Abstract
The transcription factors Mixer and Sox17β have well characterized roles in endoderm specification during Xenopus embryogenesis. In order to more thoroughly understand the mechanisms by which these endodermal regulators act, we expressed Mixer and Sox17β in naïve ectodermal tissue and, using oligonucleotide-based microarrays, compared their genomic transcriptional profile to that of unaffected tissue. Using this novel approach, we identified 71 transcripts that are upregulated by Mixer or Sox17β, 63 of which have previously uncharacterized roles in endoderm development. Furthermore, an in situ hybridization screen using antisense probes for several of these clones identified six targets of Mixer and/or Sox17β that are expressed in the endoderm during gastrula stages, providing new and regional markers of the endoderm. Our results contribute further insight into the functions of Mixer and Sox17β and bring us closer to understanding at the molecular level the pathways that regulate endoderm development.
Keywords: Mixer, Sox17, endoderm, Xenopus, microarray, Xtwik2, Borg4, March8, Gpr4, Cxcr4
Introduction
The endoderm is one of the three primary germ layers established during early vertebrate embryogenesis. The integrity of this germ layer is crucial to an organism’s survival as the cells of the endoderm will go on to form the gut epithelium and associated organs such as the liver and pancreas. The endoderm is also a source of instructive cues, providing developmental signals to structures in the embryo such as the head and heart (Nascone and Mercola, 1995; Bouwmeester et al., 1996; Beddington and Robertson, 1998; Couly et al., 2002).
Recent interest in vertebrate endoderm development has launched several studies over the past few years in which a handful of molecules involved in endoderm formation were identified (reviewed in Shivdasani, 2002; Stainier, 2002). VegT, a T-box transcription factor, is the primary maternal regulator of endoderm specification in Xenopus (Horb and Thomsen, 1997; Zhang et al., 1998; Xanthos et al., 2001). The maternal localization of VegT mRNA to the vegetal pole and the subsequent initiation of zygotic factors including Nodal-like molecules and transcription factors such as Mixer and Sox17 are the forces governing early endoderm development. Mixer is a paired-like homeodomain transcription factor identified in a functional screen for endodermal determinants in Xenopus (Henry and Melton, 1998). Sox17α and Sox17β are HMG domain-containing transcription factors identified in a subtractive PCR screen for endodermally enriched genes in Xenopus (Hudson et al., 1997). Both Mixer and Sox17β are expressed exclusively in the Xenopus endoderm, can induce endodermal cell fate in naïve ectodermal tissue and are required for proper endogenous endoderm development (Hudson et al., 1997; Henry and Melton, 1998). Mixer and Sox17 β are functionally conserved, as they are involved in Zebrafish and mouse endoderm specification (Kikuchi et al., 2000; Alexander and Stanier, 1999; Pearce and Evans, 1999; Kanai-Azuma et al., 2002). Both Mixer and Sox17β are induced by Nodal-like signals in the Xenopus embryo and have been placed in a hierarchical pathway leading to endoderm specification, with Mixer upstream of Sox17β based on its ability to induce Sox17β in naïve ectodermal explants (Henry and Melton, 1998; Shivdasani, 2002; Stainier, 2002). Genetic evidence in Zebrafish supports this epistatic relationship (Alexander et al., 1999; Alexander and Stainier, 1999; Aoki et al., 2002; Shivdasani, 2002).
Despite the extensive studies on the roles of molecules like Mixer and Sox17 β in endoderm specification, little is known about the genes that are regulated by these transcription factors. It is likely that each induces a set of molecules that define endodermal cell fate. In this report, we use oligonucleotide-based microarray technology developed by Affymetrix to identify molecules that are upregulated by Mixer, Sox17β or both. Interestingly, despite the somewhat linear progression of events thought to establish endodermal cell fate, most of the transcripts we identified were downstream of Mixer or Sox17β with few downstream of both.
Results
Experimental set-up and data interpretation
We used a genomic microarray approach to identify molecules involved in early endoderm formation. Our experimental strategy was to transform naïve ectodermal tissue into endoderm and assay the change in genome-wide gene expression (see Fig. 1). Although VegT is the farthest known upstream endoderm inducer, we did not select this molecule for our experiments because VegT can also induce mesoderm (Stennard et al., 1996). Mixer and Sox17β, on the other hand, induce endoderm in the absence of mesoderm induction and were thus selected for our experiments based on their specificity.
Fig. 1. Schematic of experimental strategy.
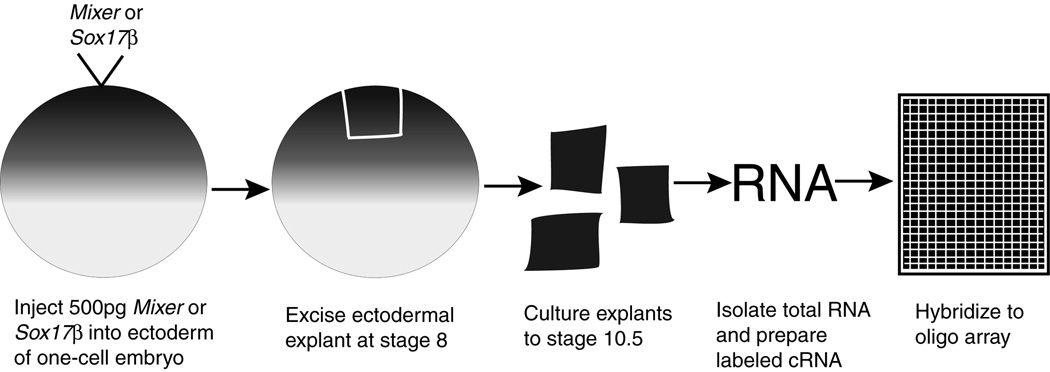
This is a simplified visualization of the experimental procedure, which begins with injection of Mixer or Sox17β into the embryo and ends with an output of data from microarrays.
To transform ectoderm into endoderm, we injected 500 pg of Mixer or Sox17β into the animal hemisphere of one-cell Xenopus embryos. At stage 8.0 (just prior to the onset of zygotic gene expression), we performed ectodermal explants and cultured the isolated tissue to stage 10.5. This is the point at which the endodermal germ layer is well established and is involuting in intact sibling embryos. We then isolated total RNA and prepared it for array hybridization (see Experimental Procedures). This entire process was repeated to generate a second set of data, which we will refer to as experiment 2.
Using the Gene Chip Operating Software provided by Affymetrix, the arrays were scanned and individual intensities for each oligo spot were assigned numerical values and averaged for each probe set. The data discussed in this publication have been deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE3334. We interpreted these values using DNA-Chip Analyzer (Li and Wong, 2001; see Experimental Procedures) and created three lists based on changes in intensities for individual probe sets within an experimental sample when compared to the uninjected control. The first list contains genes and ESTs that are induced by Mixer and Sox17β (Table 1). The second list contains genes and ESTs that are induced by Mixer, but not reliably by Sox17β (Table 2). The third list contains genes that are induced by Sox17β, but not reliably by Mixer (Table 3).
TABLE 1.
Genes induced by Mixer and Sox17β
| Name |
Accession number |
Unigene cluster |
Domains/ homology/ description |
Fold Change by Mixer |
Fold Change by Sox17β |
|---|---|---|---|---|---|
| FoxA1 | BC047130 | Xl.888 | Forkhead box A1, (Xfkh2, Hnf3 β) | 2.0, 2.6 | 5.0, 4.0 |
| Cxcr4 | BC044963 | Xl.11336 | C-X-C motif chemokine receptor 4 | 2.4, 2.6 | 2.2, 3.3 |
| Frzb-1 | U68059 | Xl.212 | Frizzled-related protein precursor | 5.0, 7.1 | 3.9, 3.0 |
| Cpeb | U14169 | Xl.984 | Cytoplasmic polyadenylation element binding protein | 2.7, 8.0 | 5.0, 26.5 |
| X-msr | U72029 | Xl.23649 | Angiotensin receptor related protein (AKA Xangio1) | 3.6, 10.6 | 2.6, 5.5 |
| p30 B9.10 | X73317 | Xl.1244 | Maternal B9.10 protein | 2.3, 2.0 | 3.5, 3.1 |
| EST-1 | CA986927 | Xl.8458 | similar to human March8 | 1.8, 4.0 | 2.0, 3.7 |
| EST-2 | BJ048011 | Xl.16876 | similar to human March8 | 2.5, 4.0 | 2.2, 3.3 |
| EST-3 | AW199159 | Xl.25556 | similar to human c-Myc target JPO1 | 2.9, 5.7 | 4.6, 15.2 |
| EST-4 | CA790591 | Xl.21726 | frog gene | 6.3, 11.1 | 3.2, 4.7 |
TABLE 2.
