Introduction
Facioscapulohumeral muscular dystrophy (FSHD [MIM 158900]) is the third most common inherited muscular dystrophy, with an estimated frequency of 1 in 20,000 (Padberg 1982). The disease is foremost characterized by a progressive and often asymmetrical weakness and wasting of the facial, shoulder, and upper-arm muscles. Generally, FSHD displays a characteristic gradual spread of muscle involvement, starting in the face and slowly progressing to the shoulder and upper-arm musculature and to the abdominal and foot-extensor muscles. Extramuscular involvement is also reported for FSHD, since approximately half of the patients present with subclinical high-tone hearing loss and retinovasculopathy (Padberg 2004). In some severely affected patients of Japanese origin, mental retardation and epilepsy are reported (Funakoshi et al. 1998; Miura et al. 1998). Males are typically more severely affected than females, and there is a wide clinical inter- and intrafamilial variability of the disease, with ∼20% of patients eventually becoming wheelchair-bound and with an equal frequency of nonpenetrant gene carriers (Padberg et al. 1991; Zatz et al. 1998).
The disease displays an autosomal dominant mode of inheritance, with the vast majority of familial cases linked to a genetic lesion in the subtelomere of chromosome 4q (4qter) (Wijmenga et al. 1990, 1992). Some familial cases have been reported that do not link to 4qter, but a second locus for FSHD has yet to be identified (Gilbert et al. 1992, 1993; Tim et al. 2001). Although, in the early 1990s, FSHD was one of the first diseases to be linked to a genomic locus by use of microsatellite repeat markers, the pathogenomic mechanism underlying FSHD is still largely unresolved. In contrast to most monogenic disorders, in which the genetic lesion typically affects the structure or function of a specific disease gene, there is increasing evidence that FSHD is caused by a complex epigenetic mechanism involving the contraction of a subtelomeric macrosatellite repeat. Thus, it is likely not the structure but rather the (spatiotemporal-restricted) transcriptional control of one or more disease genes that is perturbed in FSHD as a result of repeat-contraction–mediated chromatin alterations. This review focuses on the recent advances in understanding the cause and consequence of the repeat-array contraction and the different mechanistic disease models that have emerged from these studies.
FSHD, a Macrosatellite Repeat-Contraction Disease
One of the most prominent features of the subtelomere of chromosome 4q is a large polymorphic repeat structure consisting of 1–100 KpnI units, designated “D4Z4” (van Deutekom et al. 1993; Hewitt et al. 1994). These units, each 3.3 kb in size, are ordered in a head-to-tail orientation and show very little sequence diversity. In 95% of patients with FSHD, the D4Z4 repeat is contracted to an array of 1–10 units (Wijmenga et al. 1992; van Deutekom et al. 1993), and, apparently, at least one unit of D4Z4 is required to develop FSHD, since monosomy of 4q does not cause FSHD (Tupler et al. 1996). There is a rough and inverse relationship between clinical severity and the residual repeat size, with the smallest repeats causing the most severe phenotypes (Lunt et al. 1995; Tawil et al. 1996).
D4Z4 repeats are typically visualized on Southern blots of genomic DNA after digestion with EcoRI and hybridization with probe p13E-11. Probe p13E-11 does not recognize the repeat proper but a locus (D4F104S1) just proximal to the repeat, within the D4Z4-containing EcoRI fragment (Wijmenga et al. 1992; van Deutekom et al. 1993). Because of the large size of the repeat (10–38 kb in disease alleles and >38–350 kb in healthy alleles), Southern blots after conventional linear gel electrophoresis typically only allow visualization of the disease allele, and alleles of >50 kb comigrate at the top of the gel. Therefore, for complete assessment of D4Z4 repeats, pulsed-field gel electrophoresis (PFGE) is required (Lemmers et al. 2004b).
The D4Z4 unit has been completely sequenced and shows some peculiar features (Hewitt et al. 1994; Winokur et al. 1994; Lee et al. 1995). First, D4Z4 contains an ORF encoding a putative homeobox protein called “DUX4.” Native transcripts of DUX4 have never been identified, although recent data show some evidence of the presence of a DUX-related protein, specifically in FSHD myoblasts (Hewitt et al. 1994; Lyle et al. 1995; Gabriels et al. 1999; Coppee et al. 2004). In addition, D4Z4 harbors two classes of repetitive DNA—the GC-rich low-copy repeats hhspm3 and LSau, both of which are found predominantly in heterochromatic domains of the genome (Hewitt et al. 1994). Furthermore, D4Z4 is unusually GC rich, with no fewer than 290 CpG dinucleotides and with a GpC:CpG ratio of 0.7. D4Z4 is not restricted to chromosome 4; perfect arrays of D4Z4 units can also be detected on chromosome 10q (see the “Unique Linkage of FSHD to Chromosome 4” section) (Bakker et al. 1995; Deidda et al. 1995), whereas additional sequences homologous to D4Z4 can be identified on many heterochromatic loci, such as those of the short arms of acrocentric chromosomes and the pericentromeric region of chromosome 1q, where they are often interspersed by other (satellite) repeats (Lyle et al. 1995; Ballarati et al. 2002).
Unique Linkage of FSHD to Chromosome 4
The subtelomeric location of the D4Z4 repeat adds several intriguing findings to the pathogenic mechanism of FSHD. Subtelomeres define the proterminal ends of chromosomes and are composed of patchworks of genomic segments that are spread over many nonhomologous chromosome ends as a result of ectopic recombination and duplication events (Mefford and Trask 2002). Subtelomeres may be organized in a proximal domain that is shared with a subset of nonhomologous chromosomes and a distal domain shared with almost all chromosome ends. The proximal and distal domains are often separated by an imperfect telomere repeat (Flint et al. 1997).
The D4Z4 repeat is localized in the proximal subtelomeric domain, which, as a result of an ancient duplication, is also present at the end of chromosome 10q (Bakker et al. 1995; Deidda et al. 1995). As a consequence, 10qter harbors a highly homologous and equally polymorphic repeat array at the same chromosomal position as on chromosome 4. Interestingly, 10% of chromosomes 10 also carry an FSHD-sized repeat (Bakker et al. 1995, 1996). However, in contrast to the D4Z4 repeat on chromosome 4qter, repeat contractions on 10qter have never been reported in studies of FSHD.
