Abstract
A mature appressorium cDNA library of rice blast fungus, Magnaporthe grisea, was constructed in a λTriplEx2 vector by SMART™ cDNA library containing 2.37×106 independent clones about 100% of which harbor foreign cDNA inserts with average size of 660 bp. Of 9 randomly selected clones, 2 expressed sequence tags (ESTs) sequences did not have homologous EST sequences of M. grisea in GenBank. The appressorium cDNA library is suitable for gene expression analysis and function analysis of the late stages of appressorium formation and the early stages of penetration of M. grisea.
Keywords: Magnaporthe grisea, Appressorium, cDNA library
INTRODUCTION
The rice blast fungus Magnaporthe grisea causes one of the most destructive diseases of rice. Genetic studies of this important pathogen during the past decade made it excellent for investigating fungal-plant interactions. Appressorium is an infection structure of M. grisea. Many genes expressed in appressorium, such as PMK1 (Xu and Hamer, 1996), CYP1 (Viaud et al., 2002), MAGB and MAC1 (Choi and Dean, 1997; Liu and Dean, 1997), GAS1 and GAS2 (Xue et al., 2002), and MPG1 (Lau and Hamer, 1996; Talbot et al., 1993; 1996), are required for appressorium formation and pathogenesis by the fungus. Many approaches, such as expressed sequence tags (ESTs) sequencing (Kamakura et al., 1999; Kim et al., 2001; Rauyaree et al., 2001), SAGE (Serial analysis of gene expression) (Irie et al., 2003), microarray (Takano et al., 2003), and mutants (Balhadere et al., 1999; Shi et al., 1995; Sweigard et al., 1998), have been used to identify genes already expressed in appressorium. By now, more than 10000 ESTs have been sequenced from several cDNA libraries representing different stages of fungal growth, differentiation and plant infection of the pathogen, including appressoria, conidia, and mycelia from complete and nitrogen starvation cultures (Xu and Xue, 2002). However, appressorium cDNA libraries in existence were only constructed from conidia germinated for less than 8 h on an inductive surface (Kamakura et al., 1999; Takano et al., 2003), causing a significant problem for completely evaluating gene expression during the late stages of appressorium formation. We report here the result of producing an appressorium stage cDNA library of M. grisea from mature appressoria formed for 23.5–24.5 h.
MATERIALS AND METHODS
M. grisea isolate, appressorium induction and RNA isolation
The appressorium of M. grisea GUY11 was used to isolate total RNA for construction of cDNA library.
For appressorium formation, droplets (20 μl) of conidium suspension (1×106–1.5×106 conidia/ml in distilled water) were inoculated on an inductive surface of projection membrane (Gaoke, China) for 23.5–24.5 h at 25 °C. And then appressoria were harvested and from which total RNA was isolated with Trizol (Molecular Research Center, USA) following the manufacturer’s procedure.
Appressorium cDNA synthesis and library construction
The ss cDNA, ds cDNA and library construction were carried out according to Smart cDNA Library Construction Kit User Manual (CLI, 2001). Three μl (~5.23 μg) total RNA was used to reverse transcribe to single-strand cDNA with SMART IV™ Oligonucleotide, CDS III/3′ PCR Primer and PowerScript™ Reverse Transcriptase at 42 °C for 1 h. Two μl of first-strand cDNA was used to carry out ds cDNA synthesis by 23 cycles of PCR (95 °C for 15 s, 68 °C for 6 min) with 5′ PCR primer, CDS III/3′ PCR primer and Advantage™ 2 polymerase mix. Half of the ds cDNA were digested with proteinase K and purified with phenol:chloroform: isoamylalcohol. Then all purified ds cDNA was digested with Sfi I enzyme. The reaction product was fractionated by CHROMA SPIN-400 column and the first three fractions containing ds cDNA were collected. The cDNA of all three fractions containing ds cDNA was concentrated by ethanol and resuspended in 7 μl deionized H2O. At last, these Sfi I-digested cDNAs were ligated to Sfi I-digested, dephosphorylated λTriplEx2 vector at 16 °C overnight. The ligated cDNA/λTriplEx2 vector was packed using MaxPlax™ Lambda Packaging Extracts (Epicentre, USA).
