Abstract
The anti-oxidant naphterpin is a natural product containing a polyketide-based aromatic core with an attached 10-carbon geranyl group derived from isoprenoid (terpene) metabolism1–3. Hybrid natural products such as naphterpin that contain 5-carbon (dimethylallyl), 10-carbon (geranyl) or 15-carbon (farnesyl) isoprenoid chains possess biological activities distinct from their non-prenylated aromatic precursors4. These hybrid natural products represent new anti-microbial, anti-oxidant, anti-inflammatory, anti-viral and anti-cancer compounds. A small number of aromatic prenyltransferases (PTases) responsible for prenyl group attachment have only recently been isolated and characterized5,6. Here we report the gene identification, biochemical characterization and high-resolution X-ray crystal structures of an architecturally novel aromatic PTase, Orf2 from Streptomyces sp. strain CL190, with substrates and substrate analogues bound. In vivo, Orf2 attaches a geranyl group to a 1,3,6,8-tetra-hydroxynaphthalene-derived polyketide during naphterpin biosynthesis. In vitro, Orf2 catalyses carbon–carbon-based and carbon–oxygen-based prenylation of a diverse collection of hydroxyl-containing aromatic acceptors of synthetic, microbial and plant origin. These crystal structures, coupled with in vitro assays, provide a basis for understanding and potentially manipulating the regio-specific prenylation of aromatic small molecules using this structurally unique family of aromatic PTases.
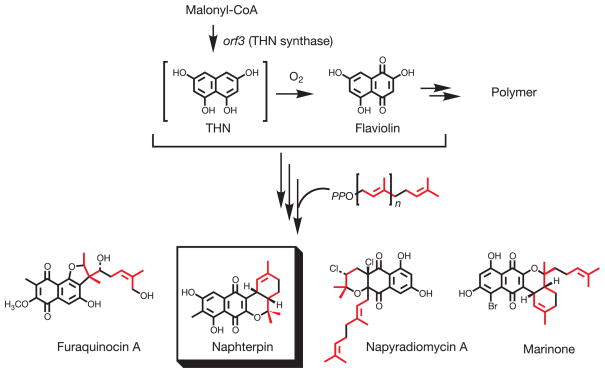
Naphterpin is produced via both mevalonate (MVA) and polyketide biosynthetic pathways. Naphterpin biosynthesis includes 1,3,6,8-tetrahydroxynaphthalene (THN), which after oxidative transformation to flaviolin, is prenylated with a 10-carbon geranyl moiety before final biosynthetic elaboration of the product3 (Fig. 1). To understand the biosynthesis of this hybrid isoprenoid-polyketide-derived natural product, the gene cluster responsible for naphterpin production was identified based upon proximity to genes encoding the MVA pathway (T. Kumano et al., unpublished data)
Figure 1. Biosynthesis of naphterpin and other hybrid isoprenoid-polyketide compounds produced by Actinomycetes.
The synthesis of naphterpin involves the prenylation of THN, flaviolin or a derived metabolite using a GPP co-substrate. The THN skeleton is further modified, prenylated and incorporated into hybrid isoprenoid-polyketides such as naphterpin, furaquinocin A, napyradiomycin A and marinone. Each isoprenoid C5 unit is shown in red.
An upstream region of the gene cluster containing the MVA pathway genes revealed three new open reading frames (ORFs) designated orf1, orf2 and orf3 (Supplementary Table S1). PSI-BLAST searches revealed strong homologies between Orf2 and three other bacterial proteins: a hypothetical protein from Streptomyces coelicolor A3(2) (HypSc, GenBank accession number AL939130) and the previously described 4-hydroxyphenylpyruvate: dimethylallyl transferase proteins encoded by the genes cloQ (accession number AF329398) and novQ (accession number AF170880) from Streptomyces roseochromogenes and Streptomyces spheroides NCIMB 11891, respectively5 (Supplementary Fig. S1). A recently identified aromatic PTase from Lyngbya majuscula (accession number AY588942) involved in lyngbyatoxin biosynthesis displays low similarity with Orf26. The Streptomyces sp. CL190 mutant strain resulting from disruption of the orf2 gene exhibited no naphterpin production. Moreover, the high degree of homology between Orf2 and the functionally characterized PTases CloQ and NovQ5, and the fact that Orf3 is a type III polyketide synthase with amino acid similarity to THN synthase (Supplementary Table S1)7,8, strongly suggest that orf2 encodes a PTase involved in geranyl group transfer to THN or a THN metabolite resulting from additional biosynthetic elaborations catalysed by Orf3 and possibly other tailoring enzymes.
