Abstract
A regulatory network of Sinorhizobium meliloti genes involved in adaptation to iron-limiting conditions and the involvement of the rhizobial iron regulator gene (rirA) were analyzed by mutation and microarray analyses. A constructed S. meliloti rirA mutant exhibited growth defects and enhanced H2O2 sensitivity in the presence of iron, but symbiotic nitrogen fixation was not affected. To identify iron-responsive and RirA-regulated S. meliloti genes, a transcriptome approach using whole-genome microarrays was used. Altogether, 45 genes were found to be jointly derepressed by mutation of rirA and under different iron-limited conditions. As expected, a number of genes involved in iron transport (e.g., hmuPSTU, shmR, rhbABCDEF, rhtX, and rhtA) and also genes with predicted functions in energy metabolism (e.g., fixN3, fixP3, and qxtAB) and exopolysaccharide production (e.g., exoY and exoN) were found in this group of genes. In addition, the iron deficiency response of S. meliloti also involved rirA-independent expression changes, including repression of the S. meliloti flagellar regulon. Finally, the RirA modulon also includes genes that are not iron responsive, including a gene cluster putatively involved in Fe-S cluster formation (sufA, sufS, sufD, sufC, and sufB).
Iron is an essential micronutrient for almost all known organisms. Due to the ready interconversion between the reduced Fe2+ ferrous form and the oxidized Fe3+ ferric form, iron is a versatile component for incorporation as a biocatalyst or electron carrier into proteins that are involved in a number of essential metabolic and enzymatic functions. In particular, rhizobia, a diverse group of symbiotic soil bacteria belonging to the genera Rhizobium, Bradyrhizobium, Mesorhizobium, Azorhizobium, and Sinorhizobium, have a special demand for iron. These so-called root nodule bacteria are known for their ability to establish nitrogen-fixing symbioses with their legume hosts. During this symbiotic interaction root nodules are formed, in which the bacteria fix atmospheric nitrogen after differentiation into bacteroids (61). Many enzymes necessary for the reduction of atmospheric nitrogen (e.g., nitrogenase and nitrogenase reductase) contain iron compounds as cofactors. In addition, the bacteroids have a very high respiratory demand, requiring an abundance of cytochromes and other ferro-proteins (14). Thus, rhizobia not only need to compete successfully with other soil organisms for iron in order to propagate in the free-living state but also need to satisfy their high iron demand during symbiosis.
Although iron is abundant in the Earth's soil, under physiological conditions it is mainly present as insoluble ferric iron. Accordingly, bacteria have evolved a number of transport systems in order to ensure a sufficient supply of iron. A common mechanism employed by most bacteria is the synthesis and secretion of siderophores, which are low-molecular-weight ligands that specifically bind ferric iron with high affinity. The ferri-siderophore complexes are then transported into the cell via cognate transporters. A number of siderophores have been characterized in different rhizobia; these include the trihydroxamate vicibactin of Rhizobium leguminosarum (10, 11), the amino polycarboxylic acid siderophore of Sinorhizobium meliloti DM4 (59), and the citrate-based dihydroxamate rhizobactin 1021 produced by S. meliloti 1021 (46). In addition to their own or exogenous ferri-siderophores, rhizobia are also known to use a wide range of other iron sources, including ferric citrate, heme, and hemoglobin (3, 11, 39, 71). This flexibility with regard to iron utilization probably reflects adaptation to complex environmental conditions and underlines the importance of iron acquisition for rhizobia.
Despite the importance of iron, high concentrations of this metal can lead to generation of hydroxyl radicals as a result of the Fenton chemistry. Therefore, iron uptake is usually strictly regulated. In many gram-negative bacteria and some gram-positive bacteria the ferric uptake regulator (Fur) has been established as a central regulator of iron-responsive genes (1, 17, 22, 23). In the sequenced genome of S. meliloti 1021, the microsymbiont of alfalfa, a Fur homologue is encoded (9, 19), but recently we demonstrated that this protein is primarily involved in the regulation of the Mn2+ transporter operon sitABCD (12), a finding that was verified independently by Platero et al. (47). A similar role of Fur was also found in R. leguminosarum (15, 70). It is therefore obvious that in S. meliloti and R. leguminosarum iron-dependent regulation is implemented very differently than it is in other gram-negative bacteria. In R. leguminosarum, Todd and colleagues have identified a regulator termed RirA (rhizobial iron regulator) which has been shown to be involved in the regulation of several iron-responsive genes (64). Additionally, proteome analyses indicated that over 100 proteins were differentially expressed in an R. leguminosarum rirA mutant, and 10 of these proteins were identified by mass spectrometry (65).
The R. leguminosarum RirA protein does not exhibit significant sequence homology to known iron-responsive regulators, such as Fur, DtxR (7), or Irr (21), and belongs to the Rrf2 family of putative transcription regulators. Other members of this protein family include NsrR, which is involved in regulating a nitrite reductase in Nitrosomonas europaea (4), Rrf2, a regulator of cytochrome synthesis in Desulfovibrio vulgaris (30), and IscR, an iron-sulfur protein involved in the regulation of Fe-S cluster formation (54). Hence, the members of this family of regulators appear to have very diverse functions in different eubacterial species.
There is an rirA homologue in S. meliloti (64), but its role in this symbiont is still unknown. In this study we analyzed the relevance of the S. meliloti rirA gene for controlling iron homeostasis during free-living growth and symbiosis. As the S. meliloti rirA gene is a likely candidate for a global iron-dependent regulator, whole-genome microarrays were utilized to identify the extent of RirA-regulated genes. These experiments were complemented with transcriptome profiling of changes in global iron-dependent expression in order to identify the mechanism with which S. meliloti adapts to iron-limiting conditions and to define the relevance of rirA for coordinating the iron deficiency response.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are shown in Table 1. Strains of Escherichia coli were routinely cultured at 37°C in antibiotic medium no. 3 (Oxoid, Wesel, Germany). S. meliloti strains were cultivated at 30°C either in tryptone yeast (TY) complex medium (6) or in Vincent minimal medium (VMM) (67). VMM was prepared without iron. A final FeCl3 concentration of 37 μM was used for iron-sufficient growth in VMM, and an FeCl3 concentration of 0.37 μM was used for iron-deficient growth in VMM. Other iron sources and the concentrations used are indicated below. Iron-limited TY medium was prepared by adding the iron chelator 2,2′-dipyridyl to a final concentration 200 μM. When appropriate, antibiotics were added at the following concentrations: neomycin, 100 μg ml−1; kanamycin, 50 μg ml−1; and streptomycin, 600 μg ml−1. Glassware was washed with 50 mM EDTA and 6 M HCl before it was thoroughly rinsed with water.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | Reference |
|---|---|---|
| Sinorhizobium meliloti strains | ||
| Rm1021 | Spontaneous mutant of wild-type strain RU47, Smr | 35 |
| Rm1021-TR2 | Rm1021 derivative, ΔrirA | This study |
| Rm1021-TR2-3 | Rm1021 derivative, ΔrirA ΔrhrA | This study |
| Escherichia coli strains | ||
| DH5αMCR | F−endA1 supE44 thi-1 λ−recA1 gyrA96 relA1 deoR Δ(lacZYA-argF)U169 φ80dlacZΔM15 mcrA Δ(mrr hsdRMS mcrBC) | 20 |
| S17-1 | E. coli 294::[RP4-2(Tc::Mu)(Km::Tn7)] pro res ΔrecA Tpr | 57 |
| Plasmid | ||
| pK18mobsacB | pUC18 derivative, sacB lacZα Kmr, mobilizable | 53 |
DNA manipulations.
The protocols of Sambrook et al. (52) were used for routine manipulations of plasmid and chromosomal DNA. Mutated DNA fragments containing either a 342-bp deletion in the rirA gene or a 810-bp deletion in the rhrA gene were constructed by gene SOEing (27). In a first PCR regions up- and downstream of the desired deletion were amplified, and then they were fused in a second PCR. The deletion constructs obtained were subsequently cloned into the suicide vector pK18mobsacB, which allows sucrose selection for vector loss (53). The resulting plasmids were conjugated into S. meliloti via E. coli S17-1 to introduce deletions by allelic exchange. Mutants were verified by PCR and Southern hybridization.
CAS siderophore assay.
Chrome azurol S (CAS) assay mixtures for siderophore detection were prepared as described by Schwyn and Neilands (55). Supernatants of S. meliloti cultures grown in VMM containing various concentrations of FeCl3 were mixed 1:1 with a CAS assay solution. After equilibrium was reached, the absorbance at 630 nm was measured. The relative siderophore activity was determined by measuring optical density ratios of different cultures.
H2O2 challenge.
To test for H2O2 sensitivity, overnight cultures of S. meliloti wild-type strain Rm1021 and rirA mutant Rm1021-TR-2 were washed, and equal amounts of the two strains were incubated for 4 h in iron-free VMM and VMM containing 60 μM FeCl3, respectively. After this, the samples were exposed to 50 mM H2O2 for 2 h. The survival rate was determined by comparing the number of CFU for H2O2-treated samples to the number of CFU for untreated control samples after incubation for 2 days on TY agar plates at 30°C.
Assays to determine nitrogen fixation efficiency in the S. meliloti-Medicago sativa symbiosis.
Nodulation tests were performed as described by Rolfe et al. (50). Solutions a to c for the plant agar plates (solution a contained 294 g/liter CaCl2 · 2H2O; solution b contained 50 g/liter KH2PO4; solution c contained 123 g/liter MgSO4, 87 g/liter K2SO4, 0.247 g/liter H3BO3, 0.288 g/liter ZnSO4, 0.1 g/liter CuSO4 · 5H2O, 0.056 g/liter CoSO4 · 7H2O, and 0.048 g/liter NaMo4 · 2H2O) were autoclaved separately. Then 0.5 ml of each solution was added to 1 liter autoclaved water containing 15 g agar. FeSO4 was added as an iron source at final concentrations of 5, 10, and 15 μM to the medium. Alfalfa (M. sativa L. cv. europe) seeds were sterilized with 32% HCl for 30 min and then washed with sterile water. After germination, seedlings were placed on nodulation plates and inoculated with equal amounts (6 × 107 CFU) of washed wild-type and mutant cells. Plants were weighed after 30 days of growth, and the nitrogen fixation activity was tested by the acetylene reduction assay (50).