Genes induced by Mixer only
| Name |
Accession number |
Unigene cluster |
Domains/ homology/ description |
Fold Change by Mixer |
Fold Change by Sox17β |
|---|---|---|---|---|---|
| Msx2a*† | AW766492 | Xl.31078 | Msh homeobox homolog 2 (Hhox-7.1') | 6.9, 8.2 | −1.5, −1.4 |
| 3.9, 2.6 | 1.1, −1.2 | ||||
| P7E4 | AB072006 | Xl.34957 | BTB (POZ) domain containing 3 | 4.7, 2.8 | −1.2, −2.0 |
| Wnt-11 | L23542 | Xl.44504 | Maternally expressed wnt gene | 3.8, 5.9 | 1.3, 1.0 |
| Sox17α | AJ001730 | Xl.3831 | SRY-related HMG-box transcription factor | 6.3, 9.7 | 1.3, 1.2 |
| Sox17-α2 | AB052691 | Xl.11957 | SRY-related HMG-box transcription factor | 7.1, 11.8 | 1.3, 1.1 |
| Sox17-β | BC070615 | Xl.44 | SRY-related HMG-box transcription factor | 13.0, 17.4 | 163.4, 280.4‡ |
| Mab-21 | AF040992 | Xl.279 | Xenopus Mab21l1 | 3.9, 7.9 | 1.4, 3.2 |
| Gata-5a | L13701 | Xl.578 | Zinc finger transcription factor | 2.0, 3.0 | −1.2, −1.7 |
| Xeel † | AB105372 | Xl.6266 | Embryonic epidermal lectin | 10.1, 21.9 | 1.3, −1.1 |
| Mig30 | AB035379 | Xl.34912 | Mixer inducible gene 30 | 2.5, 4.6 | −1.1, −1.0 |
| c-myc | M14455 | Xl.826 | Myelocytomatosis oncogene | 2.6, 3.5 | 1.1, −1.3 |
| EST-5 | BJ055630 | Xl.9623 | likely ortholog of mouse Epsin 2 | 2.2, 2.5 | −1.3, −1.2 |
| EST-6 | BG555687 | Xl.1321 | similar to Centromere/kinetochore protein Zw10 | 2.4, 7.6 | −1.1, −1.9 |
| EST-7 | CB561069 | Xl.7842 | similar to Serum/glucocorticoid regulated kinase | 3.0, 6.4 | −2.3, −1.4 |
| EST-8 | BC060483 | Xl.14214 | similar to human transmembrane protein Claudin 5 | 3.5, 2.1 | 1.0, 1.0 |
| EST-9* | BC045272 | Xl.8630 | similar to human Serum inducible kinase | 3.0, 5.6 | 1.3, 1.4 |
| 2.6, 4.7 | 1.2, 1.0 | ||||
| EST-10 | BJ046058 | Xl.9284 | similar to human GT box-binding protein Sp3 | 4.2, 2.0 | 1.4, 1.7 |
| EST-11 | BG020193 | Xl.2466 | similar to human I38026 MLN 62 protein | 2.6, 3.6 | −1.1, 1.1 |
| EST-12 | BJ087388 | Xl.16135 | similar to human K-sam precursor | 2.0, 2.0 | −1.0, −1.5 |
| EST-13 | BG486882 | Xl.4337 | similar to human Sec4 GTP binding protein | 2.1, 4.4 | 1.1, 1.0 |
| EST-14 | BC041294 | Xl.13019 | similar to human RAS-like protein | 2.3, 3.8 | −1.9, −1.3 |
| EST-15 | BJ044041 | Xl.16547 | similar to human Borg4 | 2.0, 2.8 | −1.2, −2.7 |
| EST-16 | BJ100112 | Xl.13033 | similar to human Chemokine-like super family 8 | 3.0, 9.1 | 1.1, 1.8 |
| EST-17*† | BJ085828 | Xl.3435 | frog gene | 7.2, 6.7 | −2.3, −2.1 |
| 7.3, 5.9 | −2.1, −2.0 | ||||
| EST-18 | BJ051730 | Xl.15365 | frog gene | 2.0, 4.3 | −1.7, −1.2 |
| EST-19 † | BJ056057 | Xl.10150 | frog gene | 7.1, 23.9 | 1.4, 1.0 |
| EST-20 | BJ086610 | Xl.13363 | frog gene | 3.0, 2.1 | −1.1, −1.1 |
| EST-21* | BJ075680 | Xl.15089 | frog gene | 1.6, 11.5 | 1.2, 1.1 |
| 1.5, 6.4 | 1.2, 1.2 | ||||
Indicates transcripts represented more than once on the array and meet the required standards twice in our data set for experiments 1 and 2.
Represents transcripts whose induction by Mixer was greater than 5.0 for experiments 1 and 2.
Number reflects measure of mRNA present due to injection.
TABLE 3.
Genes induced by Sox17β only
| Name |
Accession number |
Unigene cluster |
Domains/ homology/ description |
Fold Change by Mixer |
Fold Change by Sox17β |
|---|---|---|---|---|---|
| ElrD | u17599 | Xl.1036 | RNA-binding protein HuD | 1.0, 1.0 | 2.6, 10.1 |
| Clast3 | CB561662 | Xl.10510 | CD40 ligand-activated specific transcript 3 | 1.2, −1.0 | 2.6, 3.0 |
| Otx2*† | AW199379 | Xl.3004 | Orthodenticle-A like homeobox protein | 1.7, 1.2 | 6.2, 8.1 |
| 1.2, 1.1 | 2.9, 5.2 | ||||
| Irx4 † | AF338157 | Xl.12086 | Iroquois-4 homeobox transcription factor | 1.3, 1.1 | 14.5, 18.6 |
| Nr2f2 | BC044975 | Xl.14532 | Nuclear receptor subfamily 2, group F, member 2 | −1.0, 1.0 | 2.0, 2.8 |
| FoxC1 | AF116844 | Xl.180 | Winged helix transcription factor (Xfd-11) | 1.8, 1.2 | 2.5, 4.1 |
| Pdgf | BC043948 | Xl.20029 | Platelet derived growth factor receptor alpha | 1.2, 1.4 | 2.4, 2.3 |
| Crgb † | AF071563 | Xl.21441 | Crystallin, gamma B | 1.1, 1.1 | 10.1, 21.2 |
| Eomes | U75996 | Xl.373 | T-domain gene Eomesodermin | 1.5, 1.1 | 2.8, 3.0 |
| Ash1 † | M98272 | Xl.450 | Achaete-scute protein homologue | 1.3, 1.6 | 5.4, 14.3 |
| Xgam | M63446 | Xl.5871 | Gamma-tubulin | 1.3, 1.6 | 2.1, 2.9 |
| Foxd1-A | AJ011652 | Xl.66 | Forkhead box D1 (brain factor 2, Xbf-2) | 1.3, 1.2 | 2.6, 4.1 |
| Gsc*† | BJ056432 | Xl.801 | Goosecoid homeobox protein | 1.1, −1.1 | 5.0, 4.1 |
| 2.5, 1.3 | 16.3, 17.5 | ||||
| Gpr-4 | AY553187 | Xl.45565 | G-protein coupled receptor 4 | 1.3, 2.2 | 2.6, 4.3 |
| Xlim-1 | CB562197 | Xl.32655 | LIM domain-containing homeobox protein | 1.2, 1.1 | 2.4, 2.5 |
| Xtwi | M27730 | Xl.879 | bHLH Twist homolog 1 | 2.2, −1.1 | 9.3, 17.7 |
| EST-22 † | BG810694 | Xl.2565 | similar to alpha-Tubulin at 84B | −1.1, 1.0 | 84.2, 106.2 |
| EST-23 | BI478249 | Xl.19057 | similar to human amino acid transporter | −1.1, −1.1 | 2.8, 5.7 |
| EST-24 | BM191866 | Xl.7085 | similar to human hypothetical protein FLJ20511 | 1.1, 1.2 | 2.0, 2.3 |
| EST-25 | BJ044317 | Xl.1419 | similar to human Interferon regulatory factor 1 | −1.0, 1.1 | 8.0, 2.3 |
| EST-26 | BC041234 | Xl.16040 | similar to human Phosphatidylserine decarboxylase | 1.2, 1.5 | 2.0, 3.0 |
| EST-27* | BG021407 | Xl.34405 | similar to human new Ets-related factor | 1.4, 1.1 | 2.3, 3.8 |
| 1.1, 1.1 | 2.9, 4.9 | ||||
| EST-28 | BJ043709 | Xl.8559 | similar to human Protocadherin 10, isoform 1 precursor |
1.1, 1.4 | 4.6, 9.2 |
| EST-29 | CA792418 | Xl.1295 | similar to human HRAS1-related cluster protein 1 | −1.1, −1.0 | 3.7, 5.1 |
| EST-30 | BJ054524 | Xl.16561 | similar to human Chloride channel protein CLC-KA | −1.1, −1.0 | 2.9, 16.0 |
| EST-31 | BM172631 | Xl.16695 | similar to human Elongation of very long chain fatty acids |
1.3, −1.0 | 3.5, 4.4 |
| EST-32 | BI348356 | Xl.18627 | similar to human hypothetical protein DKFZp586N041.1 |
1.4, 1.5 | 2.1, 2.1 |
| EST-33 | BQ385449 | Xl.19414 | similar to human Rsu-1 homolog | 1.1, 1.1 | 2.8, 3.7 |
| EST-34 † | BJ044473 | Xl.15931 | similar to mouse Doublesex- and mab-3-related txn factor |
1.3, 1.1 | 6.3, 6.3 |
| EST-35 † | BJ048106 | Xl.23586 | likely paralog of Xtwik2 | 1.3, 1.7 | 8.5, 11.4 |
| EST-36 | BJ085642 | Xl.13426 | similar to human C2orf17 unnamed protein | 1.3, 1.1 | 2.4, 2.2 |
| EST-37 | BJ079356 | Xl.14106 | frog gene | 1.4, 2.3 | 2.7, 2.6 |
| EST-38 | BG023545 | Xl.2439 | frog gene | 1.3, 2.0 | 2.5, 5.2 |
Indicates transcripts represented more than once on the array and meet the required standards twice in our data set for experiments 1 and 2.
Represents transcripts whose induction by Sox17β was greater than 5.0 for experiments 1 and 2.
10 transcripts were induced by Mixer and Sox17β
To identify molecules that are induced by Mixer and Sox17β, we generated four initial lists of genes and ESTs from experiments 1 and 2 that had 1.8 fold or higher intensities in the Mixer and Sox17β injected samples versus control with a false detection rate (FDR) ≤ 0.10. Transcripts that appeared on all four lists were incorporated into a final list shown in Table 1. Using this screening method, six known genes and four ESTs were identified as downstream targets of both Mixer and Sox17β. Of the known genes, FoxA1 (Xfkh2, Hnf3α) has previously been shown to be expressed in the early endoderm and activated by Sox17β in ectodermal explants (Bolce et al., 1993; Sinner et al., 2004), but this is the first report of FoxA1 as a target for Mixer. The remaining five genes and four ESTs have not been characterized in terms of endoderm development.
28 transcripts were induced by Mixer
A wealth of evidence indicates that Mixer is upstream of Sox17β in the pathway of gene expression leading to endoderm specification (Henry and Melton, 1998; Alexander et al., 1999; Alexander and Stainier, 1999; Aoki et al., 2002; Shivdasani, 2002). Using data from our microarrays, we found additional molecules downstream of Mixer. We initially generated two list of genes and ESTs from experiments 1 and 2 that contained probe sets with a ≥ 2.0 increase in intensity in the Mixer samples versus controls. Transcripts that appeared on both lists with a FDR ≤ 0.10 were selected for Table 2. One exception is EST-21, which appeared twice in our data for experiment 2, with a fold change of 11.5 and 6.4 above control, but in experiment 1 had a fold change of 1.6 and 1.5. Despite the lower fold changes in experiment 1, EST-21 had a FDR of 0.01 and 0.03 and was therefore included in the list. Other transcripts that appeared more than once on our list are Msx-2a, EST-9 and EST-17 and are indicated with an asterisk in Table 2. It is also worth noting the few transcripts that had fold changes of greater than 5.0 in both experiments (indicated with a †). These include Msx-2a (6.9, 8.2), Xeel (10.1, 21.9), EST-17 (7.2, 6.7; 7.3, 6.9) and EST-19 (7.1, 23.9).