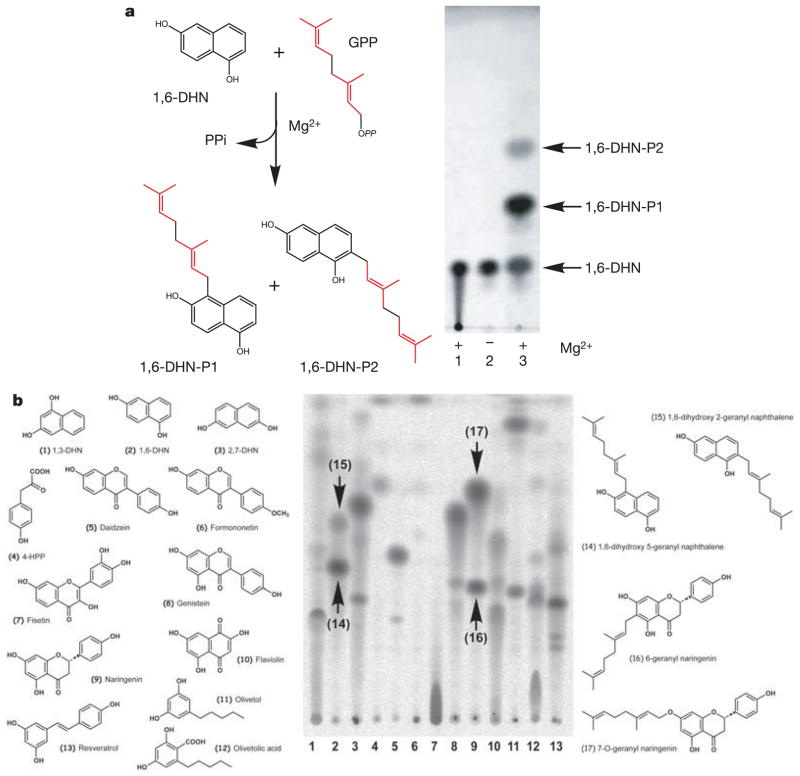
When expressed in Escherichia coli, Orf2 behaves as a soluble 33-kDa (307 residues) monomeric protein. To assess enzyme activity, purified recombinant Orf2 was incubated with putative aromatic acceptors in the presence of dimethylallyl diphosphate (DMAPP, C5), geranyl diphosphate (GPP, C10) or farnesyl diphosphate (FPP, C15). The resultant PTase activity of Orf2 displayed relaxed substrate specificity for aromatic small molecules, as prenylated products accumulate using THN analogues including 1,3-dihydroxynaphthalene (1,3-DHN), 1,6-DHN and 2,7-DHN, as well as flaviolin (Fig. 2a, b). Orf2 also catalyses the prenylation of the CloQ substrate 4-hydroxyphenylpyruvate (4-HPP) (Fig. 2b). For the isoprenoid diphosphate substrates, Orf2 exhibited no activity with DMAPP, the highest relative activity with GPP and detectable activity with FPP.
Figure 2. Enzymatic assays of Orf2.
a, Mg2+-dependent prenylation of 1,6-DHN. In lane 1 (control), Orf2 was boiled before addition. The reaction mixture analysed in lane 2 contained no MgCl2, whereas 5 mM MgCl2 was added in lane 3. b, Promiscuous activity against chemically diverse aromatic acceptors. Assays used the substrates named and numbered on the left side of the TLC. The chemical structures of four reaction products were determined by MS/NMR analyses and are shown to the right of the TLC.
