Abstract
RNA interference through the expression of small hairpin RNA (shRNA) molecules has become a very promising tool in reverse mouse genetics as it may allow inexpensive and rapid gene function analysis in vivo. However, the prerequisites for ubiquitous and reproducible shRNA expression are not well defined. Here we show that a single copy shRNA-transgene can mediate body-wide gene silencing in mice when inserted in a defined locus of the genome. The most commonly used promoters for shRNA expression, H1 and U6, showed a comparably broad activity in this configuration. Taken together, the results define a novel approach for efficient interference with expression of defined genes in vivo. Moreover, we provide a rapid strategy for the production of gene knockdown mice combining recombinase mediated cassette exchange and tetraploid blastocyst complementation approaches.
INTRODUCTION
RNA interference (RNAi) (1) is based on the introduction of double stranded RNA (dsRNA) molecules into cells, whereby one strand is complementary to the coding region of the target gene. Through pairing with the dsRNA, the target mRNAs are degraded by a cellular mechanism. Sustained gene silencing has been achieved by the intracellular transcription of small inverted repeats separated by a spacer region (2). For the expression of these short hairpin RNAs (shRNAs), the RNA polymerase III-dependent promoters U6 and H1 have been used in most cases (2,3). These promoters produce small transcripts lacking nonfunctional bases at the 5′ end and a poly-adenosine tail. The resulting shRNAs are processed in vivo into active ‘small interfering’ RNAs. Several reports describe the stable, shRNA-mediated gene silencing in mice using random transgenesis (4–7). While this demonstrates that shRNA-mediated gene knockdown in mice is feasible, the requirements with respect to copy number and integration site of the transgene for efficient gene silencing are still unclear. Since previous experiments included random transgenesis, each of the resulting mouse lines showed a unique, irreproducible shRNA expression pattern. In addition, screening of several embryonic stem cells or mouse lines with sufficient siRNA expression was required, which is laborious and time-consuming. Finally, the organ-specific activities of the most commonly used promoters for shRNA expression, U6 and H1, have not been analyzed systematically. Thus, an established protocol for simple and reproducible RNAi in mice is not available.
Here we demonstrate that a single copy shRNA transgene under the control of either the U6 or the H1 promoter mediates efficient and ubiquitous gene knockdown in mice when integrated at the rosa26 locus. By combining recombinase-mediated cassette exchange (RMCE) at the rosa26 locus and the tetraploid blastocyst complementation approach, we drastically reduced the time and effort for generating shRNA transgenic gene knockdown mice.
MATERIALS AND METHODS
Rosa targeting and exchange vectors
Rosa26 renilla luciferase (Rluc) vector
The Renilla luciferase gene (Promega) was inserted into the rosa targeting vector (8) 3′ of the splice acceptor site. shRNA cassettes were inserted into 3′ of the Renilla luciferase gene. shRNA cassettes: the U6- and the H1-promoter fragments were amplified from human genomic DNA and cloned in front of the Fluc-specific shRNA (9) and five thymidines as termination signal (shRNA). The shRNA expression cassettes were flanked by loxP-sites (lox) to allow cre-mediated deletion.
Rosa26 firefly luciferase (Fluc) vector
The firefly luciferase gene (Promega) was inserted into the rosa targeting vector (8) and the neomycin resistance gene was replaced by a PGK-hygromycin resistance gene.
Rosa26 targeting vector
The neomycin was deleted by Flp-mediated deletion in bacteria (10). The final rosa (RMCE) targeting vector (Figure 1A) was generated by standard cloning procedures and has the following order in 5′ to 3′ direction: 5′ homology arm, splice acceptor, ATG start codon, F3 site (11), zsgreen ORF (Clontech), synthetic poly(A) signal, PGK-hygro resistance gene, CAGGS-promoter, Flpe-recombinase gene (12), synthetic poly(A) signal and FRT site and the 3′ homology arm.
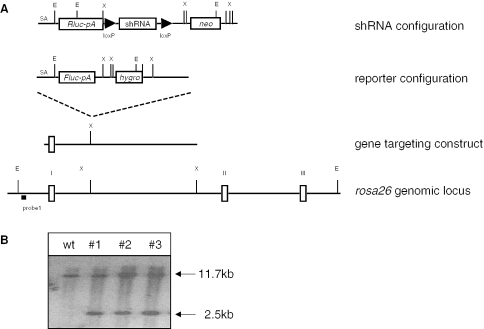
Figure 1.