Through the combination of restriction enzymes differentially recognizing chromosomes 4– and 10–derived repeat units and allele separation by PFGE (Deidda et al. 1996; Lemmers et al. 2001), the behavior of this macrosatellite repeat is now well studied in healthy and FSHD populations. Owing to subtelomeric localization, translocations of chromosome 4– and 10–derived units are encountered in the Dutch population with an equal frequency of 10% (van Deutekom et al. 1996; van Overveld et al. 2000). These translocations have been observed in other populations (Matsumura et al. 2002)—albeit at somewhat different frequencies—which suggests that de novo translocations are universal but rare. Translocated repeat arrays are not always homogeneous but are often composed of clusters of chromosome 4–derived and chromosome 10–derived repeat units (Lemmers et al. 1998; van Overveld et al. 2000). Despite this plasticity, contracted repeats on chromosome 10 have never been associated with FSHD, whereas the exact composition of the contracted repeat array on 4qter (i.e., whether it is derived from chromosome 4, 10, or both) seems to be irrelevant to the development of FSHD. It is interesting, however, that chromosome 4–derived translocated repeat arrays on chromosome 10 tend to be homogeneous, whereas the inverse translocations on chromosome 4 are almost always composed of heterogeneous clusters of chromosome 4– and 10–derived repeat units (van Overveld et al. 2000). The cause for this difference in homogeneity is currently unknown but may relate to a directional mutation mechanism.
Subtelomeric Variation Distal to D4Z4
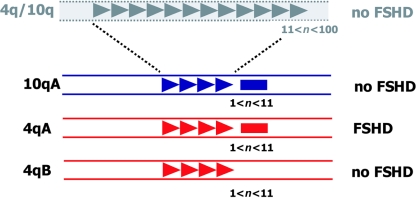
It was recently observed that two allelic variants of the 4q subtelomere exist (van Geel et al. 2002), which adds to the complexity of the pathogenesis of FSHD. Large allelic variations of subtelomeric domains may not be uncommon. For example, for chromosome 16pter, three variants have been reported that differ in size by several hundred kilobases (Wilkie et al. 1991). On the basis of some sequence variations distal to D4Z4, within different YAC clones originating from chromosome 4, two allelic variants were identified and were designated “4qA” and “4qB” (van Geel et al. 2002). Subsequent studies showed that both alleles are almost equally common in the general population but that FSHD alleles are always the 4qA type. Furthermore, it was demonstrated that both chromosomes display an equal propensity to rearrange, and it was thus hypothesized that, in addition to a contraction of D4Z4, additional elements in cis are necessary to cause FSHD (4qA) or to prevent it (4qB) (Lemmers et al. 2002). This hypothesis was recently corroborated by the demonstration that FSHD-sized repeat arrays on 4qB chromosomes, indeed, do not cause FSHD (Lemmers et al. 2004d). It is confusing that all chromosome 10 ends are of the 4qA type, making it more difficult to explain easily the unique linkage of FSHD to 4qA. Figure 1 summarizes all known genetic requirements for development of FSHD.
Figure 1.
Schematic presentation of the primary genetic defect of FSHD. Polymorphic arrays of D4Z4 units (triangles) are located in the subtelomeres of chromosomes 4q (red) and 10q (blue). On chromosome 10, this array may vary between 1 and 100 units without pathological consequences (n = number of units). On chromosome 4, however, repeat arrays in the range of 1–10 units cause FSHD, but only when the contraction is in cis with the subtelomeric variant 4qA, which contains beta satellite repeats (red rectangle), not the 4qB variant. The beta satellite repeats are also present in the 10q subtelomere (blue rectangle).
The distal ends of both alleles are only partly sequenced, and, therefore, the exact differences are unknown. The most prominent difference is the presence of a 6.2-kb beta satellite repeat directly distal to D4Z4 on 4qA chromosomes (van Geel et al. 2002). Beta satellite repeats are 68-bp repetitive elements that are found mostly on the pericentromeric domains of the acrocentric chromosomes in which they probably, like alpha satellite repeats, fulfill a structural role (Agresti et al. 1989; Vogt 1990; Lee et al. 1997). Given the epigenetic disease mechanism, it is postulated that the presence of this beta satellite repeat is essential for the disease presentation, although it does not explain why FSHD-sized repeats on chromosome 10qter with the identical beta satellite repeat are nonpathogenic.
Detailed analysis of the D4Z4 repeat on 4qA, 4qB, and 10q chromosomes showed some unexpected features. First, the size distribution of the D4Z4 repeat is not uniform but rather shows a multimodal distribution with equidistant peaks of ∼65 kb, possibly reflecting a higher-order chromatin architecture (van Overveld et al. 2000). Second, the mean length of the repeat array is different for the three chromosome ends. Whereas D4Z4 repeats on 4qA chromosomes are, on average, 136 kb in size, repeats on 4qB chromosomes are 94 kb, on average, and repeats on chromosome 10q are 75 kb, on average (van Overveld et al. 2000; Lemmers et al. 2004c). The cause of this difference in mean size is currently unknown, but it suggests that, despite the high frequency of translocated repeat arrays, interchromosomal recombination is rare.
Indeed, a detailed search for allele-specific polymorphisms revealed a PvuII polymorphism within D4Z4 that is not uniformly distributed over the repeats on 4qA and 4qB chromosomes (Lemmers et al. 2004c). Whereas approximately one-third of all most-proximal (i.e., first) repeat units on 4qB chromosomes are sensitive to PvuII, almost none of the proximal units of the array on 4qA chromosomes carry this PvuII site. However, on both alleles, the D4Z4 units after the first unit seem to be equally sensitive to PvuII. This, and other polymorphisms studied, showed that regions proximal and distal to D4Z4 are in strong linkage disequilibrium and that, although internal units of D4Z4 on 4qA and 4qB chromosomes are fairly well homogenized, recombination between 4qA and 4qB of sequences flanking D4Z4 is not frequent.
Timing and Mechanism of D4Z4 Contraction
D4Z4 displays a considerable mitotic instability. In the Dutch population, 3% of individuals are carriers of a mitotic repeat contraction or expansion on either chromosome 4 or 10 (van Overveld et al. 2000). The mechanism by which D4Z4 mitotically rearranges is well studied in de novo kindreds with FSHD. Although germline mosaicism has been only sporadically reported in studies of FSHD (Griggs et al. 1993; Weiffenbach et al. 1993), almost half of de novo FSHD cases arise through a mitotic rearrangement, either in the unaffected carrier parent of an affected nonmosaic child or in the affected individual (van der Maarel et al. 2000). In these mosaic individuals, there seems to be a relationship between the severity of the disease and the combination of the residual repeat size and the proportion of cells carrying the disease allele. The gender difference in clinical severity is well reflected in mosaic individuals. Whereas females are mostly unaffected carriers of a mosaic FSHD allele, males, when carrying a comparable mosaic allele complement, are more often affected (van der Maarel et al. 2000). This predominance of asymptomatic female carriers of a somatic contraction has been reported repeatedly (Zatz et al. 1995; Kohler et al. 1996; van der Maarel et al. 2000), although a gender difference in clinical severity could not be confirmed in a study of mosaic and nonmosaic Japanese individuals (Goto et al. 2004).