Although the true physiological substrate of Orf2 is still under investigation, significant Mg2+-dependent, in vitro activity is observed with the dihydroxy-containing THN analogues (Fig. 2a, b). Notably, the formation of two prenylated products, 1,6-DHN-P1 and 1,6-DHN-P2, was readily detected by thin-layer chromatography (TLC) when Orf2 was incubated with both 1,6-DHN and GPP. Large-scale incubations with GPP and 1,6-DHN produced a sufficient amount of both products—in a 10:1 ratio—to permit their structural elucidation using both mass spectrometry (MS) and 1H nuclear magnetic resonance (NMR) analyses (see Supplementary Data). Each compound possessed a single geranyl chain at C2 and C5, resulting in trans-2-geranyl 1,6-DHN and trans-5-geranyl 1,6-DHN, respectively (Fig. 2a).
Orf2 was next assayed against various flavonoids, isoflavonoids and related plant polyketides including resveratrol, olivetol and olivetolic acid, for which significant activity was observed (Fig. 2b). In the presence of naringenin (5,7,4′-trihydroxyflavanone) and GPP, two reaction products, 6-geranyl naringenin and 7-O-geranyl naringenin, form (Fig. 2b). 6-geranyl naringenin, also known as bonannione A9, is a prenylated flavanone displaying significant antibacterial activity10. 7-O-geranyl naringenin is, to the best of our knowledge, a novel prenylated flavonoid. Orf2 also geranylated olivetol and olivetolic acid (Fig. 2b). These two plant polyketides and their geranylated products serve as intermediates in the biosynthesis of the plant-derived polyketide-terpene natural product Δ9-tetrahydrocannabinol (Δ9-THC)11.
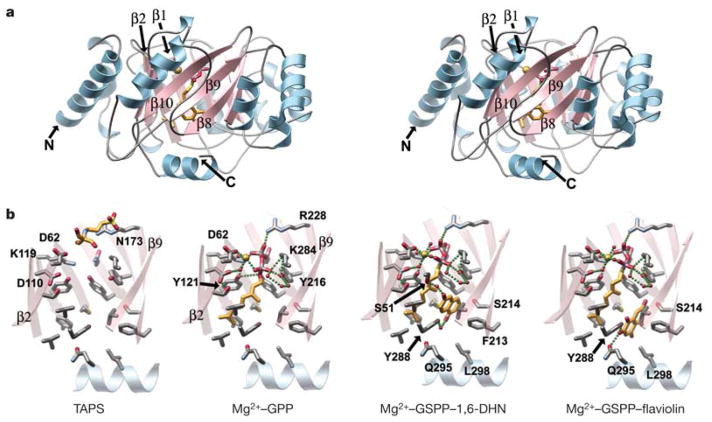
To investigate the architectural principles underlying prenyl chain-length determination, aromatic substrate selectivity and the mechanism of prenyl group transfer, we determined four X-ray crystal structures of Orf2 substrate/substrate analogue complexes, including Orf2 bound to a TAPS buffer molecule, a binary Orf2 complex containing GPP and Mg2+, a ternary Orf2 complex containing a non-hydrolysable GPP analogue, geranyl S-thiolodiphosphate (GSPP), Mg2+ and 1,6-DHN, and a ternary Orf2 complex containing GSPP, Mg2+ and flaviolin (Supplementary Table S2). The three-dimensional structure of Orf2 consists of a single domain possessing a novel barrel fold (Fig. 3). This new barrel, here termed a PT barrel, is a cylindrical β-sheet comprising ten anti-parallel β-strands arranged around a central solvent-filled core. In an arrangement reminiscent of a TIM barrel, the cylindrical β-sheet is surrounded by a ring of solvent-exposed α-helices; however, the connectivity of Orf2’s secondary structure elements is not shared with TIM barrels (Supplementary Fig. S2b).
Figure 3. Structures of Orf2’s PT barrel.
a, Stereo view of the Orf2 monomer viewed with the central barrel axis oriented vertically in the plane of the page. The cylindrical solvent-filled β-sheet and outer belt of α-helices are differentially coloured. Bound GSPP and 1,6-DHN reside within the PT barrel and are represented as colour-coded sticks. Mg2+ is shown as a gold-coloured van der Waal’s sphere. b, Orf2 substrate/substrate analogue complexes. Only the β-strands (minus β-strands 1 and 10) plus the C-terminal α-helix are displayed in the same vertical orientation as in a after a 30° anticlockwise rotation around the barrel axis. Hydrogen and coordination bonds are shown as green spheres.