Analysis of the appressorium cDNA library
The unamplified cDNA library was titered and the percentage of recombinant clones was determined by blue/white screening according to the instructions in manual of SMART™ cDNA library construction kit (Clontech, USA).
Twelve randomly selected plaques were used for the conversion of λTriplEx2 clone to pTriplEx2 plasmid according to the instructions of manual of SMART™ cDNA library construction kit. The plasmids were purified by CTAB-method DNA Mini Preparation and digested by Sfi I enzyme. The inserts were analyzed by separation on a 1% agarose/EtBr gel. And the expressed sequence tags (EST) of nine randomly selected clones were sequenced with T7 primer on an ABI 377 DNA sequencer. The sequence data were analyzed using VecScreen program (NCBI) for vector masking, and then the adaptor sequences were removed from these cDNA sequences. These EST sequences were processed using software BioEdit (Hall, 1999) for contig assembly. Process sequences were subjected to similarity searches against GenBank database using Blast 2.2.8 (Altschul et al., 1997) and against Phytopathogenic Fungi and Oomycete EST database (Version 1.4) in COGEME (http://cogeme.ex.ac.uk/index.html) and Magnaporthe grisea database (http://www.broad.mit.edu/annotation/fungi/magnaporthe/)
Appressorium cDNA library amplification and titer of the amplified library
The manual of SMART™ cDNA library construction kit suggested amplification of the unamplified cDNA library and then titering of the amplified library.
RESULTS
Appressorium induction of M. grisea
More than 96% conidia inoculated on the hydrophobic surface of a projection membrane for 23.5–24.5 h at 25 °C germinated and formed appressoria (Fig.1).
Fig. 1.
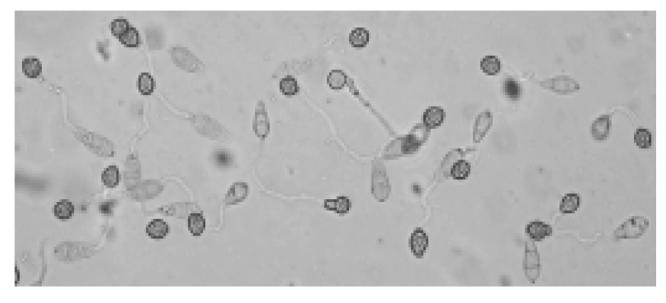
Appressoria of M. grisea GUY11 on a projection membrane inoculated for 23.5–24.5 h at 25 °C
Total RNA isolation of appressorium
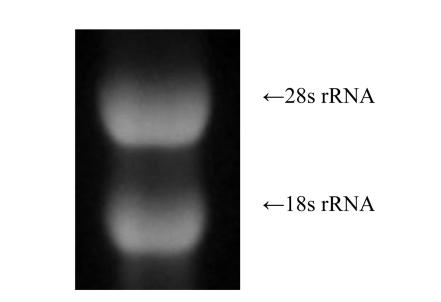
The ratio of OD260/OD280 of total RNA isolated from appressoria incubated for 23.5–24.5 h was 1.63. From the gel, total RNA appeared as two bright bands (28s rRNA and 18s rRNA) and showed intact (Fig.2).
Fig. 2.
