Abstract
Eukaryotic initiation factor (eIF) 4A unwinds secondary and tertiary structures in the 5′-untranslated region of mRNA, permitting translation initiation. Programmed cell death 4 (Pdcd4) is a novel transformation suppressor and eIF4A-binding partner that inhibits eIF4A helicase activity and translation. To elucidate the regions of eIF4A that are functionally significant in binding to Pdcd4, we generated point mutations of eIF4A. Two-hybrid analysis revealed that five eIF4A mutants completely lost binding to Pdcd4 while four eIF4A mutants retained wild-type levels of binding. The residues that, when mutated, inactivated Pdcd4 binding specified ATP binding, ATP hydrolysis, or RNA binding. With the exception of the Q-motif mutant eIF4AP56L, the eIF4A mutants inactivated for Pdcd4 binding were inactivated for binding to eIF4G (GM, GC, or both) and for enhancing translation. Several eIF4A mutants showing wild-type level binding to Pdcd4 were also inactivated for binding to eIF4G and for enhancing translation. Thus, significant dissociation of eIF4A’s Pdcd4- and eIF4G-binding regions appears to occur. Because three of the four eIF4A mutants that retained Pdcd4 binding also suppressed translation activity in a dominant-negative manner, the structure that defines the Pdcd4-binding domain of eIF4A may be necessary but is insufficient for translation. A structural homology model of eIF4A shows regions important for binding to Pdcd4 and/or eIF4G lying on the perimeters of the hinge area of eIF4A. A competition experiment revealed that Pdcd4 competes with C-terminal eIF4G for binding to eIF4A. In summary, the Pdcd4-binding domains on eIF4A impact both binding to eIF4G and translation initiation in cells.
Keywords: translation initiation, eIF4F, tumor suppressor, eIF4A homology model
INTRODUCTION
Eukaryotic translation initiation is orchestrated by a group of proteins called eukaryotic translation initiation factors (eIFs), of which eIF4A is a member. Eukaryotic translation initiation consists of three basic steps: (1) the association of the initiating tRNA, eIFs, and the 40S ribosomal subunit to form the 43S preinitiation complex; (2) binding of the 43S preinitiation complex at the 5′-cap structure of mRNA, followed by scanning along the 5′-untranslated region (5′-UTR) toward the initiation codon; and (3) binding of the 60S ribosomal subunit to the 43S preinitiation complex to form the 80S ribosome at the AUG site, whereupon protein elongation from the initiator tRNA can commence (Pain 1996). eIF4A is thought to facilitate binding of the 43S ribosomal subunit to the mRNA by catalyzing the unwinding of mRNA secondary and tertiary structure at the cap structure and 5′-UTR (Ray et al. 1985; Linder 2003), allowing scanning in a 5′-to-3′ direction toward the initiation codon and subsequent translation initiation (Kressler et al. 1997; Rogers et al. 2001; Svitkin et al. 2001).
eIF4A is a 46-kDa polypeptide that exhibits RNA-dependent ATPase and RNA helicase activities. It is part of a multi-subunit complex called eIF4F, which also includes eIF4G and eIF4E. When localized in the eIF4F complex, eIF4A is ~20-fold more efficient as an RNA helicase than when found alone (Pause and Sonenberg 1992; Rogers et al. 1999), leading to the hypothesis that functional eIF4A is associated with eIF4F (Rozen et al. 1990). eIF4A is the prototype of the DEAD-box family of proteins, with the name referring to one of the conserved domains of this family (Linder et al. 1989; Linder 2003). DEAD-box family RNA helicases typically contain seven to eight conserved domains (including the DEAD-box domain) that are involved in binding an NTP (typically ATP) and using the energy from NTP hydrolysis to unwind dsRNA (Tanner and Linder 2001; Rocak and Linder 2004). eIF4A has thus far been characterized to contain at least 10 such conserved domains (Svitkin et al. 2001; Tanner et al. 2003).
eIF4G bridges the ribosome to the mRNA. Being a scaffold protein, it binds eIF4E and eIF4A, as well as other proteins (Korneeva et al. 2000). eIF4G can be divided into three distinct functional domains: the N-terminal domain (amino acid residues 1 to 674), which contains the eIF4E-binding site; the middle domain (residues 675 to 1079), which contains the first of two eIF4A-binding sites and an RNA-binding site; and the C-terminal domain (residues 1080 to 1600), which contains the second eIF4A-binding site (Imataka and Sonenberg 1997). Studies of the eIF4G middle domain (eIF4GM) show that it is sufficient for cap-independent, internal ribosome entry site-dependent translation (Lomakin et al. 2000) and cap-independent but 5′-end-dependent translation (De Gregorio et al. 1998), whereas the eIF4G C-terminal domain (eIF4GC) serves a regulatory role during translation (Morino et al. 2000). Whether eIF4A associates with eIF4G in a 1:1 (Li et al. 2001) or 2:1 (Korneeva et al. 2001) ratio is not yet established.
Programmed cell death 4 (Pdcd4) protein was discovered by mRNA differential display analysis to be highly expressed in transformation-resistant (P−) but not in transformation-susceptible (P+) JB6 mouse epidermal cells (Cmarik et al. 1999). Overexpression of Pdcd4 in P+ cells renders them resistant to neoplastic transformation by tumor promoters (Yang et al. 2001). Pdcd4 suppresses the tumor phenotype when transfected into transformed (Tx) cells, at least in part through the inhibition of AP-1-dependent transcription (Yang et al. 2003b). Recently, overexpression of Pdcd4 in human carcinoid cells has been shown to inhibit cell proliferation (Goke et al. 2004), and to induce apoptosis in human breast cancer cells (Afonja et al. 2004).
How does Pdcd4 inhibit neoplastic transformation and tumor phenotype? Yeast two-hybrid assays have determined that Pdcd4 associates with eIF4A (Yang et al. 2003a). Through its association with eIF4A, Pdcd4 inactivates the helicase function of eIF4A and suppresses cap-dependent translation (Yang et al. 2003a). Pdcd4 mutants inactivated for eIF4A binding are also inactivated for inhibiting translation and for inhibiting the AP-1-dependent transcription required for transformation (Yang et al. 2001, 2003a). The MA-3 domains, two regions of Pdcd4 that are predicted to be mostly α-helical, are necessary for mediating this inhibitory effect of Pdcd4 (Aravind and Koonin 2000; Ponting 2000).
Pdcd4 is proposed to inhibit translation by binding to eIF4A and to eIF4GM, and by preventing eIF4A from inter-acting with eIF4GC (Yang et al. 2003a). The Pdcd4 mutants inactivated for eIF4A binding fail to compete with eIF4GC, a region of eIF4G shown to have a regulatory function in translation initiation, for binding to eIF4A. Thus, it appears that by suppressing eIF4A’s helicase activity and its interaction with eIF4GC, Pdcd4 inhibits AP-1 transactivation and thus neoplastic transformation and tumor phenotype. In this way, Pdcd4 acts as a tumorigenesis suppressor through the novel mechanism of translation regulation.
Mutation of the conserved domains in eIF4A affects its ability to bind and unwind RNA, to hydrolyze ATP, and to facilitate translation (Pause and Sonenberg 1992; Pause et al. 1993, 1994; Svitkin et al. 2001; Tanner et al. 2003). Because Pdcd4 inhibits functions inactivated by these conserved domain mutations in eIF4A, we hypothesized that such mutations may also define residues involved in Pdcd4–eIF4A interaction. Furthermore, because eIF4A activity is enhanced when bound to eIF4G in the eIF4F complex, eIF4A mutants may owe loss of translation activity to loss of binding to eIF4G. In order to determine which eIF4A residues are involved in binding to Pdcd4 and to characterize these residues for their significance in binding to eIF4GM and eIF4GC, as well as in translation initiation, we performed a mutational analysis of eIF4A. Results showed partial overlap between eIF4A regions required for Pdcd4 binding and eIF4G binding. Residues of eIF4A responsible for binding Pdcd4 are required, but not sufficient, for translation initiation, indicating that Pdcd4 contacts only some of the domains critical for translation. The ribbon and surface model reveals Pdcd4 to bind along the hinge region of eIF4A. Finally, eIF4GC and Pdcd4 compete for binding to eIF4A.