The secondary connectivity nearly conforms to a (ααββ)5 classification, but is more specifically described using the (ααββ)4-(αββ)-α nomenclature due to a helical kink between α-helices 7 and 8. Searches for structurally related proteins retrieved examples belonging to either the TIM barrel or the β-barrel structural families (Supplementary Fig. S2), both of which display barrel folds with connectivity patterns that are distinctively different from the PT barrel described here12–15 (see Supplementary Discussion). A number of hydrophobic residues located inside the PT barrel sequester the geranyl tail of GPP and GSPP, whereas the diphosphate or the thio-diphosphate head groups, respectively, point towards the ‘upper’, more polar end of the barrel where a Mg2+ ion is coordinated.
Whereas Orf2 and its structural homologue CloQ display considerable aromatic PTase activities, the primary sequence and tertiary structure of Orf2 are not shared with any known PTase or mechanistically related biosynthetic enzymes16–22. Moreover, the sequence does not contain the (N/D)DXXD signature motif indicative of Mg2+-dependent isoprenoid diphosphate recognition. Even within the Orf2 family of PTases, some members such as CloQ exhibit activity in the absence of Mg2+ ions5, whereas Orf2’s PTase activity is Mg2+ dependent (Fig. 2a). Complexes with Mg2+ and GPP, or a non-hydrolysable analogue, GSPP, precisely define the binding of GPP and the coordination of Mg2+ (Fig. 3b). Lys 119, Asn 173 and Arg 228, located near the polar open end of the barrel, tether the β-phosphate of GSPP (Fig. 4a). The α-phosphate linked to the geranyl chain forms a hydrogen bond with Tyr 216 and Lys 284, and also coordinates a Mg2+ ion. The bound Mg2+ exhibits octahedral coordination, with four equatorially arranged water molecules and two axially located oxygen atoms contributed by the side chain carboxylate of Asp 62 and a non-bridging oxygen of the aforementioned α-phosphate (Fig. 4a, b). Tyr 121 resides within hydrogen-bonding distance of the bridging atom (sulphur in GSPP and oxygen in GPP) linking the diphosphate moiety to the C10 geranyl chain. Finally, the hydrophobic geranyl chains of the GPP or GSPP molecules rest against the side chains of Val 49, Phe 123, Met 162, Tyr 175 and Tyr 216 (Fig. 4b).
Figure 4. Mg2+ dependence and substrate recognition in aromatic PTases.
a, Representative 2Fo − Fc electron density map, contoured at 1.0σ, displaying the octahedral coordination of the catalytic Mg2+. The orientation is the same as that in Figs 3b and 4b after a 180° rotation around the vertical barrel axis. b, Schematic representation of Orf2’s active site. The side chains involved in Mg2+, GSPP and 1,6-DHN binding are depicted with direct hydrogen and coordination bonds shown as green dashes and water-mediated bonds as blue dashes. Half circles depict van der Waal’s contacts with grey for residues in the back of the GSPP–1,6-DHN plane, black in the same plane and thick black in the front. c, Biochemical characterization of HypSc. PTase activity was assayed and visualized as in Fig. 2a using 1,6-DHN as a prenyl acceptor and either DMAPP or GPP as potential prenyl donors.
The ternary complexes with Mg2+, GSPP and either 1,6-DHN or flaviolin delineate the surprisingly spacious aromatic substrate binding pocket (Figs 3b and 4b), partially explaining Orf2’s relaxed specificity for aromatic small-molecule substrates. 1,6-DHN rests against the geranyl tail of GSPP, and the bicyclic aromatic rings are sequestered between the side chains of Met 162 and Phe 213. The side chains of Gln 295 and Leu 298, provided by the short carboxy-terminal helical cap of the PT barrel, line the wall of the aromatic substrate binding pocket, with additional contacts made through the side chains of Phe 213, Ser 214 and Tyr 288 (Fig. 3b). Although the aromatic planes of both 1,6-DHN and flaviolin reside in the same active site orientation, the flaviolin molecule binds in a slightly different position than 1,6-DHN, with extra pairs of hydrogen bonds formed with Ser 214, Tyr 288 and Gln 295 (Fig. 3b).
