Abstract
The N-end rule relates the in vivo half-life of a protein to the identity of its N-terminal residue. Primary destabilizing N-terminal residues (Ndp) are recognized directly by the targeting machinery. The recognition of secondary destabilizing N-terminal residues (Nds) is preceded by conjugation of an Ndp residue to Nds of a polypeptide substrate. In eukaryotes, ATE1-encoded arginyl-transferases (RD,E,C*-transferases) conjugate Arg (R), an Ndp residue, to Nds residues Asp (D), Glu (E), or oxidized Cys residue (C*). Ubiquitin ligases recognize the N-terminal Arg of a substrate and target the (ubiquitylated) substrate to the proteasome. In prokaryotes such as Escherichia coli, Ndp residues Leu (L) or Phe (F) are conjugated, by the aat-encoded Leu/Phe-transferase (L/FK,R-transferase), to N-terminal Arg or Lys, which are Nds in prokaryotes but Ndp in eukaryotes. In prokaryotes, substrates bearing the Ndp residues Leu, Phe, Trp, or Tyr are degraded by the proteasome-like ClpAP protease. Despite enzymological similarities between eukaryotic RD,E,C*-transferases and prokaryotic L/FK,R-transferases, there is no significant sequelogy (sequence similarity) between them. We identified an aminoacyl-transferase, termed Bpt, in the human pathogen Vibrio vulnificus. Although it is a sequelog of eukaryotic RD,E,C*-transferases, this prokaryotic transferase exhibits a “hybrid” specificity, conjugating Ndp Leu to Nds Asp or Glu. Another aminoacyl-transferase, termed ATEL1, of the eukaryotic pathogen Plasmodium falciparum, is a sequelog of prokaryotic L/FK,R-transferases (Aat), but has the specificity of eukaryotic RD,E,C*-transferases (ATE1). Phylogenetic analysis suggests that the substrate specificity of R-transferases arose by two distinct routes during the evolution of eukaryotes.
Keywords: proteolysis, ClpAP, ClpS, Aat, bacterial protein transferase
A protein substrate of the ubiquitin (Ub) system, which controls the levels of many intracellular proteins, is conjugated to Ub through the action of three enzymes, E1, E2, and E3. The substrate’s degradation signal (degron) is recognized by E3 (1–4). A ubiquitylated protein bears a covalently linked poly-Ub chain and is degraded by the 26S proteasome (5, 6). An essential determinant of one class of degrons, called N-degrons, is a substrate’s destabilizing N-terminal residue. The set of destabilizing residues in a given cell type yields a rule, called the N-end rule, which relates the in vivo half-life of a protein to the identity of its N-terminal residue (7–12). In eukaryotes, the N-degron consists of three main determinants, a destabilizing N-terminal residue of a substrate, its internal Lys (K) residue(s) (the site of formation of a poly-Ub chain), and a conformationally flexible region(s) in the vicinity of these determinants (7, 13, 14).
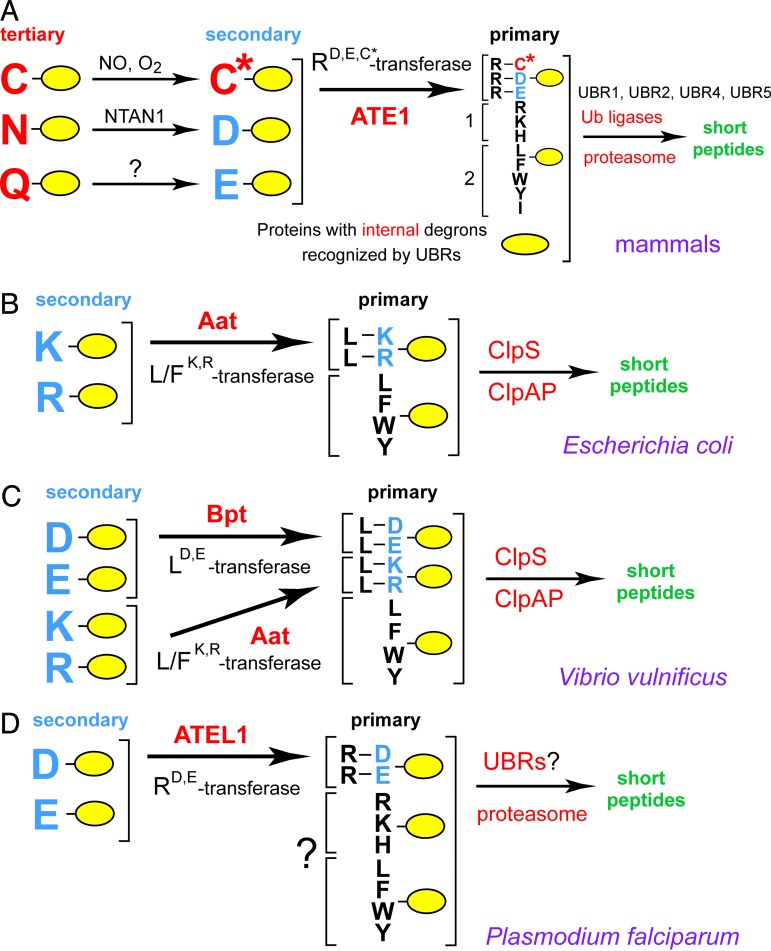
The N-end rule has a hierarchic structure (Fig. 1). In eukaryotes, N-terminal Asn (N) and Gln (Q) are tertiary destabilizing residues in that they function, through enzymatic deamidation (15, 16), to yield the secondary destabilizing N-terminal residues (Nds) Asp (D) and Glu (E) (Fig. 1A). The activity of Asp and Glu requires their conjugation, by ATE1-encoded isoforms of Arg-tRNA-protein transferase (RD,E,C*-transferase, where C* is oxidized cysteine residue), to Arg (R), one of the primary destabilizing N-terminal residues (Ndp) (11, 17, 18). The latter are recognized by the E3 Ub ligases of the N-end rule pathway, called N-recognins (Fig. 1A) (7, 9, 10, 12). In mammals, the set of destabilizing residues that function through their arginylation contains not only N-terminal Asp and Glu, but also N-terminal Cys (C), which is arginylated after its nitric oxide (NO)-dependent oxidation (Fig. 1A) (11, 18).