(A) Scheme of the targeting strategy. ShRNA and reporter constructs were independently inserted into the rosa26 locus by homologous recombination in ES cells. Genes encoding the Renilla (Rluc) and firefly luciferases (Fluc) along with an adenovirus splice acceptor sequence and a polyadenylation signal (pA) were placed downstream of the endogenous promoter of rosa26. The Fluc specific shRNA is expressed under the control of the U6- and H1-promoter, respectively, and terminated by five thymidines (shRNA). The loxP-sites flanking the shRNA expression cassettes were used to generate a negative control through cre-mediated recombination. (B) Southern blot analysis of genomic DNA from ES cells containing the targeted insertion of the shRNA (lane 1) or the target configuration (lanes 2 and 3). Homologous recombination at the rosa26 locus is detectable using EcoRV-digested genomic DNA and probe 1, resulting in a 11.7 kb band for the wild type (wt) and a 2.5 kb band for the targeted alleles. E: EcoRV; X: XbaI; neo: FRT-flanked neomycin resistance gene; hyg: FRT-flanked hygromycin resistance gene.
Exchange vector
The vector contains the F3 site and the FRT site in the same configuration as in the rosa26 targeting vector described above. The vector was generated using standard cloning procedures and has the following order in 5′ to 3′ direction: synthetic poly(A) signal, F3-site and a neomycin-resistance gene lacking the start ATG. The H1-promoter fragment was amplified from human genomic DNA with attached BbsI/AvrII sites and cloned into the basic exchange vector (pRMCE-H1). LeptinR-specific shRNA sequence [aacagtcttccactgttgctttg (sense) TTAGCACTG (loop) caaagcaacagtggaagactg (antisense)] together with five thymidines were generated by oligonucleotide annealing and inserted into pRMCE-H1 upon BbsI/AvrII restriction. Insertion at the BbsI site in pRMCE-H1 allows transcription of the shRNA starting at the first base of the hairpin sequence.
Rosa26/CAGGS-Fluc/βgal-shRNA exchange vector
The vector has the following order in 5′ to 3′ direction: synthetic poly(A) signal, F3-site, neomycin-resistance gene lacking the start ATG, U6-promoter, LacZ-specific shRNA sequence (gtggatggagccgatattggaTTCAAGAGAttcaatattggcttcatccac), five thymidines, CAGGS promoter (13), firefly luciferase gene, synthetic poly(A) signal, FRT.
Cell culture
Culture and targeted mutagenesis of Art4.12 ES cells were carried out as previously described (8,14). ES cells were cultured in ES cell medium [DMEM with 15% fetal calf serum (FCS) containing 2000 U/ml LIF (Chemicon)] on mitomycin C-treated primary feeder fibroblasts. Prior to blastocyst injection ES cells were trypsinized, resuspended in ES cell medium and pre-plated to remove feeder cells and debris.
For the Cre-mediated deletion of the shRNA cassette 1 × 107 cells were electroporated with 18 μg pCAGGScrepA (R. Kühn, unpublished) generated from pCAGGSflpe-ires-puro (15). One half of the treated cells were plated on two 10 cm dishes. At day 2 after the electroporation 1000 cells and 3000 cells were replated on 10 cm dishes. At day 8 clones were isolated on 96-well plates and analyzed by Southern blotting.
Transfection of cells with the exchange vector
One day before transfection, 2 × 105 ES cells were plated on a 3 cm dish in 2 ml medium. Before transfection 2 ml fresh medium was given to the cells. 3 μl Fugene6 Reagent (Roche; Catalog No. 1 814 443) was mixed with 100 μl serum free medium (OptiMEM 1 with Glutamax-I Invitrogen; Catalog No. 51985-035) and incubated for 5 min. 100 μl of the Fugene/OptiMEM solution was added to 2 μg circular DNA (c = 0.33 μg/μl) and incubated for 15 min. This transfection complex was added drop wise to the medium and mixed by a circuiting movement. Fresh medium was added to the transfected cells the following day. From day 2 on, the medium was changed daily, replaced by medium containing 250 μg/ml G418 (Geneticin; Invitrogen; Catalog No. 10131-019). Seven days after transfection, single clones were isolated by standard procedures as described (16).
Mice
All mice were kept in the animal facility at Artemis Pharmaceuticals GmbH in microisolator cages (Tecniplast Sealsave). B6D2F1 mice for the generation of tetraploid blastocysts were obtained from Harlan, NL.
