Abstract
Objective:
To develop a fully automated, rapid, molecular-based assay that accurately and objectively evaluates sentinel lymph nodes (SLN) from breast cancer patients.
Summary Background Data:
Intraoperative analysis for the presence of metastatic cancer in SLNs from breast cancer patients lacks sensitivity. Even with immunohistochemical staining (IHC) and time-consuming review, alarming discordance in the interpretation of SLN has been observed.
Methods:
A total of 43 potential markers were evaluated for the ability to accurately characterize lymph node specimens from breast cancer patients as compared with complete histologic analysis including IHC. Selected markers then underwent external validation on 90 independent SLN specimens using rapid, multiplex quantitative reverse transcription-polymerase chain reaction (QRT-PCR) assays. Finally, 18 SLNs were analyzed using a completely automated RNA isolation, reverse transcription, and quantitative PCR instrument (GeneXpert).
Results:
Following analysis of potential markers, promising markers were evaluated to establish relative level of expression cutoff values that maximized classification accuracy. A validation set of 90 SLNs from breast cancer patients was prospectively characterized using 4 markers individually or in combinations, and the results compared with histologic analysis. A 2-marker assay was found to be 97.8% accurate (94% sensitive, 100% specific) compared with histologic analysis. The fully automated GeneXpert instrument produced comparable and reproducible results in less than 35 minutes.
Conclusions:
A rapid, fully automated QRT-PCR assay definitively characterizes breast cancer SLN with accuracy equal to conventional pathology. This approach is superior to intraoperative SLN analysis and can provide standardized, objective results to assist in pathologic diagnosis.
Current methods of lymph node analysis are time-consuming, lack standardization, depend on subjective criteria, and risk human error. We demonstrate that a fully automated, quantitative RT-PCR assay can rapidly and objectively characterize sentinel lymph nodes for the presence of metastatic breast cancer with equivalent accuracy to complete histologic analysis.
In breast cancer and other malignancies, involvement of regional lymph nodes is a strong prognostic indicator and greatly influences staging and clinical management.1–4 One benefit from the implementation of sentinel lymph node biopsy (SLNB) techniques is the identification of metastatic foci of cancer in 10% to 15% of patients that would have been previously staged as node-negative (pN0) by conventional methods.5,6 This improved sensitivity is attributed to both the addition of immunohistochemical staining (IHC) and to an increase in sampling volume.5,6 Many of these additional positive nodes contain only micrometastatic foci of tumor. However, the clinical significance of micrometastatic disease identified by SLNB techniques is highly controversial.7–11 Nonetheless, SLNB techniques are now widely used in breast cancer and melanoma and are being applied with increasing frequency to other tumors, including colorectal, oropharyngeal, prostate, lung, and other solid organ cancers.12–15
Another clear advantage to the SLNB technique is that in the majority of breast cancer patients, SLNB safely avoids axillary lymph node dissection (ALND) and the associated morbidity when the SLN is negative for metastatic disease.16 However, rapid, frozen-section analysis of SLN for metastasis is only 50% to 70% sensitive for the detection of metastasis compared with the permanent histologic sections and IHC of the same lymph node,17,18 and complete analysis of the SLNB specimen currently requires extensive preparation and time-consuming review.8 Even in experienced hands, 10% of SLNB specimens are later found to contain metastases.19 As a result, these patients have required a second surgical procedure to complete the ALND. This is clearly an undesirable algorithm for the patient, the healthcare provider, and the healthcare payer, and this dilemma has contributed to a new controversy; if the SLNB is positive, does a completion ALND confer a therapeutic or staging benefit to the patient? Until ongoing multicenter trials determine whether completing the ALND when the SLNB is positive for metastasis benefits the patient, it is clear that the time required to accurately evaluate SLNB specimens has significant implications.20
It must also be recognized that, even with adequate time, the accurate histologic analysis of lymph nodes for metastatic disease is challenging, and discordance in the interpretation of these materials is a well-known clinical problem. Indeed, a recent study found alarming disparity among pathologists in the analysis of SLNB specimens.21 Furthermore, protocols for SLNB specimen analysis vary widely between healthcare centers.22 Thus, current methods of lymph node analysis clearly lack standardization, are dependent in part on subjective criteria, and are subject to human error.