Fig. 1.
The N-end rule pathways and the activities of aa-transferases, the Bpt LD,E-transferase of V. vulnificus (a prokaryotic pathogen) and the ATEL1 RD,E-transferase of P. falciparum (an eukaryotic pathogen). (A) The N-end rule pathway in mammals. N-terminal residues are indicated by single-letter abbreviations for amino acids. Yellow ovals denote the rest of a protein substrate. C* denotes oxidized N-terminal Cys, produced in reactions mediated by NO, with subsequent arginylation of C* by ATE1-encoded isoforms of Arg-tRNA-protein transferase (RD,E,C*-transferase) (11). The type-1 and type-2 primary destabilizing N-terminal residues are recognized by multiple E3 Ub ligases (N-recognins) of the N-end rule pathway that share the UBR motif (12). Through their other substrate-binding sites, these E3 enzymes also recognize internal (non-N-terminal) degradation signals (degrons) in other substrates of the N-end rule pathway, denoted by a larger yellow oval. (B) The E. coli N-end rule pathway (7, 25, 26). Although the purified Aat L/FK,R-transferase is capable of conjugating either L (Leu) or F (Phe), the conjugated residue in vivo (in E. coli) is largely L (26). (C) The V. vulnificus N-end rule pathway, characterized in the present work, including the aa-transferase Bpt (LD,E-transferase). ClpS was demonstrated to be the N-recognin of the E. coli N-end rule pathway (31); its similar function in V. vulnificus is an inferred one at present. (D) The putative N-end rule pathway of P. falciparum and the ATEL1 aa-transferase. A question mark denotes the expected but unproven features of this pathway in P. falciparum, where the only characterized component thus far is ATEL1.
The functions of the eukaryotic N-end rule pathway include the control of peptide import (through the conditional degradation of import’s repressor) (8, 9), the fidelity of chromosome segregation (through the degradation of a conditionally produced cohesin’s fragment) (19), the regulation of apoptosis (through the degradation of a caspase-processed inhibitor of apoptosis) (20, 21), the regulation of meiosis (10), leaf senescence in plants (22), and cardiovascular development in mammals (11, 18, 23). The latter function is likely to be mediated in part by the arginylation-dependent degradation of RGS4, RGS5, and RGS16, a set of GTPase-activating proteins that down-regulate the signaling by specific G proteins and are themselves down-regulated by the N-end rule pathway, at rates controlled by NO and oxygen (11, 18, 23). The Johanson-Blizzard syndrome, a human genetic disease, is caused by the absence of UBR1, one of the E3 N-recognins of the N-end rule pathway (Fig. 1A) (24).
Although prokaryotes lack Ub conjugation and Ub itself, they contain the (Ub-independent) N-end rule pathway, which has been so far characterized only in Escherichia coli (Fig. 1B) (25, 26). The pathway’s physiological functions in prokaryotes are unknown. In E. coli, the Ndp residues Leu (L) or Phe (F) are conjugated, by the aat-encoded Leu/Phe-tRNA-protein transferase (L/FK,R-transferase), to N-terminal Arg or Lys, which are the Nds residues in prokaryotes but Ndp residues in eukaryotes (Fig. 1 A and B) (7, 25, 26). Reporter substrates bearing the Ndp residues Leu, Phe, Trp (W), or Tyr (Y) are targeted for degradation by ClpAP (27–30), one of several proteasome-like proteases in E. coli. ClpS, a 12-kDa adaptor protein specific for ClpAP, is an essential component of the E. coli N-end rule pathway, where it functions as the N-recognin (Fig. 1B) (31).
Although eukaryotic RD,E,C*-transferases (ATE1) and prokaryotic L/FK,R-transferases (Aat) mediate analogous enzymatic reactions (Fig. 1), there is no significant sequelogy (sequence similarity) between these sets of enzymes. [In this terminology (32), sequelog and spalog denote, respectively, a sequence that is similar, to a specified extent, to another sequence, and a 3D structure that is similar, to a specified extent, to another 3D structure. These terms and their derivatives allow separate, single-word notations for sequence and spatial similarities. Their other advantage is evolutionary neutrality. The sequelog terminology complements the existing one (homolog, ortholog, paralog) by making the initial designation of a sequence similarity (sequelogy) rigorous, i.e., evolutionarily neutral, while still allowing later designations to involve homology or related terms, if their employment is justified by evidence (32).]
We describe here a prokaryotic aminoacyl-transferase (aa-transferase) termed Bpt (bacterial protein transferase) and characterized in the human pathogen Vibrio vulnificus. This Gram-negative γ-proteobacterium is naturally found in marine environments, including shellfish, and can cause gastrointestinal and bloodstream infections in humans. These infections can be severe, and occasionally fatal, in patients with compromised immune system, iron overload, or hepatic disease (ref. 33 and references therein). In contrast to E. coli Aat, the Bpt aa-transferase is a sequelog (32) of eukaryotic RD,E,C*-transferases (ATE1), but is shown here to exhibit a “hybrid” specificity: it conjugates the Ndp residue Leu to Nds residues Asp or Glu (Fig. 1C). Yet another transferase, termed ATEL1, of the eukaryotic pathogen Plasmodium falciparum (malaria parasite), is a sequelog of the prokaryotic L/FK,R-transferase (Aat), but is shown to have the specificity of the eukaryotic RD,E,C*-transferase (ATE1) (Fig. 1D). We discuss the evolution of the Ub system and possible functions of aa-transferases and the prokaryotic N-end rule pathway.
Results and Discussion
An aa-Transferase in V. vulnificus.