Production of ES mice by tetraploid embryo complementation
The production of mice by tetraploid embryo complementation was essentially performed as described (17). Briefly, embryo culture was carried out in microdrops on standard bacterial Petri dishes (Falcon) under mineral oil (Sigma). Modified CZB media (18) was used for embryo culture unless otherwise noted. HEPES buffered CZB was used for room temperature operations. After administration of hormones, superovulated B6D2F1 females were mated with B6D2F1 males. Fertilized zygotes were isolated from the oviduct and any remaining cumulus cells were removed with hyaluronidase. After overnight culture, two-cell embryos were electrofused using a CF-150B fusion chamber (BLS Ltd., Budapest, Hungary) to produce tetraploid embryos. Embryos that had not undergone membrane fusion within 1 h were discarded. Embryos were then cultured in vitro to the blastocyst stage. For microinjection, 10–20 blastocysts were placed in a drop of DMEM with 15% FCS under mineral oil. A flat tip, piezo actuated microinjection-pipette with an internal diameter of 12–15 μm was used to inject 20 ES cells into each blastocyst. After recovery, 10 injected blastocysts were transferred to each uterine horn of 2.5 days post coitum, pseudopregnant NMRI females. Embryos were either delivered naturally or were recovered from recipient mothers by C-section and subsequently cross-fostered.
Luciferase detection
Organs were homogenized at 4°C in lysis buffer (0.1 M KH2PO4, 1 mM DTT, 0.1% Triton X-100) using a tissue grinder and centrifuged for 5′ at 2000 g (4°C). Supernatant was assayed for Fluc activities using the Dual Luciferase Assay (Promega, Inc.) according to the manufacturer's protocol. The luminescence was detected using a Lumat LB 9507 (EG&G Berthold). Relative values of Fluc activity in different organs are given as indicated. All values of Fluc activity were normalized by using the Rluc activity for reference.
Histochemical detection of β-galactosidase activity
For preparation of histological sections, organs were embedded in OCT tissue freezing medium (Leica&Jung No. 020108926), frozen on dry ice and sectioned using a Leica CM3050 Cryostat. Sections were mounted onto slides and dried for 1–4 h at room temperature. For histochemical detection of β-galactosidase activity, sections were fixed for 5 min at room temperature using 0.2% glutaraldehyde, 5 mM EGTA and 2 mM MgCl2 in phosphate buffer (80 mM K2HPO4, 20 mM KH2PO4, pH 7.3). Sections were washed three times in washing buffer (2 mM MgCl2, 0.02% Nonidet-40, 80 mM K2HPO4, 20 mM KH2PO4, pH 7.3) and finally incubated in X-Gal staining solution [5 mM K3(Fe(CN)6), 5 mM K4(Fe(CN)6), 2 mM MgCl2, 1 mg/ml X-Gal in washing buffer] for 12 h at 37°C. Sections were washed twice in phosphate-buffered saline and counterstained for 10 min with Nuclear Fast Red. Sections were subsequently washed in distilled water, dehydrated using graded ethanol concentrations (70, 90, 96 and 100% ethanol; 5 min incubation for each concentration) and finally mounted using Entellan (Merck No. 1.07961.0100). Images were taken using a Hitachi HVC20M camera and the Diskus imaging program (C. Hilgers, Königswinter, Germany).
Real-time PCR
After tissue homogenization, RNA extraction was performed with the RNeasyTM Mini Kit (Qiagen). Due to the usage of exon-spanning TaqmanTM probes DNAse I digestion was not necessary. First Strand cDNA synthesis was done using Oligo-dT primers and SuperscriptTM II RT (Invitrogen). For Taqman analysis ∼50 ng cDNA of each sample was used as template. The analysis was performed on an ABI PRISMTM 7700 sequence detection system and quantification of relative cDNA transcript amounts was calculated by the ΔΔCt-method (19). For leptin receptor detection the primers and probes were used from Applied Biosystems (assay ID 0440188). Each sample was analyzed in duplicates and normalized on TATA-box binding protein representing the endogenous control. Standard Curves had an R 2 ≥ 0.997.
RESULTS
Activity of shRNA transgenes upon insertion into the rosa26 locus
To investigate the effect of different promoters on the level and tissue specificity of shRNA-mediated gene inactivation, we decided to test both, the H1 and U6 promoter in a particular chromosomal locus. We performed a side-by-side comparison of a firefly luciferase-specific shRNA transgene under the control of the human H1 and U6 promoter, respectively. A single copy of either of these constructs was inserted into the rosa26 locus of ES cells to exclude variable position effects (Figure 1). We have chosen rosa26 for this purpose, since it has been described as being accessible for transcription in all tissues (8,20). A dual reporter system was established by the expression of both, the firefly luciferase (Fluc) as a test substrate and the Renilla luciferase (Rluc) as a reference to quantify the degree of shRNA-mediated silencing. A similar Fluc/Rluc system has been previously applied for the transient assay of siRNA in vitro and in mice (21,22). We adapted this system for our purpose by expressing both reporter genes under the control of the ubiquitous rosa26 promoter. Each of the luciferases along with a splice acceptor sequence (23) was inserted downstream of the promoter of rosa26 through homologous recombination in ES cells (Figure 1). The negative control was generated by cre-mediated deletion of the loxP-flanked shRNA expression cassettes in ES cells (data not shown).