We and others have shown that a real-time, quantitative RT-PCR (QRT-PCR) analysis of lymph nodes can be more accurate than conventional histologic analysis in predicting prognosis for solid organ malignancies,23,24 and we have previously reported our development of a rapid QRT-PCR procedure that can be completed (including RNA isolation) in less than 25 minutes.25 Importantly, a revolutionary tool for molecular-based assays called the GeneXpert (Cepheid, Sunnyvale, CA) is in the final stages of development. This instrument fully automates sample preparation, RNA isolation and purification, and QRT-PCR in a single-use, cartridge-based format that removes the major obstacles limiting routine use of molecular-based assays. Thus, we aimed to determine if a rapid, fully automated, internally controlled QRT-PCR assay performed by a prototype GeneXpert instrument could produce equivalent results to histologic techniques of SLNB analysis, including IHC. This assay could produce significant benefits to patients and healthcare providers by definitively analyzing lymph nodes using objective criteria in a time frame that allows intraoperative use, reduces patient psychologic distress, and improves standardization between healthcare centers.
MATERIALS AND METHODS
Experimental Design
Literature and database surveys identified potential mRNA markers for detection of breast cancer metastases. A primary screen analyzed the expression of 43 markers in 6 primary breast carcinomas and 10 benign lymph nodes from patients without cancer. Six markers showed good characteristics for lymph node metastasis detection and entered a secondary screening phase where expression was analyzed in 25 primary tumors, 27 lymph nodes histologically positive for cancer, and 21 benign lymph nodes (73 individual patients). Based on classification characteristics, 4 markers were selected for an external validation study of 90 SLN from 90 individual breast cancer patients using a rapid, multiplex real-time PCR assay. Finally, 9 histologically negative and 9 histologically positive lymph nodes were analyzed using a completely automated and rapid RNA isolation and QRT-PCR assay on the GeneXpert instrument.
Source of Tissues
All tissues were collected through IRB approved protocols. Tissues for marker screening and GeneXpert studies were obtained from tissue banks at the University of Pittsburgh Medical Center and SLN for the marker validation study were obtained from the Minimally Invasive Molecular Staging of Breast Cancer Trial.
Tissue Preparation and Histologic Analysis
All tissues were snap-frozen in liquid nitrogen and stored at −80°C until use, when they were embedded in OCT compound for frozen sectioning on a cryostat. For the marker screening and GeneXpert studies, 45-μm sections were cut for RNA isolation. Additional sections from the beginning, middle, and end of the sections for RNA isolation were cut for hematoxylin and eosin and IHC (AE1/AE3 pancytokeratin antibodies) analysis. All slides from each specimen were independently reviewed by 2 pathologists.
For the validation study, 115 sequentially obtained SLN specimens from individual patients were identified. Five-micron serial sections were cut from each SLN, and the initial and final 2 tissue sections were mounted on slides for histologic analysis with hematoxylin and eosin staining and pancytokeratin IHC. The intervening sections were distributed 4:1:4:1:4 etc., such that 4 sections were immediately placed in chaotropic lysis buffer for RNA isolation and every fifth section was mounted on a slide for histologic review (total of 50–60 sections). Review to confirm adequate preservation of histology for pathologic analysis resulted in the exclusion of 25 specimens, principally due to freeze-drying artifact. For the remaining 90 SLNs, sections from 3 levels underwent pathologic review with both hematoxylin and eosin and IHC staining, and remaining slides were reviewed as needed.
All specimens were independently evaluated by 2 pathologists with extensive experience interpreting breast cancer specimens. The pathologists determined the presence or absence of tumor, the percentage of tumor present, and the presence of any contaminating tissues (eg, normal breast tissue). A single discordantly interpreted specimen was observed and was reviewed by both pathologists with a consensus reached that a single focus of metastatic tumor was present on only 2 of the 10 sections examined for that SLN.
RNA Isolation
For the screening and validation studies, RNA was isolated using the RNeasy minikit (Qiagen, Valencia, CA) as described by the manufacturer. The only modification was that the volume of lysis reagent was doubled and loaded on the column in 2 steps. All RNAs were DNAse treated using the DNA-free Kit from Ambion.
Quantitative RT-PCR Analysis
For the marker screening study, cDNA was synthesized using random hexamers as previously described.26 Quantitative real-time PCR was performed on the ABI Prism 7700 Sequence Detection instrument and expression of each marker gene was measured relative to the endogenous control gene β-glucuronidase using ΔCt calculations.23,27 The primary screen was performed using quantification with SYBR green.26 In the secondary screen, 5′ nuclease hybridization probes were used to increase assay specificity.26 All assays were designed using the ABI Primer Express Version 2.0 software and, where possible, amplicons spanned exon junctions to provide cDNA specificity. Negative controls were included in each PCR plate. A mixture of Universal Human Reference RNA (Stratagene, La Jolla, CA) and RNAs from human placenta, thyroid, heart, colon, PCI13 cell line, and SKBR3 cell line served as a universal positive expression control for all the genes in the marker screening process.