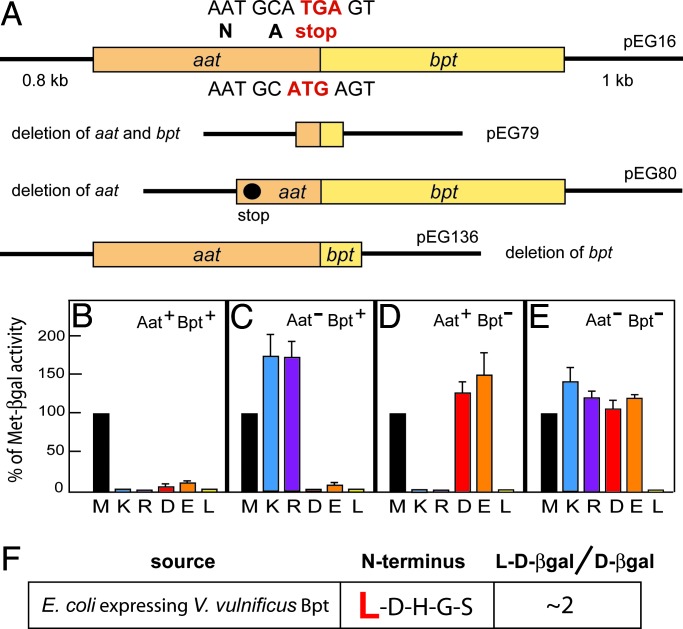
Through searches in databases we found that some prokaryotes, other than E. coli, contain proteins that are sequelogs of eukaryotic ATE1 (see Introduction for the terms and notations used). We denoted the corresponding genes as bpt to distinguish them from eukaryotic transferase genes (ATE1) (11, 18) and prokaryotic aat genes (25, 26). Remarkably, the sequenced genome of the human prokaryotic pathogen V. vulnificus (NC_004459 and NC_004460) was found to encode Aat (L/FK,R-transferase) and Bpt in a two-transferase operon (NC_004459; Vv12124), in which the ORF of bpt begins within the stop codon of the preceding aat ORF (Fig. 2A). Partial overlaps of adjacent ORFs in an operon were observed with other prokaryotic genes as well (34).
Fig. 2.
The N-end rule pathway in V. vulnificus and E. coli. (A) The V. vulnificus aat-bpt operon and its derivatives. The ORF encoding the Bpt aa-transferase begins within the stop codon of the preceding aat ORF. The nucleotide sequence of the bpt ORF (including its start codon, in red) is shown below the diagram. Shown above is the 3′ end of aat ORF, including its TGA stop codon, in red. Also indicated are the lengths of flanking V. vulnificus DNA fragments in the plasmid insert. (B) Relative enzymatic activities of βgal in Aat-lacking E. coli (TS351) expressing S. cerevisiae UBP1, both aa-transferases of V. vulnificus (WT), and either Met-βgal (M), Lys-βgal (K), Arg-βgal (R), Asp-βgal (D), Glu-βgal (E), or Leu-βgal (L), produced from the corresponding Ub fusions. (C) The same as B but in the absence of V. vulnificus Aat. (D) The same as B but in the absence of V. vulnificus Bpt. (E) The same as B but in the absence of V. vulnificus Aat and Bpt. One hundred percent is the (averaged) activity of Met-βgal (M). (F) Determination, through Edman degradation, of the N-terminal sequence of Asp-βgal (derived from Ub-Asp-βgal) from E. coli KPS18 that lacked both Aat and ClpA and expressed V. vulnificus Bpt. The indicated ratio of Leu-Asp-βgal/Asp-βgal refers to the incomplete in vivo leucylation of (overexpressed) Asp-βgal by V. vulnificus Bpt in E. coli, as detected by Edman sequencing.
To examine the recognition/conjugation specificity of V. vulnificus Bpt, an Aat-lacking strain of E. coli (aat::minitet, TS351) (26) was transformed with plasmids expressing the yeast deubiquitylating enzyme UBP1 and either the intact V. vulnificus aat-bpt operon or its derivatives with deletions (Fig. 2A) (see Materials and Methods and Supporting Text, which is published as supporting information on the PNAS web site). By cleaving, nearly cotranslationally, after the last residue of Ub in a Ub fusion, the UBP1 enzyme enabled the Ub fusion technique in E. coli (25, 35), thereby making possible the in vivo production of X-β-galactosidase (βgal) reporter proteins in which N-terminal X was a desired residue.
Bpt Transferase Has a Hybrid Recognition/Conjugation Specificity.
Asp-βgal and Glu-βgal are normally long-lived in E. coli, because N-terminal Asp and Glu are stabilizing residues in this prokaryote, in that they are not recognized by either the Aat L/FK,R-transferase or the rest of the E. coli N-end rule pathway (25, 26, 31). Previous work (13, 17, 36) has shown that the steady-state level of an X-βgal reporter is a sensitive measure of its metabolic stability. Remarkably, Asp-βgal and Glu-βgal became short-lived in Aat-lacking E. coli that expressed V. vulnificus Bpt, irrespective of the presence or absence of V. vulnificus Aat (Fig. 2 B–E). This result indicated that V. vulnificus Bpt, similarly to eukaryotic ATE1 (and in contrast to prokaryotic Aat) recognized N-terminal Asp and Glu. This result also indicated that an amino acid conjugated by V. vulnificus Bpt could not be Arg, but was expected, instead, to be either Leu, Phe, Trp, or Tyr. The reason for this conclusion is that any one of the latter four residues, if made N-terminal, would be directly recognized as a Ndp residue by the ClpS–ClpAP targeting/proteolytic complex (Fig. 1B) (31). Because the degradation of Asp-βgal and Glu-βgal, the substrates of V. vulnificus Bpt, was not affected by the absence of V. vulnificus Aat, and because a converse omission of V. vulnificus Bpt from E. coli expressing V. vulnificus Aat did not perturb the degradation of Aat substrates Arg-βgal and Lys-βgal (Fig. 2 B–E), a physical interaction between Bpt and Aat, even if it normally occurred, would not be required for the activities of these enzymes. Neither coimmunoprecipitation tests nor a cAMP-based two-hybrid in vivo assay in E. coli (37) suggested a physical interaction between V. vulnificus Bpt and Aat (Fig. 5, which is published as supporting information on the PNAS web site, and Supporting Text).