RESULTS
Discrete motifs of eIF4A specify interaction with Pdcd4, eIF4GM, and eIF4GC
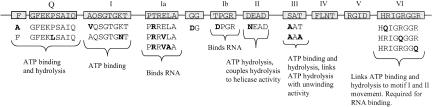
Both Pdcd4 (Yang et al. 2003a) and eIF4G (Imataka and Sonenberg 1997) have been shown to interact with eIF4A. Likewise, the regions of Pdcd4 (Yang et al. 2004) and eIF4G (Morino et al. 2000) that interact with eIF4A (the MA-3 domains) have been identified and their functions analyzed. However, the regions of eIF4A that bind to Pdcd4 and to eIF4G have not been characterized. Mutation of eIF4A has revealed the location of motifs important for translation initiation (Pause and Sonenberg 1992; Pause et al. 1993, 1994; Svitkin et al. 2001; Tanner et al. 2003). The known functions of some of the conserved eIF4A domains are shown in Figure 1 ▶. Motifs I (AQSGTGKT) and II (DEAD) are required for ATP binding and hydrolysis, motifs Ia (PTRELA) and Ib (TPGR) bind RNA, and motifs III (SAT) and VI (HRIGRGGR) link ATP hydrolysis to RNA unwinding and binding activity, respectively. Additionally, a postulated conserved Q motif is required for ATP binding and hydrolysis (Tanner et al. 2003). Because Pdcd4 inhibits translation, we hypothesized that Pdcd4 binds to eIF4A residues important for some of the translation functions. We thus performed site-directed mutagenesis of eIF4A to create mutations identical to those known to inactivate translation (Fig. 1 ▶). Wild-type (WT) and all eIF4A mutants were expression-verified in JB6 RT101 cells (figure not shown). Mammalian two-hybrid analysis used the NF-κB activation domain (pCMV-AD) fused to wild-type or mutant eIF4A cDNA and the Gal4 DNA-binding domain (pCMV-BD) fused to pdcd4, eIF4GM, or eIF4GC cDNA. Both pCMV-AD and pCMV-BD vectors contain the nuclear localization signal (NLS), enabling them to translocate into the nucleus. These AD and BD fusion constructs and Gal4-luciferase reporter gene were cotransfected into RT101 cells. After 48 h, the cells were lysed and luciferase activity was assayed as a measure of eIF4A interaction with Pdcd4 or eIF4G.
FIGURE 1.
The 15 point mutations of eIF4A, shown in bold letters where an amino acid has been changed. At the top is shown wild-type eIF4A with its 10 highly conserved domains.
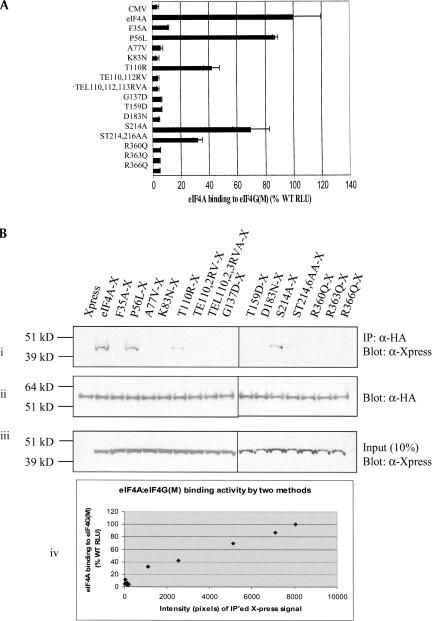
As shown in Figure 2A ▶, the Pdcd4-binding activity of wild-type eIF4A was designated as 100% when WT-Pdcd4 (bait) and WT-eIF4A (prey) were cotransfected into RT101 cells at a ratio of 1:1. The mutants eIF4AF35A, eIF4AA77V, eIF4AT110R, and eIF4AR363Q retained wild-type or greater interaction with Pdcd4. The eIF4A mutants having >100% binding activity may have a higher than wild-type affinity for Pdcd4. The mutants eIF4AP56L, eIF4AK83N, eIF4AG137D, eIF4AT159D, and eIF4AR360Q showed only background-level luciferase activity, indicating that these eIF4A mutants do not bind or weakly bind to Pdcd4. The mutants eIF4ATE110,112RV, eIF4ATEL110,112,113RVA, eIF4AD183N, eIF4AS214A, eIF4AST214,216AA, and eIF4AR366Q showed partial inactivation of eIF4A binding to Pdcd4.
FIGURE 2.
Mutational analysis of eIF4A/Pdcd4 interaction. (A) Mammalian two-hybrid analysis of wild-type and mutant eIF4A interaction with Pdcd4. Two-hundred-fifty nanograms of wild-type/mutant pCMV-AD-eIF4A and 250 ng of pCMV-BD-Pdcd4 were transfected into RT101 cells, alongside 25 ng of Gal4-luciferase reporter gene and 5 ng of thymidine kinase-Renilla luciferase gene. Values (relative light units, or RLUs) are corrected for Renilla signal and provided as percentages of wild-type (WT) eIF4A interaction with Pdcd4. These experiments were repeated three times in sextuplicate, and representative data are shown. The results are expressed as the mean ± standard deviation. (B) Pull-down verification of eIF4A/mutant-xpress interaction levels with Pdcd4. (i) One-hundred micrograms of GST-Pdcd4 were added to 100 μg of RT101 cell lysates, which were then blotted with xpress antibody. (ii) Blots were stripped and probed with GST antibody to confirm equal affinity of glutathione-Sepharose beads for GST-Pdcd4. (iii) One-hundred microgram amounts of RT101 cell lysates were blotted with xpress antibody to show equal expression and loading of xpress-labeled protein. (iv) Densitometry readings were taken of each band in panel Bi, and the values were plotted against the RLU values in A.
To confirm these interactions, we performed GST pull-down assays (Fig. 2Bi ▶). RT101 cells were transiently transfected with the xpress-tagged wild-type or mutant eIF4A vectors. After lysis and addition of GST-Pdcd4 and gluthathione-Sepharose beads, the bound proteins were analyzed by SDS-PAGE. The band intensity of xpress-tagged wild-type eIF4A binding to Pdcd4 was designated as 100%, and the mutants eIF4AF35A, eIF4AA77V, eIF4AT110R, and eIF4AR363Q bound Pdcd4 at or near wild-type level. In contrast, eIF4AP56L, eIF4AG137D, eIF4AT159D, and eIF4AR360Q showed little to no binding to Pdcd4. It should be noted that JB6 RT101 cells contain little endogenous Pdcd4, so that any loss of interaction between these eIF4A mutants and Pdcd4 is not due to interference by endogenous Pdcd4. The mutant eIF4AK83N (Fig 2Bi ▶), which had been shown not to interact with Pdcd4 in the mammalian two-hybrid assays, showed a low level of binding to Pdcd4 in the GST pull-down assays. The mutants eIF4A, eIF4ATEL110,112,113RVA, eIF4AD183N, eIF4AS214A, eIF4AST214,216AA, and eIF4AR366Q showed ~50% of the wild-type eIF4A level of binding to Pdcd4. Overall, the results of these pull-down assays were in agreement with the results of the mammalian two-hybrid assays, confirming the binding capacity of wild-type and mutant eIF4A to Pdcd4.
SDS-PAGE, followed by Western blot analysis with xpress antibody, was performed on 10% input amounts of all transfected RT101 cell lysates. All lysates showed an equal level of wild-type or mutant xpress-tagged eIF4A expression, confirming equal transfection efficiency and lane loading (Fig. 2Biii ▶). When densitometry readings were taken of the GST pull-down assays (Fig. 2Bi ▶) and compared against RLU values for the two-hybrid assays (Fig. 2A ▶), an approximate linear relationship between the two sets of measurements was evidenced (Fig. 2Biv ▶). These results indicate that wild-type eIF4A interacts with Pdcd4 both in vivo and in vitro, and that amino acid substitutions within the eIF4A protein sequence alter the ability of eIF4A to bind Pdcd4.
To determine the relationship between eIF4A residues important for binding to Pdcd4 and residues important for binding to eIF4GM, another mammalian two-hybrid analysis of eIF4A mutants was performed. Plasmids pCMV-BD-eIF4GM(bait) and pCMV-AD-WT/mutant eIF4A (prey) were cotransfected into RT101 cells with the Gal4-luciferase reporter gene. In order to increase the sensitivity of binding detection, the ratio of eIF4GM to eIF4A was set at 1:8 in-stead of the 1:1 ratio used with Pdcd4 and eIF4A. Wild-type eIF4A binding to eIF4GM was designated as 100%, and the mutant eIF4AP56L showed wild-type-level binding to eIF4GM (Fig. 3A ▶). The mutants eIF4AT110R and eIF4AS214A showed ~45% and ~70% of wild-type eIF4A binding, respectively, to eIF4GM, and eIF4AST214,216AA ~showed 30% of wild-type eIF4A-binding activity to eIF4GM. All other eIF4A mutants showed background level luciferase activity, i.e., complete inactivation of binding to eIF4GM.
FIGURE 3.