S. meliloti transcript profiling using the genome-wide SM6kOligo microarray.
For identification of the RirA modulon, S. meliloti wild-type strain Rm1021 and rirA mutant Rm1021-TR2 were cultivated in VMM containing 37 μM FeCl3. For transcriptome profiling of iron-regulated genes, the S. meliloti wild-type Rm1021 strain was grown in either TY medium (iron-sufficient complex medium), TY medium containing 200 μM 2,2′-dipyridyl (iron-limited complex medium), VMM containing 37 μM FeCl3 (iron-sufficient minimal medium), or VMM containing 0.37 μM FeCl3 (iron-limited minimal medium). All cultures were incubated in 250-ml Erlenmeyer flasks with shaking at 150 rpm at 30°C until the optical density at 580 nm was 0.9 before they were harvested. Cells were centrifuged (10,000 × g, 1 min, 4°C) and then immediately frozen in liquid nitrogen. For total-RNA isolation an RNeasy mini kit (QIAGEN, Hildesheim, Germany) was used. Cells were disrupted in the RLT buffer provided in the kit in Fast Protein tubes (Qbiogene, Carlsbad, CA) using a Ribolyser (30 s; speed, 6.5; Hybaid, Heidelberg, Germany) before RNA isolation using the RNeasy mini kit RNA purification protocol. Fluorescent labeling of cDNA by amino-allyl dye coupling was performed as described by de Risi (http://www.microarrays.org/protocols.html). For this study the Sm6kOligo microarrays described by Krol and Becker (32) were used. Hybridization and image acquisition of the microarrays were performed as described previously (5, 51). For acquisition of the mean signal and mean local background intensity for each spot of the microarray, the ImaGene 5.0 software (Biodiscovery Inc., Los Angeles, CA) for spot detection, image segmentation, and signal quantification was used. The log2 value of the intensity ratios (Mi) was calculated for each spot as follows: Mi = log2(Ri/Gi), where Ri = Ich1i − Bgch1i and Gi = Ich2i − Bgch2i (Ich1i and Ich2i are the intensities of spots in channels l and 2, respectively, and Bgch1i and Bgch2i are the background intensities of spots in channels 1 and 2, respectively). The mean intensity (Ai) was calculated for each spot as follows: Ai = log2(RiGi)0.5. A normalization method based on local regression that accounts for intensity spatial dependence in dye biases was used (73). Normalization and t-statistics were carried out using the Emma 1.1 microarray data analysis software (16). The expression of a gene was considered significantly different if the P value was ≤0.05, the log2 ratio of the intensities (M value) was ≥1 or ≤−1, and the mean intensity (A value) was ≥7. The microarray results were verified for specific genes (rhbA, shmR, and hmuS) by quantitative reverse transcription-PCR using a QuantiTect SYBR Green reverse transcription-PCR kit (QIAGEN, Hildesheim, Germany) according to the manufacturer's instructions. The measurements were obtained with a LightCycler instrument (Roche, Mannheim, Germany). The rhbA and shmR genes were chosen since previous reports showed that there was iron-responsive regulation of these genes (3, 34). Iron-dependent regulation of the hmuS genes was confirmed by glucuronidase assays (data not shown).
RESULTS AND DISCUSSION
Mutational analysis of the S. meliloti rirA gene suggested that it is involved in the regulation of iron metabolism.
One of the objectives of this work was to elucidate iron-dependent regulation in S. meliloti. As we demonstrated previously that the S. meliloti fur gene is not involved in iron-dependent regulation, we focused on the rhizobial iron regulator gene rirA. The rirA gene was initially identified in R. leguminosarum as a possible regulator of iron-responsive genes (64). In S. meliloti a gene (SMc00785) homologous to the R. leguminosarum rirA gene was identified, which has been annotated to encode a hypothetical protein. The deduced protein had a predicted molecular mass of 17 kDa and exhibited very high levels of homology to the RirA repressor of R. leguminosarum (84% identity, 93% positive). Consequently, we renamed the corresponding S. meliloti open reading frame (SMc00785) rirA. Other close homologues of the S. meliloti RirA protein occur in the closely related plant pathogen Agrobacterium tumefaciens, Mesorhizobium loti, and different Brucella and Bartonella species. In none of these proteins could a significant helix-turn-helix motif, which is typical of many bacterial transcriptional regulators, be identified by bioinformatic approaches. The S. meliloti rirA gene is flanked downstream by genes encoding a putative dipeptide transporter (dppA1-F1) and upstream by a gene coding a putative periplasmic iron binding protein (SMc00784).
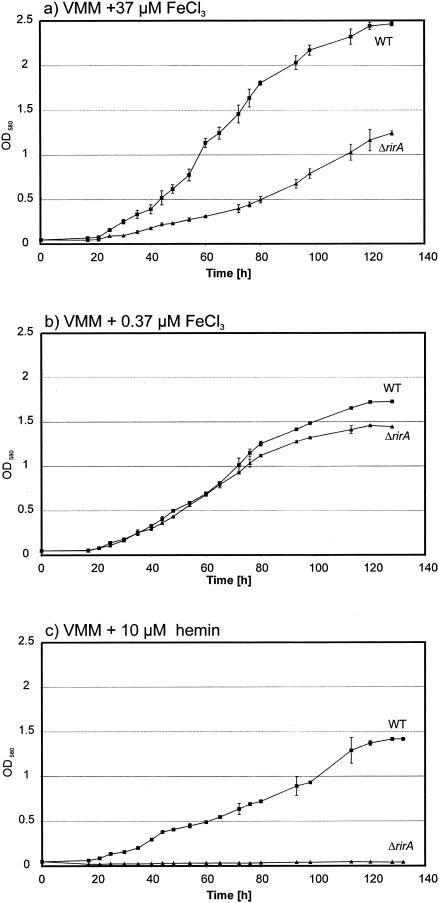
In order to assess the role of the S. meliloti rirA gene in regulation of global iron metabolism, a marker-free S. meliloti rirA deletion mutant designated Rm1021-TR2 (Table 1) was constructed and used for further studies. As we hypothesized that the rirA gene might be the central regulator of iron metabolism, we tested the influence of iron status on the growth of the rirA mutant. To do this, S. meliloti wild-type strain Rm1021 and rirA mutant Rm1021-TR2 were grown in VMM containing different iron sources. In medium containing 37 μM FeCl3 as the sole iron source, the rirA mutant exhibited a reduced growth rate (Fig. 1a). This phenotype was partially recovered in minimal medium containing only 0.37 μM FeCl3 (Fig. 1b). This reduction in growth of the rirA mutant was apparently connected to iron availability and may have been caused by oxidative stress due to derepressed iron uptake. With hemin as the iron source, virtually no growth of the rirA mutant was observed (Fig. 1c). Supplementation with other iron sources did not restore growth if hemin was present (data not shown), indicating that deregulated accumulation of this potentially toxic compound (63), rather than the inability of the rirA mutant strain to utilize hemin as an iron source, was responsible for the growth defects.
FIG. 1.
Effects of the rirA mutation on growth rates of S. meliloti strains. Overnight cultures of S. meliloti wild-type strain Rm1021 (WT) and rirA mutant Rm1021-TR2 (ΔrirA) were washed and diluted in fresh minimal medium containing 37 μM FeCl3 (a), 0.37 μM FeCl3 (b), or 10 μM hemin (c) as the sole iron source. Samples were taken, and their optical densities at 580 nm (OD580) were determined. The error bars indicate the standard deviations calculated from six independent cultures.
Further evidence for the involvement of the rirA gene in regulating iron acquisition was obtained by semiquantitative liquid CAS assays in which the siderophore concentrations in supernatants of the rirA mutant were compared to those in wild-type cultures. When cultivated under low-iron conditions (no FeCl3), both strains exhibited siderophore production, but the amount in the rirA mutant was ∼125-fold higher. Addition of 37 μM FeCl3 to the culture medium abolished siderophore production in the wild type but did not affect siderophore accumulation in the rirA mutant (data not shown). Thus, the sensitivity of the rirA mutant to high iron concentrations might be at least partially caused by derepression of siderophore-dependent iron uptake mechanisms.
Since oxidative stress caused by the Fenton reaction in the presence of iron can be enhanced by low doses of H2O2, we tested the effect of the rirA mutation on H2O2 sensitivity in the presence of FeCl3 to ascertain whether the growth deficits of the rirA mutant were a result of enhanced oxidative stress. When preincubated in medium containing no added iron sources before challenge with H2O2, the rirA mutant showed only a slightly lower survival ratio (46% ± 3%) than the wild type (58% ± 4%). However, the survival ratio of rirA mutant cells which were preincubated in iron-containing medium was reduced to only 4% ± 0.5%, whereas the viability of the wild type remained largely unchanged (40% ± 2%).
In conclusion, the bioassays indicated that a mutation in rirA leads to harmful accumulation of FeCl3 or hemin, resulting in reduced viability due to oxidative stress. In other gram-negative bacteria, like E. coli (66), Yersinia pestis (62), and Pseudomonas aeruginosa (24), a fur mutation led to iron overload, resulting in iron-dependent growth phenotypes similar to those exhibited by the S. meliloti rirA mutant. Our data therefore suggested that the importance of the S. meliloti rirA gene for maintaining the intracellular iron balance below toxic levels is similar to the importance of fur in other bacteria. It is interesting that in R. leguminosarum an rirA mutation resulted in an iron-independent growth phenotype (64), indicating that there may be a different mode of rirA-mediated regulation and oxidative stress avoidance in R. leguminosarum.