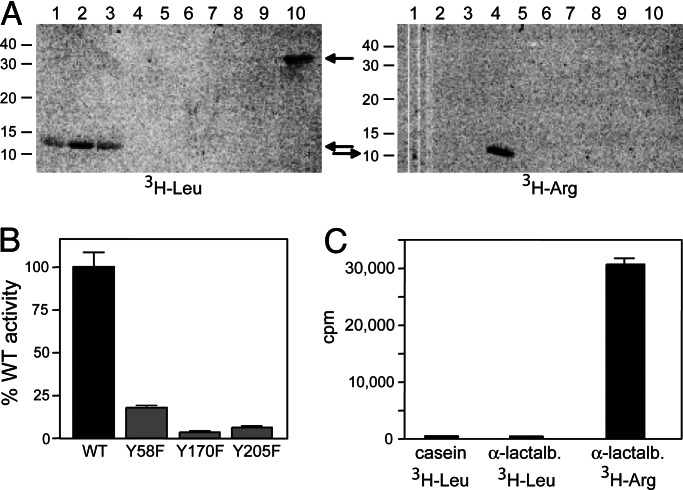
To identify directly the amino acid residue(s) conjugated by Bpt to its substrate, Asp-βgal was isolated from E. coli (KPS18) that lacked both Aat and ClpA and expressed V. vulnificus Bpt, followed by N-terminal (Edman) sequencing. It revealed the N-terminal sequence of leucylated Asp-βgal, Leu-Asp-βgal (Fig. 2F). Thus, V. vulnificus Bpt, although it recognizes N-terminal Asp and Glu (similarly to eukaryotic ATE1), conjugates Leu (Ndp in both prokaryotes and eukaryotes) to these residues, in contrast to eukaryotic ATE1, which conjugates Arg (Fig. 1 A and C). The recognition/conjugation specificity of V. vulnificus Bpt was independently confirmed by using in vitro conjugation assays with either 3H-Leu or 3H-Arg and either α-casein (bearing N-terminal Lys) or bovine α-lactalbumin (bearing N-terminal Glu) as reporters (Fig. 3A). In addition, the enzymatic specificity of strong sequelogs (32) of V. vulnificus Bpt from other prokaryotes, such as the plant pathogen Agrobacterium tumefaciens and the plant symbyont Mesorhizobium loti was found to be indistinguishable from that of V. vulnificus Bpt (Fig. 3A). We conclude that Bpt proteins are a distinct class of prokaryotic aa-transferases, termed LD,E-transferases, which exhibit a hybrid (prokaryotic/eukaryotic) specificity. It is likely (but remains to be determined) that a Bpt LD,E-transferase can also leucylate an oxidized N-terminal Cys, analogously to the arginylation of this residue by RD,E,C*-transferases (Fig. 1 A and C) (11, 18).
Fig. 3.
Enzymatic specificities of V. vulnificus Bpt and P. falciparum ATEL1 aa-transferases. (A) (Left) 3H-Leu: lane 1, purified V. vulnificus Bpt was incubated with 3H-Leu, other components of the aa-transferase assay, and the 14-kDa bovine α-lactalbumin (bearing N-terminal Glu) as a reporter. Lanes 2–5, the same as lane 1 but with A. tumefaciens Bpt, M. loti Bpt, S. cerevisiae ATE1, and E. coli Aat, respectively. Lanes 6–10, the same as lanes 1–5 but with the 24-kDa bovine α-casein (bearing N-terminal Lys) as a reporter. (Right) 3H-Arg: the same as Left but with 3H-Arg instead of 3H-Leu. Arrows indicate the bands of 3H-lactalbumin and 3H-casein, and numbers to the left indicate molecular masses, in kDa, of protein standards. (B) Relative enzymatic activities of V. vulnificus Bpt in which specific Tyr residues were converted to Phe. Purified mutant and WT V. vulnificus Bpt (0.2 μg each) were assayed in vitro with 3H-Leu as described in Supporting Text, using α-lactalbumin as a reporter. 3H was measured in triplicate samples, and the data were corrected by subtracting 3H incorporation in the control assay lacking Bpt. (C) aa-transferase assay with purified (0.25 μg) P. falciparum ATEL1 enzyme in the presence of either α-casein or α-lactalbumin and either 3H-Leu or 3H-Arg as indicated.
Conserved Tyrosines and Cysteines Essential for Bpt Activity.
Sequence alignments of Bpt proteins revealed strong conservation of specific residues (Fig. 6, which is published as supporting information on the PNAS web site, and Supporting Text). We mutated the conserved Tyr-58, Tyr-170, and Tyr-205 in V. vulnificus Bpt to Phe residues. The resulting Bpt-Y58F, Bpt-Y170F, and Bpt-Y205F were produced in E. coli KPS22 (lacking Aat and LacZ) and purified, followed by tests in in vitro conjugation assays with 3H-Leu and α-lactalbumin. Bpt-Y58F lost 70–80% of wild-type Bpt activity (Fig. 3B). Both Bpt-Y170F and Bpt-Y205F, while retaining detectable activity, lost ≈90% of it (Fig. 3B). CD spectra of the mutant proteins were similar to that of wild-type Bpt (data not shown), indicating the absence of strong folding defects. Thus, at least one of the above Tyr residues may be a part of the Bpt’s active site.
Mutants of Saccharomyces cerevisiae ATE1 that lack N-terminus-proximal Cys residues are inactive in vitro (38). Two of these Cys residues of ATE1 were also conserved among Bpt aa-transferases (Fig. 6A). In agreement with S. cerevisiae results (38) and the above sequelogy-based deductions, the Cys-18 → Ser mutant of V. vulnificus Bpt was found to be inactive as well (Fig. 6B). The similarity between prokaryotic Bpt proteins and eukaryotic ATE1 proteins involves a number of other conserved residues as well (Fig. 6 A and D).
ATEL1 of Malaria Parasite Is a Sequelog of Prokaryotic Aat Transferases, but Its Specificity Is That of Eukaryotic ATE1 Transferases.
Another kind of aa-transferase identified in the present work is encoded by the genome of P. falciparum. This eukaryote is an obligatory intracellular parasite and the cause of malaria in humans (39). blastp with E. coli Aat as a query predicted, with a moderate but statistically significant E-value of 7e-4, that P. falciparum may encode an aa-transferase (NP_473045.1) that is sequelogous to prokaryotic Aat transferases (L/FK,R-transferases). In contrast, the sequenced genomes of mammalian and arthropod species (including the hosts of P. falciparum) lack significant sequelogs of Aat (data not shown). Sequelogs of prokaryotic Aat are also present in other Plasmodium species (P. yoelii yoelii, P. chabaudi, and P. berghei), with comparable but stronger E-values (4e-7, 1e-6 and 2e-6, respectively, using E. coli Aat as a query).