We expected to see several of the genes in Table 2, including Sox17α, Sox17α2, Sox17β, Gata-5a and Mig30. All of these are known targets of Mixer or have been previously characterized for their roles in endoderm formation. All three Sox17-like transcripts have large fold change values ranging from 6.3 to 17.4. The remaining genes and ESTs in Table 2 have not been investigated for their role in endoderm formation.
33 transcripts were induced by Sox17β
For several years after Sox17β was discovered, the only known transcriptional targets for Sox17β during gastrula stage were Endodermin and Hnf1β (Tcf2; Hudson et al., 1997; Clements et al., 2003). Recently investigators used a candidate gene approach to screen for Sox17β targets and identified nine additional downstream genes (Sinner et al., 2004). In our array experiment, we identified 34 potential targets for Sox17β using the same selection criteria we described for identifying Mixer targets (Table 3). Of the 16 known genes identified, Otx2 and Goosecoid have been shown to be targets for Sox17β (Chiao et al., 2005; Sinner et al., 2004). In fact, genetic experiments in the mouse suggest a role for Otx2 in early endoderm development (Perea-Gomez et al., 2001). Otx2 and Goosecoid appear twice in each data set and have fold changes greater than 5.0. Other transcripts that have fold changes ≥ 5.0 include Irx4 (14.5, 18.6), Crgb (10.1, 21.2), Ash1 (5.4, 14.3), Xtwi (9.3, 17.7), EST-22 (84.2, 106.2), EST-35 (8.5, 11.4) and EST-34 (6.3, 6.3). EST27 appears twice in this data set. All of the genes and ESTs in Table 3, except for Goosecoid and Otx2, are novel targets for Sox17β and do not have a known role in endoderm development.
6 transcripts exhibit novel expression in the early endoderm
We have identified 71 targets for Mixer and/ or Sox17β, 63 of which may have a completely novel role in endoderm development. To determine whether these are targets for Mixer and Sox17β within the endoderm, we analyzed the expression pattern at gastrulation and later stages for 34 of the most promising transcripts identified. During gastrulation, six of the transcripts were expressed in the endoderm, two in the mesoderm, and eleven were ubiquitous. The remaining fifteen did not have detectable expression (see Table 4). Several of these clones had unique and detectable patterns of expression at later developmental times (Fig. 2). Since the molecules expressed within the endoderm during gastrulation were more likely to be endogenous targets of Mixer and Sox17β, we investigated these further and describe them in detail below (Fig. 3). They include the following molecules: Cxcr4, EST1 (March8), EST15 (Borg4), EST-21, Gpr-4 and EST35 (Xtwik2). The sequence conservation between several of these ESTs and proteins in the mouse and human database strongly suggest that they are the correct homologs. Therefore, from this point forward we will refer to EST1 as March8, EST15 as Borg4 and EST35 as Xtwik2.
TABLE 4.
In situ hybridization results
| Name |
Accession number |
Expression |
Figure Reference |
||
|---|---|---|---|---|---|
| St 10.5 | St 15 | St 30 | |||
| Cxcr4 | BC044963 | endo | X* | X* | 2 A,C |
| Cpeb | U14169 | ubiq | |||
| p30 B9.10 | X73317 | none | |||
| March8 | CA986927 | endo | 2 E,G | ||
| EST-2 | BJ048011 | ubiq | |||
| EST-3 | AW199159 | none | |||
| EST-4 | CA790591 | ubiq | |||
| Xeel | AB105372 | ubiq | |||
| EST-8 | BC060483 | none | X | X | 4 D |
| EST-9 | BC045272 | none | X | 4 H | |
| EST-10 | BJ046058 | none | X | X | 4 B |
| EST-11 | BG020193 | meso | X | X | 4 A |
| EST-14 | BC041294 | meso | X | 4 C | |
| Borg4 | BJ044041 | endo | 2 I,K | ||
| EST-16 | BJ100112 | ubiq | |||
| EST-17 | BJ085828 | ubiq | |||
| EST-19 | BJ056057 | none | X | 4 G | |
| EST-20 | BJ086610 | ubiq | |||
| EST-21 | BJ075680 | endo | X | 2 M,O; 3 A | |
| Gpr-4 | AY553187 | endo | X | X | 2 Q,S; 3 C,E |
| EST-22 | BG810694 | ubiq | |||
| EST-23 | BI478249 | ubiq | |||
| EST-24 | BM191866 | ubiq | |||
| EST-25 | BJ044317 | none | |||
| EST-26 | BC041234 | none | |||
| EST-27 | BG021407 | none | |||
| EST-28 | BJ043709 | none | X | 4 F | |
| EST-30 | BJ054524 | none | |||
| EST-33 | BQ385449 | none | |||
| EST-34 | BJ044473 | none | X | X | 4 E |
| Xtwik-2 | BJ048106 | endo | 2 U, W | ||
| EST-36 | BJ085642 | none | |||
| EST-37 | BJ079356 | none | |||
| EST-38 | BG023545 | ubiq | |||
Expression at these stages previously determined (Moepps et al., 2000).
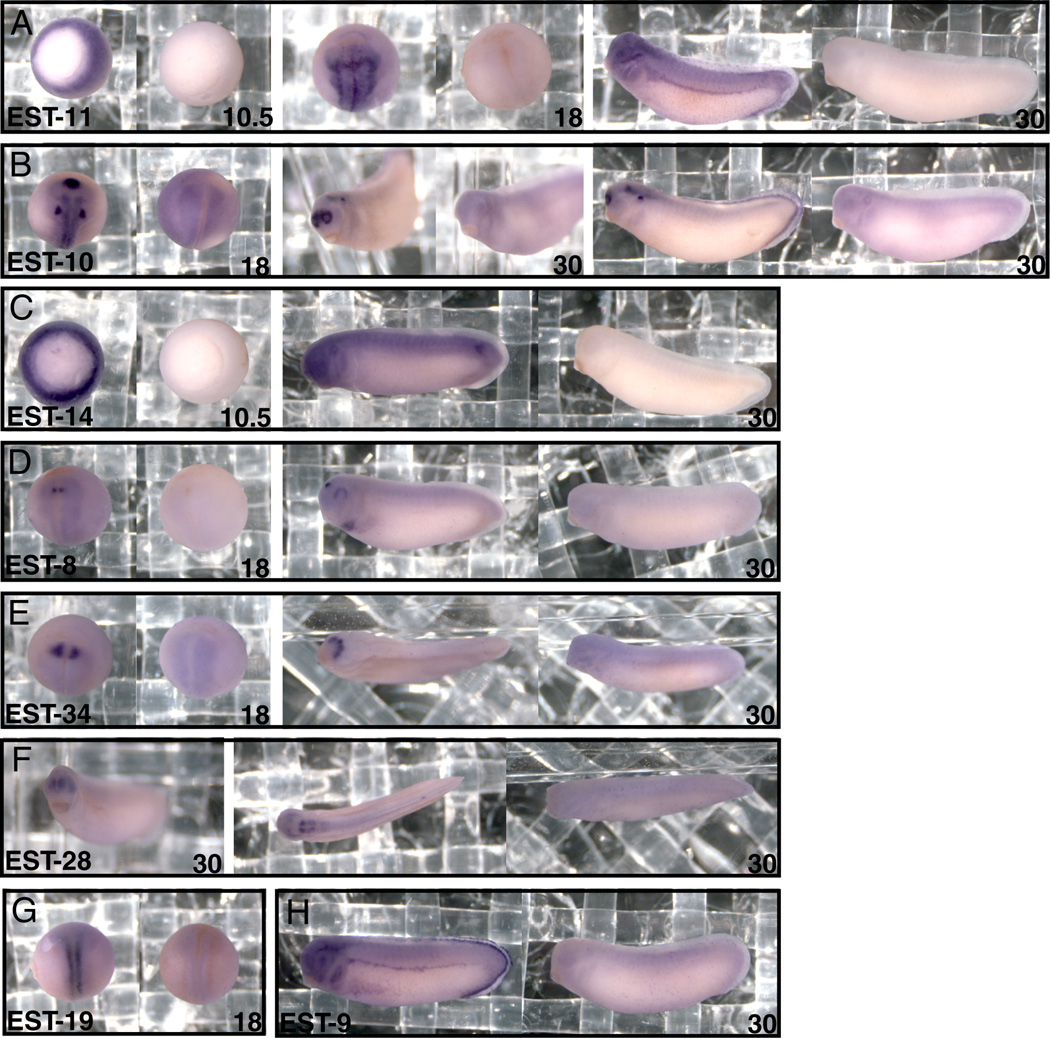
Fig. 2. In situ hybridization screen reveals additional expression patterns.
A: Embryos at gastrula (10.5), neurula (15) and tailbud (30) stage are stained with antisense (left panel) and sense (right panel) probes for EST-11 showing expression in the mesoderm, neural tube, neural crest and pronephros. B: Embryos at neurula and tailbud stage are stained with antisense and sense probes for EST-10 demonstrating expression in cement gland, neural tube nasal placode, otic placod and forebrain. C: Embryos at gastrula and tailbud stage are stained for antisense and sense probes for EST-14 demonstrating expression in mesoderm and in a single posterior somite. D: Embryos at neurula and tailbud stage are stained with antisense and sense probes for EST-8, indicating expression within forebrain and heart. E: Embryos at neurula and tailbud stage are stained with antisense and sense probes for EST-34, showing expression in the placodes. F: Embryos at tailbud stage are stained with antisense and sense probes for EST-28 showing expression in nasal placodes and hindbrain. G: Embryos at neurula stage are stained with antisense and sense probes for EST-19, indicating expression within the neural tube. H: Embryos at tailbud stage are stained with antisense and sense probes for EST-9, demonstrating expression within pronephros and neural crest.