Since deregulation of iron uptake caused by the rirA mutation had strong effects on the viability of S. meliloti, we speculated that the symbiotic properties might also be affected. To test this, M. sativa seedlings were inoculated with either the wild-type strain or the rirA mutant on plant medium containing FeSO4 at concentrations between 5 and 15 μM. However, no significant differences in nodulation efficiency, foliage fresh weight, or nitrogen fixation rate between plants inoculated with the wild type and plants inoculated with the mutant were found (data not shown). In conclusion, our results indicated that rirA is essential for maintaining a balanced iron content in the cell during iron-sufficient free-living growth, but not during symbiosis. It is therefore possible that in planta iron uptake is regulated independent of rirA; alternatively, in nodules the iron availability might be limited, or iron could be provided in a form that does not require strict regulation by rirA.
Transcriptional profiling of the S. meliloti rirA mutant and the wild-type strain supported the hypothesis that the rirA gene has a regulatory function in iron metabolism.
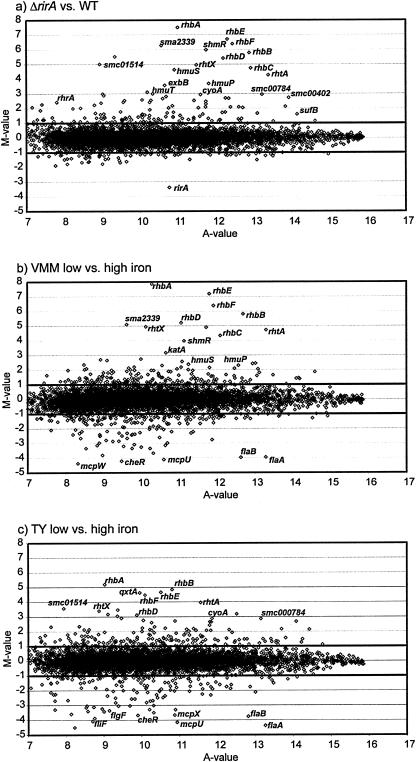
In order to define the RirA modulon, transcriptome studies utilizing Sm6kOligo microarrays (32) were performed. The complete set of genes affected by the rirA mutation was identified by comparing the transcriptomes of the rirA mutant Rm1021-TR2 and the wild-type strain Rm1021 cultivated in iron-sufficient VMM. Duplicate samples were taken from three independent cultures of each strain, which yielded a total of six replicates for each experimental microarray set. RNA isolation, labeling, hybridization, and data analysis were performed as described in Materials and Methods. Only genes with statistically significant (P ≤ 0.05) changes in expression of twofold or more were considered. Using this approach, the expression of 195 genes was found to be significantly changed by the rirA mutation. Of these 195 genes, 132 were induced, whereas only 63 genes were repressed in the rirA mutant compared to the wild type. Figure 2a shows the results of microarray experiments expressed by plotting the log2 expression ratio (M value) versus the mean signal intensity (A value) for each gene. As expected, many of the most highly induced genes (induction levels, 8- to 181-fold) belonged to the previously characterized rhizobactin 1021 synthesis and uptake cluster (including rhbABCDEF and rhtX) (34), further confirming the role of rirA as a repressor of iron uptake genes. In contrast, the only highly repressed gene that was found was the rirA gene itself, whose expression was reduced fourfold or more, an obvious result of deletion of this gene.
FIG. 2.
Scatter plots of the microarray-based analysis of S. meliloti gene expression affected by iron limitation and the rirA mutation. The plots show the logarithmic mean signal ratio (M-value) versus the logarithmic mean signal intensity (A-value) obtained by comparison of the transcriptomes of S. meliloti rirA mutant Rm1021-TR2 (ΔrirA) and S. meliloti wild-type strain Rm1021 (WT) (a), S. meliloti wild-type cells grown in iron-limited VMM and cells grown in iron-sufficient VMM (b), and S. meliloti wild-type cells incubated in iron-limited TY medium and cells grown in iron-sufficient TY medium (c). A number of genes with the greatest changes in mRNA abundance are indicated. The functions of the genes indicated are shown in Tables 2 and 3.
Transcriptome profiling of the S. meliloti wild-type strain grown with high and low iron concentrations defined the iron deficiency stimulon.
Assuming that RirA is a global regulator of iron-responsive genes, a large overlap between genes affected in expression by the rirA mutation and genes affected in expression by iron limitation was expected. Accordingly, two sets of microarray experiments were performed under iron-limited conditions to assess the role of rirA in coordinating gene expression in response to iron availability and also to identify other rirA-independent iron-regulated genes. Two different types of media were used for these experiments, defined VMM and complex TY medium, both either iron deficient or iron sufficient, in order to distinguish between iron-specific and medium-specific expression changes. The optimal culture conditions under which an iron deficiency response was elicited but which still allowed consistent growth were ascertained by monitoring the viability of S. meliloti cells incubated either in TY medium with increasing concentrations of the iron chelator 2,2′-dipyridyl or in VMM with various concentrations of FeCl3 as the sole iron source. At the same time the siderophore production was measured by using a semiquantitative CAS assay as a marker for an iron deficiency response (data not shown). Consequently, we decided to compare the transcriptome of S. meliloti wild-type cells grown in VMM containing 0.37 μM FeCl3 (iron-limited VMM) to the transcriptome of cells cultivated in VMM containing 37 μM FeCl3 (iron-sufficient VMM); likewise, the gene expression of cultures grown in TY medium containing the iron chelator 2,2′-dipyridyl at a concentration of 200 μM (iron-limited TY medium) was compared to the gene expression of cells grown in TY medium without additives.
In iron-limited VMM the expression patterns of 378 genes were altered compared to the patterns observed for growth in iron-sufficient VMM. A total of 199 of these genes were induced by iron limitation, and 179 were repressed. An overview of genes with the greatest increases in mRNA abundance in iron-limited VMM, as shown in a scatter plot (Fig. 2b), revealed a significant consensus for the most highly induced genes in the S. meliloti rirA mutant (Fig. 2a). In contrast, the most highly repressed genes during growth in iron-limited VMM were involved in chemotaxis and cell motility, which was not apparent in the rirA mutant.
Finally, with 2,2′-dipyridyl-induced iron limitation in TY medium 318 genes were found to be differentially expressed (Fig. 2c). Of these 318 genes, 184 were upregulated, and 134 were downregulated. The scatter plots derived from these microarray results demonstrated that there was a high correlation between the most highly induced and repressed genes under both sets of iron-limited conditions, indicating that a comparison of these conditions is suitable for elucidating the transcriptome response of S. meliloti for dependence on iron availability.
Venn mapping identified the S. meliloti genes whose expression was affected by iron availability and/or the rirA mutation.
A complete overview of all genes with significant changes in expression is provided in Table S1 in the supplemental material. Tables 2 and 3 list genes which are differentially expressed under at least two different conditions. For a closer analysis of these data sets, genes that were differentially expressed during growth in iron-limited VMM, in iron-limited TY medium, or in the rirA mutant were plotted in a Venn diagram (Fig. 3).
TABLE 2.
S. meliloti genes significantly induced under iron-limiting conditions and in the rirA mutanta
| Open reading frame | Gene and/or description | Expression ratiob
|
||
|---|---|---|---|---|
| VMM | TY medium | ΔrirA | ||
| SMa0002 | fdoG, probable FdoG formate dehydrogenase-O alpha subunit | −0.42 | 2.13 | 1.05 |
| SMa0142 | Possible protease | 1.60 | 1.29 | 1.08 |
| SMa0172 | Conserved hypothetical protein | 1.27 | 1.17 | 0.96 |
| SMa0312 | Hypothetical protein | 2.43 | 1.74 | 2.09 |
| SMa0314 | Hypothetical protein | 1.24 | 0.92 | 1.21 |
| SMa0316 | Conserved hypothetical protein | 2.31 | 0.90 | 1.56 |
| SMa0320 | Putative dehydrogenase | 1.81 | 0.49 | 1.68 |
| SMa0612 | fixN3, cytochrome c oxidase subunit 1 | 1.97 | 1.61 | 1.26 |
| SMa0617 | fixP3, cytochrome c oxidase membrane-anchored subunit | 1.27 | 1.05 | 1.62 |
| SMa0621 | fixI2, E1-E2-type cation ATPase | 1.00 | 0.52 | 1.11 |
