Abstract
The genetic diversity of Cylindrospermopsis strains (cyanobacteria) was examined using mainly the 16S-23S internally transcribed spacer (ITS1) sequences. Strains were grouped in three clusters: (i) America, (ii) Europe, and (iii) Africa and Australia. These results suggested a recent spread of Cylindrospermopsis across the American and European continents from restricted warm refuge areas instead of exchanges between continents. On the other hand, they also suggested a recent colonization of Australia by African strains.
Cylindrospermopsis Seenayya et Subba Raju 1972 (25) is a filamentous, heterocystous freshwater cyanobacterium genus belonging to the Nostocales order. The type species Cylindrospermopsis raciborskii was first identified in a sample collected in Java island (27) and further described as a typical tropical cyanobacterium (23). The geographical distribution of this toxic cyanobacterium has now extended from tropical to temperate areas (5-8, 17, 23). Past studies on phytoplankton diversity have indicated that C. raciborskii appeared in Europe during the 1930s. In addition, recent reviews (16, 23) clearly have shown a progressive colonization by C. raciborskii from Greece and Hungary towards Northern latitudes near the end of the 20th century. Two possible explanations of the spread of C. raciborskii towards temperate areas have been proposed by Padisák (23): either a primary radiation from Africa or the dispersal of adapted strains from Australia to the Eurasian and eventually American continents.
To test the hypotheses on the origin of European C. raciborskii strains and more globally on the phylogeography of this species, 16 Cylindrospermopsis strains and two Raphidiopsis strains isolated from freshwater lakes and reservoirs in Africa, America, Australia, and Europe were characterized genetically. The genotypic characterization was based on sequencing of the 16S-23S internally transcribed spacer (ITS1) of the ribosomal operon and completed for some of them by the sequencing of fragments of the 16S rRNA, rpoC1, and nifH genes.
The 18 strains used in this study are listed in Table 1. Based on morphological criteria, two of them were identified as Raphidiopsis sp. Fritsch et Rich 1929 (12). In contrast with the 16 other strains, these two strains lack the ability to differentiate heterocysts when grown in a nitrogen-free medium and to fix nitrogen, since their cultures in this condition turned from green to yellow within a week. However, the partial 16S rRNA gene sequences (348 bp) of the two Raphidiopsis strains, using the cyano-specific primers (21), were identical to those of seven Cylindrospermopsis strains of this study (Table 1) and of 25 Cylindrospermopsis strains available in the GenBank database (data not shown). The clonal isolates were grown in Z8 or Z8X (without nitrogen) medium at 25°C in Erlenmeyer flasks, illuminated at 25 μmol of photons · m−2 · s−1 with a 16:8-h light-dark photoperiod. For the genetic analysis, 2 ml of culture in the early growth phase was centrifuged, washed once in sterile water, and frozen in liquid nitrogen. The ITS1 was amplified using the primers 322 and 340 (15), whereas the rpoC1 and nifH genes were amplified using the primers rpoC1f (5′-ACCATTAACTACCGCACCCT-3′) and rpoC1r (5′-TTGTCAATTACCCGCAGACG-3′) and nifHf (5′-CGTAGGTTGCGACCCTAAGGCTGA-3′) and nifHr (5′-GCATACATCGCCATCATTTCACC-3′). All amplifications were performed in a volume of 50 μl containing 2 μl of clonal culture, 200 μM deoxynucleoside triphosphate, 20 μM (each) primer, and 5 μl of 10× PCR buffer, using the same PCR conditions as described by Gugger et al. (13). The ITS PCR products were cloned prior to sequencing, while the rpoC1 and nifH PCR products were directly sequenced using the same primers as for amplification. Sequencing was performed using the Applied Biosystems 373 automated DNA sequencer (Perkin-Elmer, Foster City, Calif.) according to the manufacturer's instructions. The sequences, obtained independently for both strands, were aligned using the PILEUP module of the GCG package (Genetics Computer Group, Inc., Madison, Wis.) and edited manually with GeneDoc (20). Phylogenetic trees were constructed by using the neighbor-joining method on Jukes-Cantor pairwise distances by maximum parsimony and by maximum-likelihood analyses, using the PHYLIP software package (11).
TABLE 1.