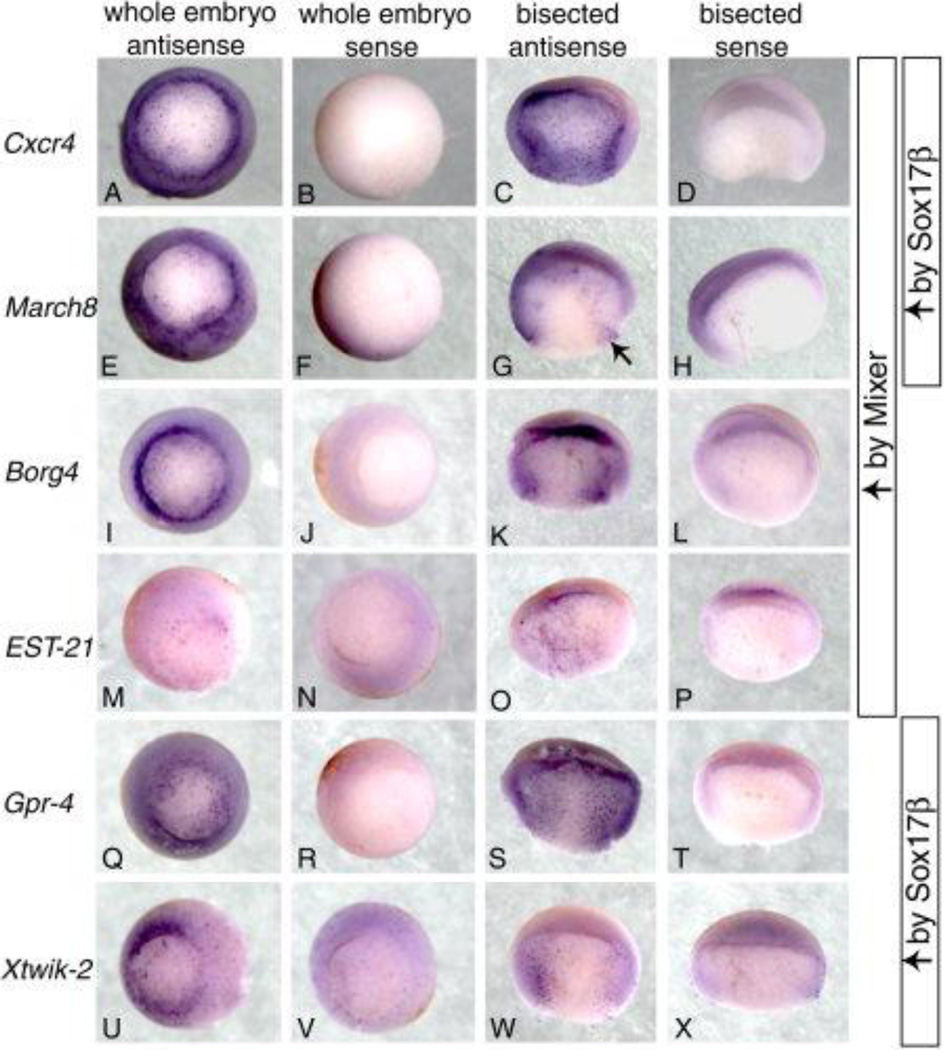
Fig. 3. Six transcripts are expressed in the early endoderm.
The first two columns display stage 10.5 embryos, vegetal view, stained with antisense and sense probes for Cxcr4 March8 Borg4 EST-21 Gpr-4 and Xtwik-2. The last two columns display hemisected stage 10.5 embryos, lateral view, stained with antisense and sense probes for the same transcripts above. The descriptions along right side of figure indicate which transcripts were upregulated with either Mixer, Sox17β or both. Arrow points to the deeper cells adjacent to blastopore ring expressing March8.
Cxcr4
In our array experiments, we observed an increase in the expression of the C-X-C motif chemokine receptor 4 (Cxcr4) in ectoderm expressing Mixer or Sox17β. Our analysis of Cxcr4 expression in Xenopus at stage 10.5 identified novel endodermal expression. In Fig. 3A, we observe Cxcr4 expression scattered throughout the yolky vegetal cells and a high level of expression in the endodermal cells most proximal to the blastopore ring. In the lateral view of a hemisected stage 10.5 embryo stained with Cxcr4 probe (Fig. 3C), we observe Cxcr4 expression throughout the deep endodermal cells and a high level of expression in the cells lining the endoderm/ mesoderm boundary extending down to the involuting cells of the blastopore ring. We also observed a low level of scattered expression throughout the ectoderm (data not shown). Cxcr4 is a member of the superfamily of heterotrimeric-G-protein-coupled receptors originally identified in leukocytes and also known to act as a co-receptor for the entry of HIV into CD4+ lymphocytes (Murphy, 1996). A role for Cxcr4 in the development of the hematopoietic system and vascularization of the gastrointestinal tract has been characterized in mice (Tachibana et al., 1998; Zou et al., 1998). In Xenopus, analysis of Cxcr4 expression from stage 13 and beyond revealed a potential role for Cxcr4 in embryonic neural development and adult B cell differentiation (Moepps et al., 2000).
March8
We identified another transcript that was increased in the presence of either Mixer or Sox17β, which we refer to as March8. Our analysis of March8 revealed expression in the superficial cells of the endoderm (Fig. 3E). We also detected transcript in the deeper cells adjacent to the blastopore ring (Fig. 3G). Some expression of March8 was observed in the ectoderm (data not shown). The predicted amino acid sequence for this transcript is 86% identical to human cellular modulator of immune recognition (c-MIR or March8) and is likely the Xenopus ortholog of this gene. Human March8 was identified based on its secondary structure similarity to two related proteins, MIR1 and 2, encoded by Kaposi’s sarcoma associated-herpes virus (Goto et al., 2003). This protein functions as a membrane-bound E3 ubiquitin ligase and contains a BKS-PHD catalytic domain responsible the E3 mediated ubiquitination and degradation of immune recognition-related molecules.
Borg4
Our analysis of Borg4, which is induced by Mixer but not by Sox17β, revealed expression in the outer endodermal cells of the blastopore ring (Fig. 3I). Significant expression in the deep endoderm cells beyond the blastopore ring was not observed (Fig. 3K). The predicted amino acid sequence of Borg4 shares 55% identity with the human Cdc42 effector protein, binder of Rho GTPase 4 (Borg4). Borg4 was identified in a two-hybrid screen of a mouse embryo library for molecules that bound to the TC10 GTPase (Joberty et al., 1999). The Rho family of GTPases regulates multiple biological processes including cell motility, morphogenesis, protein kinase cascades, gene expression and cell cycle progression. GTPase specificity is thought to be regulated by downstream effector proteins such as those of the Borg family, which are proposed negative regulators of Rho GTPase signaling (Joberty et al., 1999). The mouse homolog of Borg4 is ubiquitously expressed in adult tissue (Osada et al., 2000).
EST-21
A second transcript we identified as a target for Mixer is EST-21. Our analysis of EST-21 revealed only selective staining of cells in the deep endoderm domain (Fig. 3M, O). This transcript does not share sequence homology with any clone in the database and may be a frog-specific gene.
Gpr-4
In addition to targets for Mixer, we also identified several transcripts induced only by Sox17β. One of these is the Xenopus ortholog of the human G protein-coupled receptor 4 (Gpr-4). Our analysis of Gpr-4 in Xenopus revealed expression in the inner cells of the superficial endoderm layer with little or no expression around the blastopore ring region (Fig. 3Q). In the hemisected embryos, we observed scattered expression throughout the deep endodermal tissue (Fig. 3S). G protein-coupled receptors are known to transduce numerous extracellular signals into cells and regulate various aspects of cell proliferation (Marinissen and Gutkind, 2001). Recently, Gpr-4 was identified in a separate microarray experiment as a target gene for FGF signaling in Xenopus (Chung, et al., 2004). In this study, Gpr-4 transcript was also observed in the endoderm and functional analysis of this gene revealed a potential role for Gpr-4 in regulating gastrulation movements (Chung et al., 2004). Gpr-4 also has oncogenic properties and has been shown to regulate pH and ERK activity (Sin et al., 2004; Ludwig et al., 2003; Bektas et al., 2003). Expression of human Gpr-4 has been observed in the kidney, heart and lung (Mahadevan et al., 1995). Interestingly, Gpr-4 shares some sequence homology with the angiotensin receptor, a gene we found to be induced by Mixer and Sox17β (see Table 1).
Xtwik-2
A second transcript we identified as a potential target for Sox17β is Xtwik-2. Our analysis of Xtwik-2 revealed expression in the endodermal cells surrounding the blastopore ring (Fig. 3U). In the hemisected embryos, we observed expression in the cells proximal to the adjacent mesoderm, extending from the blastopore ring up to the blastocoel floor with much fewer cells expressing Xtwik-2 in the innermost endoderm tissue (Fig. 3W). The predicted amino acid sequence of this clone is nearly identical Xtwik-2. However, the nucleotide sequence is more divergent and thus may be a close paralog of Xtwik-2. Human Twik-2 is known to regulate cell electrogenesis and is expressed in the pancreas, stomach, spleen and uterus (Chavez et al., 1999; Medhurst et al., 2001).
EST-21 and Gpr-4 have novel expression patterns beyond stage 10.5
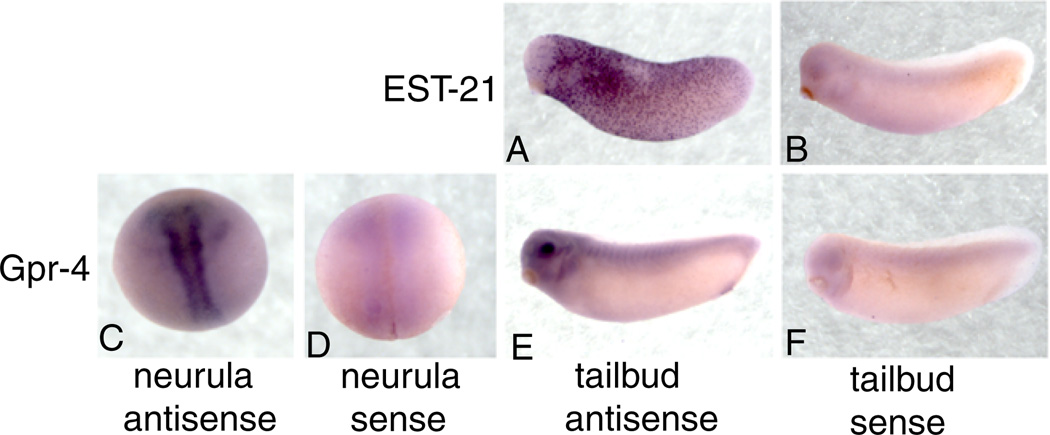
We analyzed the expression patterns of the six clones expressed in the early endoderm at later stages. Cxcr4 displayed expression patterns identical to those published (Moepps et al., 2000). EST-1, EST-15 and EST-35 did not have specific expression patterns at stages beyond 10.5. At tailbud stage, EST-21 is expressed throughout the epidermis in a scattered punctate pattern with less staining in the head region (Fig. 4A). Gpr-4 is expressed along the neural tube in neurula stage embryos and in the eye and brachial arches in tailbud stage embryos (Fig. 4C, E).