To determine the conjugation/recognition specificity of the putative aa-transferase of P. falciparum, it was produced in E. coli (lacking aat) and purified, followed by in vitro conjugation assays with either 3H-Leu or 3H-Arg and appropriate reporters. The specificity of the P. falciparum aa-transferase was found to be indistinguishable from that of “canonical” eukaryotic ATE1s (Figs. 1 and 3C), despite the absence of significant sequelogy to ATE1s. The P. falciparum aa-transferase is thus a RE-transferase. We denoted it, and its strong sequelogs in other species, as ATEL1 (“ATE-like”), to distinguish this group of R-transferases from ATE1s and their strong sequelogs Bpt aa-transferases. In vivo degradation assays with X-βgal reporters in E. coli expressing P. falciparum ATEL1 confirmed its specificity for N-terminal Glu, and in addition indicated that it arginylates N-terminal Asp (data not shown). Thus, P. falciparum ATEL1 is a RD,E-transferase (Fig. 1D).
Identical Specificities of ATE1 and ATEL1 May Be the Result of Convergent Evolution.
psi-blast with S. cerevisiae ATE1 as a query did not retrieve sequelogs of ATE1 in Plasmodium and related genomes. A converse psi-blast, with P. falciparum ATEL1 as a query, did not retrieve sequelogs in eukaryotic genomes known to encode ATE1 enzymes. Strong sequelogs of Bpt LD,E-transferases were found only in prokaryotes, where Bpt proteins are frequent in proteobacteria, except for enterobacteria and other prokaryotes that have undergone genome reduction as a result of their obligatory parasitism or symbiosis. Eukaryotes are the only organisms that contain strong sequelogs of mouse/yeast ATE1s, in that prokaryotic Bpt aa-transferases (LD,E-transferases) were much less sequelogous (32) to ATE1s of eukaryotes than the latter were among themselves, in addition to Bpt proteins being ≈2-fold smaller than eukaryotic ATE1s (for details, see the legend to Fig. 7, which is published as supporting information on the PNAS web site).
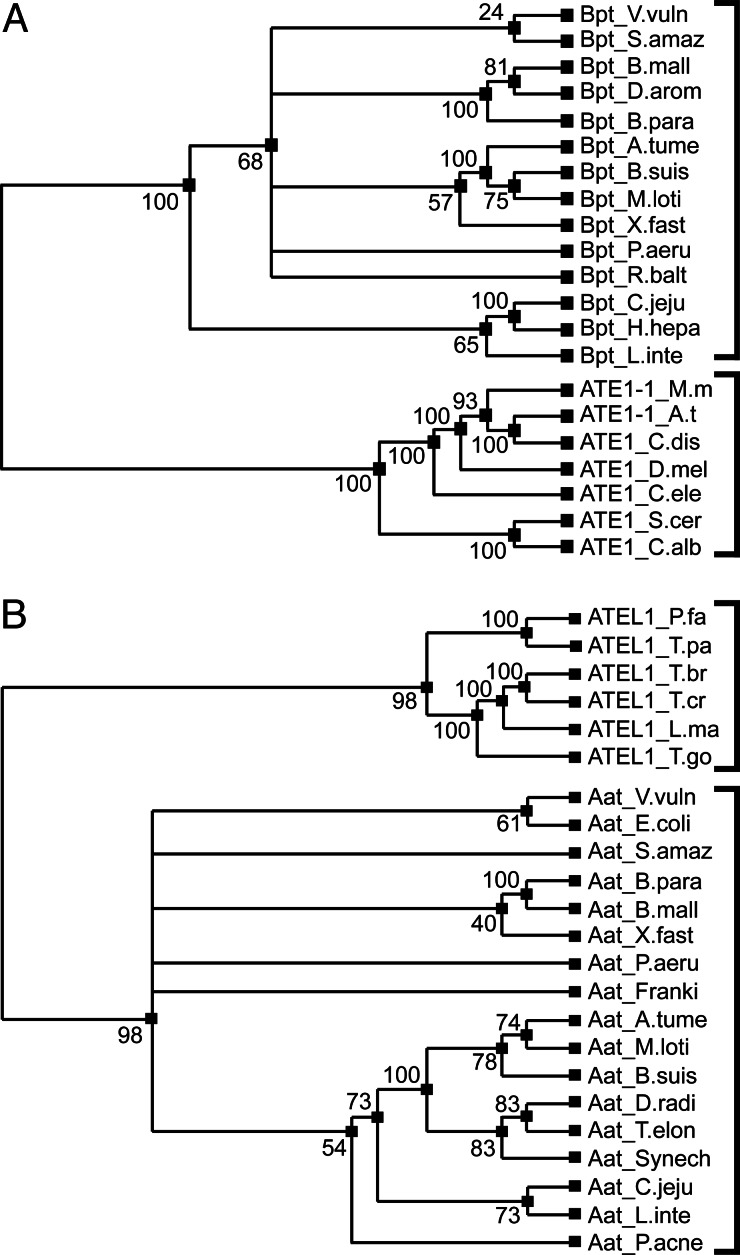
To clarify evolutionary links among aa-transferases, we carried out phylogenetic analyses of ATE1, ATEL1, Aat, and Bpt (Figs. 4 and 7 and Supporting Text), using both parsimony and distance analyses. The resulting phylogenetic trees were consistent with each other (Figs. 4 and 7). We also tested the Aat/ATEL1 family and the Bpt/ATE1 family for sequelogy (32) to each other (Fig. 6), because even marginally significant sequelogy between them would be consistent with their (remote) homology. Prokaryotic Bpt LD,E-transferases and eukaryotic ATE1 RD,E,C*-transferases (Fig. 1) were clearly sequelogous (Fig. 6 A and D), strongly suggesting their homology. However, we could not detect sequelogy between prokaryotic Aat L/FK,R-transferases and Bpt LD,E-transferases (data not shown), even though both Aat and Bpt conjugate Leu to their substrates’ N-terminal residues (Fig. 1 B and C). In contrast, the prokaryotic/eukaryotic Aat/ATEL1 aa-transferases and, separately, the prokaryotic/eukaryotic Bpt/ATE1 aa-transferases form two strongly coherent phylogenetic groups within each protein family (Figs. 4 and 7). Thus, phylogenetic analyses suggest that ancestor proteins of P. falciparum (eukaryotic) ATEL1 were derived from prokaryotic Aat aa-transferases rather than from ancestors of eukaryotic ATE1s, and that the identical substrate specificities of the contemporary ATE1s and ATEL1s (Fig. 1 A and D) may be the result of convergent evolution. Examples of such evolution, where nonsequelogous, nonspalogous (32) enzymes (see Introduction for definitions of terms) have similar enzymatic properties and physiological functions, include arsenate reductases (40) and vitamin B6-dependent enzymes (41).