Recombinant ES cells were injected into blastocysts and chimeric mice were obtained upon transfer of blastocysts into pseudopregnant females using standard protocols (16). Figure 2A shows the activity of Fluc in different organs in shRNA transgenic animals that were obtained from breeding of chimeras. Expression of the shRNA under the control of both, the U6 as well as the H1 promoter resulted in efficient and ubiquitous repression of Fluc activity, ranging between 70 and 95% reduction in all organs except spleen and testis. This result was reproducible in several, independent experiments using mice ranging in age from 6 to 25 weeks (Figure 2).
Figure 2.
Efficiency of shRNA-mediated Fluc knockdown in mice. (A) Silencing of Fluc under the control of the endogenous rosa26 promoter. Each configuration (control, H1shRNA, U6shRNA) was analyzed using three mice at the age of 6, 12 and 25 weeks, respectively. The efficiency of luciferase knockdown appeared not to be influenced by the age of the animal. Percentages of U6- (white bars) and H1-shRNA-mediated repression of Fluc activity (grey bars) ± standard error of the mean are shown. In the negative controls (black bars), the shRNA expression cassettes were removed through cre-mediated recombination. Relative values of Fluc activity in different organs are given as indicated. All values of Fluc activity were normalized by using the Rluc activity for reference. (B) Comparison of firefly luciferase expression driven by the CAGGS (black bars) and the endogenous rosa26 promoter (white bars), respectively. All values are normalized by measuring the protein content using Bradford assay for reference. (C) Silencing of Fluc under the control of the CAGGS promoter. Percentages of U6-shRNA-mediated repression of Fluc activity (white bars) ± standard error of the mean are shown. In the negative controls (black bars), the shRNA expression cassettes were removed through cre-mediated recombination. Relative values of Fluc activity in different organs are given as indicated. All values of Fluc activity were normalized by using the Rluc activity for reference.
RMCE strategy for the insertion of shRNA transgenes into the rosa26 locus
To facilitate the introduction of shRNA transgenes into the genome we adapted recombinase—mediated cassette exchange (RMCE) to the rosa26 locus. The targeting vector to prepare the rosa26 locus for RMCE is depicted in Figure 3. The vector carries a FLPe expression cassette to provide the recombinase for RMCE. The hygromycin resistance gene was used for positive selection of homologous recombinant clones. In addition, a zsGreen gene was placed between the FRT and F3 sites to allow for the identification of recombinant clones that have not undergone RMCE following secondary transfection of the exchange vector. The splice acceptor site (SA) and the ATG start codon should facilitate expression of the truncated neomycin resistance gene (Δ5′neoR) on the exchange vector by employing the endogenous rosa26 promoter following RMCE.
Figure 3.
RMCE targeting system for the rosa26 locus. (A) The rosa26 targeting vector comprising zsgreen, PGK-Hyg and CAGGS-FLP was inserted into the rosa26 locus via homologous recombination in ES cells. The F3/FRT sites are oriented in opposite direction to each other. ‘X’ marks the insertion point within the rosa26 locus. The modified locus carrying the RMCE acceptor is called rosa26(RMCE). (B) RMCE by Flpe-mediated recombination generates the rosa26(RMCE) allele. The exchange vector carries the shRNA expression cassette under the control of the H1 promoter, the F3/FRT pair and a truncated neoR gene for positive selection. A polyA signal is included to prevent expression of the truncated neoR gene at random integration sites. (C) Southern blot analysis of genomic DNA from ES cells. The sizes of wt, rosa26(RMCE) and rosa26(RMCE exchanged) are 4.4, 3.9 and 5.8 kb, respectively. In clones 1–3 and 5–9 successful RMCE had occurred. Genomic DNA was digested with HindIII and analyzed using probe 1. X: XbaI, H: HindIII.
We used the hybrid ES cell line ART4.12 ([C57BL/6 ×129S6/SvEvTac] F1) for homologous recombination, since these lines result in ES cell derived mice (ES mice) through tetraploid blastocyst complementation (8). ART4.12 cells were transfected with the rosa26 targeting vector and cultured in medium containing hygromycin B. Recombinant ES cell clones [rosa26(RMCE)] were obtained at a frequency of 2% as verified by Southern blot analysis (Figure 3C, first and second lanes). For unknown reasons, the zsgreen gene turned out to be non functional. The detection of green fluorescence was therefore not used in our screening setup.