Analysis in the marker validation study was performed using rapid, multiplex (endogenous control gene and target gene) QRT-PCR on the SmartCycler (Cepheid) as previously described.25,28 RNA input for each lymph node sample was 50 to 200 ng per QRT-PCR reaction, and all reactions were performed in duplicate. Each reaction incorporated an internal positive control oligonucleotide to demonstrate adequate assay sensitivity in the case of negative results.28 Gene specific reverse transcription primer sequences and PCR primer and probe sequences are published as supplemental data on-line at http://www.mssm.edu/labs/godfrt01/publications/index.html.
GeneXpert Analysis
Twenty-four, 5-μm sections of OCT-embedded tissue were sectioned into 800 mL of GeneXpert lysis buffer (Cepheid). The lysis buffer was filtered through a 0.22-μm syringe filter (Osmonics Inc, West Borough, MA) and loaded into a GeneXpert cartridge. The automated processes of RNA isolation, reverse transcription, and QRT-PCR on the GeneXpert are described elsewhere (Raja S et al Clin Chem. in press).
Statistical Analyses
The characteristics used to evaluate markers were sensitivity, specificity, classification accuracy and negative and positive predictive values. The evaluation included characterizing the distributions of the markers and testing the fit of the data to the log-normal distribution. For individual markers, a cutoff value was determined that maximized the classification accuracy. In cases where classification accuracy was 100%, the cutoff was set at the midpoint between the highest expressing benign node and the lowest expressing histologically positive node. Markers were also evaluated in paired combinations and a linear prediction rule was generated for each pair. The rule was equivalent to the linear predictor that equalized the fitted probabilities above and below the linear boundary. That is, points on the boundary line had a predicted probability midway between the numeric scores assigned to positive and negative nodes.
Properties of single and paired marker prediction rules were also investigated by examining the distributional properties of the expression levels and by applying parametric bootstrap validation. Data were simulated from the lognormal and bivariate lognormal distributions using moment estimators for mean, variance, and correlation between marker pairs. A total of 500 parametric samples of the original data were obtained and the prediction for each bootstrap sample was applied to the original data. Using Efron's improved bootstrap for prediction error,29 the difference between the observed classification accuracy and the average bootstrap classification accuracy was used to estimate the optimism in the resubstitution prediction rules. The single marker and double marker decision rules were then applied to data from the marker validation study and classification characteristics were calculated.
Prediction characteristics of marker combinations were also determined by generating equal probability contours. In this method, the joint distributions of marker pairs were assumed to follow a bivariate log-normal distribution. From estimates of the means, variances, and covariances of benign nodes, equal probability contours were constructed around the estimated mean values obtained for relative level of expression in benign lymph nodes. Observed values were then plotted against these equal probability ellipsoids and compared with contours for the more extreme quantiles, including the 95th, 99th, and 99.9th percentiles. This method of analyzing the data attaches a value to each point that is the approximate probability that the plotted node is benign.
RESULTS
Primary Marker Screen
Median relative expression in primary tumors and in benign lymph nodes was calculated for all 43 potential markers included in the primary screen (Table 1). In addition, we also calculated the ratio between the median expression in tumors and the highest expressing benign node and between the lowest expressing tumor and the highest expressing benign node. When using median expression in the tumors as the numerator, 4 genes, TACSTD1, cytokeratin 7 (CK7), cytokeratin 19 (CK19), and mammoglobin 1 (MGB1), stood out as having tumor/benign node ratios greater than 1000. Thus, these 4 markers were selected for further evaluation. Mammoglobin 2 (MGB2) and prolactin inducible protein (PIP) were also selected based on the primary screen data as well as previously published data regarding these markers.30 The other 37 markers were excluded from further evaluation.
TABLE 1. Relative Expression in Primary Tumor and Benign Lymph Node From Primary Screening
Secondary Marker Screen
Histologic evaluation of the 25 primary breast cancer specimens used in the secondary screen revealed a median tumor percentage of 75% (range, 5%–95%). The median tumor percentage in the 27 histologically positive nodes was 80% (range, 5%–95%). The relative expression of the 6 markers included in the secondary screen in breast tumors, positive lymph nodes, and benign lymph nodes are shown in Figure 1A. The classification characteristics of each marker (compared with pathology review) are summarized in Table 2. The observed classification accuracies ranged from 89.6% (MGB1 and PIP) to 100% (TACSTD1). Parametric bootstrap analysis of these data is also shown in Table 2, and the estimates of classification bias ranged from 2% (TACSTD1) to 6% (MGB1). Thus, the relative expression level cutoffs established for each individual marker in the screening set should accurately characterize subsequently analyzed lymph nodes.