Fig. 4. EST-21 and Gpr-4 have additional patterns during later embryonic stages.
A and B: Embryos at tailbud stage are stained with antisense and sense probes for EST-21. C and D: Neurula stage embryos are stained with antisense and sense probes for Gpr-4. E and F: Tailbud stage embryos are stained with antisense and sense probes for Gpr-4.
New endoderm markers respond as predicted by array to endoderm inducers
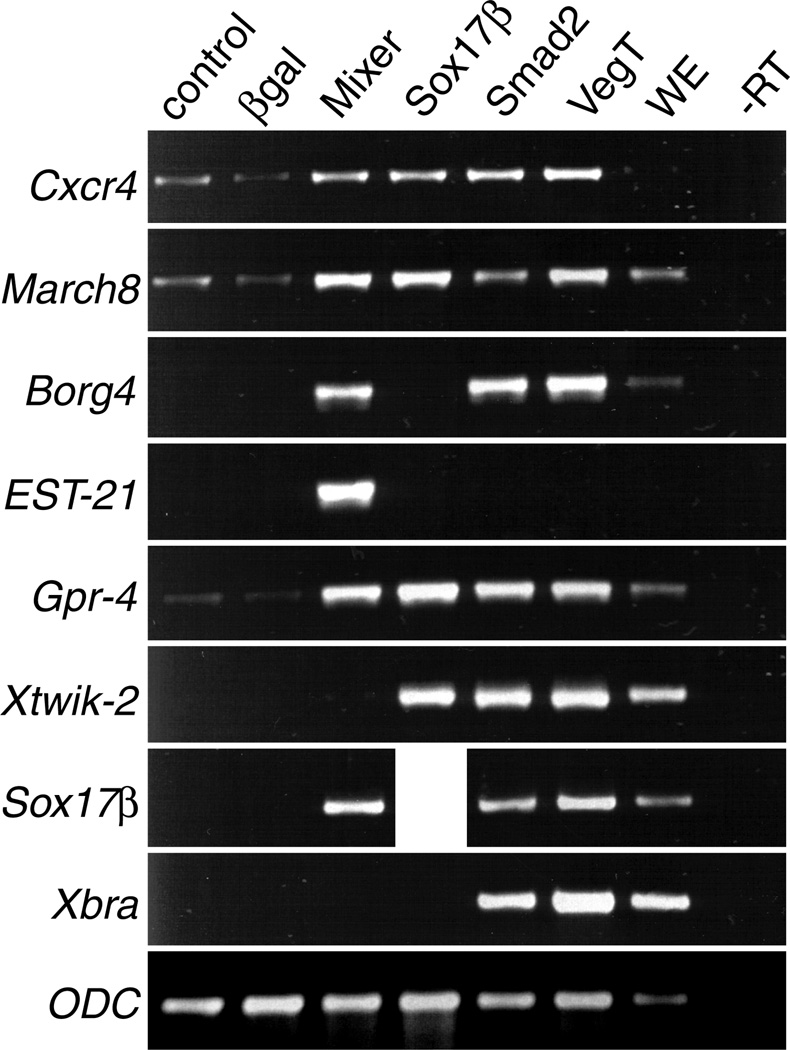
We identified six genes that have novel endoderm expression. We next examined the transcriptional response of these genes to endoderm inducers, Mixer, Sox17β, Smad2 and VegT in ectoderm via RT-PCR in order to confirm the array data and test whether these genes also respond to other known inducers of endodermal cell fate. Based on our array data, we expected to see increased expression of Cxcr4, March8, Borg4 and EST-21 in ectoderm expressing Mixer and increased expression of Cxcr4, March8, Gpr-4 and Xtwik-2 in ectoderm expressing Sox17β. As shown in Fig. 5, all six genes responded to Mixer and Sox17β as predicted. We also observed induction of Gpr-4 by Mixer, which is reflected in the microarray data for experiment 2 (2.2 fold increase), but below our threshold in experiment 1 (1.3 fold increase). Because this fold change did not meet our standards for induction by Mixer, we placed Gpr-4 in Table 3 instead of Table 1.
Fig. 5. Semiquantitative RT-PCR analysis confirms array data and supports an endogenous role for six new molecules in the endoderm pathway.
RT-PCR was performed on cDNA synthesized from ectoderm explants expressing Mixer, Sox17β, Smad2 or VegT with primers for Cxcr4 March8 Borg4 EST-21 Gpr-4 and Xtwik-2 β-gal was injected as a control. Primers for Sox17β and Xbra were used as positive controls. ODC was used as a loading control. WE, whole embryo; -RT, minus reverse transcriptase.
We also examined the transcriptional response of the six new endoderm genes to other endoderm inducers such as Smad2 and VegT. Fig. 5 shows induction or increased expression of Cxcr4, March8, Gpr-4 and Xtwik-2 by Smad2, a subtle increase in March8 expression by Smad2 and no induction of EST-21 by Smad2. VegT increased the expression of Cxcr4, March8 and Gpr-4 above background and clearly induced expression of Borg4 and Xtwik-2. Like Smad2, VegT did not induce EST-21, indicating that it is a Mixer specific target.
Discussion
Aside from adopting a cellular fate capable of becoming the epithelium and organ system of the gastrointestinal tract, the Xenopus endoderm cell has several functions in the early embryo. These include nourishing the embryo through its yolk protein stores, acting as a signaling source to instruct the overlying mesoderm to adopt specific cell fates and migrating to the interior of the embryo while establishing an anterior posterior axis during gastrulation. All of these functions require a unique set of molecules that define endodermal cell fate. Using microarrays, we identified 71 transcripts that are upregulated during the transformation of ectoderm to endoderm. Eight of these had previously described roles during endoderm development providing evidence that we were identifying known targets.
This is the first report that utilizes microarray technology to identify molecules specifically involved in endoderm formation. Many of the known genes that comprise the endoderm pathway have been identified through functional and differential expression screens, candidate gene approaches or genetics (Jones et al., 1995; Baker and Harland, 1996; Lustig et al., 1996; Hudson et al., 1997; Joseph and Melton, 1997; Henry and Melton, 1998; Lemaire et al., 1998; Sun et al., 1999; Yasuo and Lemaire, 1999; Weber et al., 2000; Xanthos et al., 2001; Afouda et al., 2005). Through these various studies, many of the important players in endoderm specification were identified including VegT, Mixer, Sox17α/β, Gata4, 5 and 6, Mix.1, and various components of the Nodal-like TGFβ signaling pathway (Jones et al., 1995; Baker and Harland, 1996; Lustig et al., 1996; Hudson et al., 1997; Joseph and Melton, 1997; Henry and Melton, 1998; Lemaire et al., 1998; Sun et al., 1999; Yasuo and Lemaire, 1999; Weber et al., 2000; Xanthos et al., 2001; Shivdasani, 2002; Stainier, 2002; Afouda et al., 2005). Many of these genes affect the transcription of one another and, perhaps more importantly, an undefined number of downstream target genes (Hudson et al., 1997; Henry and Melton, 1998; Clements et al., 1999; Yasuo and Lemaire, 1999; Engleka et al., 2001; Shivdasani, 2002; Stainier, 2002; Loose and Patient, 2004). It is this collection of unknown target genes that ultimately defines the early endoderm cell. Using microarrays, we were able to take a genome-wide snapshot of the changes in gene expression for a group of cells instructed to become endoderm. We identified 71 potential targets for Mixer and/ or Sox17β, 63 of which may have a completely novel role in defining endodermal cell fate.
The collection of signaling molecules and transcription factors, including Mixer and Sox17, that lead to endoderm specification is often depicted as a linear progression of inductive events (Yasuo and Lemaire, 1999; Shivdasani, 2002; Stainier, 2002). Therefore, we expected to see most of the genes induced by Sox17β also induced by Mixer, given that Mixer is known to induce Sox17β. However, we observed most genes in our array experiments to be downstream of Mixer or Sox17β, with only ten transcripts upregulated by both. Our results may simply reflect the limitations within the assay, with induction of transcripts by Sox17β directly being more robust than the induction of these same transcripts by Mixer via Sox17β. However, we believe our results reflect a non-linear branching of the pathway and highlight the distinct functions of Mixer and Sox17β as they lead to endoderm specification.
We identified six genes that are expressed in the early endoderm through an in situ hybridization screen of 34 transcripts found by our microarray analysis: an 18% success rate. Criticisms will be raised that this is considered a low hit rate due to experimental error from Microarray data. A closer look at the data suggests otherwise. First, of the transcripts that produced expression patterns, 32% were expressed within the endoderm. This is because of the 34 transcripts analyzed, 15 had no detectable expression at gastrula stages. Five of these did demonstrate expression in later staged embryos, suggesting that the probe could detect transcript. The other ten transcripts were not detected at any embryonic stage suggesting that the lack of signal was due to probe failure. Due to penetration problems, insitu hybridizations are less effective in the endoderm than elsewhere in the embryo and extensive troubleshooting is sometimes required to identify the best sequence from each probe for hybridizations. Therefore, we are left questioning whether there are more endoderm specific genes to be uncovered within this grouping. Second, we found 11 transcripts to be ubiquitously expressed. This does not rule out any of these genes as downstream targets of Sox17β or Mixer. Many important regulators of specific tissues are expressed ubiquitously and activated specifically in particular cell types. Whether these eleven are true targets, we have yet to know, but certainly, although they are not exclusively expressed within the endoderm, they cannot be ruled out.