| SMa0994 | Hypothetical protein | 1.37 | 1.92 | 1.08 |
| SMa1461 | Putative muconate cycloisomerase | 1.13 | 0.66 | 1.47 |
| SMa1683 | Putative arylsulfatase | 2.57 | 1.02 | 1.74 |
| SMa1686 | Putative two-component response regulator | 1.05 | 1.09 | 0.54 |
| SMa1745 | Putative iron ABC transporter permease | 1.09 | 0.51 | 1.39 |
| SMa1746 | Putative iron ABC transporter periplasmic binding protein | 1.98 | 1.96 | 2.41 |
| SMa1749 | Putative transcriptional regulator | 0.47 | 1.16 | 1.23 |
| SMa1860 | Putative ABC transporter periplasmic binding protein | 2.13 | 1.05 | 1.02 |
| SMa1985 | Hypothetical protein | 1.09 | 0.83 | 1.56 |
| SMa2259 | Hypothetical protein | 1.25 | 0.77 | 1.36 |
| SMa2294 | mrcA2, probable penicillin binding protein | 1.35 | 1.47 | |
| SMa2337 | rhtX, rhizobactin 1021 transporter | 4.95 | 3.39 | 4.96 |
| SMa2339 | Putative siderophore biosynthesis protein | 5.09 | 3.17 | 6.28 |
| SMa2400 | rhbA, rhizobactin 1021 biosynthesis protein | 7.86 | 5.20 | 7.50 |
| SMa2402 | rhbB, rhizobactin 1021 biosynthesis protein | 5.81 | 4.85 | 5.78 |
| SMa2404 | rhbC, rhizobactin 1021 biosynthesis protein | 4.34 | 2.02 | 4.74 |
| SMa2406 | rhbD, rhizobactin 1021 biosynthesis protein | 5.21 | 3.11 | 5.40 |
| SMa2408 | rhbE, rhizobactin 1021 biosynthesis protein | 7.19 | 4.66 | 6.70 |
| SMa2410 | rhbF, rhizobactin 1021 biosynthesis protein | 6.38 | 4.47 | 6.38 |
| SMa2412 | rhrA, transcriptional activator | 1.40 | 1.75 | 2.42 |
| SMa2414 | rhtA, rhizobactin 1021 receptor | 4.72 | 3.96 | 4.27 |
| SMb20022 | Conserved hypothetical protein | 1.22 | 1.23 | 1.36 |
| SMb20072 | Putative rhizopine binding protein | 1.06 | 3.46 | 0.37 |
| SMb20099 | Putative trehalose synthase protein | 1.81 | 1.17 | 1.18 |
| SMb20127 | Hypothetical protein | 1.36 | 0.56 | 2.88 |
| SMb20203 | cbbR, probable transcriptional regulator protein | 1.07 | 0.63 | 1.38 |
| SMb20545 | Hypothetical protein | 1.19 | 0.15 | 1.31 |
| SMb20600 | Hypothetical protein | 1.73 | 1.43 | 0.75 |
| SMb20755 | pccB, putative propionyl-coenzyme A carboxylase beta chain protein | 1.18 | 2.07 | 0.24 |
| SMb20757 | bhbA, methylmalonyl-coenzyme A mutase protein | 1.40 | 2.07 | 0.52 |
| SMb20924 | abfA, putative alpha-L-arabinofuranosidase protein | 1.76 | 0.70 | 1.44 |
| SMb20946 | exoY, galactosyltransferase protein | 2.06 | 1.43 | 1.18 |
| SMb20949 | exoV, putative pyruvyltransferase protein | 1.46 | 0.90 | 1.04 |
| SMb20959 | exoO, glucosyltransferase protein | 0.70 | 1.12 | 1.37 |
| SMb20960 | exoN, UDP-glucose pyrophosphorylase protein | 1.72 | 1.41 | 1.41 |
| SMb20993 | Putative monooxygenase protein | 1.05 | 0.45 | 1.09 |
| SMb21285 | Conserved hypothetical protein | 1.57 | 1.65 | 2.05 |
| SMb21337 | Putative oxidoreductase | 1.43 | 1.48 | 0.65 |
| SMb21432 | Putative iron ABC transporter periplasmic solute binding protein | 1.35 | 2.18 | 2.10 |
| SMb21456 | Hypothetical protein | 1.88 | 1.33 | 0.60 |
| SMb21464 | Putative transcriptional regulator GntR family protein | 1.07 | 0.10 | 1.42 |
| SMb21487 | cyoA, putative cytochrome o ubiquinol oxidase chain II protein | −0.51 | 2.88 | 2.94 |
| SMb21488 | cyoB, putative cytochrome o ubiquinol oxidase chain I protein | −0.29 | 2.76 | 2.55 |
| SMb21489 | cyoC, putative cytochrome o ubiquinol oxidase chain III protein | −0.09 | 2.64 | 2.08 |
| SMb21690 | exoW, glucosyltransferase protein | 2.07 | 1.22 | 1.13 |
| SMc00062 | Hypothetical protein | 1.05 | 0.88 | 1.63 |
| SMc00086 | cycG, putative diheme cytochrome c type signal peptide protein | 1.90 | −0.01 | 1.52 |
| SMc00136 | Putative oxidoreductase protein | 1.39 | 1.37 | 0.60 |
| SMc00235 | trpD, probable anthranilate phosphoribosyltransferase protein | 1.44 | −0.06 | 1.74 |
| SMc00301 | Conserved hypothetical protein | 0.40 | 1.02 | 1.31 |
| SMc00338 | Conserved hypothetical protein | 1.78 | 1.23 | 0.82 |
| SMc00371 | Conserved hypothetical protein | 1.09 | 1.11 | 0.27 |
| SMc00401 | Conserved hypothetical protein | 1.72 | 1.70 | 2.99 |
| SMc00402 | Putative iron-regulated protein A | 1.44 | 1.49 | 2.72 |
| SMc00512 | Conserved hypothetical protein | 1.07 | 1.07 | 0.29 |
| SMc00537 | Putative transport protein | 1.15 | 0.23 | 1.43 |
| SMc00591 | Hypothetical/unknown signal peptide protein | 0.21 | 2.24 | 1.77 |
| SMc00592 | Hypothetical transmembrane protein | 0.76 | 1.81 | 1.34 |
| SMc00764 | Hypothetical or unknown transmembrane protein | 1.23 | 1.43 | 0.63 |
| SMc00784 | Putative iron ABC transporter periplasmic binding protein | 1.42 | 2.86 | 2.94 |
| SMc00819 | katA, catalase | 3.17 | −0.02 | 1.16 |
| SMc00830 | Hypothetical protein | −0.32 | 1.20 | 1.08 |
| SMc00832 | glcD, probable glycolate oxidase subunit protein | 1.14 | 0.56 | 1.32 |
| SMc00922 | Putative transporter transmembrane protein | 1.20 | 0.55 | 1.06 |
| SMc01016 | Hypothetical protein | 1.09 | 1.75 | 0.71 |
| SMc01022 | Putative cytochrome transmembrane protein | 2.16 | 1.00 | 1.28 |
| SMc01095 | mexF1, probable multidrug efflux system transmembrane protein | 0.21 | 1.04 | 2.81 |
| SMc01169 | ald, probable alanine dehydrogenase oxidoreductase protein | 1.23 | 1.48 | 0.56 |
| SMc01266 | Conserved hypothetical protein | 1.48 | 1.48 | 0.98 |
| SMc01267 | Conserved hypothetical protein | 0.91 | 1.31 | 1.13 |
| SMc01471 | senC, putative cytochrome c oxidase assembly factor transmembrane protein | −0.13 | 1.47 | 1.29 |
| SMc01489 | Hypothetical protein | 2.09 | 1.30 | 1.04 |
| SMc01512 | hmuT, putative hemin ABC transporter periplasmic binding protein | 1.33 | 1.32 | 3.00 |
| SMc01513 | hmuS, Putative hemin ABC transporter ATPase | 2.39 | 3.04 | 4.62 |
| SMc01514 | Conserved hypothetical protein | 2.20 | 3.56 | 5.01 |
| SMc01516 | Conserved hypothetical protein | 1.34 | 1.77 | 2.54 |
| SMc01517 | Conserved hypothetical protein | 0.53 | 1.18 | 2.58 |
| SMc01658 | Putative transport protein | 1.36 | 0.98 | 3.11 |
| SMc01659 | Putative iron ABC transporter periplasmic binding protein | 1.21 | 1.87 | 5.51 |
| SMc01718 | Hypothetical transmembrane protein | 1.64 | 1.29 | 0.66 |
| SMc01747 | hmuP, putative hemin transport protein | 2.30 | 1.52 | 3.68 |
| SMc01765 | Hypothetical transmembrane protein | 0.90 | 1.43 | 1.04 |
| SMc01788 | Hypothetical protein | 2.45 | 1.92 | 1.21 |
| SMc01814 | Probable glutamate synthase small-chain protein | 0.15 | 2.15 | 1.17 |
| SMc02047 | gcvT, probable aminomethyltransferase | 1.58 | 2.68 | 0.24 |
| SMc02075 | Conserved hypothetical protein | 1.06 | 1.12 | −0.01 |
| SMc02084 | exbD, putative polymer transport protein | 1.21 | 1.45 | 1.99 |
| SMc02085 | exbB, putative polymer transport protein | 1.96 | 2.60 | 3.57 |
| SMc02156 | Conserved hypothetical protein | 1.86 | 1.34 | 0.96 |
| SMc02179 | Conserved hypothetical protein | 1.34 | 1.05 | 0.44 |
| SMc02254 | qxtB, putative quinol oxidase subunit II | 1.09 | 2.66 | 1.08 |
| SMc02255 | qxtA, putative quinol oxidase subunit I | 1.77 | 4.62 | 2.37 |
| SMc02266 | Conserved hypothetical protein | 1.37 | 1.48 | −0.14 |
| SMc02708 | Conserved hypothetical protein | 1.04 | 1.05 | 0.38 |
| SMc02726 | shmR, hemin binding outer membrane receptor | 3.98 | 2.90 | 5.98 |
| SMc02727 | Hypothetical protein | 1.98 | 0.46 | 2.23 |
| SMc02887 | Hypothetical protein | 2.19 | 0.78 | 2.17 |
| SMc03149 | Hypothetical protein | 0.34 | 1.38 | 1.58 |
| SMc03787 | Hypothetical protein | 1.64 | 3.19 | 2.70 |
| SMc03971 | mexF2, putative multidrug efflux system protein | 1.19 | 1.01 | 1.10 |
| SMc04049 | Putative sulfite oxidase protein | 1.11 | 1.03 | 0.21 |
| SMc04162 | Putative transcription regulator protein | 1.39 | −0.03 | 1.69 |
| SMc04164 | Hypothetical protein | 1.40 | 1.56 | 0.92 |
| SMc04206 | Putative hemolysin type calcium binding protein | 4.90 | 1.93 | 2.43 |
Genes were considered differentially expressed when according to t-statistics the P value was <0.05 with an induction level of >twofold. Only genes differentially expressed during at least two sets of microarray experiments were considered.
The expression ratios are expressed as log2 values.
TABLE 3.