Mutational analysis of eIF4A/eIF4GM(497–974) interaction. (A) Mammalian two-hybrid analysis of wild-type (WT) and mutant eIF4A interaction with eIF4G(497–974). Here 50 ng of wild-type/mutant pCMV-AD-eIF4A and 400 ng of pCMV-BD-eIF4G(497–974) were transfected into RT101 cells, alongside 25 ng of Gal4-luciferase reporter gene and 5 ng of thymidine kinase-Renilla luciferase gene. Values (relative light units, or RLUs) are corrected for Renilla signal and provided as percentages of wild-type eIF4A interaction with eIF4G(497–974). These experiments were repeated three times in sextuplicate, and representative data are shown. The results are expressed as the mean ± standard deviation. (B) Immunoprecipitation of eIF4A/mutant-xpress with eIF4G(497–974). (i) Here 2 μg of HA antibody was added to 100 μg of RT101 cell lysates, which were then blotted with xpress antibody. (ii) Blots were stripped and probed with HA antibody to confirm equal G-Sepharose bead affinity for HA antibody. (iii) One-hundred microgram amounts of RT101 cell lysates were blotted with xpress antibody to show equal expression and loading of xpress-labeled protein. (iv) Densitometry readings were taken of each band in panel Bi, and the values were plotted against the RLU values in A.
The binding of eIF4A mutants to eIF4GM was further confirmed by immunoprecipitation. RT101 cell lysates, containing transiently transfected HA-tagged eIF4GM and xpress-tagged wild-type or mutant eIF4A, were incubated with HA antibody followed by protein G-Sepharose beads. The bound proteins were analyzed by SDS-PAGE. The band intensity of xpress-tagged wild-type eIF4A binding to eIF4GM was designated as 100% (Fig. 3Bi ▶). The mutant eIF4AP56L bound eIF4GM at nearly the same level as wild-type eIF4A, and the mutant eIF4AS214A also showed a high level of interaction with eIF4GM. The mutants eIF4AT110R and eIF4AST214,216AA showed an intermediate level of binding to eIF4GM, with the eIF4AST214,216AA interaction level being slightly lower than that evidenced in the mammalian two-hybrid analysis. Overall, the results of the immunoprecipitation assays were in agreement with those of the mammalian two-hybrid assays, confirming the level of wild-type and mutant eIF4A binding to eIF4GM. All lysates showed equal levels of xpress-tagged wild-type or mutant eIF4A expression, confirming equal transfection efficiency and lane loading (Fig. 3Biii ▶). Furthermore, den-sitometry readings of the immunoprecipitation assays (Fig. 3Bi ▶) compared against RLU values for the two-hybrid assays (Fig. 3A ▶) showed an approximate linear relationship between the two methods (Fig. 3Biv ▶). These results indicate that wild-type eIF4A interacts with eIF4GM both in vivo and in vitro, and that amino acid substitutions that alter eIF4A’s binding to Pdcd4 in four of five cases also alter its ability to bind eIF4GM.
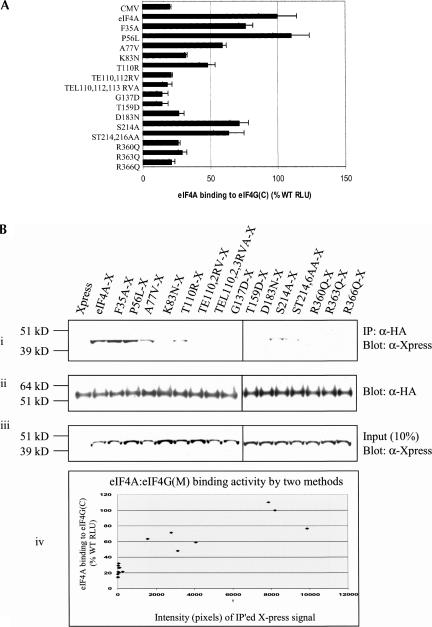
Interaction of wild-type or mutant eIF4A with eIF4GC was similarly determined by mammalian two-hybrid analysis. Plasmids pCMV-BD-eIF4GC (bait) and pCMV-AD-WT/mutant eIF4A (prey) were cotransfected into RT101 cells with Gal4-luciferase reporter gene. As in the mammalian two-hybrid assays of eIF4GM and eIF4A, the ratio of eIF4GC to eIF4A was set at 1:8 in order to increase sensitivity of binding detection. The mutant eIF4AP56L bound eIF4GC at the wild-type eIF4A level (Fig. 4A ▶). The mutants eIF4AF35A, eIF4AA77V, eIF4AK83N, eIF4AT110R, eIF4AS214A, and eIF4AST214,216AA bound eIF4GC at levels ranging from ~35%–70% of wild-type eIF4A, and the mutants eIF4ATE110,112RV, eIF4ATEL110,112,113RVA, eIF4AG137D, eIF4AT159D, eIF4AD183N, eIF4AR360Q, eIF4AR363Q, and eIF4AR366Q interacted with eIF4GC at or near background level.
FIGURE 4.
(A) Mammalian two-hybrid analysis of wild-type (WT) and mutant eIF4A interaction with eIF4G(924–1444). Fifty nanograms of wild-type/mutant pCMV-AD-eIF4A and 400 ng of pCMV-BD-eIF4G(924–1444) were transfected into RT101 cells, alongside 25 ng of Gal4-luciferase reporter gene and 5 ng of thymidine kinase-Renilla luciferase gene. Values (relative light units, or RLUs) are corrected for Renilla signal and provided as percentages of wild-type eIF4A interaction with eIF4G(924–1444). These experiments were repeated three times in sextuplicate, and representative data are shown. The results are expressed as the mean ± standard deviation. (B) Immunoprecipitation of eIF4A/mutant-xpress with eIF4G(924–1444). (i) Two micrograms of HA antibody was added to 100 μg of RT101 cell lysates, which were then blotted with xpress antibody. (ii) Blots were stripped and probed with HA antibody to confirm equal G-Sepharose bead affinity for HA antibody. (iii) One-hundred microgram amounts of RT101 cell lysates were blotted with xpress antibody to show equal expression and loading of xpress-labeled protein. (iv) Densitometry readings were taken of each band in panel Bi, and the values were plotted against the RLU values in A.
These results were confirmed by immunoprecipitation experiments. RT101 cell lysates, which contained transiently transfected HA-tagged eIF4GC (Imataka and Sonenberg 1997) and xpress-tagged wild-type or mutant eIF4A, were incubated with HA antibody and protein G-Sepharose beads. The bound proteins were analyzed by SDS-PAGE (Fig. 4Bi ▶). The eIF4A mutants eIF4AF35A and eIF4AP56L bound eIF4GC at the same level as wild-type eIF4A, while the mutants eIF4AA77V, eIF4AT110R, eIF4AS214A, and eIF4AST214,216AA bound eIF4GC at 50% of wild-type eIF4A binding to eIF4GC. The mutants eIF4AK83N, eIF4ATE110,112,RV, eIF4ATEL110,112,113RVA, eIF4AG137D, eIF4AT159D, eIF4AD183N, eIF4AR360Q, eIF4AR363Q, and eIF4AR366Q all had background level interaction with eIF4GC. These immunoprecipitation results confirmed the mammalian two-hybrid results for wild-type and mutant eIF4A interaction with eIF4GC and showed that three of the five eIF4A mutants inactivated for Pdcd4 binding were also inactivated for binding to eIF4GC.
All lysates showed equal levels of xpress-tagged wild-type or mutant eIF4A expression, confirming equal transfection efficiency and lane loading (Fig. 4Biii ▶). Likewise, densitometry readings of the immunoprecipitation assays (Fig. 4Bi ▶) compared against RLU values for the two-hybrid assays (Fig. 4A ▶) showed an approximately linear relationship between the two methods (Fig. 4Biv ▶). These results indicate that wild-type eIF4A interacts with eIF4GC both in transfected cells and in vitro, and that its ability to interact with eIF4GC can be altered by specific amino acid substitutions within its protein sequence.
The Q, I, Ia, GG, Ib, II, and VI conserved domains contribute to the translation-required regions of eIF4A
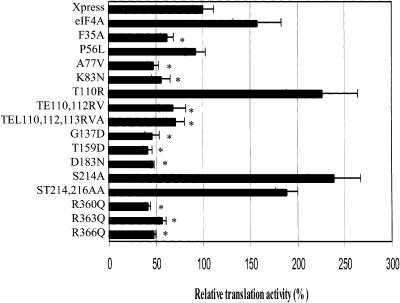
Mutation of specific eIF4A residues had been previously shown to inhibit the ability of eIF4A to initiate in vitro translation in a dominant-negative manner (Pause et al. 1994). To determine if the eIF4A mutants were similarly inactivated for in vivo translation initiation, we transfected RT101 cells with xpress-tagged wild-type or mutant eIF4A and the stem–loop structured luciferase reporter gene. The mRNA of the stem–loop structured luciferase reporter gene contains a 24-bp stem–loop structure at the 5′-untranslated region (UTR) with a free energy of -44.8 kcal/mol (Yang et al. 2004). This stem–loop structured luciferase mRNA is much less efficiently translated than an unstructured luciferase mRNA, and because it requires greater eIF4A activity, is more sensitive to inhibition by Pdcd4 (Yang et al. 2004). Thus, eIF4A must be functional in order for luciferase activity to be evidenced. The RT101 cells transfected with control vector alone showed a luciferase expression level designated as 100%, representing activity due to endogenous eIF4A (Fig. 5 ▶). Wild-type eIF4A enhanced stem–loop luciferase signal to ~1.6-fold that of endogenous background level. The mutants eIF4AT110R, eIF4AS214A, and eIF4AST214,216AA all showed translation activity greater than that of wild-type eIF4A. The eIF4A mutants eIF4AF35A, eIF4AA77V, eIF4AK83N, eIF4ATE110,112RV, eIF4ATEL110,112,113,RVA, eIF4AG137D, eIF4AT159D, eIF4AD183N, eIF4AR360Q, eIF4AR363Q, and eIF4AR366Q all showed luciferase expression levels significantly below that of control vector, suggesting a dominant-negative effect on translation activity. Although the relative concentrations of endogenous wild-type and introduced mutant eIF4A are not readily ascertainable, it is reasonable to assume from the abundance of eIF4A that mutant to wild-type ratios are less than or equal to one, thus indicating a dominant-negative effect when translation was significantly inhibited. The mutant eIF4AP56L showed no activity for enhancing translation activity.