Caveats exist within these interpretations and there are several reasons why not all of the targets identified in this screen will be endogenous players. First, certainly overexpression of molecules as potent as Sox17β and Mixer may lead to non-specific effects by promiscuously activating promoters containing HMG box or homeodomain motifs. Second, indirect inductions may occur between the start of zygotic transcription (stage 8) and the time of analysis (st10.5). In order to obtain direct targets a temporally activated system would need to be employed. Third, our FDR cut-off was < 0.10, indicating that 10% of the transcripts analyzed are indeed background. Be that as it may, we used Microarray technology as a first pass screen to obtain candidate endoderm specific genes and used the secondary in situ screen to validate. This approach effectively identified six uncharacterized endoderm genes, which display regional specific patterns. One of the great difficulties in studying endoderm has historically been a dearth of markers. Here we present the community with six new ones, more than doubling the current collection.
The endoderm specific genes identified in this screen are expressed in discrete locations within the endoderm, representing either subblastoporal or marginal regions (Fig. 3 and Fig. 6). Interestingly, both Mixer and Sox17β induce genes that are expressed in each region. For example, Mixer induces specifically Borg4, which is expressed strongly in the boundary with the mesoderm, and EST21, which is restricted to the subblastoporal cells. Although both genes are induced selectively by Mixer, they display non-overlapping patterns of expression within the endoderm. This indicates that Mixer can induce may complementary cell types within the endoderm and further reflects that endoderm is a heterogeneous population of cells, comprising at least several distinguishable cell types.
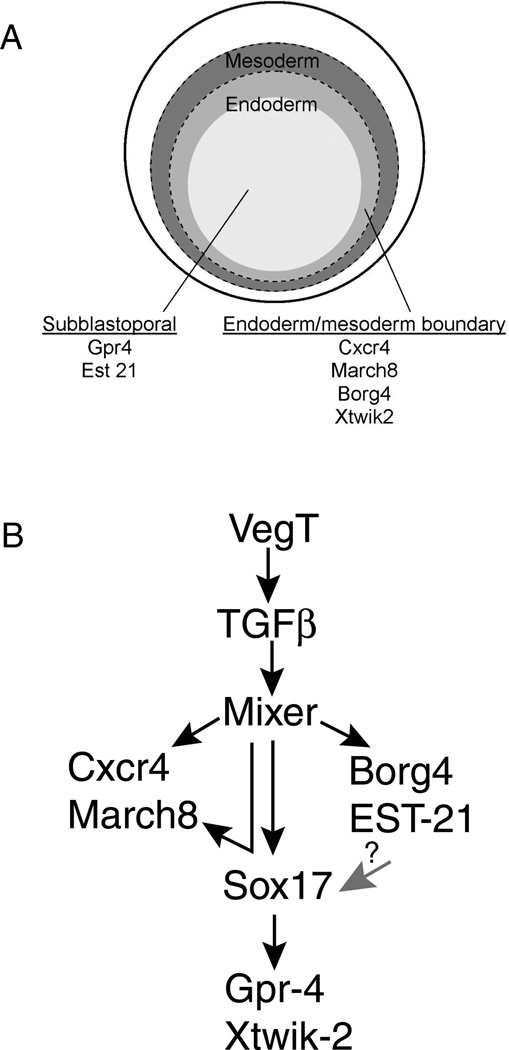
Fig. 6. Six new genes are expressed in discrete regions of the endoderm and may play roles in the endoderm pathway.
A. This cartoon illustrates the vegetal hemisphere of a Xenopus embryo during gastulation. The most vegetal cells (off white) are referred to as subblastoporal endoderm and express Gpr4 and Est21. A population of endoderm (light grey) lies adjacent to the mesoderm and expresses Cxcr4 March8, Borg4 and Xtwik2. Mesoderm is depicted in dark grey. B. A simplified version of the endoderm pathway is diagramed hypothesizing how the endoderm specific targets may be involved. Cxcr4 and March8 are placed downstream of Mixer and Sox17β. Borg4 and EST-21 are placed downstream of Mixer. It is unknown whether Borg4 and EST-21 activate Sox17β (gray arrow with ?). Gpr-4 and Xtwik-2 are placed downstream of Sox17β.
The proposed locations of these six genes within the endoderm pathway is summarized in Fig. 6. Borg4 and EST-21 are induced by Mixer, but not by Sox17β. It is possible that these genes may also have a function within the pathway and serve as intermediates leading to the induction of Sox17β or other downstream endoderm molecules (Fig. 6, gray arrow). Gpr-4 and Xtwik2 are induced by Sox17β, but not by Mixer, and are therefore placed farthest downstream. Cxcr4 and March8 are induced by both Mixer and Sox17β. The upregulation of Cxcr4 and March8 may be due independent inductive events by Mixer and Sox17β, or may simply reflect a by-product of Sox17β initiation by Mixer.
We can begin to make hypotheses about the roles of some of these genes in endoderm development based on their structure and specific expression pattern. Four of the six endodermally expressed genes are either known receptors (Cxcr4 and Gpr-4) or membrane spanning proteins (March8 and Xtwik2) (Chavez et al., 1999; Moepps et al., 2000; Goto et al, 2003; Chung et al, 2004). Borg4 may be a member of a signal transduction cascade (Joberty et al., 1999). These proteins may play a role in transducing extracellular signals secreted from within the endoderm itself or from the neighboring mesoderm. We observed a heavy concentration of expression around the blastopore ring for Cxcr4, March8, Borg4 and Xtwik-2. These may be involved in integrating the instructional cues for endoderm cells to begin involution during gastrulation. Cxcr4, for example, has been characterized for its role in neural crest migration (Moepps et al., 2000) and may operate through a similar mechanism to direct endodermal cell movements during gastrulation. Borg4 may regulate cell motility within the endoderm through its association with Rho GTPases, integrins and the extracellular matrix.
The putative downstream targets of Mixer and Sox17β described in this paper may also be transcriptionally upregulated by other inducers of endoderm. We found that four of the six endodermally expressed clones were induced by VegT, and all six were induced by Smad2. Recently, Cxcr4 and Borg4, in addition to c-myc, Xmsr, FoxA1 and FoxC1, were identified in a cDNA-based microarray experiment as possible targets of VegT (Taverner et al., 2005). Xmsr was found to be a direct target of VegT (Taverner et al., 2005), and although its transcription is induced by Mixer and Sox17β, it is unknown whether Xmsr is a direct target of these transcription factors. It will be interesting to sort out the hierarchy of induction for these molecules during endoderm formation.
We have laid the foundation for many future studies with our list of 63 potentially novel regulators of endoderm formation, especially the six we found to be expressed in the endoderm. At this point we can only speculate what these molecules are doing in the context of endoderm development. Overexpression analysis of these genes will reveal possible inductive roles in endoderm specification. These studies would be complemented by morpholino or dominant-negative loss of function analyses to identify molecules within our list that are required for the formation of endoderm. The ultimate goal will be to explore epistatic and biochemical relationships between these genes and other known components of the endoderm pathway.
Experimental Procedures
mRNA synthesis and injection for array samples
DNA plasmid constructs for Mixer (pCS300) (Henry and Melton, 1998) and Sox17β (pSPJC2L) (Hudson et al., 1997) were linearized with AscI and XhoI, respectively. SP6 transcription of mRNA was performed using mMESSAGE machine (Ambion, Austin, TX). Female frogs were primed for ovulation with human chorionic gonadotropin (Condie and Harland, 1987). Embryos were collected into 0.1× MR solution, fertilized in vitro, and de-jellied with 2.5% cysteine, pH 8.0. Embryos were transferred onto mesh grid plates containing 1/3× MR with 2.5% ficoll for injection. Embryos were injected with 500 pg of Mixer or Sox17β mRNA at the one-cell stage in the presumptive ectoderm.
Ectodermal explants
Injected and control uninjected embryos were cultured to stage 8 and transferred to agarose coated dishes containing 3/4× NAM solution (Peng, 1991) for tissue excision. 90 explants per sample were performed; 80 to be used for the array experiment and 10 for a control RT-PCR assay. Explants were harvested at stage 10.5.
Total RNA isolation
Stage 10.5 explants were homogenized in lysis buffer (0.5% SDS, 5 mM EDTA, 50 mM Tris pH 7.5, 50 mM NaCl) containing 0.2 mg/ml proteinase K and incubated at 42°C for 30 minutes. Samples were then extracted with equal volumes of phenol:chloroform:IAA, ethanol precipitated (with 0.1 volume 3 M NaOAc, 2.5 volume ethanol and 1 µl glycogen), washed and resuspended in 15 µl DEPC water. DNAse treatment (added to samples): 2.5 µl 10× DNase buffer, 0.4 µl DNase1 (Ambion, Austin, TX), 1.25 µl 20 mM DTT, 0.5 µl RNase inhibitor, 5.4 µl water. Samples were incubated at 37°C for 30 minutes, then brought up to a volume of 100 µl with water. Phenol extraction, ethanol precipitation (minus glycogen) and wash were performed as above. Samples were resuspended in 100 µl water. A second ethanol precipitation was performed with 10 µl 3 M NaOAc, 1 µl glycogen and 250 µl ethanol and incubated overnight at −20°C. Samples were centrifuged for 20 minutes at 4°C. Pellets were washed twice with 80% ethanol followed by a 5 minute centrifugation, then dried and resuspended in 12 µl DEPC water.
cDNA synthesis
1 µl T7(dT) 24 primer (100 pmol/µl) (Affymetrix, Santa Clara, CA) was added to 10 µg total RNA and incubated at 70°C for 10 minutes. First and second strand cDNA synthesis was performed using SuperScript Double-Stranded cDNA Synthesis kit (Invitrogen, Carlsbad, CA). cDNA was then phenol chloroform extracted using Phase Lock Gel tubes (Eppendorf, Hamburg, Germany); equal volume of phenol chloroform was added to cDNA and centrifuged for 2 minutes. Upper phase was added to 0.75 volume 5 M NH4OAc and 2.5 volume cold ethanol, vortexed and centrifuged for 20 minutes. Pellets were washed twice in 80% cold ethanol followed by a 5 minute centrifugation, dried and resuspended in 12 µl DEPC water.