Initially, somatic mosaicism was only described in unaffected carrier parents of nonmosaic affected children (e.g., see Upadhyaya et al. [1995] and Kohler et al. [1996]). However, with the increased resolution of PFGE, somatic mosaicism was also detected—with equal frequency—in affected de novo patients. Indeed, it was recently demonstrated that the conventional linear gel-based Southern blot detection for FSHD alleles, a standard procedure in most diagnostic centers, often fails to detect somatic mosaicism for the disease allele in mosaic patients, since it relies fully on a (minor) decreased signal intensity for mosaic alleles and not on the presence of >4 alleles (Lemmers et al. 2004a).
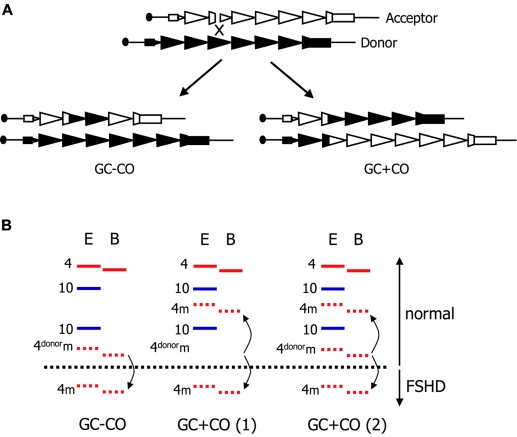
The presence of only five alleles in the genomes of mosaic individuals suggests that most mitotic rearrangements occur by a single gene-conversion event without crossover. Such mitotic gene conversion, by definition, results in the presence of two cell populations: one carrying the unchanged parental donor allele and one carrying a rearranged acceptor allele. The exact mechanism by which the D4Z4 repeat mitotically contracts was studied in further detail in a cohort of mosaic Dutch kindreds with FSHD. This demonstrated that, most likely, D4Z4 rearranges by a synthesis-dependent strand-annealing mechanism without crossover. However, gene conversion with crossover was also detected, with an unexpectedly high frequency. In three-quarters of the mosaic individuals, two cell populations were identified by the presence of five alleles (two alleles derived from chromosome 10, one nonrearranged allele derived from chromosome 4, and two alleles with reduced intensity derived from chromosome 4 prior to and after gene conversion without crossover). However, the remainder of mosaic individuals had two cell populations carrying distinct mitotically rearranged alleles derived from gene conversions with crossover. These individuals sometimes have, in addition, a cell population that carries the unchanged alleles prior to rearrangement (fig. 2). On the basis of these studies, it was suggested that mitotic contractions of D4Z4 likely occur within the first few cell divisions after fertilization. Further analysis of polymorphisms within, proximal to, and distal to D4Z4 showed that the preferred partner for these gene conversions is the sister chromatid rather than the homologous chromosome (Lemmers et al. 2004c). This finding was rather unexpected, since, in a previous study, it was shown that mosaic individuals have an increased frequency of translocated chromosome 4–derived repeats on chromosome 10, suggesting that the presence of supernumerary identical—but not homologous—repeats adds to the mitotic instability of D4Z4 (van der Maarel et al. 2000). Similar findings were recently confirmed in another population (Wu et al. 2004). In line with this model, there seems to be a slight but significant increased pairing frequency, at interphase, of 4qter and 10qter domains in cultured lymphocytes from patients with FSHD (Stout et al. 1999).
Figure 2.
Mechanism of mitotic contractions of D4Z4 in FSHD. A, Generally, repeat rearrangements are induced by double-strand breaks, in this case within D4Z4 (triangles). Break repair through gene conversion with crossover (GC+CO) or without crossover (GC−CO) requires the pairing of the damaged DNA strand (acceptor indicated by unblackened triangles) with the homologous donor (blackened triangles). In FSHD, the sister chromatids are likely the preferred partners in this process. By unequal pairing, the repair of repetitive DNA by GC+CO or by GC−CO is associated with an expansion or contraction of the repeat. B, Schematic representation of somatic mosaicism encountered in FSHD. D4Z4 alleles are visualized in DNA digested by EcoRI (E) and hybridized by probe p13E-11. Chromosome 4 alleles (red) are distinguished from chromosome 10 alleles (blue) by their resistance to BlnI (B). Chromosome 4–derived EcoRI/BlnI fragments are 3 kb smaller because of a single internal BlnI site. GC−CO causes the contraction (or expansion) of a single allele, and the donor alleles remain unchanged. Mosaic (m) alleles are recognized by their reduced signal intensity (dashed lines). During GC+CO, the size of the donor and acceptor alleles are changed. When crossover occurs before the first zygotic division, this yields two cell populations with the newly formed donor and acceptor allele (GC+CO 1). When it occurs in zygotic divisions after the first division, a population of cells, in addition to the two newly formed alleles, will be present carrying the ancestral allele (GC+CO 2).
Interestingly, it was recently described that the subtelomeric domains of chromosomes 4 and 10 occupy distinct territories in the nucleus. Of all subtelomeres studied, 4qter displayed the highest preference for the nuclear periphery, whereas the 10q subtelomeres were much more localized to the interior of the nucleus (Masny et al. 2004). This finding was recently corroborated by another study (Tam et al. 2004), in which the preferential localization of 4q35 in the heterochromatic nuclear periphery was suggested to be related to gene expression profiles along chromosome 4, with the subtelomere of chromosome 4q being substantially poorer in transcriptional activity than 4p, which was located more interiorly. Apart from being the first structural (and perhaps first functional) difference between chromosomes 4 and 10, this observation may provide an explanation for the preferred partner for recombination given the increased frequency of 4-derived repeats on chromosome 10 in mosaic individuals. It is becoming increasingly evident that chromosomes occupy distinct territories in the mammalian nucleus, and there is some evidence that the spatial organization is largely retained after mitosis (Gerlich et al. 2003). Could it be possible that the presence of translocated chromosome 4–derived D4Z4 repeats on chromosome 10 has a subtle impact on integrity and faithful transmission during early embryogenesis? Clearly, the issue is still largely unsettled, but, as a first step, it would be interesting to investigate whether the presence of translocated repeats on either chromosome influences their nuclear localization and their relative positions, with respect to each other, in dividing and nondividing cells. Tam et al. (2004) demonstrated that the preference for the nuclear periphery is an intrinsic property of 4qter, since the presence of 4 Mb of 4qter sequences on a derivative chromosome X in a cell line with a 4;X translocation causes a more peripheral localization of this chromosome. Nevertheless, both studies suggested a region directly proximal to D4Z4 as responsible for the perinuclear localization (Masny et al. 2004; Tam et al. 2004). Moreover, it should be realized that, although mitotic rearrangements of D4Z4 are fairly common in the general population, in individuals they seem to be restricted to a single event, most likely during early embryogenesis. Therefore, it is tempting to speculate that specific characteristics of early cell division, such as the spatial nuclear separation of paternal and maternal haploid genomes (Mayer et al. 2000), are contributing to the mitotic D4Z4 instability.