At least two distinct catalytic mechanisms can be considered for prenylation of aromatic substrates. One reaction invokes carbon-mediated nucleophilic attack on C1 of GPP, with the diphosphate moiety stabilized by Mg2+ coordination and the basic character of the diphosphate binding site serving as a leaving group. This SN2-like mechanism has been described for protein farnesyltransferase (PFTase)17. A second mechanism invokes carbocation-mediated electrophilic capture, as proposed for the trans-prenyltransferase reaction of FPP synthase21 and numerous terpene synthases (cyclases) of secondary metabolism22 (see Supplementary Discussion). The distance between the site of C-prenylation on 1,6-DHN and C1 of GSPP is 4 Å. Because C1 of GPP is spatially constrained by covalent attachment to the electrostatically coordinated diphosphate moiety, PTase-catalysed cleavage of the carbon–oxygen bond before prenylation of the aromatic acceptor is more consistent with an electrophilic aromatic substitution reaction in Orf2 (Supplementary Fig. S3). Ongoing kinetic studies using 2-fluoro-substituted GPP (2F-GPP) will provide an experimental means to ascertain the most likely mechanism.
In order to decipher prenyl diphosphate chain-length selectivity, aromatic substrate recognition and divalent cation dependence, homology modelling of CloQ, NovQ and HypSc was carried out using Orf2 as a structural template (Supplementary Fig. S4). Asp 62, involved in the diphosphate binding site of Orf2 via a coordinated Mg2+, is replaced in HypSc by Asn 63 and in CloQ and NovQ by Ser 64. As a possible explanation for the Mg2+-independent PTase activity of CloQ and NovQ, Ser 51 in Orf2 is replaced by Lys 54 in CloQ and NovQ, and Arg 51 in HypSc. These basic residues can be positioned over the Mg2+ observed in the Orf2 active site. These notable active site differences in CloQ/NovQ and HypSc possibly serve as catalytic surrogates for Mg2+-mediated α-phosphate binding and catalysis, supporting the Mg2+-independent activity previously observed for CloQ and NovQ (ref. 5), and suggestive of a Mg2+-independent mechanism in HypSc. Also, in both the CloQ/NovQ and HypSc models, residues equivalent to Ser 64 and Gly 286 in Orf2 are replaced by Arg and Glu residues, respectively (Supplementary Fig. S4). The presence of putative salt bridges across these two positions in the prenyl diphosphate binding sites of CloQ, NovQ and HypSc appear to prevent the binding of a prenyl donor with an isoprenoid chain longer than dimethylallyl, again supporting the observed selectivity of CloQ and NovQ for the five-carbon DMAPP substrate5.
These models not only explain the chain-length selectivity and Mg2+-independent catalytic activity measured for CloQ and NovQ but suggest that HypSc should exhibit DMAPP specificity and Mg2+-independent PTase activity as well. To test these hypotheses, HypSc was overexpressed in E. coli as an octahistidine-tagged protein and assayed either in the absence or presence of MgCl2 using DMAPP and GPP as prenyl donors and 1,6-DHN as an aromatic acceptor. Measurable PTase activity was detected when using DMAPP and 1,6-DHN as substrates in the absence or presence of Mg2+, supporting the chain-length selectivity and Mg2+-independent catalytic activity of HypSc first suggested from its homology model described above (Fig. 4c).
The present work describes the identification of two novel aromatic prenyltransferases: Orf2 from Streptomyces sp. CL190 and HypSc from Streptomyces coelicolor A3(2), each possessing PTase activity against aromatic small molecule substrates. Owing to Orf2’s relaxed specificity for aromatic substrates, it can be used as an efficient biological catalyst for the regio-specific prenylation of aromatic small molecules. Moreover, the experimental crystal structures of Orf2 and the homology models of phylogenetically related PTases serve as starting points for rationally engineering the in vitro and in vivo prenylation of natural products of both microbial and plant origin.