FIGURE 5.
The Q, I, Ia, GG, Ib, II, and VI conserved domains contribute to the translation-required regions of eIF4A. RT101 cells were transfected with 400 ng of wild-type (WT)/mutant eIF4A, 25 ng of stem–loop luciferase reporter, and 5 ng of thymidine kinase-Renilla luciferase gene. Values (relative light units) are corrected for Renilla signal and provided as percentages of wild-type eIF4A translational activity. These experiments were repeated three times in sextuplicate, and representative data are shown. The results are expressed as the mean ± standard deviation. (*) Significant difference compared with the control (xpress), as determined by the Student’s t-test (*,<0.0001).
In total, 11 of the 15 eIF4A mutants that were transfected into RT101 cells with stem–loop structured lu-ciferase reporter gene exhibited a dominant-negative effect on in vivo translation of luciferase. These 11 mutants were generated in the eIF4A conserved Q (eIF4AF35A) and GG (eIF4AG137D) domains, as well as conserved domains I (eIF4AA77V and eIF4AK83N), Ia (eIF4ATE110,112RV and eIF4ATEL110,112,113,RVA), Ib (eIF4AT159D), II (eIF4AD183N), and VI (eIF4AR360Q, eIF4AR363Q, and eIF4AR366Q). The data from the eIF4A mutants showing a dominant-negative effect on translation were statistically analyzed using a two-tailed Student’s t-test; the P-value of all 11 mutants was found to be <0.0001 compared to control vector. In summary, the Q, I, Ia, GG, Ib, II, and VI conserved domains contribute to the translation-required regions of eIF4A (Fig. 1 ▶).
Mutant eIF4A associates with the eIF4F complex
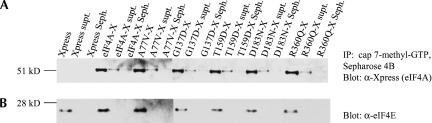
Several of the eIF4A mutants were found to be inactivated for binding to eIF4GM and eIF4GC, yet were capable of inhibiting translation of stem–loop structured luciferase gene in a dominant-negative manner (Fig. 5 ▶). Because eIF4A helicase activity is dependent on its association with the eIF4F complex, a complex containing eIF4E, eIF4A, and eIF4G, this posed the question of what mutant eIF4A might interact with to inhibit translation. To test whether mutant eIF4A can still bind to the eIF4F complex, RT101 cells were transfected with xpress-tagged eIF4AWT, eIF4AA77V, eIF4AG137D, eIF4AT159D, eIF4AD183N, or eIF4AR360Q, and the eIF4F complex was pulled down by cap 7-methyl-GTP Sepharose 4B beads (Polunovsky et al. 2000). Both wild-type and mutant eIF4A were found associated with the purified cap 7-methyl-GTP beads at similar levels, as determined by Western blot analysis with xpress antibody (Fig. 6A ▶). These aforementioned mutants had all been shown to inhibit translation in a dominant-negative manner and to be inactivated for binding to eIF4GM, and four of them had also been found to be inactivated for binding to eIF4GC. Both wild-type and mutant eIF4A showed a background level of association with the supernatant fractions of the RT101 cell lysates following spin-down of the cap 7-methyl-GTP bead mixtures (Fig. 6A ▶) and the capless Sepharose 4B bead mixtures (Fig. 6A ▶). When blotted with eIF4E antibody, RT101 cell lysates found positive for association of wild-type and mutant eIF4A with cap 7-methyl-GTP were also found positive for the presence of eIF4E (Fig. 6B ▶). These findings suggest that both wild-type and mutant eIF4A are found in the eIF4F complex, and that mutant eIF4A inhibits translation via its association with eIF4F, despite its inability to interact with the eIF4GM fragment.
FIGURE 6.
Pull-down of eIF4A/mutant-xpress with 7-methyl-GTP. (A) Thirty micrograms of cap 7-methyl-GTP Sepharose 4B were added to 100 μg of RT101 cell lysates transfected with eIF4A/mutant-xpress, spun down, and the SDS extract subjected to PAGE and immunoblotted with xpress antibody. Supernatant fractions, following spin-down of lysates with cap 7-methyl-GTP, were also blotted with xpress antibody. Prior to pull-down with 7-methyl-GTP, Sepharose 4B (50 μg) was added to 100 μg of RT101 cell lysates transfected with eIF4A/mutant-xpress, then blotted with xpress antibody. (B) Membranes were incubated with eIF4E antibody as a loading control for eIF4F complex.
Pdcd4 and the C-terminal region of eIF4G compete for binding to eIF4A
We have previously demonstrated that addition of Pdcd4 effectively competed with eIF4GC for binding to eIF4A (Yang et al. 2003a). It can be hypothesized that competition between Pdcd4 and eIF4GC involves their mutually conserved MA-3 domains. Further studies of this competition were initiated, using Pdcd4 mutants Pdcd4D253 and Pdcd4D418A, both of which contain point mutations in their MA-3 N-terminal or C-terminal domains. These Pdcd4 point mutants were found incapable of competing with eIF4GC for binding to eIF4A, confirming the importance of the MA-3 domains in binding to eIF4A (Yang et al. 2004). Likewise, when eIF4GCE1329Kand eIF4GC D1333A, both of which contain point mutations in their MA-3 domains, were tested for interaction with eIF4A, both mutants bound eIF4A with only 10% of wild-type eIF4A binding activity (Yang et al. 2004). Because Pdcd4 has been found to have discrete regions specifying competition for eIF4A, we asked if corresponding regions could be identified on eIF4A. Figures 2 ▶ and 4 ▶ show that the mutant eIF4AP56L has background level interaction with Pdcd4 but wild-type level interaction with eIF4GC, respectively. The mutant eIF4A F35A has wild-type-level interaction with both Pdcd4 (Fig. 2 ▶) and (Fig. 4 ▶). We therefore performed a competition eIF4GC assay using transfection of vectors containing the NLS, enabling all fusion proteins to translocate into the nucleus. To ensure that Pdcd4 would be capable of translocating to the nucleus, it was inserted into the vector pCMV-AD. However, to make certain that Pdcd4 did not interfere with eIF4A-AD and eIF4GC-BD binding, the NF-κB activation domain (AD) portion of the vector was deleted, creating a vector we termed pCMV-NLS-Pdcd4. RT101 cells were transfected with pCMV-AD-eIF4A (or pCMV-AD-eIF4AF35A or pCMV-AD-eIF4AP56L), pCMV-BD-eIF4GC, and increasing amounts of pCMV-NLS-Pdcd4 and Gal4-luciferase reporter gene. After 48 h, cells were lysed and measured for luciferase activity. The level of luciferase activity was taken as a measure of eIF4A–eIF4GC binding with respect to cotransfected Pdcd4 concentration. As shown in Figure 7 ▶, the level of luciferase expression was significantly reduced when eIF4A or eIF4AF35A was cotransfected with eIF4GC and Pdcd4. When compared with eIF4AP56L–eIF4GC-binding activity at 0 ng of CMV-Pdcd4 using a two-tailed Student’s t-test, the P-value of the mutant eIF4AF35A, at 200 ng (ratio of 0.5) of CMV-Pdcd4, was found to be 0.013. As the ratio of Pdcd4 to eIF4A increased from 0 to 0.5, eIF4A–eIF4GC-binding activity decreased from 100% to 68%, and eIF4AF35A–eIF4GC-binding activity decreased from 100% to 73%. When JB6 RT101 cells were transfected with pCMV-AD-eIF4AP56L (inactivated for binding to Pdcd4), pCMV-BD-eIF4GC, and increasing amounts of pCMV-NLS-Pdcd4, luciferase expression decreased only slightly, from 100% to 93% as the ratio of Pdcd4 to eIF4A increased from 0 to 0.5. Thus, eIF4AP56L binding to eIF4GC was much less inhibited by the addition of Pdcd4 than was eIF4A or eIF4AF35A binding to eIF4GC. Because Pdcd4 did not efficiently compete with eIF4GC for binding to eIF4AP56L and because eIF4AP56L had been shown to be inactivated for binding to Pdcd4 (Fig. 2 ▶) but not to eIF4GC (Fig. 4 ▶), this supports the proposed model of Pdcd4 and eIF4Gc competing for binding to eIF4A.