S. meliloti genes significantly repressed under iron-limiting conditions and in the rirA mutanta
| Open reading frame | Gene and/or description | Expression ratiob
|
||
|---|---|---|---|---|
| VMM | TY medium | ΔrirA | ||
| SMa0249 | Conserved hypothetical protein | −1.12 | −1.06 | −0.07 |
| SMa0252 | Conserved hypothetical protein | −2.32 | −1.09 | −0.40 |
| SMa1289 | Hypothetical protein | −1.68 | −1.53 | −0.10 |
| SMa2032 | Putative nonheme chloroperoxidase | −1.70 | −1.92 | −1.11 |
| SMb20293 | Hypothetical protein | −1.47 | −1.21 | −0.03 |
| SMb20397 | Putative oxidoreductase protein | −2.85 | −1.69 | −0.47 |
| SMb20649 | nadE1, putative NH3-dependent NAD+ synthetase protein | −1.09 | −1.62 | −0.02 |
| SMb20650 | Putative long-chain fatty acid coenzyme A ligase protein | −1.18 | −1.71 | −0.02 |
| SMb20651 | Hypothetical protein | −2.30 | −1.96 | 0.10 |
| SMb20764 | phnL, putative phosphonate uptake ABC transporter ATP binding protein | −2.76 | −2.31 | −0.29 |
| SMb20986 | narB, putative nitrate reductase large-subunit protein | −1.61 | −1.39 | 0.39 |
| SMb21170 | Putative guanylate kinase | −1.08 | −1.41 | −0.25 |
| SMb21174 | phoT, phosphate uptake ABC transporter permease protein | −1.07 | −1.75 | −0.11 |
| SMb21175 | phoE, phosphate uptake ABC transporter permease protein | −1.70 | −1.90 | −0.31 |
| SMb21177 | phoC, phosphate uptake ABC transporter ATP binding protein | −1.36 | −1.31 | −0.15 |
| SMc00158 | Hypothetical protein | −1.41 | −2.25 | −0.22 |
| SMc00159 | Hypothetical protein | −1.70 | −2.12 | 0.12 |
| SMc00283 | Putative transcription regulator protein | −3.82 | −3.05 | −0.64 |
| SMc00335 | rpsA, 30S ribosomal protein s1 | −0.44 | −1.14 | −1.05 |
| SMc00565 | rplI, probable 50S ribosomal protein 19 | −0.63 | −1.12 | −1.05 |
| SMc00578 | Hypothetical protein | −0.47 | −2.05 | −1.20 |
| SMc00765 | mcpZ, probable methyl-accepting chemotaxis protein | −3.68 | −3.29 | −0.23 |
| SMc00887 | Hypothetical protein | −3.35 | −3.43 | −0.50 |
| SMc00888 | Hypothetical protein | −3.08 | −3.01 | −0.08 |
| SMc00975 | mcpU, probable chemoreceptor (methyl-accepting chemotaxis) transmembrane protein | −4.11 | −4.14 | −0.81 |
| SMc00986 | Hypothetical protein | −1.83 | −1.79 | −0.45 |
| SMc00998 | Conserved hypothetical signal peptide protein | −1.63 | −1.19 | 0.08 |
| SMc01104 | mcpX, probable chemoreceptor (methyl-accepting chemotaxis) transmembrane protein | −2.98 | −3.67 | −0.36 |
| SMc01160 | Putative transcriptional regulator protein | −2.51 | −0.47 | −1.17 |
| SMc01319 | rplJ, probable 50S ribosomal protein | −1.33 | −1.10 | −1.51 |
| SMc01326 | tufB, probable elongation factor Tu protein | −1.04 | −0.42 | −1.25 |
| SMc01469 | mcpW, probable methyl-accepting chemotaxis transmembrane protein | −4.38 | −3.41 | −0.63 |
| SMc01842 | Putative methyltransferase transcription regulator protein | −0.49 | −2.05 | −1.00 |
| SMc01847 | Putative methyltransferase protein | −1.40 | −1.25 | −0.92 |
| SMc02392 | Hypothetical protein | −1.39 | −1.81 | −0.76 |
| SMc02634 | Hypothetical transmembrane protein | −2.60 | −2.12 | −0.91 |
| SMc03004 | mcpE, putative chemoreceptor (methyl-accepting chemotaxis protein) | −1.36 | −1.44 | −0.57 |
| SMc03005 | Conserved hypothetical protein | −1.21 | −1.85 | −0.30 |
| SMc03007 | cheA, chemotaxis protein (sensory transduction histidine kinase) | −2.80 | −3.04 | −0.57 |
| SMc03008 | cheW1, chemotaxis protein | −2.04 | −2.29 | −0.37 |
| SMc03009 | cheR, chemotaxis protein methyltransferase | −4.21 | −3.68 | −0.43 |
| SMc03012 | cheD, chemotaxis protein | −2.18 | −2.00 | −0.63 |
| SMc03013 | Conserved hypothetical protein | −2.07 | −2.03 | −0.52 |
| SMc03017 | Conserved hypothetical protein | −1.61 | −1.80 | −0.84 |
| SMc03021 | fliM, flagellar motor switch transmembrane protein | −2.41 | −2.83 | −0.71 |
| SMc03022 | motA, chemotaxis (motility protein A) | −2.66 | −2.81 | −0.80 |
| SMc03023 | Conserved hypothetical protein | −2.42 | −2.69 | −0.76 |
| SMc03024 | flgF, flagellar basal body rod protein | −3.14 | −3.36 | −1.00 |
| SMc03027 | flgB, flagellar basal body rod protein | −3.25 | −3.50 | −0.84 |
| SMc03028 | flgC, flagellar basal body rod protein | −1.35 | −1.74 | −0.59 |
| SMc03029 | fliE, flagellar hook basal body complex protein | −3.20 | −2.72 | −0.79 |
| SMc03030 | flgG, flagellar basal body rod protein | −3.35 | −3.30 | −0.89 |
| SMc03031 | flgA, flagellar precursor transmembrane protein | −1.16 | −1.88 | −0.69 |
| SMc03034 | flgH, flagellar 1 ring protein precursor | −1.62 | −1.97 | −0.56 |
| SMc03035 | fliL, flagellar transmembrane protein | −2.66 | −3.11 | −1.19 |
| SMc03037 | flaA, flagellin protein | −3.95 | −4.37 | −0.11 |
| SMc03038 | flaB, flagellin protein | −3.97 | −3.76 | −0.42 |
| SMc03040 | flaC, flagellin protein | −2.28 | −2.36 | −0.45 |
| SMc03042 | motB, chemotaxis protein | −1.84 | −2.00 | −0.83 |
| SMc03043 | motC, chemotaxis protein precursor | −1.92 | −2.49 | −0.96 |
| SMc03044 | motD, chemotaxis protein | −2.97 | −3.04 | −0.82 |
| SMc03045 | Hypothetical transmembrane protein | −4.50 | −1.41 | |
| SMc03047 | flgE, flagellar hook protein | −1.66 | −1.61 | −0.50 |
| SMc03048 | flgK, putative flagellar hook-associated protein | −2.50 | −1.90 | −0.81 |
| SMc03049 | flgL, putative flagellar hook-associated protein | −3.14 | −2.25 | −0.85 |
| SMc03051 | flbT, putative flagellin synthesis repressor protein | −2.21 | −1.90 | −0.68 |
| SMc03052 | flgD, putative basal body rod modification protein | −1.94 | −1.71 | −0.64 |
| SMc03057 | Conserved hypothetical transmembrane protein | −1.57 | −2.56 | −0.74 |
| SMc03071 | Hypothetical protein | −1.68 | −2.06 | −0.56 |
| SMc03072 | Conserved hypothetical protein | −3.35 | −2.48 | −0.62 |
| SMc03242 | Probable GTP binding protein | −0.21 | −1.10 | −1.61 |
| SMc03857 | ffh, probable signal recognition particle protein | −0.84 | −1.16 | −1.64 |
| SMc04007 | Conserved hypothetical protein | −1.18 | −0.86 | −1.19 |
| SMc04009 | Conserved hypothetical protein | −0.85 | −1.03 | −1.14 |
| SMc04059 | Hypothetical protein | −2.86 | −3.23 | −0.66 |
| SMc04300 | afuC, probable iron ABC transporter ATPase | −0.75 | −1.40 | −1.39 |
| SMc04317 | afuA, probable iron ABC transporter periplasmic binding protein | −1.52 | −1.55 | −0.90 |
Genes were considered differentially expressed when according to t-statistics the P value was <0.05 with a repression level of >two fold. Only genes differentially expressed during at least two sets of microarray experiments were considered.
The expression ratios are expressed as log2 values.
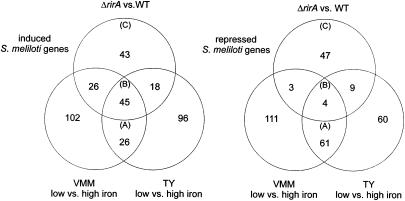
FIG. 3.
Venn mapping of S. meliloti genes with significantly altered expression obtained from three microarray experiments. The amounts of significantly induced genes (left) and repressed genes (right) were derived from the microarray-based global profiling of iron-responsive and RirA-regulated gene expression in S. meliloti. Of special interest were genes that were substantially up- or downregulated under both low- and high-iron conditions but not in the rirA mutant (subset A), during all three microarray experiments (subset B), and only in the rirA mutant compared to the wild type (WT) (subset C). The Venn diagrams are based on the microarray results shown in Table S1 in the supplemental material.
This led to identification of 136 genes (71 induced genes and 65 repressed genes) that were differentially expressed in both iron-limited media compared to growth under iron-sufficient conditions (Fig. 3, subsets A and B). These up- and downregulated genes are likely to be generally involved in the adaptation of S. meliloti to low-iron conditions and were considered to represent the S. meliloti iron deficiency stimulon. Of the 71 induced genes of the iron deficiency stimulon, 45 were also found to be induced in the rirA mutant (Fig. 3, subset B), thus supporting the hypothesis that rirA has a role as a major repressor of iron-responsive genes. In contrast, only 4 of the 65 repressed genes belonging to the iron deficiency stimulon were found to be repressed in the rirA mutant and under iron-limiting conditions, further indicating that rirA is involved primarily in repression rather than in induction of genes with dependence on iron.
An additional 87 genes of the iron deficiency stimulon (26 upregulated genes and 61 downregulated genes) (Fig. 3, subset A) were not affected by the rirA mutation, indicating that other regulatory mechanisms are involved during adaptation of S. meliloti to growth under low-iron conditions. Finally, 90 genes were found to be differentially expressed exclusively in the rirA mutant, independent of iron availability (Fig. 3, subset C). Several explanations might account for these transcriptome changes: (i) the rirA gene might also exert regulatory control on genes in an iron-independent way, (ii) the iron limitation used in this study might not have been sufficient to change the expression above the threshold value that we considered significant, and (iii) the deregulated iron uptake in the rirA mutant and the resulting changes in the intracellular iron content might have led to secondary transcriptome changes independent of actual regulation mediated by rirA.
S. meliloti genes involved in iron acquisition, energy metabolism, and exopolysaccharide production are induced in the rirA mutant, as well as under iron-limiting conditions.