Cyanobacterial strains used in this study
| Species | Straina | Geographic origin | Locus sequencedb
|
||||
|---|---|---|---|---|---|---|---|
| 16S | ITS1-S | ITS1-L | rpoCl | nifH | |||
| Cylindrospermopsis raciborskii | PMC98.14 | Viry-ChÂtillon, France | + | + | + | + | + |
| PMC99.12 | Chanteraînes, France | + | + | + | + | + | |
| PMC114.02 | Courneuve, France | + | + | ||||
| ACT-9502 | Balaton, Hungary | + | + | + | |||
| CYLI 53 | Lake Melangsee, Germany | + | + | ||||
| PMC99.06 | Epazote, Mexico | + | + | ||||
| PMC99.08 | Jabali, Mexico | + | + | ||||
| PMC00.01 | Jucazinho, Brazil | + | + | + | |||
| ITEP-A3 | Riacho do Pau, Brazil | + | + | ||||
| ITEP-018 | Tabocas reservoir, Brazil | + | + | ||||
| CYP-030B | Bourke, NSW,c Australia | + | + | + | |||
| CYP-023 | Bourke, NSW, Australia | + | |||||
| CYP-026J | Bourke, NSW, Australia | + | + | + | |||
| PMC118.02 | Guiers, Senegal | + | + | + | |||
| PMC117.02 | Guiers, Senegal | + | + | + | |||
| Cylindrospermopsis africana Raphidiopsis sp. | PMC115.02 | Guiers, Senegal | + | + | + | ||
| ITEP-005 | Tapacurá, Brazil | + | + | + | + | NO | |
| ITEP-007 | Ingazeira, Brazil | + | + | + | |||
Culture collections: PMC, Paris Museum Collection, C. Bernard of Museum National d'Histoire Naturelle, Paris, France; CYLI, J. Fastner of Technical University Berlin, Berlin, Germany; ACT, A. Kovacs of Institute of the Hungarian Academy of Sciences, Tihany, Hungary; ITEP, R. Molica of Instituto Tecnologico do Estado de Pernambuco, Recife, Brazil; CYP, P. Baker of Australian Water Quality Center, Salisbury, Australia.
+, sequence obtained; NO, sequence not obtained for this isolate.
NSW, New South Wales.
ITS1 rRNA analysis.
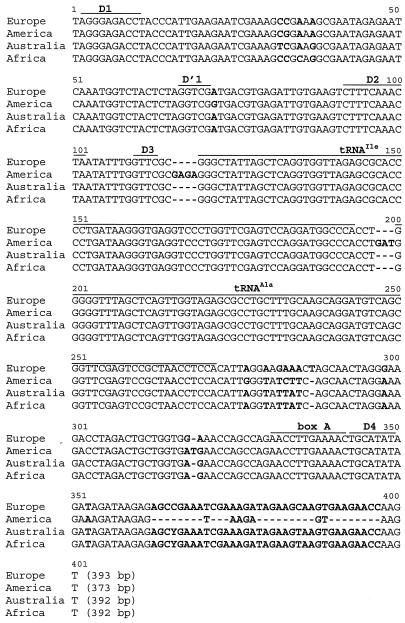
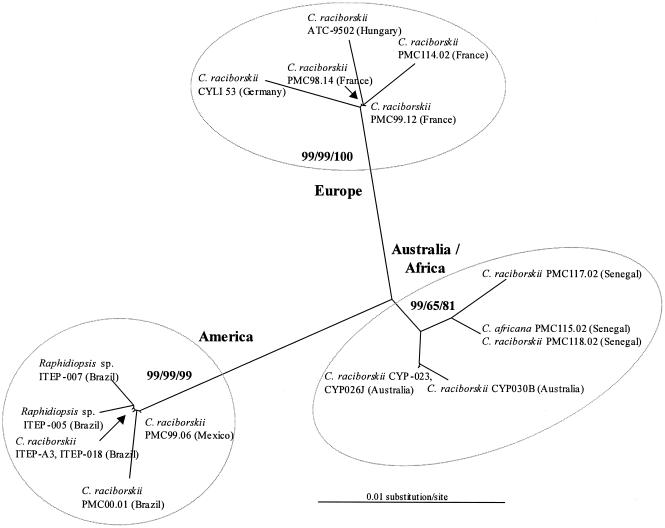
Two amplified products, ranging from 408 to 435 bp and from 569 to 593 bp, respectively, corresponding to short and long ITS1 with flanking regions, were obtained from each strain. The short (ITS1-S) and long (ITS1-L) ITS1 products were sequenced. The ITS1-L sequences contained two tRNA genes and were more informative than the ITS1-S sequences, because the flanking regions of each tRNA gene were polymorphic (Fig. 1). The ITS1-L sequences of the Cylindrospermopsis and Raphidiopsis American strains were about 20 bp shorter than the ones from other continents (Fig. 1). A very high sequence similarity (>99%) was revealed within European strains, American strains, and African and Australian strains, whereas less than 91% similarity was found between these three groups of strains. The phylogenetic analysis was consistent with these findings and identified three clusters highly supported by the bootstrap analysis, whatever the phylogenetic algorithm used (Fig. 2).