Fig. 4.
Parsimony-based phylogenetic analyses of selected Bpt/ATE1 and Aat/ATEL1 aa-transferases. (A) Phylogeny of Bpt and ATE1. (B) Phylogeny of Aat and ATEL1. Numbers by the phylogenetic branchpoints give their statistical strength, with 100 being a maximum score. See the text and Supporting Text for details, including the complete names of the indicated organisms.
The distribution of Aat/ATEL1 and Bpt/ATE1 proteins in extant organisms is striking in its unevenness. Although many proteobacterial genomes encode both Aat and Bpt aa-transferases, other bacteria have only Aat or only Bpt, and no detectable Aat or Bpt exist in archaea. ATE1 genes are present in a broad set of “better-known” eukaryotes, such as fungi, plants, animals, and protists. However, we found that ATEL1-like proteins, while apparently absent from the above eukaryotes, are also distributed broadly among other eukaryotes, such as apicomplexans, leishmanias, trypanosomes, cnidaria, diatoms, dinoflagellates, and oomycetes (see Fig. 7 for details). These patterns are consistent with the hypothesis that aa-transferases arose twice, independently, in a common ancestor of proteobacteria and eukaryotes, and underwent two kinds of evolutionary changes in the eukaryotic lineage. First, ATE1 and ATEL1 independently evolved to function as R-transferases (rather than L-transferases, their presumed ancestral states). Second, either ATE1 or ATEL1 were lost from various deep lineages of eukaryotes, so that extant eukaryotes contain one of them, but apparently not both. Larger sets of sequenced genomes, particularly of underexplored unicellular eukaryotes, and specificity comparisons of their aa-transferases would be required to verify the above scenario.
On the Functions of Prokaryotic N-End Rule Pathways.
In addition to proteomic and genetic approaches to the pathway’s function(s) in prokaryotes, one might extrapolate from its known functions in eukaryotes. For example, in S. cerevisiae (and possibly also in multicellular eukaryotes) the N-end rule pathway is the controller of peptide import, through the regulated degradation of CUP9, a transcriptional repressor of PTR2, which encodes the main importer of dipeptides and tripeptides in yeast (8, 9). The discovery of this positive-feedback circuit, which takes advantage of the multiple substrate-binding sites of UBR1, yielded a plausible scenario of how the “peptide-sensing” properties of the N-end rule pathway could either coevolve with peptide-import systems or be “recruited” by them in the course of evolution (8, 9). It remains to be determined whether prokaryotic N-end rule pathways are involved in regulating transmembrane import or export of specific compounds.
Another function of the eukaryotic N-end rule pathway is fidelity of chromosome segregation, through the degradation of a conditionally produced fragment of a subunit of cohesin, an oligomeric protein whose chromosome-associated molecules hold together sister chromatids (19, 42). Might the segregation of prokaryotic chromosomes and/or plasmids also involve a cleavage event that produces a protein fragment whose degradation by the N-end rule pathway is functionally relevant? To the best of our knowledge, no evidence either suggests or precludes this possibility in prokaryotes.
Many protein-size and peptide-size toxic polypeptides that are capable of entering eukaryotic or prokaryotic cells bear destabilizing N-terminal residues and would be, thus, potential N-end rule substrates that might be modified by aa-transferases and/or degraded by the N-end rule pathway. These polypeptides include, in particular, bacteriocins such as colicins, eukaryotic toxins such as, for example, ricin, and some of the yeast killer toxins, as well as a large variety of naturally toxic peptide-size polypeptides that can enter the cytosol of prokaryotic or eukaryotic cells (43, 44).
Concluding Remarks.
The hierarchic organization of N-end rules, i.e., their tertiary, secondary, and primary destabilizing N-terminal residues, is a feature more conserved in evolution than either the Ub dependence of N-end rule pathways or the specificity of enzymatic reactions that modify destabilizing residues (Fig. 1) (7, 11, 25). For example, the identities of Nds can be different between prokaryotic and eukaryotic N-end rule pathways (Fig. 1 A and B). The apparent confinement of Leu-conjugating aa-transferases (Bpt and Aat) to prokaryotes and Arg-conjugating aa-transferases (ATE1 and ATEL1) to eukaryotes (Fig. 1) suggests that Arg became a Ndp residue late in evolution of the N-end rule pathway, after the emergence of eukaryotes. The absence of sequelogy (32) between the yeast (S. cerevisiae) NTA1 N-terminal amidase (Nt-amidase) and the mammalian Asn-specific NTAN1 NtN-amidase (15, 16) suggests that the tertiary destabilizing N-terminal residues Asn and Gln (Fig. 1A) became part of the N-end rule still later, after the divergence of fungal and metazoan lineages.
ClpS, a 12-kDa adaptor protein specific for the prokaryotic ClpAP protease (27, 28, 30), contains a region of sequelogy to a conserved sequence of the much larger UBR1 and other E3 Ub ligases (N-recognins) of the eukaryotic N-end rule pathway (12, 45). ClpS is the N-recognin of the E. coli N-end rule pathway, where it binds to the Ndp residues Leu, Phe, Trp, or Tyr (31). ClpS also binds to the ClpA subunits of ClpAP and thereby functions as an adaptor in mediating the processive degradation of N-end rule substrates by ClpAP (31). The targeting layout of prokaryotic N-end rule substrates (specifically, the N-recognin ClpS and its ligand the ClpAP protease) is thus analogous (distantly homologous?) to that of eukaryotic N-end rule substrates, specifically, the N-recognin Ub ligase UBR1 and the 26S proteasome, including the UBR1–proteasome interaction (46). But the former machine, ClpS–ClpAP, does not involve Ub (Fig. 1B).