Irrespective of the exact mechanism, partial deletions of the D4Z4 repeat array are not always confined to the repeat proper. In 1% of sporadic and familial patients, the partial D4Z4 deletion extends in the proximal direction and includes the probe region D4F104S1 (Lemmers et al. 1998, 2003). Proximal deletions can include up to 60 kb proximal to D4Z4, and the inverted D4Z4 repeat unit (D4S2463) 40 kb proximal to D4Z4 might play a role in the mutational mechanism. Typically, patients with these deletions present a classic FSHD phenotype. Therefore, with the standard DNA diagnosis of FSHD performed by use of probe p13E-11, the absence of an FSHD-sized chromosome 4–derived D4Z4 fragment does not necessarily exclude the involvement of this region in these cases of FSHD. With the recognition of proximal extended deletions, additional diagnostic protocols have been developed to address this issue, including the dosage test that quantifies specific chromosome 4– and chromosome 10–derived restriction fragments (van der Maarel et al. 1999), PFGE, and the use of a distal 4qA probe as an alternative to p13E-11 (Lemmers et al. 2003).
Epigenetic Consequences of D4Z4 Contractions
Because contractions of D4Z4 did not seem to disturb the structure of a specific disease gene, it was soon hypothesized that chromatin conformational changes could underlie the disease mechanism of FSHD. These conformational changes, in turn, would lead to inappropriate downregulation or upregulation of one or more genes within or in the close vicinity of the D4Z4 repeat.
Since C5 methylation of cytosine is the most common modification of mammalian DNA and is known to be involved in development, X-chromosome inactivation, imprinting, and gene silencing (Robertson and Wolffe 2000), efforts were undertaken to investigate the DNA methylation of D4Z4. In an initial survey focusing on several different methylation sites in the repeat, D4Z4 was found to be highly methylated in a limited set of normal and FSHD lymphoblasts, as well as in somatic tissues, including skeletal muscle (Tsien et al. 2001). In a more recent study that analyzed the DNA methylation in lymphoblast DNA from a much larger cohort of patients and controls by a different strategy, D4Z4 was shown to be significantly hypomethylated at the disease allele in patients with FSHD (van Overveld et al. 2003). While, in control individuals, D4Z4 methylation levels of ∼50% were detected at each CpG dinucleotide tested, these methylation levels were reduced by half in disease alleles of patients and nonpenetrant gene carriers. This observation was confirmed in a limited set of control and FSHD muscle DNA. Importantly, in a small set of patients with an FSHD phenotype but no contraction of the D4Z4 repeat (non–4q-linked FSHD), D4Z4 was also hypomethylated, suggesting that, in non–4q-linked FSHD, the disease mechanism also acts through D4Z4, albeit unrelated to the contraction. In contrast to 4q-linked patients in whom only the contracted allele was hypomethylated, non–4q-linked patients showed hypomethylated D4Z4 on both chromosome 4q ends. The different outcomes of both methylation studies may be explained by differences in sample size, the method employed, and the methylation-sensitive restriction enzymes used.
Interestingly, strong hypomethylation of D4Z4 was first reported in immunodeficiency–centromeric instability–facial anomalies syndrome (ICF syndrome [MIM 242860]), an autosomal recessive disorder characterized by immunodeficiency, facial anomalies, and subtle developmental delay (Kondo et al. 2000). ICF syndrome is a very rare disorder (∼35 patients, primarily European, have been described) that invariably presents with abnormalities of the juxtacentromeric heterochromatin of chromosomes 1 and 16 in mitogen-stimulated lymphocytes. These chromosomes have long juxtacentromeric heterochromatin regions of satellite DNA. ICF is caused by mutations in the catalytic domain of the DNMT3B gene that reduce, but do not abolish, its DNA methyltransferase activity (Xu et al. 1999). As a result, DNA of patients with ICF shows reduced methylation at specific repetitive DNA sequences, including the pericentromeric Sat2, Sat3, and NBL2 repeats at the aforementioned chromosomes (1 and 16) and D4Z4 repeats at the subtelomeres of chromosomes 4q and 10q (reviewed by Ehrlich [2003]). Although hypomethylation of D4Z4 is observed in 4q-linked and non–4q-linked FSHD, ICF does not present with a myopathic phenotype. It may be argued that the lack of muscular dystrophy in patients with ICF may be due to the severity of the disorder and, accordingly, to the relative young age at which these patients usually die, whereas FSHD typically starts in the 2nd decade of life. However, two observations argue against this explanation for the absence of myopathy in ICF: the occurrence of the infantile form of FSHD, starting in the 1st decade of life and the absence of any subclinical myopathic features in ICF, even in relatively old patients with unusual mild ICF syndrome due to high residual DNA methyltransferase activity of DNMT3B.
One obvious explanation for the lack of muscular dystrophy in patients with ICF may relate to the allelic variation of 4qter distal to D4Z4 and by the absence of 4qA alleles in patients with ICF syndrome. However, analysis of a limited number of patients also showed the presence of 4qA in some of these patients (authors' unpublished results).
In patients with ICF, the D4Z4 hypomethylation is much more pronounced than in patients with 4q-linked FSHD and, rather than hypomethylation being restricted to a single chromosome, the alleles on chromosomes 4 and 10 are equally hypomethylated. Although similar low levels of methylation were found at both chromosome 4q ends, and possibly at chromosome 10, in patients with non–4q-linked FSHD, these patients do not suffer from a defect in DNMT3B (authors' unpublished results).
The proposed heterochromatic nature of 4qter was further studied by means of chromatin immunoprecipitation assays. Analysis of the histone H4 acetylation levels for a region immediately adjacent to D4Z4 demonstrated that 4qter had properties of unexpressed euchromatin, rather than constitutative heterochromatin, in fibroblasts, lymphoblastoid cell lines, and mononuclear blood cells of controls and FSHD cases (Jiang et al. 2003). Moreover, on the basis of histone acetylation studies and semiquantitative expression analyses of genes on chromosome 4qter, there was no evidence of a spreading of heterochromatinization emanating from the D4Z4 repeat. In a follow-up study, other markers of heterochromatin were evaluated by cytogenetic and immuno-FISH analysis of healthy and FSHD myoblasts. This again suggested that 4qter did not resemble constitutive heterochromatin, in either patient or control cells. More specifically, the 4qter region did not seem to colocalize with 4′-6-diamidino-2-phenylindole (DAPI)–bright foci, or with regions enriched in heterochromatin protein-1α or histone H3 trimethylated at lysine 9. Finally, the replication timing of 4qter was found to be very close to that of unexpressed euchromatin, and there was no indication of a change in replication timing in FSHD cells (Yang et al. 2004).
