2.Caugant, D. A., L. O. Froholm, K. Bovre, E. Holten, C. E. Frasch, L. F. Mocca, W. D. Zollinger, and R. K. Selander. 1986. Intercontinental spread of a genetically distinctive complex of clones of Neisseria menigitidis causing epidemic disease. Proc. Natl. Acad. Sci. USA 83:4927-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Combet, C., C. Blanchet, C. Geourjon, and G. Deléage. 2000. NPS@: Network Protein Sequence Analysis. Trends Biochem. Sci. 25:147-150. [DOI] [PubMed] [Google Scholar]
- 4.Cornelissen, C. N., J. E. Anderson, and P. F. Sparling. 1997. Characterization of the diversity and the transferrin-binding domain of gonococcal transferrin-binding protein 2. Infect. Immun. 65:822-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danve, B., L. Lissolo, M. Mignon, P. Dumas, S. Colombani, A. B. Schryvers, and M.-J. Quentin-Millet. 1993. Transferrin-binding proteins isolated from Neisseria meningitidis elicit protective and bactericidal antibodies in laboratory animals. Vaccine 11:1214-1220. [DOI] [PubMed] [Google Scholar]
- 6.De Loof, H., M. Rossenau, R. Brasseur, and J. M. Ruysschaert. 1986. Use of hydrophobicity profiles to predict receptor binding domains on apolipoprotein E and the low-density lipoprotein apolipoprotein B-E receptor. Proc. Natl. Acad. Sci. USA 83:2295-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisenberg, D., R. M. Weiss, and T. C. Terwilliger. 1982. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature 299:371-374. [DOI] [PubMed] [Google Scholar]
- 8.Eisenberg, D., E. Schwarz, M. Komaromy, and R. Wall. 1984. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179:125-142. [DOI] [PubMed] [Google Scholar]
- 9.Fuller, C. A., R. Yu, S. W. Irwin, and A. B. Schryvers. 1998. Biochemical evidence for a conserved interaction between bacterial transferrin-binding protein A and transferrin-binding protein B. Microb. Pathog. 24:75-87. [DOI] [PubMed] [Google Scholar]
- 10.Gallet, X., B. Charloteaux, A. Thomas, and R. Brasseur. 2000. A fast method to predict protein interaction sites from sequences. J. Mol. Biol. 302:917-926. [DOI] [PubMed] [Google Scholar]
- 11.Gray-Owen, S. D., and A. B. Schryvers. 1996. Bacterial transferrin and lactoferrin receptors. Trends Microbiol. 14:843-850. [DOI] [PubMed] [Google Scholar]
- 12.Holdgate, G. A. 2001. Making cool drugs hot: the use of isothermal titration calorimetry as a tool to study binding energetics. BioTechniques 31:164-184. [PubMed] [Google Scholar]
- 13.Krell, T., G. Renauld-Mongenie, M.-C. Nicolai, S. Fraysse, M. Chevalier, Y. Berard, J. Oakhill, R. W. Evans, A. Gorringe, and L. Lissolo. 2003. Insight into the structure and function of the transferrin receptor from Neisseria meningitidis using microcalorimetric techniques. J. Biol. Chem. 278:14712-14722. [DOI] [PubMed] [Google Scholar]
- 14.Legrain, M., V. Mazarin, S. W. Irwin, B. Bouchon, M.-J. Quentin-Millet, E. Jacobs, and A. B. Schryvers. 1993. Cloning and characterization of Neisseria meningitidis genes encoding the transferrin-binding proteins Tbp1 and Tbp2. Gene 130:73-80. [DOI] [PubMed] [Google Scholar]
- 15.Legrain, M., A. Findeli, D. Villeval, M.-J. Quentin-Millet, and E. Jacobs. 1996. Molecular characterization of hybrid Tbp2 proteins from Neisseria meningitidis. Mol. Microbiol. 19:159-169. [DOI] [PubMed] [Google Scholar]
- 16.Legrain, M., B. Rokbi, D. Villeval, and E. Jacobs. 1998. Characterization of genetic exchanges between various highly divergent tbpBs having occurred in Neisseria meningitidis. Gene 208:51-59. [DOI] [PubMed] [Google Scholar]
- 17.Lissolo, L., G. Maitre-Wilmotte, P. Dumas, S. Colombani, A. B. Schryvers, and M.-J. Quentin-Millet. 1995. Evaluation of transferrin-binding protein 2 within the transferrin-binding protein complex as a potential antigen for future meningococcal vaccines. Infect. Immun. 63:884-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marrec-Fairley, M., A. Piette, X. Gallet, R. Brasseur, H. Hara, C. Faipont, J.-M. Ghysen and J.-M. Nguyen-Distèche. 2000. Differential functionalities of amphiphilic peptide segments of the cell-septation penicillin-binding protein 3 of Escherichia coli. Mol. Microbiol. 37:1019-1031. [DOI] [PubMed] [Google Scholar]