The exchange vector (Figure 3B) carries the FRT and F3 sites together with a truncated neoR gene for positive selection of RMCE. The upstream poly(A) signal was included to prevent expression of the truncated neoR gene in ES cells carrying randomly integrated vectors. ES cells were transfected with the exchange vector and selected in medium containing G418. Southern blot analysis of G418 resistant colonies revealed that successful RMCE had occurred in >95% of clones (Figure 3C and data not shown).
Silencing of a highly expressed firefly luciferase transgene
To determine whether the concentration of mRNA is critical for the degree of shRNA-mediated RNAi, we used the strong CAGGS promoter (13) for expression of the Fluc gene. A CAGGS-Fluc expression cassette was inserted into the ES cell genome using RMCE at the rosa26 locus to allow a side-by-side comparison with the Fluc target under the control of the endogenous rosa26 promoter. Recombinant ES cells were injected into blastocysts and mice were obtained upon transfer of blastocysts into pseudopregnant females. Comparison of Fluc activities in various organs showed that the CAGGS promoter conferred up to ∼100-fold higher expression level of the Fluc transgene compared with the endogenous rosa26 promoter (Figure 2B). The Fluc-specific shRNA gene under the control of the human U6 promoter and the CAGGS-Fluc transgene were combined through breeding of mice. Measurement of luciferase activity in protein extracts from various organs revealed a roughly similar pattern of CAGGS-Fluc repression compared with the shRNA-mediated knockdown of rosa26-Fluc in all organs except liver (Figure 2C). In liver, the degree of Fluc silencing was less pronounced compared with the data from low expressing Fluc mice (51 versus 82% knockdown).
Analysis of shRNA-mediated knockdown at the single cell level using lacZ
We next introduced a lacZ specific shRNA under the control of the human U6 promoter through RMCE in ART4.12/rosa26(RMCE) ES cells. Recombinant ES cells were injected into tetraploid blastocysts and ES cell obtained mice were derived as described (8). A highly expressed β-galactosidase gene was provided through breeding using a mouse strain carrying lacZ under the control of the ubiquitous CAGGS promoter (13), that had been placed into the Rosa26 promoter. In this strain, β-galactosidase is ubiquitously expressed at high levels in every single cell of adult mice (S. Andreas and N. Faust, unpublished data). X-Gal staining on tissue sections revealed a strong, uniform expression of lacZ under the control of the CAGGS promoter in every single cell, whereas the presence of the shRNA construct resulted in marked reduction of β-galactosidase activity in the vast majority of cells (Figure 4).
Figure 4.
Analysis of shRNA-mediated β-galactisidase silencing at the single cell level. Histological sections of organs from wt and CAGGS-lacZ transgenic mice, as well as two CAGGS-lacZ/shRNA transgenic animals are shown as indicated. Cryosections were incubated with X-gal (blue staining) and counterstained with Nuclear Fast Red (red staining). Magnification: 40×.
Generation and analysis of leptin receptor knockdown mice
The RMCE strategy was used for targeted insertion of a shRNA sequence directed against all known splice variants of the leptin receptor mRNA. We have chosen this target for proof of concept, since the phenotype of leptin receptor deficiency in db/db mice represents a well characterized disease model, encompassing hyperphagia, obesity and insulin resistance (24). The shRNA expression cassette was placed 3′ of the neoR gene on the exchange vector. Upon transfection, recombinase mediated integration of the exchange vector at the rosa26 locus was observed in >90% of G418 resistant colonies (Figure 3C). The activity of the inserted shRNA expression cassette in the surviving ES cell clones was tested using real-time PCR analysis. The presence of leptin receptor specific shRNA transgene resulted in a >70% reduction of leptin receptor mRNA. In contrast, a p53-specific shRNA construct in a similar configuration did not affect the level of leptin receptor mRNA (data not shown), confirming the specificity of the selected shRNA.
ShRNA transgenic ES cells were injected into tetraploid blastocysts and ES cell derived mice were obtained three weeks later at a frequency of 3%. Real-time PCR analysis of 15-week-old mice indicated a >80% reduction of leptin receptor mRNA in most organs, reaching 95–99% in heart, brain, muscle, pancreas and white adipose tissue (Figure 5). In liver, the degree of leptin receptor repression reached 65%. Initial inspection of leptin receptor knockdown mice revealed a strongly increased food intake and 30% increased body weight when fed with normal diet (data not shown).
Figure 5.
TaqMan analysis of leptin receptor mRNA in shRNA transgenic versus wild-type control mice. Percentages of leptin receptor mRNA expression in shRNA transgenic mice (grey bars) in comparison with expression levels found in wild-type mice (black bars) ± standard error of the mean are shown. All assays were performed with groups of 4–5 mice at age of 4 months from different organs as indicated.