FIGURE 7.
Wild-type but not eIF4A mutants inactivated for binding to Pdcd4 compete for interaction with eIF4GC. RT101 cells were transfected with 50 ng of pCMV-BD-eIF4G (924–1444), 400 ng of pCMV-AD-eIF4A or the mutants pCMV-AD-eIF4AF35A or pCMV-AD-eIF4AP56L, 25 ng of Gal4-luciferase reporter gene, and 5 ng of thymi-dine kinase-Renilla luciferase gene, with 0–200 ng of pCMV-Pdcd4. Values (relative light units) are corrected for Renilla signal and expressed as the mean ± standard deviation. Experiments were repeated three times in sextuplicate, and representative data are shown.
The double globular structure of eIF4A reveals that several residues involved in binding to Pdcd4, eIF4GM, and eIF4GC are buried inside of the protein
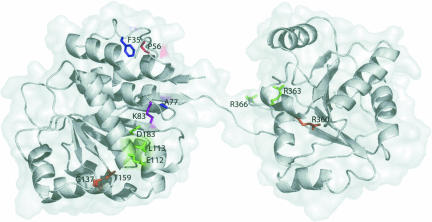
In order to determine which regions of the three-dimensional structure of eIF4A are important for binding to Pdcd4, eIF4GM, and eIF4GC, we mapped our mutations to a homology model of mouse eIF4A constructed based on the crystal structure of yeast eIF4A (Caruthers et al. 2000). Using the Deep View Swiss-PdbViewer program (Guex and Peitsch 1997), we made an alignment of the sequence of yeast eIF4A protein with the amino acid sequence of mouse eIF4A, in which our mutations were made. Since the yeast and mouse eIF4A are 63% identical, the homology model is expected to adequately represent the mouse protein. The ribbon diagram superimposed on a transparent surface representation in Figure 8 ▶ (created using Pymol; http://www.pymol.org) depicts the mutated residues labeled according to the mouse eIF4A sequence. Amino acids that, when mutated, disrupted binding were highlighted as follows: blue (F35, A77), eIF4GM interaction; red (P56), Pdcd4 interaction; purple (K83), eIF4GM and Pdcd4 interaction; green (D183, E112, L113, R363), eIF4GM and eIF4GC interaction; and orange (G137, T159, R360), eIF4GM, eIF4GC, and Pdcd4 interaction. E112, L113, G137, T159 R363, and R366, all of which have exposed side chains, are candidates for direct interactions with binding proteins. D183 is only partially exposed and so the explanation of its effect, either through disruption of direct protein–protein interaction or local structure, is uncertain. F35, P56, A77, K83, and R360 are all buried and thus are expected to affect a localized region of the structure. Because the loss of binding activity between eIF4A–Pdcd4, eIF4A–eIF4GM, or eIF4A–eIF4GC was expected to be the result of surface–surface interaction loss, and not the action of amino acids buried deep within the eIF4A protein structure itself, we substituted the native amino acids with their mutant counterparts and studied what effect these changes might have on the eIF4A protein structure. We found that the mutated amino acids could have three possible effects on eIF4A interaction with Pdcd4, eIF4GM, and eIF4GC: (1) the mutation could disrupt direct surface–surface interaction, as seen with the amino acids located on the outside of the structure; (2) cause local unfolding of the eIF4A structure in a region important for binding to Pdcd4, eIF4GM, or eIF4GC; or (3) the mutation could affect the catalytic (ATPase and/or helicase) activity of eIF4A, thus also affecting the ability of eIF4A to bind Pdcd4 eIF4GM or eIF4GC. One notable exception to these three possibilities was the eIF4AG137D point mutation: the substitution of an aspartic acid for a glycine places a side chain in an area where there was none, leading to potential steric interference with interactions of proteins in the region of T159. We believe that no mutant caused the unfolding of the overall structure of eIF4A, because many of our mutants inactivated for binding to Pdcd4, eIF4GM, or eIF4GC inhibited translation in a dominant-negative manner. This effect would not be seen if eIF4A were completely unfolded and thus nonfunctional. Furthermore, deletion mutation studies on eIF4A had shown the protein to be uniquely sensitive to any loss of its amino acids, and to result in incorrect conformational folding, which leads to rapid degradation in vivo (data not shown).
FIGURE 8.
Homology model of mouse eIF4A. A ribbon representation is superimposed on a transparent surface depiction of a homology model of mouse eIF4A. Specific amino acid residues are colored as follows: blue (F35, A77) = eIF4GM interaction; red (P56) = Pdcd4 interaction; purple (K83) = eIF4GM and Pdcd4 interaction; green (E112, L113, D183, R363) = eIF4GM and eIF4GC interaction; and orange (G137, T159, R360) = eIF4GM, eIF4GC, and Pdcd4 interaction. The figure was created using Pymol (http://www.pymol.org).
DISCUSSION
The present study establishes that the Pdcd4-binding region of eIF4A partially overlaps with its translation “enabling” region. Four of the five eIF4A mutants inactivated for Pdcd4 binding (eIF4AK83N, eIF4AG137D, eIF4AT159D, and eIF4AR360Q) were also inactivated for supporting translation, and four of the 11 mutants inactivated for translation were also inactivated for Pdcd4 binding (Table 1 ▶). Parts of the eIF4A: Pdcd4 interaction region may even be completely dissociated from the eIF4A:eIF4GM interaction region, as exhibited by the mutants eIF4AF35A and eIF4AP56L (eIF4AF35A binds Pdcd4, not eIF4GM, while eIF4AP56L binds eIF4GM, not Pdcd4). The residues important for Pdcd4 binding lie along the hinge region of eIF4A and reside in both N-terminal and C-terminal globular domains.
TABLE 1.
Summary of eIF4A mutant characterization
| Region | Translation | eIF4G(M) | eIF4G(C) | Pdcd4 | Change | Locality | |
| eIF4A | N/A | ↑ | + | + | + | N/A | N/A |
| F35A | Q | ↓ | - | + | + | Structure | Inside |
| P56L | Q | = | + | + | - | Structure | Inside |
| A77V | I | ↓ | - | +/− | + | Structure | Inside |
| K83N | I | ↓ | - | +/− | - | Charge | Inside |
| T110R | Ia | ↑ | + | +/− | + | Structure/charge | Outside |
| TE110,112RV | Ia | ↓ | - | - | +/− | Structure/charge | Outside |
| TEL110,112,113RVA | Ia | ↓ | - | - | +/− | Structure | Outside |
| G137D | GG | ↓ | - | - | - | Structure/charge | Outside |
| T159D | Ib | ↓ | - | - | - | Charge | Outside |
| D183N | II | ↓ | - | - | +/− | Charge | Inside |
| S214A | III | ↑ | + | +/− | +/− | Charge | Inside |
| ST214,216AA | III | ↑ | +/− | +/− | +/− | Charge | Outside |
| R360Q | VI | ↓ | - | - | - | Charge | Inside |
| R363Q | VI | ↓ | - | - | + | Charge | Outside |
| R366Q | VI | ↓ | - | - | +/− | Charge | Unknown |
Wilde-type and all 15 point mutations of eIF4A, shown with their effects on in cell cap-dependent translation and their in cell interaction with eIF4GM, eIF4GC, and Pdcd4. The up and down arrows signify activation above or below the translational activity level seen with wild-type eIF4A, respectively. + and - indicators signify wild-type or background interaction, respectively, with eIF4GM, eIF4GC, or Pdcd4. Locality data are taken from Figure 8 ▶.
The mammalian two-hybrid analysis and translation initiation activity experiments reported here show that the 11 eIF4A mutants that lost binding to eIF4GM also showed a dominant-negative effect on cellular translation, repressing expression levels of structured stem–loop luciferase below that of background (Table 1 ▶; Figs. 3A ▶, 5 ▶). This was not the case with eIF4A mutants that lost interaction with either Pdcd4 or eIF4GC. Some mutants retained interaction with Pdcd4, eIF4GC, or both, yet still lost translation initiation activity, depending only on their loss of interaction with eIF4GM. These results show that some of the residues of eIF4A responsible for binding Pdcd4 and/or eIF4GC are required but insufficient, for translation. However, all residues of eIF4A that bind eIF4GM appear essential for translation initiation activity.
It is interesting to note that of the four eIF4A mutants that retained binding to eIF4GM (Table 1 ▶; Fig. 3A ▶), only two (eIF4AP56L and eIF4AS214A) retained almost wild-type-level binding to eIF4GM, yet at least three were capable of initiating wild-type or greater translation initiation activity. This suggests that translation initiation is dependent on a certain threshold level of functional eIF4A, and may not require wild-type levels.