In vitro transcription (IVT) and fragmentation of cRNA
Biotin-labeled cRNA was synthesized using BioProbe T7 RNA Transcript Labeling kit (ENZO Biochem Inc., Farmingdale, NY) and purified using RNeasy Mini Protocol for RNA Cleanup (Qiagen, Valencia, CA). An adjusted cRNA yield was calculated using the following formula: adjusted cRNA yield = µg cRNA after IVT- (µg total RNA used initially)(fraction of cDNA reaction used in IVT). 20 µg (adjusted value) cRNA was incubated in fragmentation buffer (0.2 M Tris-acetate 8.1, 0.5 M KOAc, 0.15 M MgOAc) at 94°C for 35 minutes.
Array hybridization
Hybridization of biotin-labeled cRNA to Xenopus laevis Genome Arrays (Affymetrix, Santa Clara, CA) was performed using procedures previously described (Wodicka et al., 1997).
Array analysis
Arrays were scanned and data was imported using Gene Chip Operating Software version 1.0 (Affymetrix, Santa Clara, CA). Data was analyzed using DNA-Chip Analyzer (dChip) (Li and Wong, 2001; www.dchip.org). Data from six arrays (duplicated experiments from control explants and explants injected with Mixer or Sox17β) was normalized to the baseline array containing the median probe intensity (Sox17β, experiment 2). Data was analyzed based on the Perfect Match-only model based expression index (Li and Wong, 2001). Low values were truncated to an intensity of 33.22 (10th percentile of expressions called “A”). Samples were compared based on fold increases in intensity for individual probe sets in experimental samples (Mixer and Sox17β) versus baseline (control). False detection rates for each list were generated using the rank products (RP) method (Breitling et al., 2004).
In situ hybridization screen
Dioxygenin-labeled anti-sense probes were generated from clones obtained from Open Biosystems (www.openbiosystems.com) and NIBB (xenopus.nibb.ac.jp). Xenopus embryos were developed to various stages between mid-gastrulation (10.5) and early tadpole (30) and fixed in MEMFA. For bi-sected embryos: Embryos were harvested at stage 10.5, fixed in MEMFA, and dehydrated in MeOH. Embryos were then bisected with a scalpel in MeOH and subsequently processed by In situ hybridization. In situ analysis was performed as described (Harland, 1991).
mRNA synthesis and injection for RT-PCR analysis
Sox17β and Mixer were cut and transcribed as above. VegT (cs105) (Zhang et al., 1998) and Smad2 (cs105) (Baker and Harland, 1996) were linearized with AscI and transcribed with SP6 as above. 500 pg Sox17β, Mixer, VegT or Smad2 was injected into the presumptive ectoderm of one-cell embryos. Explants were performed as above (20 per sample).
RT-PCR
Explants were cultured in 3/4× NAM to stage 10.5. RNA was isolated and cDNA was synthesized as previously described (Wilson and Melton, 1994). 48 µl PCR reactions were assembled with the following ingredients: 34.5 µl dH2O, 5 µl 10X PCR buffer, 4 µl 25 mM MgCl2, 1 µl 10mM dNTP mix, 1 µl cDNA (from 20 µl reaction), 1 µl each 0.1 µg /µl primer, 0.5 µl Taq Polymerase (Applied Biosystems, Foster City, CA). PCR was performed using the following parameters: denature 94°C 2 minutes, 24–28 cycles of (denature 94°C 1 minute, anneal 62–64°C 1.5 minutes, elongate 72°C 1.5 minutes), final elongation 72°C 5 minutes. The following primers were used:
| Gene | Primer Sequence | Reference |
|---|---|---|
| ODC | F-CAGCTAGCTGTGGTGTGG | (Agius et al., 2000) |
| R-CAACATGGAAACTCACACC | ||
| Sox17β | F-AACTCCCACCAGCAGGCTACTTTG | (Myers et al., 2004) |
| R-TGTCAATGTCACTCTCCAGATGTCC | ||
| Xbra | F-AACTGGTCTACCCTTCAAATGCC | (New) |
| R-CGTGACATCATACTGGTTTTCTGC | ||
| March8 | F-TCCTCGGACATCAGTGACTCCATC | (New) |
| R-AAGAACATACAGGGACCAAACGAC | ||
| Cxcr4 | F-GGCTATCAAAAGAAATCCAGGACC | (New) |
| R-GCAGGAATCTAAACCCAAACAGTC | ||
| Borg4 | F-CGGGTGATGCCTTTGGAGATAC | (New) |
| R-GGAACAGTTGCTGGACTTGAGC | ||
| Gpr-4 | F-AGGGAAACATCTTGGGCATCTAC | (New) |
| R-TCCTTGAACGGAGTGGGAAAAC | ||
| EST-21 | F-ACACTTCACCACAATACCAGGGAG | (New) |
| R-CTTTTCCATCGGGGCTCAAG | ||
| Xtwik-2 | F-GGAAGCAGAACACAGTAACAATCCG | (New) |
| R-CACAAGTAGCGTGAGTAACAGCCAG |
Acknowledgements
We thank Dr. Gil Chu and Dr. Laura Attardi for their assistance in adapting the Affymetrix protocol to Xenopus and acknowledge Andy Hufton and Suzanne Jacobs for technical advice. We thank Dr. Hugh Woodland for supplying the Sox17β (pSPJC2L) plasmid and Dr. Doug Melton for supplying the Mixer (pCS300) plasmid. We thank Dr. Cheng Li and Dr. Wing Wong for providing dChip software and advice. We also thank NIBB for their generous contribution of EST clones. This paper was supported by NIH R01 HD 41557 and a generous gift from the Baxter Foundation.
References
- Afouda BA, Ciau-Uitz A, Patient R. GATA4, 5 and 6 mediate TGFbeta maintenance of endodermal gene expression in Xenopus embryos. Development. 2005;132:763–774. doi: 10.1242/dev.01647. [DOI] [PubMed] [Google Scholar]
- Agius E, Oelgeschlager M, Wessely O, Kemp C, De Robertis EM. Endodermal Nodal-related signals and mesoderm induction in Xenopus. Development. 2000;127:1173–1183. doi: 10.1242/dev.127.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J, Stainier DY. A molecular pathway leading to endoderm formation in zebrafish. Curr Biol. 1999;9:1147–1157. doi: 10.1016/S0960-9822(00)80016-0. [DOI] [PubMed] [Google Scholar]
- Alexander J, Rothenberg M, Henry GL, Stainier DY. casanova plays an early and essential role in endoderm formation in zebrafish. Dev Biol. 1999;215:343–357. doi: 10.1006/dbio.1999.9441. [DOI] [PubMed] [Google Scholar]
- Aoki TO, David NB, Minchiotti G, Saint-Etienne L, Dickmeis T, Persico GM, Strahle U, Mourrain P, Rosa FM. Molecular integration of casanova in the Nodal signalling pathway controlling endoderm formation. Development. 2002;129:275–286. doi: 10.1242/dev.129.2.275. [DOI] [PubMed] [Google Scholar]
- Baker JC, Harland RM. A novel mesoderm inducer, Madr2, functions in the activin signal transduction pathway. Genes Dev. 1996;10:1880–1889. doi: 10.1101/gad.10.15.1880. [DOI] [PubMed] [Google Scholar]
- Beddington RS, Robertson EJ. Anterior patterning in mouse. Trends Genet. 1998;14:277–284. doi: 10.1016/s0168-9525(98)01499-1. [DOI] [PubMed] [Google Scholar]
- Bektas M, Barak LS, Jolly PS, Liu H, Lynch KR, Lacana E, Suhr KB, Milstien S, Spiegel S. The G protein-coupled receptor GPR4 suppresses ERK activation in a ligand-independent manner. Biochemistry. 2003;42:12181–12191. doi: 10.1021/bi035051y. [DOI] [PubMed] [Google Scholar]
- Bolce ME, Hemmati-Brivanlou A, Harland RM. XFKH2, a Xenopus HNF-3 alpha homologue, exhibits both activin-inducible and autonomous phases of expression in early embryos. Dev Biol. 1993;160:413–423. doi: 10.1006/dbio.1993.1317. [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Kim S, Sasai Y, Lu B, De Robertis EM. Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann's organizer. Nature. 1996;382:595–601. doi: 10.1038/382595a0. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Chavez RA, Gray AT, Zhao BB, Kindler CH, Mazurek MJ, Mehta Y, Forsayeth JR, Yost CS. TWIK-2, a new weak inward rectifying member of the tandem pore domain potassium channel family. J Biol Chem. 1999;274:7887–7892. doi: 10.1074/jbc.274.12.7887. [DOI] [PubMed] [Google Scholar]
- Chiao E, Leonard J, Dickinson K, Baker JC. High-throughput functional screen of mouse gastrula cDNA libraries reveals new components of endoderm and mesoderm specification. Genome Res. 2005;15:44–53. doi: 10.1101/gr.2993405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HA, Hyodo-Miura J, Kitayama A, Terasaka C, Nagamune T, Ueno N. Screening of FGF target genes in Xenopus by microarray: temporal dissection of the signalling pathway using a chemical inhibitor. Genes Cells. 2004;9:749–761. doi: 10.1111/j.1356-9597.2004.00761.x. [DOI] [PubMed] [Google Scholar]
- Clements D, Friday RV, Woodland HR. Mode of action of VegT in mesoderm and endoderm formation. Development. 1999;126:4903–4911. doi: 10.1242/dev.126.21.4903. [DOI] [PubMed] [Google Scholar]
- Clements D, Cameleyre I, Woodland HR. Redundant early and overlapping larval roles of Xsox17 subgroup genes in Xenopus endoderm development. Mech Dev. 2003;120:337–348. doi: 10.1016/s0925-4773(02)00450-1. [DOI] [PubMed] [Google Scholar]
- Condie BG, Harland RM. Posterior expression of a homeobox gene in early Xenopus embryos. Development. 1987;101:93–105. [PubMed] [Google Scholar]
- Couly G, Creuzet S, Bennaceur S, Vincent C, Le Douarin NM. Interactions between Hox-negative cephalic neural crest cells and the foregut endoderm in patterning the facial skeleton in the vertebrate head. Development. 2002;129:1061–1073. doi: 10.1242/dev.129.4.1061. [DOI] [PubMed] [Google Scholar]
- Engleka MJ, Craig EJ, Kessler DS. VegT activation of Sox17 at the midblastula transition alters the response to nodal signals in the vegetal endoderm domain. Dev Biol. 2001;237:159–172. doi: 10.1006/dbio.2001.0366. [DOI] [PubMed] [Google Scholar]