Appressorium total RNA (5 μl) on a 1.2% denaturing formaldehyde/agarose gel
cDNA synthesis and library construction quality analysis
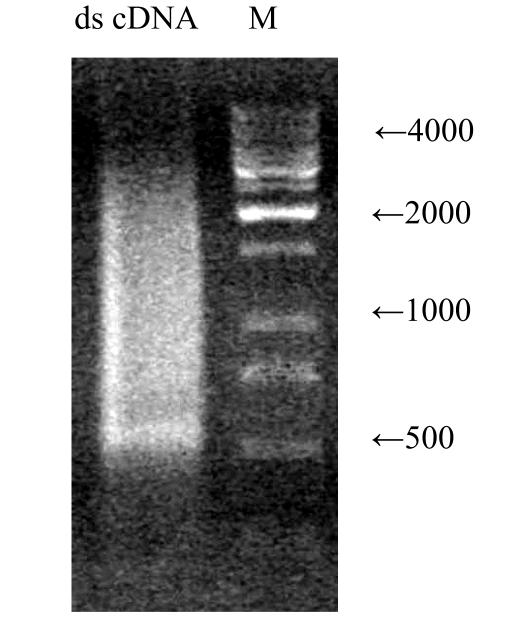
The sizes of the ds cDNA produced from the 24 h appressorium mRNA ranged from 200 bp to 6 kb, mainly between 500 bp and 5 kb on a 1.1% agarose gel (Fig.3). After fractionation, the first three fraction containing cDNA (8, 9 and 10 fractions) were collected and used for ligation to vector.
Fig. 3.
Ten μl ds cDNA product synthesized from appressorium RNA electrophoresed on a 1.1% agrose/EtBr gel
Lane M, 1-kb DNA size marker (Fermentas, Lithuania) (0.2 μg loaded)
The cDNA was ligated unidirectionally into λTriplEx2 vector by three ligation reactions. From the three ligations combined, a primary library of 2.37×106 independent clones was obtained. The result of blue/white screening of the primary cDNA library in E. coli XL1-Blue showed that the ratio of white (recombinant) to blue (nonrecombinant) in two LB/MgSO4 plates containing IPTG and X-gal were 684/0 and 931/0 respectively. So, the recombination efficiency of cDNA ligation reached 100%. After amplification, the amplified cDNA library had a titer of 2.86×109 pfu/ml.
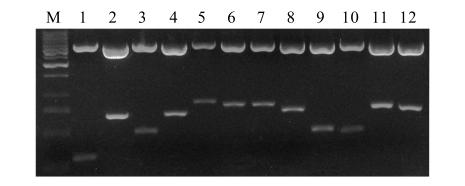
Twelve randomly selected lambda plaques of the primary library were converted to plasmids by in vivo conversion and were analyzed for insert size. The plasmids were purified with CTAB method and digested by Sfi I. All 12 plasmids showed that the insert sizes ranged from 300 bp to 900 bp, average of 660 bp (Fig.4).
Fig. 4.
Sfi I-digested plasmids of 12 clones on a 1% agrose/EtBr gel
Lane M, 1-kb DNA marker size (Fermentas, Lithuania). 1~12: clones 1~12
Nine randomly selected cDNA clones were partially sequenced using T7 primers. Analysis of the 9 cDNA sequences using BioEdit software (Hall, 1999) generated 8 nonredundant EST sequences. All 8 unique EST sequences were subjected to similarity searches against GenBank (Blastn 2.2.8 and Blastx 2.2.8) (Altschul et al., 1997), COGEME Db and Magnaporthe grisea Database. Six of them had homologous sequences of M. grisea in dbEST and the other 2 clones’ EST sequences were found to be inhomologous sequences (Table 1) until submission. Clone C02 was homologous to putative protein of cDNA of Neurospora crassa (GenBank Accn: AW723279, Expect value=3e−53). As for clone C04, no homologous putative protein sequence or EST sequence hits with E-value less than e−3 were found in NCBI blast. These sequences have been deposited in the dbEST of GenBank database (GenBank Accn: CK725218~CK725225).
Table 1.