The ancient origins of the N-end rule pathway (it appears to predate the Ub system, of which it is a part in eukaryotes), its presence in both eukaryotes and prokaryotes, the code-like nature of the N-end rule, and the diversity of proteins that either produce or target specific N-degrons may make this pathway especially useful in phylogenetic analyses, including reconstructions of early cellular life. Being a “bridge” between eukaryotes and prokaryotes, the N-end rule pathway (Fig. 1) may also help understand the unexplained contrast between the massive size (>1,000 genes in a mammal) and broad functions of the Ub system in eukaryotes and the absence of this system in prokaryotes. The latter contain proteins with the characteristic Ub fold but lack the isopeptide bond-mediated ubiquitylation and Ub itself. The resulting dichotomy is striking, because it does not result from any obvious deficiency in prokaryotic protein degradation, in that the processive proteolysis in prokaryotes is mechanistically sophisticated and efficacious (27–31).
The origins of Ub and ubiquitylation in eukaryotes remain unclear. One idea (A.V., unpublished work) is that primordial Ub (a protein containing the Ub fold) emerged (before the appearance of eukaryotes) as an N-terminal cotranslational chaperone of very early (suboptimally folding) proteins, playing a role analogous to the one that modern Ub appears to play as a cotranslational chaperone of two specific ribosomal proteins (48). The chaperone function of primordial Ub might account for its initial spread through positive selection, before the appearance of Ub-specific enzymes. Apart from the known similarities between the eukaryotic 26S proteasome and its prokaryotic counterparts such as ClpAP (5, 27, 28, 30), the functional analogy (as well as sequelogy) between the prokaryotic N-recognin ClpS and the N-recognins (Ub ligases) of the eukaryotic N-end rule pathway (31) is unique so far, to the best of our knowledge, in linking the processive proteolysis in prokaryotes and the Ub system in eukaryotes.
Materials and Methods
Construction of E. coli Mutants.
The strains and plasmids used, as well as their construction, are described in Supporting Text and Tables 1 and 2, which are published as supporting information on the PNAS web site.
Cloning and Alterations of the V. vulnificus aat-bpt Operon.
The plasmid pEG16 (which contained the aat-bpt operon) and its deletion derivatives were constructed as described in Supporting Text.
Plasmids Expressing X-βgal Reporters and UBP1.
pUB23-X plasmids encoding Ub-X-βgal fusion proteins (35, 49) were converted into plasmids that coexpressed these fusions and the S. cerevisiae UBP1 deubiquitylating enzyme, as described in Supporting Text.
Assays for βgal Activity and N-Terminal Sequencing of X.
These procedures were carried out as described in Supporting Text.
Expression and Purification of aa-Transferases.
See Supporting Text for description of the cloning of the E. coli aat, V. vulnificus bpt, M. loti bpt, A. tumefaciens bpt, and P. falciparum ATEL1 genes, their expression in specific E. coli strains, and purification of the corresponding enzymes.
Site-Directed Mutagenesis of V. vulnificus Bpt.
Mutated alleles of the V. vulnificus bpt gene were obtained as described in Supporting Text, followed by expression and purification of the mutant Bpt proteins.
In Vitro aa-Transferase Assays.
The reporter substrates were either α-casein (with N-terminal Lys, a substrate of E. coli Aat) or bovine α-lactalbumin (with N-terminal Glu, a substrate of S. cerevisiae ATE1). Specific aa-transferases were incubated with substrates in the presence of either3H-Leu or 3H-Arg and components (including ATP, tRNAs and aminoacyl-tRNA synthetases) that enabled the amino acid–protein conjugation, followed by SDS/PAGE and autoradiography.
Phylogenetic Analysis.
Searches for sequelogs (32) of specific aa-transferases were carried out with blastp, tblastn, or psi-blast. Multiple sequence alignments were produced by using probcons. Phylogenetic analyses were carried out with phylip. See Supporting Text for references and details.
Supplementary Material
Acknowledgments
We thank J. D. Oliver (University of North Carolina, Charlotte) for the V. vulnificus C7184 translucent strain, F. Wellmer (California Institute of Technology) for the A. tumefaciens strain C58, T. Kaneko (Kazusa DNA Research Institute, Chiba, Japan) for genomic DNA of M. loti, L.-I. Hor (National Cheng-Kung University, Taiwan) for the plasmid pJRD215, the Malaria Research and Reference Reagent Resource Center (MR4; Manassas, VA) for P. falciparum 3D7 genomic DNA, which was contributed to MR4 by D. J. Carucci (National Institutes of Health, Bethesda), and D. Ladant (Institut Pasteur, Paris) for the E. coli-based two-hybrid system. This study was supported by National Institutes of Health Grants DK39520 and GM31530 (to A.V.). E.G. and K.P. were supported, respectively, by the International Human Frontier Science Program and Colvin postdoctoral fellowships.