FIG. 1.
Alignment of the consensus sequences obtained for Cylindrospermopsis and Raphidiopsis ITS1-L from each continent. Europe: ITS-L from six Cylindrospermopsis strains; America: ITS-L from six Cylindrospermopsis/Raphidiopsis strains; Australia: ITS-L from three Cylindrospermopsis strains; Africa: ITS-L from three Cylindrospermopsis strains.
FIG. 2.
Maximum-likelihood unrooted tree based on Cylindrospermopsis and Raphidiopsis ITS1-L sequences. Neighbor-joining, maximum-parsimony, and maximum-likelihood bootstrap values >70%, respectively, are given at the nodes (500 bootstrap resampling).
rpoC1 and nifH gene analysis.
In order to compare some of our strains to Cylindrospermopsis sequences available in the GenBank database, a 380-bp fragment rpoC1 gene was obtained from 10 strains originating from the four continents (Table 1). The rpoC1 gene did not yield much information due to the low level of polymorphism (36 out of 380 positions). There was a high homology between the European and the African and Australian Cylindrospermopsis sequences (98 to 100% identity) and between the sequences of the Raphidiopsis strain from Brazil and the Cylindrospermopsis strain from Mexico (96% identity). The phylogenetic analysis (data not shown) allowed only the differentiation of the American strains from all the other strains. A 297-bp fragment of the nifH gene from three African, three French, and two Mexican Cylindrospermopsis strains (Table 1) was sequenced for comparison with 19 sequences of this locus available in the GenBank database (10). No amplification of the nifH gene was obtained for the Raphidiopsis strains, whereas amplicons were retrieved with these primers for other nostocalean strains, such as Anabeana and Aphanizomenon strains (data not shown). In Cylindrospermopsis strains, this locus was highly conserved (>98% similarity). The nifH sequences of the 10 European strains were identical, and the 3 African sequences were similar to those of the 6 Australian strains. Finally, the eight American sequences were divergent by only five nucleotide substitutions. Four sites allowed the distinction of three groups of strains on the basis of the three continental areas: Africa and Australia, America, and Europe.
The ITS1 and rpoC1 loci showed that Cylindrospermopsis and Raphidiopsis strains from the same continent were more closely related to each other than the Cylindrospermopsis strains originating from different continents. Thus, it would be very interesting to perform a more complete study using a polyphasic approach in order to investigate the question of the taxonomic validity of these two genera.
Previous studies based on 16S rRNA gene, cpcBA-IGS, and nifH sequences (10, 19) had revealed a separation of the Australian, European, and American strains. But these loci contained too few polymorphic sites to distinguish clusters supported by the bootstrap analyses. Therefore, the ITS1 fragment is, so far, the only genetic marker providing robust phylogenetic inferences between Cylindrospermopsis strains. This marker was previously used to study subgeneric phylogenetic relationships in Microcystis (14, 22), Arthrospira (1, 24), and Microcoleus (2).
Our findings with ITS1 suggest that the recent invasion of Europe and of Central and North America by Cylindrospermopsis did not result from recent colonization events by African or Australian isolates as proposed by Padisák (23). In view of the history of the earth's climatic changes and of biogeographic evidence for numerous plant and animal models (e.g., see references 18 and 26), we can propose the following hypothesis. First, the multiple glaciations or dry climatic conditions, during the Pleistocene age, for example, could have led to the extinction of Cylindrospermopsis in most of its geographical distribution areas and allowed it to survive only in warm refuge areas on each continent. More recently, the elevation of temperatures has allowed the colonization of more and more septentrional areas from these warm refuge areas on the European and American continents. This hypothesis is in agreement with our ecophysiological study with the same strains, showing that the light and temperature optima for the growth of the European isolates are the same as those for tropical ones (3, 4) and, therefore, that these European strains could have originated in a warm area located on the Eurasian continent.
To test this alternative hypothesis on the spread of C. raciborskii from warm refuge areas, it would be very interesting to evaluate the phylogenetic relationship between strains from Europe and strains from such potential warm refuge areas on the Eurasian continent. For example, the Indonesian Peninsula could be examined, because in this region the climate has been consistently tropical over the past 100 million years. But other possibilities also exist, the investigation of which would require extensive sampling on the whole Eurasian continent.
Finally, concerning the close relationship between African and Australian strains, more ITS1 sequences are needed from strains isolated on each of these continents in order to confirm a recent colonization of Australia by strains originating from Africa. Since Australia has been subjected to periods of cooling during the earth's history, this could have led to the local extinction of the species in this isolated continent and to the need of a recent colonization, in relation with human activities, for example.