DISCUSSION
In this report, we describe a defined, single copy shRNA configuration that mediates efficient and ubiquitous gene knockdown in mice. We provide the first side-by-side comparison of the most commonly used promoters for shRNA expression, U6 and H1. ShRNA expression cassettes were inserted into the rosa26 locus through targeted transgenesis in ES cells. Both, the U6 and the H1 promoter, showed a comparably broad activity in this configuration, resulting in 70–95% silencing of the luciferase reporter gene in most organs. Single cell analysis of tissue sections demonstrated that the degradation of target mRNAs is achieved in the vast majority rather than in just a fraction of cells (Figure 4). We did not observe a strong impact of the mRNA level, as the comparison of the Fluc target expressed at different levels did not significantly affect the degree of knockdown (Figure 2). Also, the organ-specific pattern was highly reproducible using different reporter targets. The reason for the lower degree of RNAi observed in lymphocytes, testes, and, in some cases, in liver remains unclear. It is unlikely that a limited activity of RNA polymerase III-dependent promoters accounts for these differences as Oberdoerffer et al. (25) did not find any obvious correlation of shRNA expression level and RNAi efficiency. We rather suspect that the degree of RNAi is affected by other factors, such as the availability of RISC and/or dicer proteins.
We confirmed the activity of our configuration by introducing a leptin receptor specific shRNA construct under the control of the H1 promoter at the rosa26 locus. Here, repression of the endogenous target ranged between 95–99% in brain, heart muscle, pancreas and white adipose tissue. Gross inspection of the phenotype revealed a significantly increased food intake and body weight when fed with normal diet (data not shown), reproducing aspects of the phenotype found in db/db mice. A detailed characterization of the strain is ongoing and will be published elsewhere.
Sustained, shRNA-mediated RNAi in mice had been demonstrated in recent publications using random transgenesis (4–7,26,27). Several technical approaches have followed: pronuclear injection and transfection of plasmid-based shRNA constructs, as well as lentiviral infection. Pronuclear injection of transgenes resulted in individual founder lines, each with a unique and irreproducible pattern of shRNA expression (27,28). Depending on the site of transgene integration, the efficiency of gene silencing in those animals was variable ranging from undetectable levels to >90%. Rosenquist et al. failed to produce any distinct phenotype when shRNA constructs directed against seven known targets were introduced via the standard transgenesis approach (28). To circumvent this problem, transfected ES cells were screened for proper shRNA expression prior to the generation transgenic mouse lines through blastocyst injection (4,28). While the severity of the observed in vivo phenotypes correlated roughly with the strength of shRNA expression in ES cells, the level and pattern of gene silencing was still variable in these experiments. Thus, a thorough characterization of the resulting mouse lines is still required to determine the level of RNAi in all relevant organs. Moreover, the approach is just applicable for a fraction of target genes that are expressed in ES cells. Lentivirus-based vectors were used for the introduction of shRNA transgenes into primary cells, ES cells and embryos (5–7). Again, only a fraction of infected cells showed detectable RNAi at various levels (7), indicating that the activity of lentiviral vectors is subjected to position dependent modulation. Taken together, all previous reports describing shRNA-mediated gene knockdown in mice employed random transgenesis, requiring time-consuming and laborious screening of embryonic stem cell or mouse lines due to the inherent problem of position-dependent effects on transgene expression.
Our targeted transgenesis strategy for the insertion of shRNA expression cassettes has several advantages over the current approaches of random transgenesis: The configuration at the rosa26 locus mediates efficient RNAi in most organs. The shRNA expression pattern in mice is predictable and not affected by variable chromosomal position effects. Therefore, the generation and time-consuming characterization of multiple transgenic strains for a given shRNA transgene is not required. Furthermore, a single copy of the shRNA gene is inserted into the genome, avoiding modulation of shRNA expression by secondary rearrangement of multiple copy arrays or segregation of transgenes integrated at different chromosomal loci.
To speed up the generation of shRNA transgenic animals, we adapted recombinase-mediated cassette exchange (RMCE) to the rosa26 locus. So far, only few examples of successful RMCE in ES had been described (15,26,29–32). In these experiments, random integration of the exchange vector as well as incomplete recombination frequently produced unwanted transgene configurations. The efficiency of RMCE varied strongly, depending on the choice of recombination sites, the selection strategy and the chromosomal target. The following features where combined in our RMCE strategy to address these problems.
We utilized Flp-mediated RMCE using a wild type Flp target site (FRT) in combination with an inverted F3 site (11). The F3/F3 couple is recombined by FLP with the same efficiency as two wild type recombinase recognition sites (RRS) whereas recombination of a FRT/F3 pair is not catalyzed (33). This characteristic contrasts other pairs of wild type and mutant RRS, such as loxp/lox511, that was found to mediate a residual recombination activity (34,35).