METHODS
Protein expression and purification
The orf2 gene from Streptomyces sp. strain CL190 (GenBank accession AB187169) was amplified by polymerase chain reaction (PCR) from total genomic DNA using oligonucleotides designed for ligation into the E. coli expression vector pQE30 (Qiagen), generating pQEORF2. PCR amplification using pQEORF2 and oligonucleotides for ligation into the E. coli expression vector pHIS8 (ref. 23) was carried out with the forward primer 5′-GGGGGGGGATCCTCCGAAGCCGCTGATGTCG-3′ (BamHI site underlined) and the reverse primer 5′-GGGGGGGAATTCTCAGTCCTCCAGC GAGTCG-3′ (EcoRI site underlined) to generate pHIS8ORF2. Constructs of pHIS8ORF2 were transformed into E. coli BL21 (DE3). Recombinant Orf2 protein was obtained and purified using a protocol previously described23. Seleno-methionine (Se-Met)-substituted protein was obtained from E. coli grown in M9 minimal medium using the methionine pathway inhibition approach24, and purified as described for the native protein. HypSc (GenBank accession number AB187170) was subcloned from Streptomyces coelicolor A3(2) genomic DNA using the forward primer 5′-GGGGGGCCATGGCCCTACTGG TAGAACGACGG-3 ′ (NcoI site underlined) and the reverse primer 5 ′-GGGGGGGGATCCTCAGCGGTTCCAGTAGCCG-3′ (BamHI site underlined), inserted into the NcoI and BamHI sites of pHIS8, overexpressed in E. coli as an octahistidine-tagged protein and purified by Ni2+-chelation chromatography and gel filtration chromatography, as described23.
Crystallization and data collection
Crystals were obtained by vapour diffusion at 4 °C. Two-microlitre hanging drops containing a 1:1 mixture of 15 mg ml−1 protein with crystallization buffer (28% (w/v) PEG 4000, 0.3 M magnesium nitrate, 2 mM DL-dithiothreitol (DTT), 0.1 M TAPS, pH 8.5) equilibrated over a 500μl reservoir of the same solution produced small diffracting crystals overnight. Larger crystals were obtained by macro-seeding into the same conditions. Crystals were stabilized by soaking briefly in a cryoprotectant solution (30% (w/v) PEG 4000, 15% (v/v) glycerol, 0.3 M magnesium nitrate, 2 mM DTT, 0.1 M TAPS, pH 8.5), and flash frozen in liquid nitrogen before data collection. Orf2 crystals belong to the P21212 space group with average unit cell dimensions of a = 71 Å, b = 92 Å, c = 48 Å, α = β = γ = 90°, contain one monomer per asymmetric unit and have a solvent content of 45%. Se-Met-substituted crystals were obtained as described for native crystals. Substrate and substrate analogue complexes were obtained by soaking Orf2 crystals in stabilization solutions containing 5 mM GPP, or 10 mM GSPP and 40 mM 1,6-DHN, or 10 mM GSPP and 10 mM flaviolin (GPP and GSPP were purchased from Echelon Biosciences).
Structure determination and refinement
A multi-wavelength anomalous dispersion (MAD) data set was collected at the selenium edge on a Se-Met Orf2 crystal at the Brookhaven National Laboratory (BNL) on beamline X8C. Data were processed with HKL2000 (ref. 25) and reduced to a unique set of indexed intensities to a resolution of 1.6 Å. Single-wavelength data sets were collected at the Salk Institute for Biological Studies, BNL (beamline X6A), the European Synchrotron Radiation Facility (ESRF, beamline FIP/BM30A) and the Stanford Synchrotron Radiation Laboratory (SSRL, beamline 9.1) on substrate and substrate analogue complexes (Supplementary Table S2). Phasing, density modification and automatic model building were carried out with the program suite Solve/Resolve26 using seven identified Se sites. Additional rounds of building and refinement were carried out with O27 and CNS28, respectively. Subsequent complexes were solved by molecular replacement with AMoRe29.