The fact that translation inhibition occurred in a dominant-negative manner corroborates structural modeling showing that mutant eIF4A is correctly folded and capable of competing with wild-type eIF4A for assembly into the translation-initiation complex (Morino et al. 2000). However, considering that the 11 translation-inhibiting eIF4A mutants are not directly associating with eIF4GM, how can their dominant-negative effect on translation be explained? One possibility is that these mutant eIF4As are interacting with intact eIF4G and/or with other members of the eIF4F complex but not with fragments of eIF4G. Our results from cap pull-down assays (Fig. 6A,B ▶) support this possibility. All five of the tested mutants are inactivated for binding to eIF4GM, and all of them act dominant-negatively to inhibit translation initiation in cells. However, all of these eIF4A mutants appear in the eIF4F complex. Thus, eIF4A mutants may be inhibiting translation initiation by interacting with the eIF4F complex of proteins, despite their inability to interact with fragments of eIF4G.
With respect to eIF4GC and Pdcd4, our results suggest that these two proteins compete for eIF4A via their mutual MA-3 domains. Wild-type eIF4A, or the mutant eIF4AF35A, both of which interact equally with eIF4GC and Pdcd4, were susceptible to competition between eIF4GC and Pdcd4. The mutant eIF4AP56L, which interacts with eIF4GC but not Pdcd4, was not susceptible to this competition. The MA-3 domain is found in the C-terminal domain of human DAP-5/NAT1/p97 as well as the second eIF4A-binding domain of eIF4G (Yang et al. 2004). Interestingly, C-terminal DAP-5/ NAT1/p97 does not bind eIF4A because several critical amino acids are not conserved (Yang et al. 2004). Due to this lack of MA-3 conservation, we were previously able to generate Pdcd4 mutants inactivated for eIF4A binding based on the MA-3 domain of DAP-5/NAT1/p97 protein (Yang et al. 2003a, 2004).
Because MA-3 domains are found in two eIF4A-binding proteins, namely, Pdcd4 and eIF4GC, we predicted that it is the MA-3 domain that binds eIF4A directly. Pdcd4 mutants containing deletions at either the N-terminal or C-terminal MA-3 regions showed complete loss of binding to eIF4A, confirming this hypothesis (Yang et al. 2004). This inactivation of binding to eIF4A was further seen with the Pdcd4 mutants Pdcd4E249K, Pdcd4D253A, Pdcd4D414K, and Pdcd4D418A, all containing mutations in specific amino acids of the N-terminal or C-terminal MA-3 regions (Yang et al. 2003a, 2004).
As illustrated in Table 1 ▶, the mutants eIF4AG137D, eIF4AT159D, and eIF4AR360Q are inactivated for binding to both Pdcd4 and eIFGC. Because the domains GG and TPGR lie in proximity to each other, it is likely that they constitute the actual site of competition between Pdcd4 and eIF4GC for binding to eIF4A. The observation that other eIF4A mutants lost binding to either Pdcd4 or eIF4GC, but not both, suggests that eIF4A contains a wide area of overlap for binding to these two proteins. Thus, the eIF4A conserved domains GG and TPGR are possible regions of competition between Pdcd4 and eIF4GC.
Svitkin et al. (2001) reported that two mutants, namely, eIF4AG137D and eIF4AT159D, did not inhibit cap-dependent translation, thereby not displaying a dominant-negative effect on translation. The explanation for this apparent discrepancy in our findings may lie in the fact that Svitkin et al. conducted translation experiments in vitro, while ours were performed in transfected cells. Likewise, Svitkin et al. (2001) showed that two other mutants, eIF4ATEL110,112,113RVA and eIF4AR363Q, are capable of interacting with eIF4G via stable interaction with the eIF4F complex, while the same mutants, according to our observations, were initially found not capable of interacting with eIF4GM and/or eIF4GC. However, immunoprecipitation with cap 7-methyl-GTP revealed that eIF4A mutants do interact with full-length eIF4G (and intact eIF4F complex).
Our eIF4A structure model shows that most of the mutations inactivated for binding to Pdcd4 and/or eIF4G lie on the perimeters of the hinge area of eIF4A. This suggests that Pdcd4 and/or eIF4G interact with eIF4A by “locking in” at its hinge region, where many of the conserved domains are located. The results also confirm our previously proposed eIF4A–Pdcd4 interaction model, wherein the two MA-3 domains of Pdcd4 bind to the two globular domains of eIF4A to achieve maximal binding capacity (Yang et al. 2004). In addition, our structural homology model indicates that Pdcd4 could, indeed, be binding eIF4A at both of its globular domains, as evidenced by the residues P56, K83, G137, and T159, all of which are located on the N-terminal globule of eIF4A and all of which are important for Pdcd4 binding, and the residue R360, which is located on the C-terminal globule of eIF4A and is also important for binding to Pdcd4. Although in our structural model the binding surfaces for Pdcd4 appear to be discontinuous, the flexible hinge region between the two globular domains of eIF4A would allow for movement of one globule relative to the other, potentially bringing those binding surfaces closer together in space. In addition, conformational changes in eIF4A that reorient one globule relative to another may occur in the presence of other binding partners, such as ATP.
In summary, the eIF4A residues important for Pdcd4 binding specify ATP binding, ATP hydrolysis, or RNA binding and are to a considerable extent dissociated from those important for binding to eIF4GM. This is not so for the eIF4GC- and Pdcd4-binding domains, which are in direct competition with each other for eIF4A. These data support a proposed model of competition between eIF4GC and Pdcd4, and a partial overlap between residues binding Pdcd4 and eIF4GC along the hinge region of eIF4A. Mutant eIF4A also does not need to be in direct contact with Pdcd4 to inhibit translation, suggesting eIF4A mutant-induced structural changes or the involvement of other, yet unknown, factors. Further discovery of the structure–function outcomes of eIF4A–Pdcd4 binding will greatly increase our understanding of protein expression, and of how the same mechanisms that bring about its initiation can also mediate cell transformation and cancer.
MATERIALS AND METHODS
Plasmids
Plasmids pCMV-BD-Pdcd4, middle domain pCMV-BD-eIF4G(497–974), or C-terminal pCMV-BD-eIF4G(924–1444) were used for mammalian two-hybrid analysis with pCMV-AD-eIF4A or its mutants (Fig. 1 ▶). Pdcd4, eIF4G(497–974), and eIF4G(924–1444) cDNAs were inserted into the EcoRI and XbaI sites of the pCMV-BD vector (Stratagene), while eIF4A or its mutant cDNAs were inserted into the BamHI and XhoI sites of the pCMV-AD vector (Stratagene). For competition assays, pCMV-BD-eIF4G(924–1444) and pCMV-AD-eIF4A, or the mutants pCMV-AD-eIF4AF35A and pCMV-AD-eIF4AP56L, were used, along with pCMV-pdcd4. The pCMV-pdcd4 vector was created by digesting pCMV-AD with restriction enzymes ClaI and BamHI to remove the p65 activation domain, followed by blunting with T4 DNA polymerase and ligation. The vector was digested with XbaI and T4 DNA polymerase, again added to synthesize blunt ends. Finally, the vector was restriction-digested with EcoRI to remove the MCS fragment. Pdcd4 was ligated into the pCMV vector after having been restriction-digested with ApaI, blunted with T4 DNA polymerase, and restriction-digested with EcoRI. For the translation assays, stem–loop structured luciferase vector (Yang et al. 2003a) was transfected with eIF4A or its mutants. eIF4A or its mutants were inserted into the BamHI and XhoI sites of the xpress vector pcDNA4/HisMAX C (Invitrogen) after being restriction-digested from the BamHI and XhoI sites of pCMV-AD. For all GST pull-downs and immunoprecipitations, eIF4A and its mutants were used in the xpress vector (Invitrogen), and for some of the immunoprecipitations, pcDNA3HA-eIF4G(497–974) and pcDNA3HA-eIF4G(924–1444) (Imataka and Sonenberg 1997) were used as well.