We added a constitutive FLPe expression cassette on the targeting vector to provide sufficient recombinase activity until successful RMCE of the exchange vector. Thus, incomplete recombination intermediates as observed by Belteki et al. (29) should be avoided.
A 5′ truncated neomycin phosphotransferase gene including a splice acceptor site but lacking a functional promoter was inserted into the exchange vector. Expression of the selection marker should therefore be mediated by the endogenous rosa26 promoter following successful RMCE, but not through random integration of the exchange vector.
We found that our RMCE strategy facilitates the insertion of shRNA transgenes into the rosa26 locus with high efficiencies, consistently reaching a frequency of >90% correct clones. Given the simple cloning strategy for the insertion of shRNA transgenes into the RMCE exchange vector and the limited screening efforts, our RMCE approach largely facilitates the generation of shRNA transgenic mice. By employing the tetraploid blastocyst complementation approach for the direct generation of ES cell derived mice, we further reduced the production time to less than 2 months (1 week for cloning of the RMCE exchange vector, 3 weeks for generating targeted ES cell clones, 3 weeks for the generation of newborn mice through tetraploid blastocyst injection).
Taken together, we describe a novel approach to efficiently knock down genes in mice in a much easier, faster and more predictable manner than achieved by conventional techniques. Combined with engineered H1 or U6 promoters allowing the temporal control of shRNA expression (7,36,37) our strategy may provide a general tool for rapid gene function analysis in adult animals.
Acknowledgments
We thank Sandra Niehaves, Maria da Silva and Isolde Falkner for excellent technical assistance, and Dave Lewis for advice. This work was supported by Artemis Pharmaceuticals GmbH. Funding to pay the Open Access publication charges for this article was provided by Artemis Pharmaceuticals GmbH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 3.Tuschl T. Expanding small RNA interference. Nat. Biotechnol. 2002;20:446–448. doi: 10.1038/nbt0502-446. [DOI] [PubMed] [Google Scholar]
- 4.Kunath T., Gish G., Lickert H., Jones N., Pawson T., Rossant J. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. Nat. Biotechnol. 2003;21:559–561. doi: 10.1038/nbt813. [DOI] [PubMed] [Google Scholar]
- 5.Rubinson D.A., Dillon C.P., Kwiatkowski A.V., Sievers C., Yang L., Kopinja J., Rooney D.L., Ihrig M.M., McManus M.T., Gertler F.B., et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nature Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 6.Tiscornia G., Singer O., Ikawa M., Verma I.M. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc. Natl Acad. Sci. USA. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ventura A., Meissner A., Dillon C.P., McManus M., Sharp P.A., Van Parijs L., Jaenisch R., Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl Acad. Sci. USA. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seibler J., Zevnik B., Kuter-Luks B., Andreas S., Kern H., Hennek T., Rode A., Heimann C., Faust N., Kauselmann G., et al. Rapid generation of inducible mouse mutants. Nucleic Acids Res. 2003;31:e12. doi: 10.1093/nar/gng012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paddison P.J., Caudy A.A., Bernstein E., Hannon G.J., Conklin D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchholz F., Angrand P.O., Stewart A.F. A simple assay to determine the functionality of Cre or FLP recombination targets in genomic manipulation constructs. Nucleic Acids Res. 1996;24:3118–3119. doi: 10.1093/nar/24.15.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlake T., Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry. 1994;33:12746–12751. doi: 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- 12.Buchholz F., Angrand P.O., Stewart A.F. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- 13.Okabe M., Ikawa M., Kominami K., Nakanishi T., Nishimune Y. Green mice as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 14.Eggan K., Rode A., Jentsch I., Samuel C., Hennek T., Tintrup H., Zevnik B., Erwin J., Loring J., Jackson-Grusby L., et al. Male and female mice derived from the same ES cell clone by tetraploid embryo complementation. Nat. Biotechnol. 2002;20:455–459. doi: 10.1038/nbt0502-455. [DOI] [PubMed] [Google Scholar]