Prenylation assays and product identification
The reaction conditions for prenylation of 1,6-DHN using either Orf2 or HypSc consisted of 50 mM HEPES (pH 7.5), either no MgCl2 (minus) or 5 mM MgCl2 (plus), 5 mM 1,6-DHN, and 5 mM DMAPP, GPP or FPP in a final volume of 20μl. Reactions were initiated with 20μg of Orf2 or 20μg HypSc. After incubation at 25 °C for 4 h (Orf2) or overnight (HypSc), reactions were dried and spotted on a silica gel TLC plate. TLC plates were developed with a chloroform/methanol (20:1) solvent mixture. 1,6-DHN and reaction products were detected at 254 nm. The chemical analyses of the two HPLC-purified products obtained from Orf2 incubations were accomplished by MS and 1H NMR (Supplementary Data). Reactions measuring the activity of Orf2 against various synthetic and natural aromatic molecules (see Fig. 2b) consisted of 50 mM HEPES (pH 7.5), 5 mM MgCl2, 0.1 mM GPP, 0.009 mM [14C]GPP, 0.1 mM each aromatic substrate, 30μg of Orf2 in a final volume of 20μl. After incubation at 25 °C for 6 h, the mixtures were dried, spotted on a silica gel TLC plate and developed with a chloroform/methanol (15:1) solvent mixture. Products were detected using a [14C] imaging plate (Fuji Photo Film).
Database searches
Searches were performed with PSI-BLAST and VAST (http://www.ncbi.nlm.nih.gov), SSM and DALI (http://www.ebi.ac.uk/msd-srv/ssm), CE (http://cl.sdsc.edu/ce.html) and DEJAVU (http://portray.bmc.uu.se/), available through the Protein Data Bank (http://www.rcsb.org/pdb/), the Structural Classification of Proteins (SCOP; http://scop.mrc-lmb.cam.ac.uk/scop), and the CATH Protein structure classification (http://www.biochem.ucl.ac.uk/bsm/cath) websites.
Supplementary Material
Acknowledgments
We thank F. Taura, S. Morimoto and Y. Shoyama for providing olivetol and olivetolic acid, as well as N. Funa and S. Horinouchi for the flaviolin sample. We also thank the staff of beamlines X8C and X6A at BNL, BL9.1 at SSRL and BM30A at ESRF. Work performed at SSRL was supported by grants from the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program and the Department of Energy, Office of Biological and Environmental Research. This work was supported by grants from the National Institutes of Health (J.P.N) and a Grant-in-Aid of Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (T.K.).
Footnotes
Author Contributions T.K., J.P.N. and S.B.R. conceived and designed the experiments; T.K. and S.B.R. performed the experiments; T.K., J.P.N. and S.B.R. analysed the data, provided analysis tools and contributed materials; and T.K., J.P.N. and S.B.R. wrote the paper.
Author Information The atomic coordinates and structure factors of Orf2 in complex with TAPS, Mg2+–GPP, Mg2+–GSPP–1,6-DHN and Mg2+–GSPP–flaviolin have been deposited in the Protein Data Bank under accession codes 1ZDY, 1ZCW, 1ZB6 and 1ZDW, respectively. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Shinya K, et al. Isolation and structural elucidation of an antioxidative agent, naphterpin. J Antibiot (Tokyo) 1990;43:444–447. doi: 10.7164/antibiotics.43.444. [DOI] [PubMed] [Google Scholar]
- 2.Shinya K, et al. Biosynthetic studies of naphterpin, a terpenoid metabolite of. Streptomyces Tetrahedr Lett. 1990;31:6025–6026. [Google Scholar]
- 3.Seto H, Watanabe H, Furihata K. Simultaneous operation of the mevalonate and non-mevalonate pathways in the biosynthesis of isopentenyl diphosphate in Streptomyces aeriouvifer. Tetrahedr Lett. 1996;37:7979–7982. [Google Scholar]
- 4.Botta B, et al. Prenylated flavonoids: pharmacology and biotechnology. Curr Med Chem. 2005;12:717–739. doi: 10.2174/0929867053202241. [DOI] [PubMed] [Google Scholar]
- 5.Pojer F, et al. CloQ, a prenyltransferase involved in clorobiocin biosynthesis. Proc Natl Acad Sci USA. 2003;100:2316–2321. doi: 10.1073/pnas.0337708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards DJ, Gerwick WH. Lyngbyatoxin biosynthesis: sequence of biosynthetic gene cluster and identification of a novel aromatic prenyltransferase. J Am Chem Soc. 2004;126:11432–11433. doi: 10.1021/ja047876g. [DOI] [PubMed] [Google Scholar]