Disease-Mechanism Models for FSHD
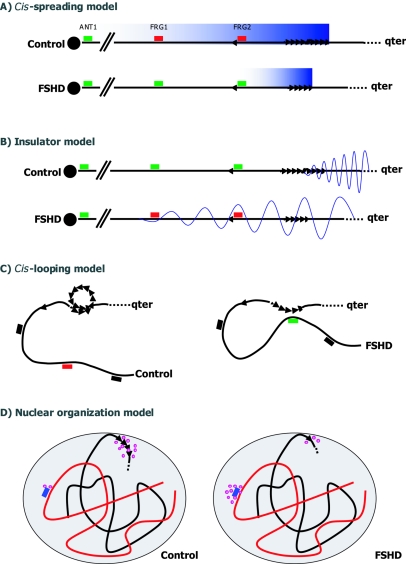
Over the past decade, several models have been put forward to explain the disease mechanism in FSHD. These are schematically represented in figure 3. Originally, when it became clear that the structure of the disease gene was not compromised and that D4Z4 shared properties of heterochromatic sequences, it was postulated that D4Z4 and surrounding sequences would be packed as heterochromatin and that partial loss of the D4Z4 repeat would lead to local chromatin relaxation (i.e., loss of heterochromatinization) and, consequently, to the transcriptional upregulation of genes near D4Z4, possibly in a distance-related manner (cis-spreading model in fig. 3A) (Hewitt et al. 1994; Winokur et al. 1994). Alternatively, it was proposed that D4Z4 acts as an insulator, separating heterochromatic telomeric sequences distal to D4Z4 from euchromatic sequences more upstream (van Deutekom 1996). Contracted arrays would not be able to completely separate both domains, and, consequently, heterochromatic spreading into proximal sequences would silence subtelomeric genes in cis (insulator model in fig. 3B). Although recent molecular data do not fully support either of these models, it is too early to completely refute these possibilities.
Figure 3.
Different models to explain the epigenetic disease mechanism of FSHD. The location of the most well-studied genes (FRG1, FRG2, and ANT1) are indicated. A, In the cis-spreading model, the long D4Z4 repeat (triangles) and nearby sequences share features of heterochromatin. Upon contraction, a local chromatin relaxation (blue gradiation) causes the transcriptional upregulation of 4qter genes (red boxes for downregulated and green boxes for upregulated genes) in a distance-dependent manner, possibly through the action of the D4Z4 repressor complex. B, In the insulator model, D4Z4 acts as a spacer between heterochromatic sequences distal to the D4Z4 repeat and proximal euchromatic sequences. Upon contraction, this insulator function is incomplete, allowing heterochromatinization (wave) of proximal sequencing and transcriptional downregulation of genes within this region. C, The cis-looping model postulates that normally intra-array loops in arrays >11 units prevent the interaction of D4Z4 with genes in cis at a large distance. Disruption of this interaction may cause inappropriate gene expression by long-range interaction with D4Z4. D, Finally, the nuclear organization model predicts that the interaction of 4qter with the nuclear lamina, in which chromatin and transcription factors (purple circles) are tethered, is perturbed in FSHD. This perturbation, in turn, may lead to a misbalance of chromatin and transcription factors at 4qter and unrelated loci.
Evidence for the cis-spreading model was provided by the identification of a repressor complex that binds to a specific sequence in D4Z4 (Gabellini et al. 2002). In muscle of patients with FSHD, a distance-dependent transcriptional upregulation was found for three 4qter genes (FRG2, FRG1, and ANT1). Moreover, the magnitude of upregulation of the gene closest to the D4Z4 repeat seemed to be inversely related to the residual size of the D4Z4 repeat in FSHD muscle. This repressor complex consists of YY1, a transcription factor that can act as transcriptional activator or repressor; the chromatin architectural protein HMGB2; and the RNA-binding protein nucleolin, which is involved in transcriptional control of ribosomal RNA and in ribogenesis. In cell culture, depletion of any of these components of the repressor complex was shown to upregulate FRG2, the gene closest to D4Z4. However, in several independent follow-up studies, the upregulation of these 4qter genes in FSHD muscle could not be confirmed by semiquantitative real-time PCR or array studies (Jiang et al. 2003; Winokur et al. 2003; R. J. L. F. Lemmers, T. Rijkers, R. R. Frants, and S. M. van der Maarel, unpublished results).
In the meantime, other models have been put forward to explain FSHD. First, contraction of D4Z4 may lead to inappropriate gene expression by a cis-looping model (fig. 3C) (Jiang et al. 2003). According to this model, long-distance loops between D4Z4 and its target gene(s) occur only when the formation of normal D4Z4 intra-array loops are impaired by chromatin constraints due to D4Z4 contraction. The observation that the size distribution of D4Z4 repeats is multimodal, with equidistant peaks 65 kb apart, may support a model in which normal-sized D4Z4 repeats form intra-array loops (van Overveld et al. 2000). Moreover, somewhat similar observations have already been reported for the regulation of the β-globin gene cluster by its locus control region and by the observation of loop-like chromatin-chromatin interactions of several megabases by FISH analysis (Volpi et al. 2000).
The most recent model is one of disturbed nuclear localization (nuclear organizing model in fig. 3D) and is not necessarily mutually exclusive of the cis-looping model (Masny et al. 2004). Expression analysis of 4qter genes remains controversial, and no consistent transcriptional upregulation of 4qter genes has been identified by array and semiquantitative RT-PCR studies to support a cis-spreading effect (Jiang et al. 2003; Winokur et al. 2003; R. J. L. F. Lemmers, T. Rijkers, R. R. Frants, and S. M. van der Maarel, unpublished results). Therefore, since the mammalian nucleus is highly compartmentalized, with individual chromosomes occupying distinct territories most likely reflecting their gene density, transcriptional activity, replication timing, and chromosome size (Sun et al. 2000; Tanabe et al. 2002), the organization of 4qter in the nucleus was studied. As described above, 4qter largely occupies a peripheral territory in the nucleus, independent of cell type and chromosome-territory effects. Importantly, this localization seems to be dependent on the integrity of the nuclear lamina, since this peripheral localization of 4qter is lost in cells deficient of lamin A/C (Masny et al. 2004). Interestingly, several neuromuscular disorders, the so-called laminopathies, arise from deficiencies in nuclear lamina proteins, including lamin A/C and emerin (reviewed by Maraldi et al. [2004]). By contrast, 10qter is more localized to the interior of the nucleus, suggesting that these chromosome ends are functionally different. Moreover, it is not D4Z4 itself but rather sequences proximal to D4Z4—which are not present on chromosome 10—that mediate the interaction with the nuclear lamina (Masny et al. 2004). This may explain the chromosome 4 specificity of FSHD. In this study (Masny et al. 2004), no disturbed localization was observed for the disease allele in myoblasts of patients with FSHD. Since no difference was observed in the localization of healthy and disease chromosomes, it was postulated that the interaction between 4qter and the nuclear envelope is different because of an altered recruitment of chromatin and transcription factors at the nuclear lamina. Independently, Tam et al. (2004) reported largely similar observations. They demonstrated that intrinsic properties of 4qter are necessary and sufficient to localize this subtelomeric domain in the nuclear periphery and suggested a role for D4Z4 in regulating the local heterochromatic state (Tam et al. 2004). According to this study, D4Z4 may operate as a silencer or insulator separating distal heterochromatin from proximal genes.