Site-directed mutagenesis of eIF4A
Point mutants of eIF4A were generated by subjecting the pCMV-AD-eIF4A vector to mutagenesis using the GeneTailor Site-Directed Mutagenesis System (Invitrogen). The following mutagenic oligomers were used (with mutation codons in bold):
for pCMV-AD-eIF4AF35A, 5′-AATGAAATTGTTGATAACGCTGATGATATG-3′and 5′-GTTATCAACAATTTCATTCCAGT TGCTCTC-3′;\
for pCMV-AD-eIF4AP56L, 5′-GCATATGGTTTTGAGAAGCTTTCAGCTATT-3′ and 5′-CTTCTCAAAACCATATGCATAGAT GCCTCG-3′;
for pCMV-AD-eIF4AA77V, 5′-TATGATGTGATTGCTCAAGTTCAGTCAGGT-3′ and 5′-TTGAGCAATCACATCATACCCTT TAATACA-3′;
for pCMV-AD-eIF4AK83N, 5′-CAGTCAGGTACTGGCAATACAGCCACATTT-3′ and 5′-GCCAGTACCTGACTGAGCTTGAG CAATCAC-3′;
for pCMV-AD-eIF4AT110R, 5′-CTAGTATTGGCCCCCAGAAGAGAACTGGCT-3′ and 5′-GGGGGCCAATACTAGTGCTTGG GTCTCCTT-3′;
for pCMV-AD-eIF4ATE110,112RV, 5′-TTGGCCCCCAGAAGAGTACTGGCTCAACAG-3′ and 5′-TCTTCTGGGGGCCAATAC TAGTGCTTGGGT-3′;
for pCMV-AD-eIF4ATEL110,112,113RVA, 5′-GCCCCCAGAAGAGTAGCTGCTCAACAGATC-3′ and 5′-TACTCTTCTGGGGGC CAATACTAGTGCTTG-3′;
for pCMV-AD-eIF4AG137D, 5′-ACTTGTCATGCTTGCATTGATGGAACAAATGTT-3′ and 5′-AATGCAAGCATGACAAGT TGCTCCCATATA-3′;
for pCMV-AD-eIF4AT159D, 5′-CCTCACATTGTTGTTGGTGATCCAGGGAGA-3′ and 5′-ACCAACAACAATGTGAGGGGCTT CAGCCTG-3′;
for pCMV-AD-eIF4AD183N, 5′-ATCAAAATGTTCGTTTTGAACGAAGCAGAT-3′ and 5′-CAAAACGAACATTTTGATCCATT TTGGAGA-3′;
for pCMV-AD-eIF4AS214A, 5′-CAGGTTGTGTTGCTTGCCGCCACAATGCCA-3′ and 5′-AAGCAACACAACCTGAATGCTT GTATTTAA-3′;
for pCMV-AD-eIF4AST214,216AA, 5′-GTGTTGCTTGCCGCCGCCATGCCAACTGAT-3′ and 5′-GGCGGCAAGCAACACAACC TGAATGCTTGT-3′;
for pCMV-AD-eIF4AR360Q, 5′-CGTGAAAACTATATTCACCAAATTGGCAGA-3′ and 5′-GTGAATATAGTTTTCACGATTG GTAGGTAG-3′;
for pCMV-AD-eIF4AR363Q, 5′-TATATTCACAGAATTGGCCAAGGGGGTCGA-3′ and 5′-GCCAATTCTGTGAATATAGTTTT CACGATT-3′; and
for pCMV-AD-eIF4AR366Q, 5′-AGAATTGGCAGAGGGGGTCAATTTGGGAGG-3′ and 5′-ACCCCCTCTCCGAATTCTGT GAATATAGTT-3′.
All mutants were verified by sequencing.
Mammalian two-hybrid assay of protein–protein binding
In the mammalian two-hybrid assay, a luciferase reporter becomes activated when a DNA-binding domain (BD) fusion protein binds to an activation domain (AD) fusion protein. RT101 cells (1.5 × 104) were seeded in 24-well plates in Eagle’s minimal essential medium (EMEM) with 4% fetal bovine serum (FBS). After 16 h, cells were transfected with 50 ng of pCMV-BD-eIF4G(497–974) [or pCMV-BD-eIF4G(924–1444)], 400 ng of pCMV-AD-eIF4A (or its mutants), 25 ng of Gal4-luciferase reporter gene, and 5 ng of thymidine kinase-Renilla luciferase gene in 2 μL of Lipofectamine (Invitrogen) for 4 h. Alternatively, RT101 cells were transfected with 250 ng of pCMV-BD-pdcd4 and 250 ng of pCMV-AD-eIF4A (or its mutants) with 25 ng of Gal4-luciferase reporter gene and 5 ng of thymidine kinase-Renilla luciferase gene. Cells were then incubated with fresh EMEM containing 4% FBS. At 48 h, cells were lysed in 100 μL of 1× passive lysis buffer (Promega). Then 25-μL aliquots of the lysates were analyzed for luminescent signal with a Microtiter Plate Luminometer (Dynex Technologies).
Expression and purification of recombinant GST-Pdcd4
GST-Pdcd4 was expressed in insect Sf9 cells. Sf9 cells, having been grown to a concentration of 2 × 106 cells/mL in Sf-900 II SFM (Invitrogen), were infected with recombinant baculovirus encoding GST-Pdcd4 at a multiplicity of infection of 5 and cultured for 48 h at 27°C. Then, 2 × 107 cells/mL of insect cell lysis buffer (10 mM TrisCl at pH 7.5, 130 mM NaCl, 1% Triton X-100, 10 mM NaF, 10 mM sodium phosphate at pH 7.5, 10 mM sodium pyrophosphate, and 1× protease inhibitor cocktail) were lysed on ice for 45 min. After centrifugation, 1 mL of glutathioneagarose beads was added to the 50-mL supernatant total for 30 min at 4°C. After centrifugation, the glutathioneagarose beads were washed with PBS four times. The recombinant GST-Pdcd4 was eluted by 5 mM reduced glutathione in Tris-HCL (pH 8.0). All elutions were tested for GST-Pdcd4 expression via SDS-PAGE and stored at −80°C.
Preparation of cell lysates and Western blot
RT101 cells (4 × 105 cells/100 mm plate) were transfected with 8 μg of the vector pCMV-AD (Invitrogen), into which eIF4A or its mutants had been inserted, with 32 μL of Lipofectamine (Invitrogen). After 4 h, cells were incubated with fresh EMEM containing 4% FBS. At 48 h, cells were lysed with 300 μL of nondenaturing lysis buffer (20 mM HEPES KOH at pH 7.6, 100 mM KCl, 0.5 mM EDTA, 20% glycerol, 0.5% Triton X-100, 50 μg/mL RNAse A, 1× protease inhibitor cocktail) for 30 min at 4°C. The cells were scraped from the 100-mm plates, sheared by passage through a 22-gauge needle, frozen at -80°C, and spun at 20,000 rpm for 20 min to remove the nuclear pellet (these steps were critical for efficient solubilization of eIF4A mutant proteins). A 10-μg aliquot of each lysate was separated by 4%–12% SDS-PAGE and transferred to a nitrocellulose membrane. The protein-bound membrane was blotted with goat eIF4AII polyclonal antibody (Santa Cruz Biotech.) or mouse monoclonal B-actin (Sigma) antibody, followed by ECL (Amersham Biosciences).
GST pull-down and immunoprecipitation assays
RT101 cells (4 × 105 cells/100 mm plate) were transfected with 8 μg of xpress-tagged wild-type/mutant eIF4A vector (Invitrogen) or 857 ng of pcDNA3HA-eIF4G(497–974)/pcDNA3HA-eIF4G(924–1444) and 7 μg of xpress-tagged wild-type/mutant eIF4A vector (Invitrogen), with 32 μL of Lipofectamine (Invitrogen). After 4 h, cells were incubated with fresh EMEM containing 4% FBS. At 48 h, cells were rinsed in PBS and lysed with 300 μL of nondenaturing lysis buffer (described previously) for 30 min at 4°C. The cells were scraped from the 100-mm plates and sheared by passage through a 22-gauge needle as described above. Then, 100 μg of GST-Pdcd4, 2 μg of HA antibody (12CA5; Roche), or 50 μg of Sepharose 4B beads were added to 100 μg of each lysate and the mixtures were rotated for 2 or 4 h at 4°C. After being washed with lysis buffer, 10 μL of glutathione-Sepharose beads, 50 μL of protein G-Sepharose beads (Sigma), or 10 μL of cap 7-methyl-GTP Sepharose 4B beads (Amersham Pharmacia Biotech), respectively, were added and the lysates were rotated overnight at 4°C. Lysates were loaded onto filter columns, spun down, and supernatants were discarded or stored for analysis by Western blot. The remaining beads were washed twice with 750 μL of lysis buffer and spun. The beads were taken off the column with 24 μL of lysis buffer and placed into tubes, to which 6 μL of loading dye was added. The beads were heated to 95°C for 5 min and analyzed by 4%–12% SDS-PAGE. After protein transfer, protein levels were measured using mouse monoclonal xpress antibody (Invitrogen), followed by ECL. Verification of equal glutathione-Sepharose or protein G-Sepharose bead affinity for GST-Pdcd4 or HA antibody was performed by incubating the nitrocellulose membranes in 15 mL of Restore Western Blot Stripping Buffer (Pierce), followed by blotting with rabbit polyclonal GST (Santa Cruz Biotech.) or HA antibody (12CA5). Alternatively, membranes used for cap 7-methyl-GTP analysis were blotted with mouse monoclonal eIF4E (Santa Cruz Biotech.) following incubation in the stripping buffer. In all, 10 μg of all lysates were separately analyzed by 4%–12% SDS-PAGE, followed by blotting with xpress antibody, to confirm equal protein input for all pull-down and immunoprecipitation experiments.