- 7.Izumikawa M, et al. Expression and characterization of the type III polyketide synthase 1,3,6,8-tetrahydroxynaphthalene synthase from Streptomyces coelicolor A3(2) J Ind Microbiol Biotechnol. 2003;30:510–515. doi: 10.1007/s10295-003-0075-8. [DOI] [PubMed] [Google Scholar]
- 8.Funa N, et al. A new pathway for polyketide synthesis in microorganisms. Nature. 1999;400:897–899. doi: 10.1038/23748. [DOI] [PubMed] [Google Scholar]
- 9.Bruno M, et al. New flavonoids from Bonannia-Graeca (L) halacsy. Heterocycles. 1985;23:1147–1153. [Google Scholar]
- 10.Schutz BA, et al. Prenylated flavanones from leaves of Macaranga pleiostemona. Phytochemistry. 1995;40:1273–1277. [Google Scholar]
- 11.Taura F, Morimoto S, Shoyama Y. Purification and characterization of cannabidiolic-acid synthase from Cannabis sativa L. Biochemical analysis of a novel enzyme that catalyzes the oxidocyclization of cannabigerolic acid to cannabidiolic acid. J Biol Chem. 1996;271:17411–17416. doi: 10.1074/jbc.271.29.17411. [DOI] [PubMed] [Google Scholar]
- 12.Gerlt JA, Raushel FM. Evolution of function in β/α8-barrel enzymes. Curr Opin Chem Biol. 2003;7:252–264. doi: 10.1016/s1367-5931(03)00019-x. [DOI] [PubMed] [Google Scholar]
- 13.Sacchettini JC, Gordon JI, Banaszak LJ. Crystal structure of rat intestinal fatty-acid-binding protein. Refinement and analysis of the Escherichia coli-derived protein with bound palmitate. J Mol Biol. 1989;208:327–339. doi: 10.1016/0022-2836(89)90392-6. [DOI] [PubMed] [Google Scholar]
- 14.Xu Z, Bernlohr DA, Banaszak LJ. The adipocyte lipid-binding protein at 1.6 Å resolution. Crystal structures of the apoprotein and with bound saturated and unsaturated fatty acids. J Biol Chem. 1993;268:7874–7884. [PubMed] [Google Scholar]
- 15.Branden CI, Tooze J. Introduction to Protein Structure. 2. Garland; New York: 1999. [Google Scholar]
- 16.Park HW, et al. Crystal structure of protein farnesyltransferase at 2.25 angstrom resolution. Science. 1997;275:1800–1804. doi: 10.1126/science.275.5307.1800. [DOI] [PubMed] [Google Scholar]
- 17.Long SB, Casey PJ, Beese LS. Reaction path of protein farnesyltransferase at atomic resolution. Nature. 2002;419:645–650. doi: 10.1038/nature00986. [DOI] [PubMed] [Google Scholar]
- 18.Koyama T, et al. Identification of significant residues in the substrate binding site of Bacillus stearothermophilus farnesyl diphosphate synthase. Biochemistry. 1996;35:9533–9538. doi: 10.1021/bi960137v. [DOI] [PubMed] [Google Scholar]
- 19.Kharel Y, Koyama T. Molecular analysis of cis-prenyl chain elongating enzymes. Nat Prod Rep. 2003;20:111–118. doi: 10.1039/b108934j. [DOI] [PubMed] [Google Scholar]
- 20.Liang PH, Ko TP, Wang AH. Structure, mechanism and function of prenyltransferases. Eur J Biochem. 2002;269:3339–3354. doi: 10.1046/j.1432-1033.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 21.Tarshis LC, et al. Crystal structure of recombinant farnesyl diphosphate synthase at 2.6- Å resolution. Biochemistry. 1994;33:10871–10877. doi: 10.1021/bi00202a004. [DOI] [PubMed] [Google Scholar]
- 22.Cane DE, editor. Comprehensive Natural Products Chemistry. Elsevier; London: 1998. Isoprenoids, Including Carotenoids and Steroids. [Google Scholar]
- 23.Jez JM, et al. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry. 2000;39:890–902. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- 24.Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger TC. Automated structure solution, density modification and model building. Acta Crystallogr D. 2002;58:1937–1940. doi: 10.1107/s0907444902016438. [DOI] [PubMed] [Google Scholar]
- 27.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr D. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.