Conclusions
During the past 5 years, our understanding of FSHD has accelerated enormously. Regardless of the exact molecular mechanism, the unifying theme that emerges is a D4Z4 contraction–mediated chromatin conformational change at 4qter as the primary pathogenic mechanism. In turn, this conformational change may cause the inappropriate expression of one or more disease genes, not necessarily in cis with the contracted D4Z4 repeat. Several genes have been proposed on the basis of their localization, evolutionary conservation, and function, including FRG1, FRG2, ANT1, and ALP, but conclusive evidence for their involvement is not currently available. The development of cellular and animal models for this disease will undoubtedly prove to be of critical importance in further understanding this enigmatic disease.
Acknowledgments
We apologize to those whose work we could not cite because of space limitations, and we thank Richard Lemmers for critically reading the manuscript. Our FSHD research is supported by grants from the Netherlands Organisation for Scientific Research (NWO), the Prinses Beatrix Fonds, the Stichting Spieren voor Spieren, the Muscular Dystrophy Association USA, the FSH Society, the Stichting FSHD, the Shaw family, and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (National Institutes of Health).
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FSHD and ICF)
References
- Agresti A, Meneveri R, Siccardi AG, Marozzi A, Corneo G, Gaudi S, Ginelli E (1989) Linkage in human heterochromatin between highly divergent Sau3A repeats and a new family of repeated DNA sequences (HaeIII family). J Mol Biol 205:625–631 [DOI] [PubMed] [Google Scholar]
- Bakker E, van der Wielen MJ, Voorhoeve E, Ippel PF, Padberg GW, Frants RR, Wijmenga C (1996) Diagnostic, predictive, and prenatal testing for facioscapulohumeral muscular dystrophy: diagnostic approach for sporadic and familial cases. J Med Genet 33:29–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker E, Wijmenga C, Vossen RH, Padberg GW, Hewitt J, van der Wielen M, Rasmussen K, Frants RR (1995) The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve 2:S39–S44 [PubMed] [Google Scholar]
- Ballarati L, Piccini I, Carbone L, Archidiacono N, Rollier A, Marozzi A, Meneveri R, Ginelli E (2002) Human genome dispersal and evolution of 4q35 duplications and interspersed LSau repeats. Gene 296:21–27 [DOI] [PubMed] [Google Scholar]
- Coppee F, Matteotti C, Ansseau E, Sauvage S, Leclercq I, Leroy A, Marcowycz A, Gerbaux C, Figlewicz D, Ding H, Belayew A (2004) The DUX gene family and FSHD. In: Upadhyaya M, Cooper DN (eds) Facioscapulohumeral muscular dystrophy: clinical medicine and molecular cell biology. Garland Science, Oxon, pp 117–134 [Google Scholar]
- Deidda G, Cacurri S, Grisanti P, Vigneti E, Piazzo N, Felicetti L (1995) Physical mapping evidence for a duplicated region on chromosome 10qter showing high homology with the facioscapulohumeral muscular dystrophy locus on chromosome 4qter. Eur J Hum Genet 3:155–167 [DOI] [PubMed] [Google Scholar]
- Deidda G, Cacurri S, Piazzo N, Felicetti L (1996) Direct detection of 4q35 rearrangements implicated in facioscapulohumeral muscular dystrophy (FSHD). J Med Genet 33:361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M (2003) The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin Immunol 109:17–28 [DOI] [PubMed] [Google Scholar]
- Flint J, Bates GP, Clark K, Dorman A, Willingham D, Roe BA, Micklem G, Higgs DR, Louis EJ (1997) Sequence comparison of human and yeast telomeres identifies structurally distinct subtelomeric domains. Hum Mol Genet 6:1305–1314 [DOI] [PubMed] [Google Scholar]
- Funakoshi M, Goto K, Arahata K (1998) Epilepsy and mental retardation in a subset of early onset 4q35-facioscapulohumeral muscular dystrophy. Neurology 50:1791–1794 [DOI] [PubMed] [Google Scholar]
- Gabellini D, Green M, Tupler R (2002) Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110:339–348 [DOI] [PubMed] [Google Scholar]
- Gabriels J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, Belayew A (1999) Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene 236:25–32 [DOI] [PubMed] [Google Scholar]
- Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, Ellenberg J (2003) Global chromosome positions are transmitted through mitosis in mammalian cells. Cell 112:751–764 [DOI] [PubMed] [Google Scholar]
- Gilbert JR, Stajich JM, Speer MC, Vance JM, Stewart CS, Yamaoka LH, Samson F, Fardeau M, Potter TG, Roses AD (1992) Linkage studies in facioscapulohumeral muscular dystrophy (FSHD). Am J Hum Genet 51:424–427 [PMC free article] [PubMed] [Google Scholar]
- Gilbert JR, Stajich JM, Wall S, Carter SC, Qiu H, Vance JM, Stewart CS, Speer MC, Pufky J, Yamaoka LH (1993) Evidence for heterogeneity in facioscapulohumeral muscular dystrophy (FSHD). Am J Hum Genet 53:401–408 [PMC free article] [PubMed] [Google Scholar]
- Goto K, Nishino I, Hayashi YK (2004) Very low penetrance in 85 Japanese families with facioscapulohumeral muscular dystrophy 1A. J Med Genet 41:e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs RC, Tawil R, Storvick D, Mendell JR, Altherr MR (1993) Genetics of facioscapulohumeral muscular dystrophy: new mutations in sporadic cases. Neurology 43:2369–2372 [DOI] [PubMed] [Google Scholar]
- Hewitt JE, Lyle R, Clark LN, Valleley EM, Wright TJ, Wijmenga C, van Deutekom JC, Francis F, Sharpe PT, Hofker M, Frants RR, Williamson R (1994) Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Mol Genet 3:1287–1295 [DOI] [PubMed] [Google Scholar]
- Jiang G, Yang F, van Overveld PG, Vedanarayanan V, van der Maarel S, Ehrlich M (2003) Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum Mol Genet 12:2909–2921 [DOI] [PubMed] [Google Scholar]
- Kohler J, Rupilius B, Otto M, Bathke K, Koch MC (1996) Germline mosaicism in 4q35 facioscapulohumeral muscular dystrophy (FSHD1A) occurring predominantly in oogenesis. Hum Genet 98:485–490 [DOI] [PubMed] [Google Scholar]
- Kondo T, Bobek MP, Kuick R, Lamb B, Zhu X, Narayan A, Bourc’his D, Viegas-Pequignot E, Ehrlich M, Hanash SM (2000) Whole-genome methylation scan in ICF syndrome: hypomethylation of non-satellite DNA repeats D4Z4 and NBL2. Hum Mol Genet 9:597–604 [DOI] [PubMed] [Google Scholar]
- Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC (1997) Human centromeric DNAs. Hum Genet 100:291–304 [DOI] [PubMed] [Google Scholar]
- Lee JH, Goto K, Matsuda C, Arahata K (1995) Characterization of a tandemly repeated 3.3-kb KpnI unit in the facioscapulohumeral muscular dystrophy (FSHD) gene region on chromosome 4q35. Muscle Nerve 2:S6–S13 [PubMed] [Google Scholar]
- Lemmers RJLF, de Kievit P, Sandkuijl L, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM (2002) Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet 32:235–236 [DOI] [PubMed] [Google Scholar]
- Lemmers RJLF, de Kievit P, van Geel M, van der Wielen MJ, Bakker E, Padberg GW, Frants RR, van der Maarel SM (2001) Complete allele information in the diagnosis of facioscapulohumeral muscular dystrophy by triple DNA analysis. Ann Neurol 50:816–819 [DOI] [PubMed] [Google Scholar]
- Lemmers RJLF, Osborn M, Haaf T, Rogers M, Frants RR, Padberg GW, Cooper DN, van der Maarel SM, Upadhyaya M (2003) D4F104S1 deletion in facioscapulohumeral muscular dystrophy: phenotype, size, and detection. Neurology 61:178–183 [DOI] [PubMed] [Google Scholar]
- Lemmers RJLF, van der Maarel SM, van Deutekom JCT, van der Wielen MJR, Deidda G, Dauwerse HG, Hewitt J, Hofker M, Bakker E, Padberg GW, Frants RR (1998) Inter- and intrachromosomal subtelomeric rearrangements on 4q35: implications for facioscapulohumeral muscular dystrophy (FSHD) aetiology and diagnosis. Hum Mol Genet 7:1207–1214 [DOI] [PubMed] [Google Scholar]
- Lemmers RJLF, van der Wielen MJR, Bakker E, Padberg GW, Frants RR, van der Maarel SM (2004a) Somatic mosaicism in FSHD often goes undetected. Ann Neurol 55:845–850 [DOI] [PubMed] [Google Scholar]
- Lemmers RJLF, van der Wielen MJR, Bakker E, van der Maarel SM (2004b) Molecular diagnosis of FSHD. In: Upadhyaya M, Cooper DN (eds) Facioscapulohumeral muscular dystrophy: clinical medicine and molecular cell biology. Garland Science, Oxon, pp 211–234 [Google Scholar]
- Lemmers RJLF, van Overveld PGM, Sandkuijl LA, Vrieling H, Padberg GW, Frants RR, van der Maarel SM (2004c) Mechanism and timing of mitotic rearrangements in the subtelomeric D4Z4 repeat involved in facioscapulohumeral muscular dystrophy. Am J Hum Genet 75:44–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJLF, Wohlgemuth M, Frants RR, Padberg GW, Morava E, van der Maarel SM (2004d) Contractions of D4Z4 on 4qB subtelomeres do not cause facioscapulohumeral muscular dystrophy. Am J Hum Genet 75:1124–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt PW, Jardine PE, Koch MC, Maynard J, Osborn M, Williams M, Harper PS, Upadhyaya M (1995) Correlation between fragment size at D4F104S1 and age at onset or at wheelchair use, with a possible generational effect, accounts for much phenotypic variation in 4q35-facioscapulohumeral muscular dystrophy (FSHD). Hum Mol Genet 4:951–958 [DOI] [PubMed] [Google Scholar]
- Lyle R, Wright TJ, Clark LN, Hewitt JE (1995) The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics 28:389–397 [DOI] [PubMed] [Google Scholar]
- Maraldi NM, Squarzoni S, Sabatelli P, Capanni C, Mattioli E, Ognibene A, Lattanzi G (2004) Laminopathies: involvement of structural nuclear proteins in the pathogenesis of an increasing number of human diseases. J Cell Physiol (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Masny PS, Bengtsson U, Chung SA, Martin JH, van Engelen B, van der Maarel SM, Winokur ST (2004) Localization of 4q35.2 to the nuclear periphery: is FSHD a nuclear envelope disease? Hum Mol Genet 13:1857–1871 [DOI] [PubMed] [Google Scholar]
- Matsumura T, Goto K, Yamanaka G, Lee J, Zhang C, Hayashi YK, Arahata K (2002) Chromosome 4q;10q translocations: comparison with different ethnic populations and FSHD patients. BMC Neurol 2:7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T (2000) Embryogenesis—demethylation of the zygotic paternal genome. Nature 403:501–502 [DOI] [PubMed] [Google Scholar]
- Mefford HC, Trask BJ (2002) The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet 3:91–102 [DOI] [PubMed] [Google Scholar]
- Miura K, Kumagai T, Matsumoto A, Iriyama E, Watanabe K, Goto K, Arahata K (1998) Two cases of chromosome 4q35-linked early onset facioscapulohumeral muscular dystrophy with mental retardation and epilepsy. Neuropediatrics 29:239–241 [DOI] [PubMed] [Google Scholar]
- Padberg GW (1982) Facioscapulohumeral disease. PhD thesis, Leiden University, Leiden [Google Scholar]
- ——— (2004) Facioscapulohumeral muscular dystrophy: a clinician’s experience. In: Upadhyaya M, Cooper DN (eds) Facioscapulohumeral muscular dystrophy: clinical medicine and molecular cell biology. Garland Science, Oxon, pp 41–54 [Google Scholar]
- Padberg GW, Lunt PW, Koch M, Fardeau M (1991) Diagnostic criteria for facioscapulohumeral muscular dystrophy. Neuromuscul Disord 1:231–234 [DOI] [PubMed] [Google Scholar]
- Robertson KD, Wolffe AP (2000) DNA methylation in health and disease. Nat Rev Genet 1:11–19 [DOI] [PubMed] [Google Scholar]
- Stout K, van der Maarel S, Frants RR, Padberg GW, Ropers H-H, Haaf T (1999) Somatic pairing between subtelomeric regions: implications for human genetic disease? Chrom Res 7:323–329 [DOI] [PubMed] [Google Scholar]
- Sun HB, Shen J, Yokota H (2000) Size-dependent positioning of human chromosomes in interphase nuclei. Biophys J 79:184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam R, Smith KP, Lawrence JB (2004) The 4q subtelomere harboring the FSHD locus is specifically anchored with peripheral heterochromatin unlike most human telomeres. J Cell Biol 167:269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe H, Habermann FA, Solovei I, Cremer M, Cremer T (2002) Non-random radial arrangements of interphase chromosome territories: evolutionary considerations and functional implications. Mutat Res 504:37–45 [DOI] [PubMed] [Google Scholar]
- Tawil R, Forrester J, Griggs RC, Mendell J, Kissel J, McDermott M, King W, Weiffenbach B, Figlewicz D (1996) Evidence for anticipation and association of deletion size with severity in facioscapulohumeral muscular dystrophy. The FSH-DY Group. Ann Neurol 39:744–748 [DOI] [PubMed] [Google Scholar]