In conclusion, our molecular data suggest an alternative hypothesis to that based on phenotypic and ecological data to explain the actual distribution of C. raciborskii in the world. The use of the ITS1 molecular marker on an extensive set of strains will probably provide an answer to this question. Whatever the origin of the European strains is, the spread of Cylindrospermopsis on the European continent is currently advancing rapidly, as demonstrated by the increasing occurrence of this species in several water bodies in France (9).
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited in the GenBank-EMBL database under the accession numbers AJ582102 to AJ592110 (16S rRNA gene), AJ831386 to AJ831392 (ITS1-S), AJ582268 to AJ582284 (ITS1-L), AJ582089 to AJ582093 and AJ582285 to AJ582289 (rpoC1), and AJ582094 to AJ582101 (nifH).
Acknowledgments
We thank E. Menthon and S. Lacoste for assistance with cyanobacterial cultures and M. Bouvy and G. Sarazin for sampling. We are grateful to P. Baker, J. Fastner, and A. Kovacs for kindly providing Australian, German, and Hungarian Cylindrospermopsis raciborskii strains.
This work was supported by the “Ministère de l'Ecologie et du Développement Durable” (Programme “Invasions Biologiques”). R. Molica was supported by a grant of the “Institut de Recherche pour le Développement” (IRD).
Footnotes
This work is a contribution of the French Institute for Research and Development (IRD/UR098).
REFERENCES
- 1.Baurain, D., L. Renquin, S. Grubisic, P. Scheldeman, A. Belay, and A. Wilmotte. 2002. Remarkable conservation of internally transcribed spacer sequences of Arthrospira (“Spirulina”) (Cyanophyceae, Cyanobacteria) strains from four continents and of recent and 30-year-old dried samples from Africa. J. Phycol. 38:384-393. [Google Scholar]
- 2.Boyer, S. L., J. R. Johansen, and V. R. Flechtner. 2002. Phylogeny and genetic variance in terrestrial Microcoleus (cyanophyceae) species based on sequence analysis of the 16S rRNA gene and associated 16S-23S ITS region. J. Phycol. 38:1222-1225. [Google Scholar]
- 3.Briand, J.-F., C. Leboulanger, J. F. Humbert, C. Bernard, and P. Dufour. 2004. Cylindrospermopsis raciborskii invasion at mid-latitudes: selection, wide physiological tolerance or global warming? J. Phycol. 40:231-238. [Google Scholar]
- 4.Briand, J. F., C. Robillot, C. Quiblier-Llobéras, J. F. Humbert, A. Couté, and C. Bernard. 2002. Environmental context of Cylindrospermopsis raciborskii (cyanobacteria) blooms in a shallow pond in France. Water Res. 36:3183-3192. [DOI] [PubMed] [Google Scholar]
- 5.Chapman, A. D., and C. L. Schelske. 1997. Recent appearance of Cylindrospermopsis (cyanobacteria) in five hypertrophic Florida lakes. J. Phycol. 33:191-195. [Google Scholar]
- 6.Couté, A., M. Leitao, and C. Martin. 1997. Première observation du genre Cylindrospermopsis (cyanophyceae, Nostocales) en France. Cryptogam. Algol. 18:57-70. [Google Scholar]
- 7.Couté, A., M. Leitao, and H. Sarmento. 2004. Cylindrospermopsis sinuosa spec. nova (Cyanophyceae, Nostocales), une nouvelle espèce du sud-ouest de la France. Arch. Hydrobiol. Suppl. 150:1-15. [Google Scholar]
- 8.Druart, J.-C., and J.-F. Briand. 2002. First record of Cylindrospermopsis raciborskii (Woloszyńska) Seenayya et Subba Raju (Cyanobacteria) in a lotic system in France. Ann. Limnol. 38:339-342. [Google Scholar]
- 9.Dufour, P., C. Bernard, J. F. Humbert, and C. Quiblier. 2004. Diversité génétique et épidémiologie de Cylindrospermopsis raciborskii, cyanobactérie toxique. Rapport final, Programme Invasions Biologiques. Ministère de l'Ecologie et du Développement Durable, Paris, France.