Abbreviations
- aa-transferase
aminoacyl-transferase
- Bpt
bacterial protein transferase
- Ub
ubiquitin
- Ndp
primary destabilizing N-terminal residue
- Nds
secondary destabilizing N-terminal residue
- C*
oxidized cysteine residue
- βgal
β-galactosidase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Varshavsky A. Trends Biochem. Sci. 2005;30:283–286. doi: 10.1016/j.tibs.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A., Ciechanover A., Varshavsky A. Nat. Med. 2000;10:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 3.Pickart C. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 4.Petroski M. D., Deshaies R. J. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 5.Baumeister W., Walz J., Zühl F., Seemüller E. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 6.Wolf D. H., Hilt W. Biochim. Biophys. Acta. 2004;1695:19–31. doi: 10.1016/j.bbamcr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Varshavsky A. Proc. Natl. Acad. Sci. USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner G. C., Du F., Varshavsky A. Nature. 2000;405:579–583. doi: 10.1038/35014629. [DOI] [PubMed] [Google Scholar]
- 9.Du F., Navarro-Garcia F., Xia Z., Tasaki T., Varshavsky A. Proc. Natl. Acad. Sci. USA. 2002;99:14110–14115. doi: 10.1073/pnas.172527399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon Y. T., Xia Z. X., An J. Y., Davydov I. V., Seo J. W., Xie Y., Varshavsky A. Mol. Cell Biol. 2003;23:8255–8271. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu R. G., Sheng J., Xin Q., Xu Z., Takahashi T. T., Varshavsky A. Nature. 2005;437:981–986. doi: 10.1038/nature04027. [DOI] [PubMed] [Google Scholar]
- 12.Tasaki T., Mulder L. C. F., Iwamatsu A., Lee M. J., Davydov I. V., Varshavsky A., Muesing M., Kwon Y. T. Mol. Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki T., Varshavsky A. EMBO J. 1999;18:6017–6026. doi: 10.1093/emboj/18.21.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prakash S., Tian L., Ratliff K. S., Lehotzky R. E., Matouschek A. Nat. Struct. Mol. Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 15.Baker R. T., Varshavsky A. J. Biol. Chem. 1995;270:12065–12074. doi: 10.1074/jbc.270.20.12065. [DOI] [PubMed] [Google Scholar]
- 16.Kwon Y. T., Balogh S. A., Davydov I. V., Kashina A. S., Yoon J. K., Xie Y., Gaur A., Hyde L., Denenberg V. H., Varshavsky A. Mol. Cell. Biol. 2000;20:4135–4148. doi: 10.1128/mcb.20.11.4135-4148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon Y. T., Kashina A. S., Varshavsky A. Mol. Cell Biol. 1999;19:182–193. doi: 10.1128/mcb.19.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon Y. T., Kashina A. S., Davydov I. V., Hu R.-G., An J. Y., Seo J. W., Du F., Varshavsky A. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 19.Rao H., Uhlmann F., Nasmyth K., Varshavsky A. Nature. 2001;410:955–960. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 20.Ditzel M., Wilson R., Tenev T., Zachariou A., Paul A., Deas E., Meier P. Nat. Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- 21.Varshavsky A. Nat. Cell Biol. 2003;5:373–376. doi: 10.1038/ncb0503-373. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida S., Ito M., Gallis J., Nishida I., Watanabe A. Plant J. 2002;32:129–137. doi: 10.1046/j.1365-313x.2002.01407.x. [DOI] [PubMed] [Google Scholar]
- 23.Lee M. J., Tasaki T., Moroi K., An J. Y., Kimura S., Davydov I. V., Kwon Y. T. Proc. Natl. Acad. Sci. USA. 2005;102:15030–15035. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zenker M., Mayerle J., Lerch M. M., Tagariello A., Zerres K., Durie P. R., Beier M., Hülskamp G., Guzman C., Rehder H., et al. Nat. Genet. 2005;37:1345–1350. doi: 10.1038/ng1681. [DOI] [PubMed] [Google Scholar]
- 25.Tobias J. W., Shrader T. E., Rocap G., Varshavsky A. Science. 1991;254:1374–1377. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- 26.Shrader T. E., Tobias J. W., Varshavsky A. J. Bacteriol. 1993;175:4364–4374. doi: 10.1128/jb.175.14.4364-4374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottesman S. Annu. Rev. Cell Dev. Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- 28.Piszczek G., Rozycki J., Singh S. K., Ginsburg A., Maurizi M. R. J. Biol. Chem. 2005;280:12221–12230. doi: 10.1074/jbc.M411733200. [DOI] [PubMed] [Google Scholar]
- 29.Burton B. M., Baker T. A. Protein Sci. 2005;14:1945–1954. doi: 10.1110/ps.051417505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin A., Baker T. A., Sauer R. T. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 31.Erbse A., Schmidt R., Bornemann T., Schneider-Mergener J., Mogk A., Zahn R., Dougan D. A., Bukau B. Nature. 2006;439:753–756. doi: 10.1038/nature04412. [DOI] [PubMed] [Google Scholar]
- 32.Varshavsky A. Curr. Biol. 2004;14:R181–R183. doi: 10.1016/j.cub.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 33.Kim Y. R., Kim S. Y., Kim C. M., Lee S. E., Rhee J. H. FEMS Microbiol. Lett. 2005;243:497–503. doi: 10.1016/j.femsle.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Fukuda Y., Nakayama Y., Tomita M. Gene. 2004;323:181–187. doi: 10.1016/j.gene.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 35.Varshavsky A. Methods Enzymol. 2005;399:777–799. doi: 10.1016/S0076-6879(05)99051-4. [DOI] [PubMed] [Google Scholar]
- 36.Xie Y., Varshavsky A. EMBO J. 1999;18:6832–6844. doi: 10.1093/emboj/18.23.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karimova G., Pidoux J., Ullman A., Ladant D. Proc. Natl. Acad. Sci. USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J., Pickart C. M. Biochemistry. 1995;34:15829–15837. doi: 10.1021/bi00048a028. [DOI] [PubMed] [Google Scholar]
- 39.Gardner M. J., Hall N., Fung E., White O., Berriman M., Hyman R. W., Carlton J. M., Pain A., Nelson E., Bowman S., et al. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukhopadhyay R., Rosen B. P., Phung L. T., Silver S. FEMS Microbiol. Rev. 2002;26:311–325. doi: 10.1111/j.1574-6976.2002.tb00617.x. [DOI] [PubMed] [Google Scholar]
- 41.Paiardini A., Contestabile R., D’Aguanno S., Pascarella S., Bossa F. Biochim. Biophys. Acta. 2003;1647:214–219. doi: 10.1016/s1570-9639(03)00050-5. [DOI] [PubMed] [Google Scholar]
- 42.Uhlmann F., Lottspeich F., Nasmyth K. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 43.Zasloff M. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 44.Selitrennikoff C. P. Appl. Environ. Microbiol. 2001;67:2883–2894. doi: 10.1128/AEM.67.7.2883-2894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lupas A. N., Koretke K. K. J. Struct. Biol. 2003;141:77–83. doi: 10.1016/s1047-8477(02)00582-8. [DOI] [PubMed] [Google Scholar]
- 46.Xie Y., Varshavsky A. Proc. Natl. Acad. Sci. USA. 2000;97:2497–2502. doi: 10.1073/pnas.060025497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudolph M. J., Wuebbens M. M., Rajagopalan K. V., Schindelin H. Nat. Struct. Biol. 2001;8:42–46. doi: 10.1038/83034. [DOI] [PubMed] [Google Scholar]
- 48.Finley D., Bartel B., Varshavsky A. Nature. 1989;338:394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- 49.Bachmair A., Finley D., Varshavsky A. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.