- Goto E, Ishido S, Sato Y, Ohgimoto S, Ohgimoto K, Nagano-Fujii M, Hotta H. c-MIR, a human E3 ubiquitin ligase, is a functional homolog of herpesvirus proteins MIR1 and MIR2 and has similar activity. J Biol Chem. 2003;278:14657–14668. doi: 10.1074/jbc.M211285200. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Henry GL, Melton DA. Mixer, a homeobox gene required for endoderm development. Science. 1998;281:91–96. doi: 10.1126/science.281.5373.91. [DOI] [PubMed] [Google Scholar]
- Horb ME, Thomsen GH. A vegetally localized T-box transcription factor in Xenopus eggs specifies mesoderm and endoderm and is essential for embryonic mesoderm formation. Development. 1997;124:1689–1698. doi: 10.1242/dev.124.9.1689. [DOI] [PubMed] [Google Scholar]
- Hudson C, Clements D, Friday RV, Stott D, Woodland HR. Xsox17alpha and -beta mediate endoderm formation in Xenopus. Cell. 1997;91:397–405. doi: 10.1016/s0092-8674(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Joberty G, Perlungher RR, Macara IG. The Borgs, a new family of Cdc42 and TC10 GTPase-interacting proteins. Mol Cell Biol. 1999;19:6585–6597. doi: 10.1128/mcb.19.10.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Kuehn MR, Hogan BL, Smith JC, Wright CV. Nodal-related signals induce axial mesoderm and dorsalize mesoderm during gastrulation. Development. 1995;121:3651–3662. doi: 10.1242/dev.121.11.3651. [DOI] [PubMed] [Google Scholar]
- Joseph EM, Melton DA. Xnr4: a Xenopus nodal-related gene expressed in the Spemann organizer. Dev Biol. 1997;184:367–372. doi: 10.1006/dbio.1997.8510. [DOI] [PubMed] [Google Scholar]
- Kanai-Azuma M, Kanai Y, Gad JM, Tajima Y, Taya C, Kurohmaru M, Sanai Y, Yonekawa H, Yazaki K, Tam PP, Hayashi Y. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129:2367–2379. doi: 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Trinh LA, Reiter JF, Alexander J, Yelon D, Stainier DY. The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes Dev. 2000;14:1279–1289. [PMC free article] [PubMed] [Google Scholar]
- Lemaire P, Darras S, Caillol D, Kodjabachian L. A role for the vegetally expressed Xenopus gene Mix.1 in endoderm formation and in the restriction of mesoderm to the marginal zone. Development. 1998;125:2371–2380. doi: 10.1242/dev.125.13.2371. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose M, Patient R. A genetic regulatory network for Xenopus mesendoderm formation. Dev Biol. 2004;271:467–478. doi: 10.1016/j.ydbio.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Ludwig MG, Vanek M, Guerini D, Gasser JA, Jones CE, Junker U, Hofstetter H, Wolf RM, Seuwen K. Proton-sensing G-protein-coupled receptors. Nature. 2003;425:93–98. doi: 10.1038/nature01905. [DOI] [PubMed] [Google Scholar]
- Lustig KD, Kroll KL, Sun EE, Kirschner MW. Expression cloning of a Xenopus T-related gene (Xombi) involved in mesodermal patterning and blastopore lip formation. Development. 1996;122:4001–4012. doi: 10.1242/dev.122.12.4001. [DOI] [PubMed] [Google Scholar]
- Mahadevan MS, Baird S, Bailly JE, Shutler GG, Sabourin LA, Tsilfidis C, Neville CE, Narang M, Korneluk RG. Isolation of a novel G protein-coupled receptor (GPR4) localized to chromosome 19q13.3. Genomics. 1995;30:84–88. doi: 10.1006/geno.1995.0013. [DOI] [PubMed] [Google Scholar]
- Marinissen MJ, Gutkind JS. G-protein-coupled receptors and signaling networks: emerging paradigms. Trends Pharmacol Sci. 2001;22:368–376. doi: 10.1016/s0165-6147(00)01678-3. [DOI] [PubMed] [Google Scholar]
- Medhurst AD, Rennie G, Chapman CG, Meadows H, Duckworth MD, Kelsell RE, Gloger II, Pangalos MN. Distribution analysis of human two pore domain potassium channels in tissues of the central nervous system and periphery. Brain Res Mol Brain Res. 2001;86:101–114. doi: 10.1016/s0169-328x(00)00263-1. [DOI] [PubMed] [Google Scholar]
- Moepps B, Braun M, Knopfle K, Dillinger K, Knochel W, Gierschik P. Characterization of a Xenopus laevis CXC chemokine receptor 4: implications for hematopoietic cell development in the vertebrate embryo. Eur J Immunol. 2000;30:2924–2934. doi: 10.1002/1521-4141(200010)30:10<2924::AID-IMMU2924>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Murphy PM. Chemokine receptors: structure, function and role in microbial pathogenesis. Cytokine Growth Factor Rev. 1996;7:47–64. doi: 10.1016/1359-6101(96)00009-3. [DOI] [PubMed] [Google Scholar]
- Myers AP, Corson LB, Rossant J, Baker JC. Characterization of mouse Rsk4 as an inhibitor of fibroblast growth factor-RAS-extracellular signal-regulated kinase signaling. Mol Cell Biol. 2004;24:4255–4266. doi: 10.1128/MCB.24.10.4255-4266.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascone N, Mercola M. An inductive role for the endoderm in Xenopus cardiogenesis. Development. 1995;121:515–523. doi: 10.1242/dev.121.2.515. [DOI] [PubMed] [Google Scholar]
- Osada N, Kusuda J, Suzuki Y, Sugano S, Hashimoto K. Sequence analysis, gene expression, and chromosomal assignment of mouse Borg4 gene and its human orthologue. J Hum Genet. 2000;45:374–377. doi: 10.1007/s100380070012. [DOI] [PubMed] [Google Scholar]
- Pearce JJ, Evans MJ. Mml, a mouse Mix-like gene expressed in the primitive streak. Mech Dev. 1999;87:189–192. doi: 10.1016/s0925-4773(99)00135-5. [DOI] [PubMed] [Google Scholar]
- Peng HB. Xenopus laevis: Practical uses in cell and molecular biology. Solutions and protocols. Methods Cell Biol. 1991;36:657–662. [PubMed] [Google Scholar]
- Perea-Gomez A, Lawson KA, Rhinn M, Zakin L, Brulet P, Mazan S, Ang SL. Otx2 is required for visceral endoderm movement and for the restriction of posterior signals in the epiblast of the mouse embryo. Development. 2001;128:753–765. doi: 10.1242/dev.128.5.753. [DOI] [PubMed] [Google Scholar]
- Shivdasani RA. Molecular regulation of vertebrate early endoderm development. Dev Biol. 2002;249:191–203. doi: 10.1006/dbio.2002.0765. [DOI] [PubMed] [Google Scholar]
- Sin WC, Zhang Y, Zhong W, Adhikarakunnathu S, Powers S, Hoey T, An S, Yang J. G protein-coupled receptors GPR4 and TDAG8 are oncogenic and overexpressed in human cancers. Oncogene. 2004;23:6299–6303. doi: 10.1038/sj.onc.1207838. [DOI] [PubMed] [Google Scholar]
- Sinner D, Rankin S, Lee M, Zorn AM. Sox17 and beta-catenin cooperate to regulate the transcription of endodermal genes. Development. 2004;131:3069–3080. doi: 10.1242/dev.01176. [DOI] [PubMed] [Google Scholar]
- Stainier DY. A glimpse into the molecular entrails of endoderm formation. Genes Dev. 2002;16:893–907. doi: 10.1101/gad.974902. [DOI] [PubMed] [Google Scholar]
- Stennard F, Carnac G, Gurdon JB. The Xenopus T-box gene, Antipodean, encodes a vegetally localised maternal mRNA and can trigger mesoderm formation. Development. 1996;122:4179–4188. doi: 10.1242/dev.122.12.4179. [DOI] [PubMed] [Google Scholar]
- Sun BI, Bush SM, Collins-Racie LA, LaVallie ER, DiBlasio-Smith EA, Wolfman NM, McCoy JM, Sive HL. derriere: a TGF-beta family member required for posterior development in Xenopus. Development. 1999;126:1467–1482. doi: 10.1242/dev.126.7.1467. [DOI] [PubMed] [Google Scholar]
- Tachibana K, Hirota S, Iizasa H, Yoshida H, Kawabata K, Kataoka Y, Kitamura Y, Matsushima K, Yoshida N, Nishikawa S, Kishimoto T, Nagasawa T. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 1998;393:591–594. doi: 10.1038/31261. [DOI] [PubMed] [Google Scholar]
- Taverner NV, Kofron M, Shin Y, Kabitschke C, Gilchrist MJ, Wylie C, Cho KW, Heasman J, Smith JC. Microarray-based identification of VegT targets in Xenopus. Mech Dev. 2005;122:333–354. doi: 10.1016/j.mod.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Weber H, Symes CE, Walmsley ME, Rodaway AR, Patient RK. A role for GATA5 in Xenopus endoderm specification. Development. 2000;127:4345–4360. doi: 10.1242/dev.127.20.4345. [DOI] [PubMed] [Google Scholar]
- Wilson PA, Melton DA. Mesodermal patterning by an inducer gradient depends on secondary cell-cell communication. Curr Biol. 1994;4:676–686. doi: 10.1016/s0960-9822(00)00152-4. [DOI] [PubMed] [Google Scholar]
- Wodicka L, Dong H, Mittmann M, Ho MH, Lockhart DJ. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- Xanthos JB, Kofron M, Wylie C, Heasman J. Maternal VegT is the initiator of a molecular network specifying endoderm in Xenopus laevis. Development. 2001;128:167–180. doi: 10.1242/dev.128.2.167. [DOI] [PubMed] [Google Scholar]
- Yasuo H, Lemaire P. A two-step model for the fate determination of presumptive endodermal blastomeres in Xenopus embryos. Curr Biol. 1999;9:869–879. doi: 10.1016/s0960-9822(99)80391-1. [DOI] [PubMed] [Google Scholar]
- Zhang J, Houston DW, King ML, Payne C, Wylie C, Heasman J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 1998;94:515–524. doi: 10.1016/s0092-8674(00)81592-5. [DOI] [PubMed] [Google Scholar]
- Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]