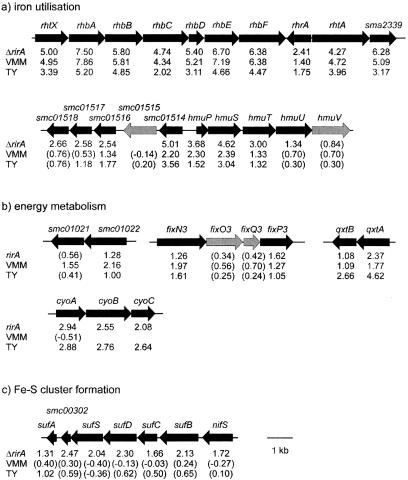
The phenotypic analyses suggested that the rirA gene is involved in the repression of iron uptake and siderophore synthesis. In agreement with this, a large number of the 45 genes induced both under iron-limiting conditions and by the rirA mutation (Fig. 3, subset B) could be connected to iron acquisition (Table 2). Some of the most highly induced genes were the complete rhbABCDEF rhizobactin 1021 synthesis operon (34) and genes encoding the cognate outer membrane receptor (rhtA) and the recently characterized inner membrane transporter (rhtX) (13) (Fig. 4a). The transcriptional activator rhrA, which was demonstrated to be involved in the induction of siderophore synthesis and uptake genes (34), is located in the same gene cluster. Since rhrA was induced by iron limitation as well as the rirA mutation, indirect regulation of the rhizobactin 1021 synthesis and uptake cluster mediated by rirA via rhrA appeared to be likely. In fact, in a constructed rhrA/rirA double mutant designated Rm1021-TR2-3, siderophore overproduction was abolished, as indicated by CAS assays (data not shown), providing evidence that the rirA-mediated regulation of siderophore synthesis is dependent on the rhrA gene. A putative siderophore synthesis gene (SMa2339) which is possibly involved in the formation of rhizobactin 1021 in a hitherto unknown way is located in the immediate vicinity of rhtA. Moreover, a number of putative siderophore type ABC transporter genes (SMa1746, SMb21432, and SMc01659) were found among the genes induced under iron-limiting conditions and in the rirA mutant. It is of interesting that according to the original annotation SMb21431 codes for the C terminus of a periplasmic binding protein, but resequencing of the corresponding DNA region showed that in the published sequence an error resulting in a frameshift is present. In fact, SMb21431 and SMb21432 constitute a single open reading frame and are referred to as SMb21432 in this report. In addition to putative siderophore type ABC transporter genes, a chromosomal region encoding a putative hemin ABC transporter was also induced by iron limitation and in the rirA mutant (Fig. 4a). This putative hemin transporter is encoded by hmuP, hmuS, hmuT, hmuU, and hmuV, which are homologous to heme uptake genes of Bradyrhizobium japonicum (38), R. leguminosarum (71), and Y. pestis (26). The expression ratios of these genes are shown in Fig. 4a. In addition to these genes, shmR, encoding an outer membrane receptor for hemin (3), was also significantly induced in the rirA mutant and under both iron-limiting conditions. The deregulation of these genes in the rirA mutant is also likely to be a major factor contributing to the observed sensitivity to hemin (Fig. 1c). Interestingly, genes coding for hypothetical proteins in the immediate vicinity of the hmu transporter genes (Fig. 4a) were also induced both under iron-limiting conditions and in the rirA mutant (SMc01514, SMc01516, SMc01517, and SMc01518). Overall, these findings suggest that the secretion and uptake of siderophores and the induction of heme utilization systems are the main strategy that S. meliloti uses to counter iron deficiency and that the repression of these systems is mediated by rirA. The only nonsiderophore/heme-type iron transporter gene found to be expressed more highly during iron limitation and in the rirA mutant was SMc00784, which encodes the periplasmic binding protein of a ferric type ABC transporter. This protein exhibits 52% identity to FbpA of Mannheimia (formerly Pasteurella) hemolytica, which is involved in iron acquisition during pathogenic host invasion (31, 56). Interestingly, no other components of an ABC transporter were located in the vicinity of SMc00784. None of these putative transporters have been characterized yet, and our results indicate that they may play a major role in the adaptation of S. meliloti to low-iron conditions. For rhbA, shmR, and hmuS the results of the microarray experiments were verified by quantitative PCR. The same tendencies were found, although the induction ratios obtained in the quantitative PCR experiments were higher than the values obtained in the microarray experiments (data not shown). This is consistent with other experiments in which the Sm6kOligo microarray was used (32).
FIG. 4.
Clustered S. meliloti genes whose expression is affected by iron availability and by the rirA mutation. (a and b) Genetic maps of the rhizobactin 1021 synthesis/uptake and putative hemin uptake clusters (a) and a selection of putative energy metabolism gene clusters which are induced in the rirA mutant and under iron-limited conditions (b). (c) Gene cluster putatively involved in Fe-S cluster formation which is induced only in the S. meliloti rirA mutant. The numbers below the genes indicate the log2 expression ratios of the genes derived from the microarray-based transcriptome analysis. Genes that were significantly induced in at least one experiment are indicated by solid arrows. The values in parentheses are expression ratios that were below the threshold used in this study.
Curiously, genes encoding the subunits of a putative ferric-type transporter (afuA and afuC) were downregulated instead of upregulated during iron-limited growth and in the rirA mutant. Recent work of Krol and Becker suggested that these genes might in fact be under the control of the phosphate-dependent regulator PhoB (32), and it is possible that despite high levels of homology to ferric-type transporters this transporter might play a different role in S. meliloti.
In addition to iron transporter genes, SMc00402, exbB, and exbD were also found to be derepressed in the rirA mutant and in either iron-limited medium. The deduced gene product of SMc00402 exhibits low levels of homology to IrpA (iron-regulated protein A) of Synechococcus sp. strain PCC7942 (48), which was proposed to be involved in iron acquisition, and a similar role for SMc00402 might be proposed for S. meliloti. The ExbB and ExbD proteins are generally involved in forming the energy-transducing TonB complex, which energizes the transport of iron substrates by specific receptors across the outer membrane (8). It is therefore likely that the S. meliloti exbB and exbD genes are involved in the transport of ferri-siderophores or heme compounds by the corresponding outer membrane receptors. We found that in the genome of S. meliloti only one tonB homologue could be detected (SMc01515), which was not differentially expressed in response to iron limitation or by the rirA mutation. An individual analysis of the microarray experiments with S. meliloti grown in TY medium or in VMM under iron-limiting conditions revealed no other genes with homologies to iron acquisition genes, demonstrating that all iron acquisition genes induced under iron-limiting conditions are also affected by the rirA mutation, which is strong evidence that the S. meliloti RirA protein is the main regulator of iron uptake.
In addition to iron acquisition genes, other genes involved in energy metabolism, noniron transport, and exopolysaccharide synthesis exhibited enhanced levels of expression during iron limitation and in the rirA mutant (Table 2). Altogether, 10 electron transport-associated genes were found to be expressed at an elevated level in the rirA mutant and under at least one type of iron-limiting growth conditions (SMc01022, fixN3, fixP3, qxtA, qxtB, cyoA, cyoB, cyoC, and cycG) (Table 2). The fix-3 gene cluster encodes a putative cytochrome oxidase and represents another iteration of the previously characterized fix-1 and fix-2 gene clusters (2). While at least either a functional fix-1 or fix-2 gene cluster is known to be essential for establishing a successful symbiosis (49), the fixN3 and fixP3 genes obviously have functions other than thsoe of their paralogues and, according to our results, are also regulated differently. Overall, the microarray analyses suggest that iron availability might influence the path of the respiratory chain in S. meliloti, which is consistent with the results of similar studies of other bacteria (40, 42, 43, 45, 58).
Three of the S. meliloti exopolysaccharide genes (exoW, exoY, and exoN) were induced under all three conditions, and two additional genes (exoO and exoV) were induced in the rirA mutant and during growth in iron-limited TY medium and VMM, respectively. These findings are in agreement with previous reports, which described enhanced production of exopolysaccharides in other bacteria as a result of nutrient stress (68, 72). In addition, katA encoding a monofunctional catalase was found to be induced in the rirA mutant and in iron-limited VMM. Previously, the S. meliloti katA gene was found to be inducible by H2O2 (25) and to be highly expressed in bacteroids (28). Our results demonstrate that the regulation of this gene is also affected by iron availability and the rirA gene. Finally, in addition to the activator gene rhrA mentioned above, a number of putative transcription regulator genes (SMa1749, SMc04162, SMc01160, and SMb21464) were differentially expressed depending on the rirA mutation and under at least one type of iron-limiting conditions. Hence, it is possible that in some cases rirA-mediated iron-dependent regulation might be exerted indirectly via these putative regulators, and it would be interesting to elucidate how these regulators contribute to the rirA-mediated regulatory network.
In R. leguminosarum iron-responsive regulators were detected, which are potential RirA binding sites (74). We conducted in silico searches in the upstream region of genes induced in the S. meliloti rirA mutant but were not able to identify any iron-responsive regulator-like sequences.
S. meliloti genes involved in cell motility are repressed under iron-limiting conditions but not in the rirA mutant.
Altogether, 87 genes were differentially expressed during growth in both iron-limited media but were not affected by the rirA mutation (Fig. 3, subset A). No function could be assigned to a large portion of these genes (57% of the induced genes and 21% of the repressed genes). A striking observation was that 30 of the downregulated genes are involved in motility. These genes represent 46% of all predicted motility genes of this bacterium. Twenty-four of these repressed motility genes are located in a cluster which represents the S. meliloti flagellar regulon (60).
These genes are known to be regulated in a hierarchical order, with VisN and VisR acting as global activators. In iron-limited VMM the visN and visR genes were also found to be significantly repressed two- to threefold (see Table S1 in the supplemental material), which probably accounts for the massive reduction of motility genes. This is also likely to be the case during growth in iron-limited TY medium, despite the fact that the visNR operon was repressed only ∼1.4-fold, which is below the threshold that we used in this study.