- 19.Mazarin, V., B. Rokbi, and M.-J. Quentin-Millet. 1995. Diversity of the transferrin-binding protein Tbp2 of Neisseria meningitidis. Gene 158:145-146. [DOI] [PubMed] [Google Scholar]
- 20.Nairn, J., N. C. Price, S. M. Kelly, D. Rigden, L. A. Fothergill-Gilmore, and T. Krell. 1996. Phosphoglycerate mutase from Schizosccharomyces pombe: development of an expression system and characterization of three histidine mutants of the enzyme. Biochim. Biophys. Acta 1296:69-75. [DOI] [PubMed] [Google Scholar]
- 21.Renauld-Mongenie, G., D. Poncet, L. von Olleschik-Elbheim, T. Cournez, M. Mignon, M. A. Schmidt, and M.-J. Quentin-Millet. 1997. Identification of human transferrin-binding sites within meningococcal transferrin-binding protein B. J. Bacteriol. 179:6400-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renauld-Mongénie, G., M. Latour, D. Poncet, S. Naville, and M.-J. Quentin-Millet. 1998. Both the full-length and the N-terminal domain of the meningococcal transferrin-binding protein B discriminate between human iron-loaded and apo-transferrin. FEMS Microbiol. Lett. 169:171-177. [DOI] [PubMed] [Google Scholar]
- 23.Retzer, M. D., R. Yu, and A. B. Schryvers. 1999. Identification of sequences in human transferrin that bind to the bacterial receptor protein, transferrin-binding protein B. Mol. Microbiol. 32:111-121. [DOI] [PubMed] [Google Scholar]
- 24.Rokbi, B., G. Renauld-Mongenie, M. Mignon, B. Danve, D. Poncet, C. Chabanel, D. A. Caugant, and M. J. Quentin-Millet. 2000. Allelic diversity of the two transferrin binding protein B gene isotypes among a collection of Neisseria meningitidis strains representative of serogroup B disease: implication for the composition of a recombinant TbpB-based vaccine. Infect. Immun. 68:4938-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rokbi, B., M. Mignon, G. Maitre-Wilmott, L. Lissolo, B. Danve, D. A. Caugant, and M. J. Quentin-Millet. 1997. Evaluation of recombinant transferrin-binding protein B variants from Neisseria meningitidis for their ability to induce cross-reactive and bactericidal antibodies against a genetically diverse collection of serogroup B strains. Infect. Immun. 65:55-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rokbi, B., V. Mazarin, G. Maitre-Willmotte, and M. J. Quentin-Millet. 1993. Identification of two major families of transferrin receptors among Neisseria meningitidis strains based on antigenic and genomic features. FEMS Microbiol. Lett. 110:51-58. [DOI] [PubMed] [Google Scholar]
- 27.Ronpirin, C., A. E. Jerse, and C. N. Cornelisson. 2001. Gonococcal genes encoding transferrin-binding proteins A and B are arranged in a bicistronic operon but are subject to differential expression. Infect. Immun. 69:6336-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schryvers, A. B., and I. I. Stojilkovic. 1999. Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 32:1117-1123. [DOI] [PubMed] [Google Scholar]
- 29.Schryvers, A. B., and L. J. Morris. 1988. Identification and characterization of the transferrin receptor from Neisseria meningitidis. Mol. Microbiol. 2:281-288. [DOI] [PubMed] [Google Scholar]
- 30.Sims, K. L., and A. B. Schryvers. 2003. Peptide-peptide interactions between human transferrin and transferrin-binding protein B from Moraxella catarrhalis. J. Bacteriol. 185:2603-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor, J. W., and E. T. Kaiser. 1987. Structure-function analysis of proteins through the design, synthesis, and study of peptide models. Methods Enzymol. 154:473-498. [DOI] [PubMed] [Google Scholar]
- 32.Tettelin, H., N. J., Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 33.Vonder Haar, R. A., M. Legrain, H. V. J. Kolbe, and E. Jacobs. 1994. Characterization of a highly structured domain in Tbp2 from Neisseria meningitidis involved in binding to human transferrin. J. Bacteriol. 176:6207-6213. [DOI] [PMC free article] [PubMed] [Google Scholar]