Homologous sequences from M. grisea of 8 EST sequences by Blastn in GenBank
| Clone noes | Putative product/function |
| C01 | Glutathione peroxidase (BU641649, e−140)* |
| C02 | Not found |
| C04 | Not found |
| C05 | Cyclophilin (peptidyl-prolyl cis-trans isomerase) (BI808723, 0.0) |
| C06 | ATP synthase subunit 9, lipid-binding protein, mitochondrial (AA415145, 0.0) |
| C09 | Unknown (CD031047, 0.0) |
| C11 | Unknown (CD036090, e−119) |
| C12 | Unknown (BM872180, 0.0) |
GenBank accession numbers and E-values for homologues are in parentheses
DISCUSSION
Although there are many M. grisea cDNA libraries in the world, mainly in Ebbole’s lab at Texas A&M University and Ralph Dean’s lab at North Carolina State Biotechnology Center (Ebbole et al., 2002; Xu and Xue, 2002), and there are five reported appressorium cDNA libraries built from germinated conidia on apprressorium-inductive surface (http://www.tigr.org/tdb/tgi/mggi/searching/xpress_search.html ), no cDNA library was build strictly from mature appressorium. The appressorium cDNA lambda expression library reported here was constructed from mature appressoria inoculated on an inductive surface for 23.5~24.5 h at 25 °C, and had 2.37×106 pfu independent clones with 100% recombinants enough to screen genes expressed in appressorium. In addition, all cDNA sequences inserted into MCS of λTriplEx2, the vector of this library, can be expressed for screening genes with protein antigens. And the vector λTriplEx2 is easily converted from phage to a plasmid vector via Cre-lox-mediated subcloning.
Two of 8 non-redundant EST sequences have no homologues with E-value less than e−3 in M. grisea ESTs available in GenBank until submission. This cDNA library probably contains many genes missed by previous M. grisea cDNA libraries. Clone C05 encoded a reported peptidyl prolyl cis-trans isomerase (cyclophilin). Cyclophilin gene CYP1 is a virulence factor in M. grisea (Viaud et al., 2002). CYP1 mutants show reduced virulence and are impaired in associated functions, such as penetration peg formation and inducement of appressorium turgor. From this cDNA library made from mature appressoria, a 3.0 kb long cDNA sequence of MGTA1 gene has been successfully cloned (1Wang, unpublished). The appressorium cDNA library ESTs can be used for identification of genes expressed in appressorium. For example, promoters of 12 genes were isolated from the rice blast fungus based on the sequences of randomly ESTs (appressorium formation stage cDNA library of Magnaporthe available from GenBank) and then these promotors can be used to identify the genes expressed during infection stages (Banno et al., 2003). So, this 24 h appressorium cDNA library is a representative cDNA library that reveales the gene expression state in appressorium and is suitable for gene expression and function analysis of M. grisea appressorium.
Footnotes
Project supported by the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry, and in part by the National Natural Science Foundation of China (No. 30270049)
Wang, J.Y., Liu, X.H., Lin, F.C., 2004. Cloning and characterization of MGTA1 gene in rice blast pathogen Magnaporthe grisea. Fungal Genetics and Biology.
References
- 1.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balhadere PV, Foster AJ, Talbot NJ. Identification of pathogenicity mutants of the rice blast fungus Magnaporthe grisea using insertional mutagenesis. Mol. Plant–Microbe Interact. 1999;12:129–142. [Google Scholar]
- 3.Banno S, Kimura M, Tokai T, Kasahara S, Higa-Nishiyama A, Takahashi-Ando N, Hamamoto H, Fujimura M, Staskawicz BJ, Yamaguchi I. Cloning and characterization of genes specifically expressed during infection stages in the rice blast fungus. FEMS Microbiol Lett. 2003;222:221–227. doi: 10.1016/S0378-1097(03)00307-0. [DOI] [PubMed] [Google Scholar]