- 15.Schaft J., Ashery-Padan R., van der Hoeven F., Gruss P., Stewart A.F. Efficient FLP recombination in mouse ES cells and oocytes. Genesis. 2001;31:6–10. doi: 10.1002/gene.1076. [DOI] [PubMed] [Google Scholar]
- 16.Hogan B., Beddington R., Costantini F., Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 17.Eggan K., Akutsu H., Loring J., Jackson-Grusby L., Klemm M., Rideout W.M., III, Yanagimachi R., Jaenisch R. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc. Natl Acad. Sci. USA. 2001;98:6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatot C.L., Lewis J.L., Torres I., Ziomek C.A. Development of 1-cell embryos from different strains of mice in CZB medium. Biol. Reprod. 1990;42:432–440. doi: 10.1095/biolreprod42.3.432. [DOI] [PubMed] [Google Scholar]
- 19.Grace M.B., McLeland C.B., Blakely W.F. Real-time quantitative RT–PCR assay of GADD45 gene expression changes as a biomarker for radiation biodosimetry. Int. J. Radiat. Biol. 2002;78:1011–1021. doi: 10.1080/09553000210158056. [DOI] [PubMed] [Google Scholar]
- 20.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 21.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 22.Lewis D.L., Hagstrom J.E., Loomis A.G., Wolff J.A., Herweijer H. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nature Genet. 2002;32:107–108. doi: 10.1038/ng944. [DOI] [PubMed] [Google Scholar]
- 23.Friedrich G., Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 24.Friedman J.G., Halaas J.L. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 25.Oberdoerffer P., Kanellopoulou C., Heissmeyer V., Paeper C., Borowski C., Aifantis I., Rao A., Rajewsky K. Efficiency of RNA interference in the mouse hematopoietic system varies between cell-types and developmental stages. Mol. Cell. Biol. 2005 doi: 10.1128/MCB.25.10.3896-3905.2005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cesari F., Rennekampff V., Vintersten K., Vuong L.G., Seibler J., Bode J., Wiebel F.F., Nordheim A. Elk-1 knock-out mice engineered by Flp recombinase-mediated cassette exchange. Genesis. 2004;38:87–92. doi: 10.1002/gene.20003. [DOI] [PubMed] [Google Scholar]
- 27.Hasuwa H., Kaseda K., Einarsdottir T., Okabe M. Small interfering RNA and gene silencing in transgenic mice and rats. FEBS Lett. 2002;532:227–230. doi: 10.1016/s0014-5793(02)03680-3. [DOI] [PubMed] [Google Scholar]
- 28.Carmell MA., Zhang L., Conklin D.S., Hannon G.J., Rosenquist T.A. Germline transmission of RNAi in mice. Nature Struct. Biol. 2003;10:91–92. doi: 10.1038/nsb896. [DOI] [PubMed] [Google Scholar]
- 29.Belteki G., Gertsenstein M., Ow D.W., Nagy A. Site-specific cassette exchange and germline transmission with mouse ES cells expressing phiC31 integrase. Nat. Biotechnol. 2003;21:321–324. doi: 10.1038/nbt787. [DOI] [PubMed] [Google Scholar]
- 30.Feng Y.Q., Seibler J., Alami R., Eisen A., Westerman K.A., Leboulch P., Fiering S., Bouhassira E.E. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J. Mol. Biol. 1999;292:779–785. doi: 10.1006/jmbi.1999.3113. [DOI] [PubMed] [Google Scholar]
- 31.Kolb A.F. Selection-marker-free modification of the murine beta-casein gene using a lox2272 site. Anal. Biochem. 2001;290:260–271. doi: 10.1006/abio.2000.4984. [DOI] [PubMed] [Google Scholar]
- 32.Seibler J., Schubeler D., Fiering S., Groudine M., Bode J. DNA cassette exchange in ES cells mediated by Flp recombinase: an efficient strategy for repeated modification of tagged loci by marker-free constructs. Biochemistry. 1998;37:6229–6234. doi: 10.1021/bi980288t. [DOI] [PubMed] [Google Scholar]
- 33.Seibler J., Bode J. Double-reciprocal crossover mediated by FLP-recombinase: a concept and an assay. Biochemistry. 1997;36:1740–1747. doi: 10.1021/bi962443e. [DOI] [PubMed] [Google Scholar]
- 34.Langer S.J., Ghafoori A.P., Byrd M., Leinwand L. A genetic screen identifies novel non-compatible loxP sites. Nucleic Acids Res. 2002;30:3067–3077. doi: 10.1093/nar/gkf421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lauth M., Spreafico F., Dethleffsen K., Meyer M. Stable and efficient cassette exchange under non-selectable conditions by combined use of two site-specific recombinases. Nucleic Acids Res. 2002;30:e115. doi: 10.1093/nar/gnf114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsukura S., Jones P.A., Takai D. Establishment of conditional vectors for hairpin siRNA knockdowns. Nucleic Acids Res. 2003;31:e77. doi: 10.1093/nar/gng077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van de Wetering M., Oving I., Muncan V., Pon Fong M.T., Brantjes H., van Leenen D., Holstege F.C., Brummelkamp T.R., Agami R., Clevers H. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]