Moreover, seven additional genes annotated to encode hypothetical proteins (SMc03005, SMc03013, SMc03017, SMc03023, SMc03057, SMc3071, and SMc3072) located in the motility cluster and four other genes which are putatively involved in chemotaxis (SMc00765, SMc01469, SMc01104, and Smc00975) but are not located in this cluster were also found to be downregulated during iron limitation. It has been reported that C, N, or phosphate starvation in S. meliloti results in a reduction in motility (69). Here we provide evidence that iron limitation also negatively affects the expression of motility genes and that this may be caused by reduced transcription of the master activators of the flagellar regulon.
S. meliloti genes involved in the biosynthesis of Fe-S clusters are differentially expressed in the rirA mutant but not under iron-limiting conditions.
Altogether, 90 genes involved in a variety of functions were differentially expressed in the rirA mutant (Fig. 3, subset C) but were not affected by iron availability. Although we are not able to easily distinguish between direct rirA-mediated regulation and secondary effects, as proposed above, in at least some cases reasonable hypotheses can be advanced to explain the observed regulation. For instance, a cluster of genes (SMc00530, SMc00531, SMc00532, SMc00533, SMc00302, and SMc00301), including a putative nifS gene, was derepressed in the S. meliloti rirA mutant background (Fig. 4 c). The products encoded by genes in this cluster were annotated as either hypothetical proteins or putative ABC transporters, but our renewed annotation showed that the products of SMc00301, SMc00530, SMc00531, SMc00532, and SMc00533 exhibited homology to the E. coli SufA, SufB, SufC, SufD, and SufS proteins, respectively, which are involved in assembly of Fe-S clusters (18). In E. coli and Erwinia chrysanthemi these genes are important for iron acquisition and were found to be induced by oxidative stress and iron starvation (33, 36, 44). Thus, it has been assumed that the suf genes are specifically adapted to synthesize Fe-S clusters when iron or sulfur metabolism is disrupted by iron starvation or oxidative stress (41). In both enterobacteria the iron-dependent regulation of the suf genes was mediated by Fur. Since we demonstrated that in S. meliloti numerous usually fur-mediated functions are fulfilled by rirA, it is resonable to hypothesize that rirA might also be involved in the regulation of the suf genes. Another possibility is that the oxidative stress generated in the rirA mutant by deregulated iron uptake might indirectly lead to the induction of the suf homologues in S. meliloti. In a recent report an suf cluster in R. leguminosarum was found to be induced in an rirA mutant, which is in accordance with our results. Interestingly, the induction was found to be highest with high iron concentrations (65), possibly indicating that iron-generated oxidative stress also contributes to the regulation of the suf genes in R. leguminosarum.
Among the downregulated genes, the most striking groups of genes (17 genes) are involved in protein biosynthesis, including the synthesis of ribosomal proteins and elongation factors (see Table S1 in the supplemental material). This observation is probably a secondary effect due to the reduced growth of the S. meliloti rirA mutant.
Conclusions.
Iron acquisition is an important aspect of the rhizobial life cycle, and iron limitation severely inhibits the effectiveness of the rhizobium-legume symbiosis (29, 37). The large number of genes differentially regulated in response to iron availability found in this study stresses the importance of this micronutrient for S. meliloti. Bioassays with the S. meliloti rirA mutant clearly demonstrated the importance of the rirA gene for maintaining intracellular iron concentrations below toxic levels under free-living conditions. This finding was supported by the results of the microarray experiments, which implied that rirA has a central role in coordinating the transcriptional response with iron availability. In the case of siderophore synthesis and the uptake cluster, we obtained evidence that the rirA-dependent regulation is exerted indirectly via the transcriptional activator gene rhrA. Furthermore, the number and variety of regulated genes indicate that regulation mediated by rirA is not limited to iron acquisition. While it was not the goal of this work to define the RirA regulon on a molecular level, our analyses identified a number of possible directly RirA-regulated genes that may be used as targets for further studies. Overall, the extent of rirA-mediated regulation and the phenotypic analyses of the S. meliloti rirA mutant revealed striking parallels to fur-mediated regulation. It appears that in S. meliloti and likely also in R. leguminosarum RirA has the role of a central coordinator of iron-dependent regulation, while the Fur proteins in these rhizobia are involved mainly in the regulation of manganese uptake (12, 15, 47). In this study a large number of genes whose expression was previously not known to be affected by iron, as well as uncharacterized genes with previously unknown functions, were identified. This clearly illustrates the need for further analyses to truly understand the iron metabolism of S. meliloti. Our study thus was the first global analysis of iron metabolism and its novel form of regulation by rirA in a rhizobial species. The full data sets for genes whose expression is affected by iron availability and the rirA mutation constitute a sound basis for targeted mutagenesis experiments.
Supplementary Material
Acknowledgments
We thank Victoria Bartelsmeier for performing the microarray experiments.
This work was supported by a scholarship from the Graduate School for Bioinformatics and Genome Research, funded by the Ministerium für Wissenschaft und Forschung (MWF), and by grant BIZ 7 from the Deutsche Forschungsgemeinschaft (DFG).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, M. J., R. F. Fisher, T. Jones, C. Komp, A. P. Abola, F. Barloy-Hubler, L. Bowser, D. Capela, F. Galibert, J. Gouzy, M. Gurjal, A. Hong, L. Huizar, R. W. Hyman, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, C. Palm, M. C. Peck, R. Surzycki, D. H. Wells, K. C. Yeh, R. W. Davis, N. A. Federspiel, and S. R. Long. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA 98:9883-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battistoni, F., R. Platero, R. Duran, C. Cervenansky, J. Battistoni, A. Arias, and E. Fabiano. 2002. Identification of an iron-regulated, hemin-binding outer membrane protein in Sinorhizobium meliloti. Appl. Environ. Microbiol. 68:5877-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaumont, H. J., S. I. Lens, W. N. Reijnders, H. V. Westerhoff, and R. J. van Spanning. 2004. Expression of nitrite reductase in Nitrosomonas europaea involves NsrR, a novel nitrite-sensitive transcription repressor. Mol. Microbiol. 54:148-158. [DOI] [PubMed] [Google Scholar]
- 5.Becker, A., H. Berges, E. Krol, C. Bruand, S. Rüberg, D. Capela, E. Lauber, E. Meilhoc, F. Ampe, F. J. de Bruijn, J. Fourment, A. Francez-Charlot, D. Kahn, H. Küster, C. Liebe, A. Pühler, S. Weidner, and J. Batut. 2004. Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol. Plant-Microbe Interact. 17:292-303. [DOI] [PubMed] [Google Scholar]
- 6.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, J., M. N. Oza, and J. R. Murphy. 1990. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. USA 87:5968-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, V., K. Gunter, and K. Hantke. 1991. Transport of iron across the outer membrane. Biol. Met. 4:14-22. [DOI] [PubMed] [Google Scholar]
- 9.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Pühler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carson, K. C., M. J. Dilworth, and A. R. Glenn. 1992. Siderophore production and iron transport in Rhizobium leguminosarum bv viciae Mnf710. J. Plant Nutr. 15:2203-2220. [Google Scholar]
- 11.Carson, K. C., S. Holliday, A. R. Glenn, and M. J. Dilworth. 1992. Siderophore and organic acid production in root nodule bacteria. Arch. Microbiol. 157:264-271. [DOI] [PubMed] [Google Scholar]
- 12.Chao, T. C., A. Becker, J. Buhrmester, A. Pühler, and S. Weidner. 2004. The Sinorhizobium meliloti fur gene regulates, with dependence on Mn(II), transcription of the sitABCD operon, encoding a metal-type transporter. J. Bacteriol. 186:3609-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuiv, P. O., P. Clarke, D. Lynch, and M. O'Connell. 2004. Identification of rhtX and fptX, novel genes encoding proteins that show homology and function in the utilization of the siderophores rhizobactin 1021 by Sinorhizobium meliloti and pyochelin by Pseudomonas aeruginosa, respectively. J. Bacteriol. 186:2996-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delgado, M. J., E. J. Bedmar, and J. A. Downie. 1998. Genes involved in the formation and assembly of rhizobial cytochromes and their role in symbiotic nitrogen fixation. Adv. Microb. Physiol. 40:191-231. [DOI] [PubMed] [Google Scholar]
- 15.Diaz-Mireles, E., M. Wexler, G. Sawers, D. Bellini, J. D. Todd, and A. W. Johnston. 2004. The Fur-like protein Mur of Rhizobium leguminosarum is a Mn2+-responsive transcriptional regulator. Microbiology 150:1447-1456. [DOI] [PubMed] [Google Scholar]
- 16.Dondrup, M., A. Goesmann, D. Bartels, J. Kalinowski, L. Krause, B. Linke, O. Rupp, A. Sczyrba, A. Pühler, and F. Meyer. 2003. EMMA: a platform for consistent storage and efficient analysis of microarray data. J. Biotechnol. 106:135-146. [DOI] [PubMed] [Google Scholar]
- 17.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flint, D. H. 1996. Escherichia coli contains a protein that is homologous in function and N-terminal sequence to the protein encoded by the nifS gene of Azotobacter vinelandii and that can participate in the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase. J. Biol. Chem. 271:16068-16074. [PubMed] [Google Scholar]
- 19.Galibert, F., T. M. Finan, S. R. Long, A. Pühler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorhölter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 20.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamza, I., S. Chauhan, R. Hassett, and M. R. O'Brian. 1998. The bacterial Irr protein is required for coordination of heme biosynthesis with iron availability. J. Biol. Chem. 273:21669-21674. [DOI] [PubMed] [Google Scholar]