- 4.Choi W, Dean RA. The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell. 1997;9:1973–1983. doi: 10.1105/tpc.9.11.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebbole DL, Yuan J, Li D, et al. Abstracts for Plant, Animal & Microbe Genomes X. San Diego, CA: 2002. Gene Discovery in the Rice Blast Fungus, Magnaporthe Grisea: Analysis of ESTs; p. 18. [Google Scholar]
- 6.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 7.Irie T, Matsumura H, Terauchi R, Saitoh H. Serial Analysis of Gene Expression (SAGE) of Magnaporthe grisea: genes involved in appressorium formation. Mol. Genet. Genomics. 2003;270:181–189. doi: 10.1007/s00438-003-0911-6. [DOI] [PubMed] [Google Scholar]
- 8.Kamakura T, Xiao J, Choi W, Kochi T, Yamaguchi S, Teraoka T, Yamaguchi I. cDNA subtractive cloning of genes expressed during early stage of appressorium formation by Magnaporthe grisea . Biosci. Biotechnol. Biochem. 1999;63:1407–1413. doi: 10.1271/bbb.63.1407. [DOI] [PubMed] [Google Scholar]
- 9.Kim S, Ahn IP, Lee YH. Analysis of genes expressed during rice–Magnaporthe grisea interactions. Mol. Plant–Microbe Interact. 2001;14:1340–1346. doi: 10.1094/MPMI.2001.14.11.1340. [DOI] [PubMed] [Google Scholar]
- 10.Lau GW, Hamer JE. Regulatory genes controlling MPG1 expression and pathogenicity in the rice blast fungus Magnaporthe grisea . Plant Cell. 1996;8:771–781. doi: 10.1105/tpc.8.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S, Dean RA. G protein α-subunit genes control growth, development and pathogenicity of Magnaporthe grisea . Mol Plant–Microbe Interact. 1997;10:1075–1086. doi: 10.1094/MPMI.1997.10.9.1075. [DOI] [PubMed] [Google Scholar]
- 12.Rauyaree R, Choi W, Fang E, Blackmon B, Dean RA. Genes expressed during early stages of rice infection with the rice blast fungus Magnaporthe grisea . Mol. Plant Pathol. 2001;2:347–354. doi: 10.1046/j.1464-6722.2001.00085.x. [DOI] [PubMed] [Google Scholar]
- 13.Shi Z, Christian D, Leung H. Enhanced transformation in Magnaporthe grisea by restriction enzyme mediated integration of plasmid DNA. Phytopathology. 1995;85:329–333. [Google Scholar]
- 14.CLI (CLONTECH Laboratories, Inc.) Smart cDNA Library Construction Kit User Manual. USA: 2001. pp. 1–51. [Google Scholar]
- 15.Sweigard JA, Carroll AM, Farrall L, Chumley FG, Valent B. Magnaporthe grisea pathogenicity genes obtained through insertional mutagenesis. Mol. Plant–Microbe Interact. 1998;11:404–412. doi: 10.1094/MPMI.1998.11.5.404. [DOI] [PubMed] [Google Scholar]
- 16.Takano Y, Choi W, Mitchell TK, Okuno T, Dean RA. Large scale parallel analysis of gene expression during infection-related morphogenesis of Magnaporthe grisea . Mol. Plant Pathol. 2003;4:337–347. doi: 10.1046/j.1364-3703.2003.00182.x. [DOI] [PubMed] [Google Scholar]
- 17.Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea . Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talbot NJ, Kershaw MJ, Wakley GE, De Vries OMH, Wessels JGH, Hamer JE. MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea . Plant Cell. 1996;8:985–999. doi: 10.1105/tpc.8.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viaud MC, Balhadere PV, Talbot NJ. A Magnaporthe grisea cyclophilin acts as a virulence determinant during plant infection. Plant Cell. 2002;14:917–930. doi: 10.1105/tpc.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu JR, Hamer JE. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea . Genes Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- 21.Xu JR, Xue CY. Time for a blast: genomics of Magnaporthe grisea . Mol. Plant Pathol. 2002;3:173–176. doi: 10.1046/j.1364-3703.2002.00108.x. [DOI] [PubMed] [Google Scholar]
- 22.Xue CY, Park G, Choi W, Zheng L, Dean RA, Xu JR. Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell. 2002;14:2107–2119. doi: 10.1105/tpc.003426. [DOI] [PMC free article] [PubMed] [Google Scholar]