- 10.Dyble, J., H. W. Paerl, and B. A. Neilan. 2002. Genetic characterization of Cylindrospermopsis raciborskii (cyanobacteria) isolates from diverse geographic origins based on nifH and cpcBA-IGS nucleotide sequence analysis. Appl. Environ. Microbiol. 68:2567-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1989. PHYLIP: phylogeny inference package. Cladistics 5:258-266. [Google Scholar]
- 12.Fritsch, F. E., and F. Rich. 1929. Contributions to our knowledge of the freshwater algae of Africa. 7. Freshwater algae (exclusive to Diatoms) from Griqualand West. Trans. R. Soc. S. Afr. 18:1-92. [Google Scholar]
- 13.Gugger, M., C. Lyra, P. Henriksen, A. Couté, J. F. Humbert, and K. Sivonen. 2002. Phylogenetic comparison of the cyanobacterial genera Anabaena and Aphanizomenon. Int. J. Syst. Evol. Microbiol. 52:1867-1880. [DOI] [PubMed] [Google Scholar]
- 14.Humbert, J. F., D. Latour-Duris, B. Le Berre, H. Giraudet, and M. J. Salençon. Genetic diversity in Microcystis populations of a French storage reservoir assessed by sequencing of the 16S-23S rRNA intergenic spacer. Microb. Ecol., in press. [DOI] [PubMed]
- 15.Iteman, I., R. Rippka, N. Tandeau de Marsac, and M. Herdman. 2000. Comparison of conserved structural and regulatory domains within divergent 16S rRNA-23S rRNA spacer sequences of cyanobacteria. Microbiology 146:1275-1286. [DOI] [PubMed] [Google Scholar]
- 16.Komárek, J., and J. Komárková. 2003. Phenotype diversity of the cyanoprokaryotic genus Cylindrospermopsis (Nostocales); review 2002. Czech Phycol. 3:1-30. [Google Scholar]
- 17.Li, R., W. W. Carmichael, S. Brittain, G. K. Eaglesham, G. R. Shaw, A. Mahakhant, N. Noparatnaraporn, W. Yongmanitchai, K. Kaya, and M. M. Watanabe. 2001. Isolation and identification of the cyanotoxin cylindrospermopsin and deoxy-cylindrospermopsin from a Thailand strain of Cylindrospermopsis raciborskii (cyanobacteria). Toxicon 39:973-980. [DOI] [PubMed] [Google Scholar]
- 18.Mayr, E., and R. J. O'Hara. 1986. The biogeographic evidence supporting the Pleistocene forest refuge hypothesis. Evolution 40:55-67. [DOI] [PubMed] [Google Scholar]
- 19.Neilan, B. A., M. L. Saker, J. Fastner, A. K. Törökné, and B. P. Burns. 2003. Phylogeography of the invasive cyanobacterium Cylindrospermopsis raciborskii. Mol. Ecol. 12:133-140. [DOI] [PubMed] [Google Scholar]
- 20.Nicholas, K. B., Nicholas, H. B., Jr., and D. W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. Version 2.6.002.
- 21.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otsuka, S., S. Suda, R. Li, M. Watanabe, H. Oyaizu, S. Matsumoto, and M. M. Watanabe. 1999. Phylogenetic relationships between toxic and non-toxic strains of the genus Microcystis based on 16S to 23S internal transcribed spacer sequence. FEMS Microb. Lett. 172:15-21. [DOI] [PubMed] [Google Scholar]
- 23.Padisák, J. 1997. Cylindrospermopsis raciborskii (Woloszyńska) Seenayya et Subba Raju, an expanding, highly adaptive cyanobacterium: worldwide distribution and review of its ecology. Arch. Hydrobiol. Suppl. 107:563-593. [Google Scholar]
- 24.Scheldeman, P., D. Baurain, R. Bouhy, M. Scott, M. Mühling, B. A. Whitton, A. Belay, and A. Wilmotte. 1999. Arthrospira (Spirulina) strains from four continents are resolved into only two clusters, based on amplified ribosomal DNA restriction analysis of the internally transcribed spacer. FEMS Microb. Lett. 172:213-222. [DOI] [PubMed] [Google Scholar]
- 25.Seenayya, G., and N. Subba Raju. 1972. On the ecology and systematic position of the alga known as Anabaenopsis raciborskii (Wolosz.) Elenk. and a critical evaluation of the forms described under the genus Anabaenopsis, p. 52-57. Taxonomy and biology of blue-green algae. University of Madras, Madras, India.
- 26.Stuart, A. 1991. Mammalian extinction in the late Pleistocene of northern Eurasia and North America. Biol. Rev. Camb. Philos. Soc. 66:453-562. [DOI] [PubMed] [Google Scholar]
- 27.Woloszyńska, J. 1912. Das Phytoplankton einiger javanischer Seen mit Berücksichtigung des Sawa-Planktons. Bull. Int. Acad. Sci. Cracovie Ser. B 6:649-709. [Google Scholar]