Cap-dependent translation assays
RT101 cells (1.5 × 104) were seeded in 24-well plates in EMEM with 4% FBS and transfected with 400 ng of xpress-tagged wild-type/mutant eIF4A vector (Invitrogen), 25 ng of stem–loop structured luciferase vector, and 5 ng of thymidine kinase-Renilla luciferase gene in 2 μL of Lipofectamine (Invitrogen). After 4 h, cells were incubated with fresh EMEM containing 4% FBS. At 48 h, cells were lysed in 100 μL of 1× passive lysis buffer (Promega). Then 25-μL aliquots of the lysates were analyzed for luminescent signal with a Microtiter Plate Luminometer (Dynex Technologies).
In vivo competition assays
RT101 cells (2 × 104) were seeded in 24-well plates in EMEM with 4% FBS and transfected with 50 ng of pCMV-BD-eIF4G(924–1444), 400 ng of pCMV-AD-eIF4A (or the eIF4A mutants pCMV-AD-eIF4AF35A or pCMV-AD-eIF4AP56L), 25 ng of Gal4-luciferase reporter gene, 5 ng of thymidine kinase-Renilla luciferase gene, and 0–200 ng of pCMV-Pdcd4 in 2 μL of Lipofectamine (Invitrogen) for 4 h. The total DNA was equalized at 680 ng by adding pcDNA3.1 (Invitrogen) in lieu of pCMV-Pdcd4. Cells were incubated with fresh EMEM containing 4% FBS. At 48 h, cells were lysed in 100 μL of 1× passive lysis buffer (Promega). Then 25-μL aliquots of the lysates were analyzed for luminescent signal with a Microtiter Plate Luminometer (Dynex Technologies).
Homology modeling of mouse eIF4A
Since the sequences of mouse and yeast eIF4A are 63% identical, a homology model of mouse eIF4A was constructed based on the crystal structure of eIF4A from yeast (PDB code 1FUU; Caruthers et al. 2000) using the program DeepView Swiss-PdbViewer and the SWISS-MODEL server (Guex and Peitsch 1997). The resulting model was further energy minimized in DeepView using Gromos 96 (Guex and Peitsch 1997).
Acknowledgments
We thank David Derse, NCI, and the members of the Gene Regulation Section, LCP, NCI, for helpful discussions.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7191905.
REFERENCES
- Afonja, O., Juste, D., Das, S., Matsuhashi, S., and Samuels, H.H. 2004. Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene 23: 8135–8145. [DOI] [PubMed] [Google Scholar]
- Aravind, L. and Koonin, E.V. 2000. Eukaryote-specific domains in translation initiation factors: Implications for translation regulation and evolution of the translation system. Genome Res. 10: 1172–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruthers, J.M., Johnson, E.R., and McKay, D.B. 2000. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl. Acad. Sci. 97: 13080–13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmarik, J.L., Min, H., Hegamyer, G., Zhan, S., Kulesz-Martin, M., Yoshinaga, H., Matsuhashi, S., and Colburn, N.H. 1999. Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation. Proc. Natl. Acad. Sci. 96: 14037–14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio, E., Preiss, T., and Hentze, M.W. 1998. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end-dependent. RNA 4: 828–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goke, R., Barth, P., Schmidt, A., Samans, B., and Lankat-Buttgereit, B. 2004. Programmed cell death protein 4 suppresses CDK1/cdc2 via induction of p21Waf1/Cip1. Am. J. Physiol. Cell Physiol. 287: C1541–C1546. [DOI] [PubMed] [Google Scholar]
- Guex, N. and Peitsch, M.C. 1997. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18: 2714–2723. [DOI] [PubMed] [Google Scholar]
- Imataka, H. and Sonenberg, N. 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 17: 6940–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneeva, N.L., Lamphear, B.J., Hennigan, F.L., and Rhoads, R.E. 2000. Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J. Biol. Chem. 275: 41369–41376. [DOI] [PubMed] [Google Scholar]
- Korneeva, N.L., Lamphear, B.J., Hennigan, F.L., Merrick, W.C., and Rhoads, R.E. 2001. Characterization of the two eIF4A-binding sites on human eIF4G-1. J. Biol. Chem. 276: 2872–2879. [DOI] [PubMed] [Google Scholar]
- Kressler, D., de la Cruz, J., Rojo, M., and Linder, P. 1997. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 7283–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Belsham, G.J., and Proud, C.G. 2001. Eukaryotic initiation factors 4A (eIF4A) and 4G (eIF4G) mutually interact in a 1:1 ratio in vivo. J. Biol. Chem. 276: 29111–29115. [DOI] [PubMed] [Google Scholar]
- Linder, P. 2003. Yeast RNA helicases of the DEAD-box family involved in translation initiation. Biol. Cell 95: 157–167. [DOI] [PubMed] [Google Scholar]
- Linder, P., Lasko, P.F., Ashburner, M., Leroy, P., Nielsen, P.J., Nishi, K., Schnier, J., and Slonimski, P.P. 1989. Birth of the D-E-A-D box. Nature 337: 121–122. [DOI] [PubMed] [Google Scholar]
- Lomakin, I.B., Hellen, C.U., and Pestova, T.V. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20: 6019–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino, S., Imataka, H., Svitkin, Y.V., Pestova, T.V., and Sonenberg, N. 2000. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20: 468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain, V.M. 1996. Initiation of protein synthesis in eukaryotic cells. Eur. J. Biochem. 236: 747–771. [DOI] [PubMed] [Google Scholar]
- Pause, A. and Sonenberg, N. 1992. Mutational analysis of a DEAD box RNA helicase: The mammalian translation initiation factor eIF-4A. EMBO J. 11: 2643–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause, A., Methot, N., and Sonenberg, N. 1993. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol. Cell. Biol. 13: 6789–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause, A., Methot, N., Svitkin, Y., Merrick, W.C., and Sonenberg, N. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13: 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polunovsky, V.A., Gingras, A.C., Sonenberg, N., Peterson, M., Tan, A., Rubins, J.B., Manivel, J.C., and Bitterman, P.B. 2000. Translational control of the antiapoptotic function of Ras. J. Biol. Chem. 275: 24776–24780. [DOI] [PubMed] [Google Scholar]
- Ponting, C.P. 2000. Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem. Sci. 25: 423–426. [DOI] [PubMed] [Google Scholar]
- Ray, B.K., Lawson, T.G., Kramer, J.C., Cladaras, M.H., Grifo, J.A., Abramson, R.D., Merrick, W.C., and Thach, R.E. 1985. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J. Biol. Chem. 260: 7651–7658. [PubMed] [Google Scholar]
- Rocak, S. and Linder, P. 2004. DEAD-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell. Biol. 5: 232–241. [DOI] [PubMed] [Google Scholar]
- Rogers Jr., G.W., Richter, N.J., and Merrick W.C. 1999. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 274: 12236–12244. [DOI] [PubMed] [Google Scholar]
- Rogers Jr., G.W., Richter, N.J., Lima, W.F., and Merrick, W.C. 2001. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 276: 30914–30922. [DOI] [PubMed] [Google Scholar]
- Rozen, F., Edery, I., Meerovitch, K., Dever, T.E., Merrick, W.C., and Sonenberg, N. 1990. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 10: 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin, Y.V., Pause, A., Haghighat, A., Pyronnet, S., Witherell, G., Belsham, G.J., and Sonenberg, N. 2001. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7: 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, N.K. and Linder, P. 2001. DExD/H box RNA helicases: From generic motors to specific dissociation functions. Mol. Cell 8: 251–262. [DOI] [PubMed] [Google Scholar]
- Tanner, N.K., Cordin, O., Banroques, J., Doere, M., and Linder, P. 2003. The Q motif: A newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell 11: 127–138. [DOI] [PubMed] [Google Scholar]
- Yang, H.-S., Jansen, A.P., Nair, R., Shibara, K., Verma, A.K., Cmarik, J.L., and Colburn, N.H. 2001. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-κB or ODC transactivation. Oncogene 20: 669–676. [DOI] [PubMed] [Google Scholar]
- Yang, H.S., Jansen, A.P., Komar, A.A., Zheng, X., Merrick, W.C., Costes, S., Lockett, S.J., Sonenberg, N., and Colburn, N.H. 2003a. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol. Cell. Biol. 23: 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H.S., Knies, J.L., Stark, C., and Colburn, N.H. 2003b. Pdcd4 suppresses tumor phenotype in JB6 cells by inhibiting AP-1 transactivation. Oncogene 22: 3712–3720. [DOI] [PubMed] [Google Scholar]
- Yang, H.S., Cho, M.H., Zakowicz, H., Hegamyer, G., Sonenberg, N., and Colburn, N.H. 2004. A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol. Cell. Biol. 24: 3894–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]