- Tim RW, Gilbert JR, Stajich JM, Rampersaud E, Viles KD, Tawil R, Padberg GW, Frants R, van der Maarel S, Bossen EH, Friedman AH, Pericak-Vance MA, Speer MC (2001) Clinical studies in non-chromosome 4-linked facioscapulohumeral muscular dystrophy. J Clin Neuromusc Dis 3:1–7 [DOI] [PubMed] [Google Scholar]
- Tsien F, Sun B, Hopkins NE, Vedanarayanan V, Figlewicz D, Winokur S, Ehrlich M (2001) Methylation of the FSHD syndrome-linked subtelomeric repeat in normal and FSHD cell cultures and tissues. Mol Genet Metab 74:322–331 [DOI] [PubMed] [Google Scholar]
- Tupler R, Berardinelli A, Barbierato L, Frants R, Hewitt JE, Lanzi G, Maraschio P, Tiepolo L (1996) Monosomy of distal 4q does not cause facioscapulohumeral muscular dystrophy. J Med Genet 33:366–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya M, Maynard J, Osborn M, Jardine P, Harper PS, Lunt P (1995) Germinal mosaicism in facioscapulohumeral muscular dystrophy (FSHD). Muscle Nerve 2:S45–S49 [PubMed] [Google Scholar]
- van der Maarel SM, Deidda G, Lemmers RJ, Bakker E, van der Wielen MJ, Sandkuijl L, Hewitt JE, Padberg GW, Frants RR (1999) A new dosage test for subtelomeric 4;10 translocations improves conventional diagnosis of facioscapulohumeral muscular dystrophy (FSHD). J Med Genet 36:823–828 [PMC free article] [PubMed] [Google Scholar]
- van der Maarel SM, Deidda G, Lemmers RJLF, van Overveld PGM, van der Wielen M, Hewitt JE, Sandkuijl L, Bakker B, van Ommen GJB, Padberg GW, Frants RR (2000) De novo facioscapulohumeral muscular dystrophy: frequent somatic mosaicism, sex-dependent phenotype, and the role of mitotic transchromosomal repeat interaction between chromosomes 4 and 10. Am J Hum Genet 66:26–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Bakker E, Lemmers RJ, van der Wielen MJ, Bik E, Hofker MH, Padberg GW, Frants RR (1996) Evidence for subtelomeric exchange of 3.3 kb tandemly repeated units between chromosomes 4q35 and 10q26: implications for genetic counselling and etiology of FSHD1. Hum Mol Genet 5:1997–2003 [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR (1993) FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet 2:2037–2042 [DOI] [PubMed] [Google Scholar]
- van Deutekom JCT (1996) Towards the molecular mechanism of facioscapulohumeral muscular dystrophy. PhD thesis, Leiden University, Leiden [Google Scholar]
- van Geel M, Dickson MC, Beck AF, Bolland DJ, Frants RR, van der Maarel SM, de Jong PJ, Hewitt JE (2002) Genomic analysis of human chromosome 10q and 4q telomeres suggests a common origin. Genomics 79:210–217 [DOI] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Deidda G, Sandkuijl L, Padberg GW, Frants RR, van der Maarel SM (2000) Interchromosomal repeat array interactions between chromosomes 4 and 10: a model for subtelomeric plasticity. Hum Mol Genet 9:2879–2884 [DOI] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM (2003) Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet 35:315–317 [DOI] [PubMed] [Google Scholar]
- Vogt P (1990) Potential genetic functions of tandem repeated DNA sequence blocks in the human genome are based on a highly conserved “chromatin folding code.” Hum Genet 84:301–336 [DOI] [PubMed] [Google Scholar]
- Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell RD, Goldsworthy M, Powis SH, Ragoussis J, Trowsdale J, Sheer D (2000) Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci 113:1565–1576 [DOI] [PubMed] [Google Scholar]
- Weiffenbach B, Dubois J, Storvick D, Tawil R, Jacobsen SJ, Gilbert J, Wijmenga C, Mendell JR, Winokur S, Altherr MR (1993) Mapping the facioscapulohumeral muscular dystrophy gene is complicated by chromsome 4q35 recombination events. Nat Genet 4:165–169 [DOI] [PubMed] [Google Scholar]
- Wijmenga C, Frants RR, Brouwer OF, Moerer P, Weber JL, Padberg GW (1990) Location of facioscapulohumeral muscular dystrophy gene on chromosome 4. Lancet 336:651–653 [DOI] [PubMed] [Google Scholar]
- Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R, van Ommen GJ, Padberg GW, Frants RR (1992) Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet 2:26–30 [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Higgs DR, Rack KA, Buckle VJ, Spurr NK, Fischel-Ghodsian N, Ceccherini I, Brown WR, Harris PC (1991) Stable length polymorphism of up to 260 kb at the tip of the short arm of human chromosome 16. Cell 64:595–606 [DOI] [PubMed] [Google Scholar]
- Winokur ST, Bengtsson U, Feddersen J, Mathews KD, Weiffenbach B, Bailey H, Markovich RP, Murray JC, Wasmuth JJ, Altherr MR (1994) The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Res 2:225–234 [DOI] [PubMed] [Google Scholar]
- Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, Tapscott SJ, van der Maarel SM, Hayashi Y, Flanigan KM (2003) Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum Mol Genet 12:2895–2907 [DOI] [PubMed] [Google Scholar]
- Wu ZY, Wang ZQ, Murong SX, Wang N (2004) FSHD in Chinese population: characteristics of translocation and genotype-phenotype correlation. Neurology 63:581–583 [DOI] [PubMed] [Google Scholar]
- Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Pequignot E (1999) Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402:187–191 [DOI] [PubMed] [Google Scholar]
- Yang F, Shao CB, Vedanarayanan V, Ehrlich M (2004) Cytogenetic and immuno-FISH analysis of the 4q subtelomeric region, which is associated with facioscapulohumeral muscular dystrophy. Chromosoma 112:350–359 [DOI] [PubMed] [Google Scholar]
- Zatz M, Marie SK, Cerqueira A, Vainzof M, Pavanello RC, Passos-Bueno MR (1998) The facioscapulohumeral muscular dystrophy (FSHD1) gene affects males more severely and more frequently than females. Am J Med Genet 77:155–161 [PubMed] [Google Scholar]
- Zatz M, Marie SK, Passos Bueno MR, Vainzof M, Campiotto S, Cerqueira A, Wijmenga C, Padberg G, Frants R (1995) High proportion of new mutations and possible anticipation in Brazilian facioscapulohumeral muscular dystrophy. Am J Hum Genet 56:99–105 [PMC free article] [PubMed] [Google Scholar]