- 22.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 23.Hantke, K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182:288-292. [DOI] [PubMed] [Google Scholar]
- 24.Hassett, D. J., P. A. Sokol, M. L. Howell, J. F. Ma, H. T. Schweizer, U. Ochsner, and M. L. Vasil. 1996. Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosa demonstrate defective siderophore-mediated iron uptake, altered aerobic growth, and decreased superoxide dismutase and catalase activities. J. Bacteriol. 178:3996-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herouart, D., S. Sigaud, S. Moreau, P. Frendo, D. Touati, and A. Puppo. 1996. Cloning and characterization of the katA gene of Rhizobium meliloti encoding a hydrogen peroxide-inducible catalase. J. Bacteriol. 178:6802-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornung, J. M., H. A. Jones, and R. D. Perry. 1996. The hmu locus of Yersinia pestis is essential for utilization of free haemin and haem-protein complexes as iron sources. Mol. Microbiol. 20:725-739. [DOI] [PubMed] [Google Scholar]
- 27.Horton, R. M. 1995. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol. Biotechnol. 3:93-99. [DOI] [PubMed] [Google Scholar]
- 28.Jamet, A., S. Sigaud, G. Van de Sype, A. Puppo, and D. Herouart. 2003. Expression of the bacterial catalase genes during Sinorhizobium meliloti-Medicago sativa symbiosis and their crucial role during the infection process. Mol. Plant-Microbe Interact. 16:217-225. [DOI] [PubMed] [Google Scholar]
- 29.Johnston, A. W., K. H. Yeoman, and M. Wexler. 2001. Metals and the rhizobial-legume symbiosis—uptake, utilization and signalling. Adv. Microb. Physiol. 45:113-156. [DOI] [PubMed] [Google Scholar]
- 30.Keon, R. G., R. Fu, and G. Voordouw. 1997. Deletion of two downstream genes alters expression of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough. Arch. Microbiol. 167:376-383. [DOI] [PubMed] [Google Scholar]
- 31.Kirby, S. D., F. A. Lainson, W. Donachie, A. Okabe, M. Tokuda, O. Hatase, and A. B. Schryvers. 1998. The Pasteurella haemolytica 35 kDa iron-regulated protein is an FbpA homologue. Microbiology 144:3425-3436. [DOI] [PubMed] [Google Scholar]
- 32.Krol, E., and A. Becker. 2004. Global transcriptional analysis of the phosphate starvation response in Sinorhizobium meliloti strains 1021 and 2011. Mol. Genet. Genomics 272:1-17. [DOI] [PubMed] [Google Scholar]
- 33.Lee, J. H., W. S. Yeo, and J. H. Roe. 2004. Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol. Microbiol. 51:1745-1755. [DOI] [PubMed] [Google Scholar]
- 34.Lynch, D., J. O'Brien, T. Welch, P. Clarke, P. O. Cuiv, J. H. Crosa, and M. O'Connell. 2001. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 183:2576-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meade, H. M., S. R. Long, G. B. Ruvkum, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nachin, L., L. Loiseau, D. Expert, and F. Barras. 2003. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 22:427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nadler, K. D., A. W. Johnston, J. W. Chen, and T. R. John. 1990. A Rhizobium leguminosarum mutant defective in symbiotic iron acquisition. J. Bacteriol. 172:670-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nienaber, A., H. Hennecke, and H. M. Fischer. 2001. Discovery of a haem uptake system in the soil bacterium Bradyrhizobium japonicum. Mol. Microbiol. 41:787-800. [DOI] [PubMed] [Google Scholar]
- 39.Noya, F., A. Arias, and E. Fabiano. 1997. Heme compounds as iron sources for nonpathogenic Rhizobium bacteria. J. Bacteriol. 179:3076-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277-1287. [DOI] [PubMed] [Google Scholar]
- 41.Outten, F. W., O. Djaman, and G. Storz. 2004. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 52:861-872. [DOI] [PubMed] [Google Scholar]
- 42.Palma, M., S. Worgall, and L. E. Quadri. 2003. Transcriptome analysis of the Pseudomonas aeruginosa response to iron. Arch. Microbiol. 180:374-379. [DOI] [PubMed] [Google Scholar]
- 43.Palyada, K., D. Threadgill, and A. Stintzi. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186:4714-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patzer, S. I., and K. Hantke. 1999. SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe-2S] FhuF protein in Escherichia coli. J. Bacteriol. 181:3307-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paustian, M. L., B. J. May, and V. Kapur. 2001. Pasteurella multocida gene expression in response to iron limitation. Infect. Immun. 69:4109-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Persmark, M., P. Pittman, J. S. Buyer, B. Schwyn, P. R. Gill, and J. B. Neilands. 1993. Isolation and structure of rhizobactin-1021, a siderophore from the alfalfa symbiont Rhizobium meliloti 1021. J. Am. Chem. Soc. 115:3950-3956. [Google Scholar]
- 47.Platero, R., L. Peixoto, M. R. O'Brian, and E. Fabiano. 2004. Fur is involved in manganese-dependent regulation of mntA (sitA) expression in Sinorhizobium meliloti. Appl. Environ. Microbiol. 70:4349-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddy, K. J., G. S. Bullerjahn, D. M. Sherman, and L. A. Sherman. 1988. Cloning, nucleotide sequence, and mutagenesis of a gene (irpA) involved in iron-deficient growth of the cyanobacterium Synechococcus sp. strain PCC7942. J. Bacteriol. 170:4466-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Renalier, M. H., J. Batut, J. Ghai, B. Terzaghi, M. Gherardi, M. David, A. M. Garnerone, J. Vasse, G. Truchet, T. Huguet, and P. Boistard. 1987. A new symbiotic cluster on the pSym megaplasmid of Rhizobium meliloti 2011 carries a functional fix gene repeat and a nod locus. J. Bacteriol. 169:2231-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rolfe, B. G., P. M. Gresshoff, and J. Shine. 1980. Rapid screening for symbiotic mutants of Rhizobium meliloti and white clover. Plant Sci. Lett. 19:277-284. [Google Scholar]
- 51.Rüberg, S., Z. X. Tian, E. Krol, B. Linke, F. Meyer, Y. Wang, A. Pühler, S. Weidner, and A. Becker. 2003. Construction and validation of a Sinorhizobium meliloti whole genome DNA microarray: genome-wide profiling of osmoadaptive gene expression. J. Biotechnol. 106:255-268. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 53.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 54.Schwartz, C. J., J. L. Giel, T. Patschkowski, C. Luther, F. J. Ruzicka, H. Beinert, and P. J. Kiley. 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. USA 98:14895-14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 56.Shouldice, S. R., D. R. Dougan, P. A. Williams, R. J. Skene, G. Snell, D. Scheibe, S. Kirby, D. J. Hosfield, D. E. McRee, A. B. Schryvers, and L. W. Tari. 2003. Crystal structure of Pasteurella haemolytica ferric ion-binding protein A reveals a novel class of bacterial iron-binding proteins. J. Biol. Chem. 278:41093-41098. [DOI] [PubMed] [Google Scholar]
- 57.Simon, R., U. B. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 58.Singh, A. K., L. M. McIntyre, and L. A. Sherman. 2003. Microarray analysis of the genome-wide response to iron deficiency and iron reconstitution in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 132:1825-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith, M. J., J. N. Shoolery, B. Schwyn, I. Holden, and J. B. Neilands. 1985. Rhizobactin, a structurally novel siderophore from Rhizobium meliloti. J. Am. Chem. Soc. 107:1739-1743. [Google Scholar]
- 60.Sourjik, V., W. Sterr, J. Platzer, I. Bos, M. Haslbeck, and R. Schmitt. 1998. Mapping of 41 chemotaxis, flagellar and motility genes to a single region of the Sinorhizobium meliloti chromosome. Gene 223:283-290. [DOI] [PubMed] [Google Scholar]
- 61.Spaink, H. P. 2000. Root nodulation and infection factors produced by rhizobial bacteria. Annu. Rev. Microbiol. 54:257-288. [DOI] [PubMed] [Google Scholar]
- 62.Staggs, T. M., J. D. Fetherston, and R. D. Perry. 1994. Pleiotropic effects of a Yersinia pestis fur mutation. J. Bacteriol. 176:7614-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stojiljkovic, I., B. D. Evavold, and V. Kumar. 2001. Antimicrobial properties of porphyrins. Expert Opin. Investig. Drugs 10:309-320. [DOI] [PubMed] [Google Scholar]
- 64.Todd, J. D., M. Wexler, G. Sawers, K. H. Yeoman, P. S. Poole, and A. W. Johnston. 2002. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology 148:4059-4071. [DOI] [PubMed] [Google Scholar]
- 65.Todd, J. D., G. Sawers, K. H., and A. W. Johnston. 2005. Proteomic analysis reveals the wide-ranging effects of the novel, iron-responsive regulator RirA in Rhizobium leguminosarum bv. viciae. Mol. Genet. Genomics 273:197-206. [DOI] [PubMed] [Google Scholar]
- 66.Touati, D., M. Jacques, B. Tardat, L. Bouchard, and S. Despied. 1995. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vincent, J. M. 1970. A manual for the practical study of root nodule bacteria. IBP handbook no. 15. Oxford University Press, Oxford, United Kingdom.
- 68.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei, X., and W. D. Bauer. 1998. Starvation-induced changes in motility, chemotaxis, and flagellation of Rhizobium meliloti. Appl. Environ. Microbiol. 64:1708-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wexler, M., J. D. Todd, O. Kolade, D. Bellini, A. M. Hemmings, G. Sawers, and A. W. Johnston. 2003. Fur is not the global regulator of iron uptake genes in Rhizobium leguminosarum. Microbiology 149:1357-1365. [DOI] [PubMed] [Google Scholar]
- 71.Wexler, M., K. H. Yeoman, J. B. Stevens, N. G. de Luca, G. Sawers, and A. W. Johnston. 2001. The Rhizobium leguminosarum tonB gene is required for the uptake of siderophore and haem as sources of iron. Mol. Microbiol. 41:801-816. [DOI] [PubMed] [Google Scholar]
- 72.Wrangstadh, M., P. L. Conway, and S. Kjelleberg. 1986. The production and release of an extracellular polysaccharide during starvation of a marine Pseudomonas sp. and the effect thereof on adhesion. Arch. Microbiol. 145:220-227. [DOI] [PubMed] [Google Scholar]
- 73.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yeoman, K. H., A. R. Curson, J. D. Todd, G. Sawers, and A. W. Johnston. 2004. Evidence that the Rhizobium regulatory protein RirA binds to cis-acting iron-responsive operators (IROs) at promoters of some Fe-regulated genes. Microbiology 150:4065-4074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.