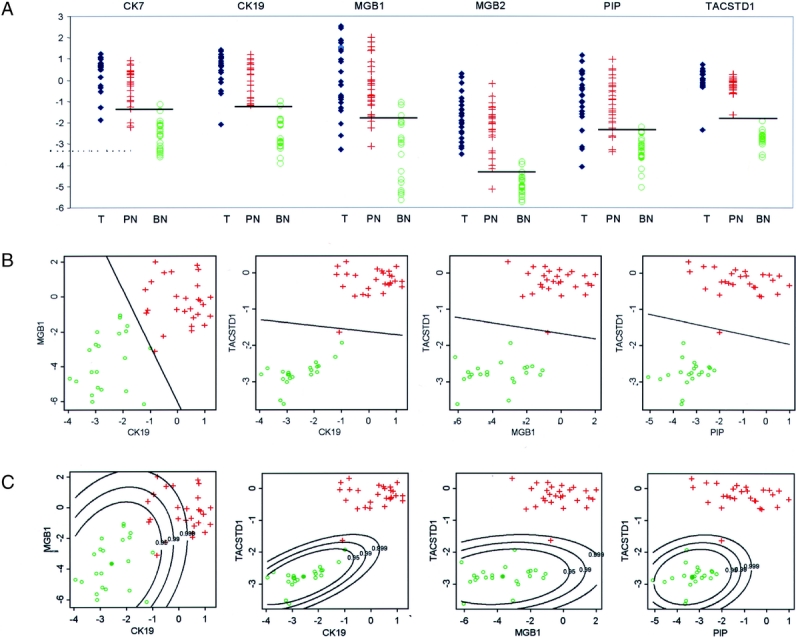
FIGURE 1. A, Data obtained from the secondary screening set of lymph nodes on individual gene expression observed in primary tumors, benign and positive lymph nodes. The horizontal line indicates the most accurate cutoff value for classifying the lymph node as positive or benign (T, tumor; PN, positive node; BN, benign node). B, Secondary screening set data on gene expression for potential 2-marker combinations using a linear discriminator decision rule. The line equalizes the predicted probability that a lymph node is positive or benign. (green o = benign lymph node; red + = histologically positive lymph node). C, Secondary screening set data on gene expression for potential 2-marker combinations applying equal probability contour statistical analysis. Equal probability curves were generated around the mean expression value observed for the 2 markers in benign lymph nodes. This demonstrates that while the marker combination of CK19 and MGB1 accurately characterizes the lymph nodes (Table 2), the wide distribution of expression observed in benign nodes diminishes their ability as accurate predictors. By this method of analysis, the marker combination of TACSTD1 and PIP more confidently characterizes the lymph nodes.
TABLE 2. Secondary Screen Marker Characteristics
We examined all possible combinations of marker pairs to determine if a 2-marker assay produces a more robust lymph node characterization. The relative expression of each possible marker pairing was analyzed using a linear decision rule that optimized characterization accuracy, and these decision rules were again internally validated using a parametric bootstrap analysis. These data are depicted in Figure 1B and summarized in Table 2. Eleven of the 15 combinations provided 100% classification accuracy in the observed data, but only 2 combinations retained 100% predicted accuracy in the bootstrap analysis. In general, the use of a pair of markers resulted in a reduction in classification bias (0%–3.2%) confirming that a 2-marker assay improved assay classification confidence compared with single markers.
Since linear classification rules are not necessarily the best method for lymph node classification in the marker combination analysis, we developed a novel classification method based on the observed distribution of expression levels for each marker in a given pair. Equal probability contours were calculated around the mean values obtained for relative expression in benign lymph nodes (Fig. 1C). This analysis demonstrates that the distribution of relative expression values obtained from benign lymph nodes impacts the confidence for classifying a positive lymph node. While CK19/MGB1 provided the best classification based on a linear prediction rule, the probability contour plot clearly shows that the wide distribution of expression for both of these markers in benign nodes negatively impacts the confidence with which a positive node can be identified. By this analysis, the combinations of TACSTD1/PIP, CK19/TACSTD1, TACSTD1/MGB1, and TACSTD1/MGB2 provide the best classification with all positive nodes correctly identified with probabilities >0.99 and in most cases >0.999. For all 4 of these combinations, all benign nodes fell within the 0.99 probability contour, and all but one was within the 0.95 probability contour. Therefore, in the screening data, 4 marker combinations were capable of providing 100% sensitivity with >99% specificity.
Validation of QRT-PCR Classification in a Rapid, Multiplex Format
To externally validate the classification accuracy of selected markers tested in the secondary screen, an independent, validation set of 90 breast cancer SLNs was prospectively analyzed (Fig. 2). Furthermore, to demonstrate the potential for intraoperative analysis, this study was performed on the SmartCycler instrument (Cepheid) using rapid, multiplex QRT-PCR. Subtle differences in calculated relative expression values were observed (data not shown) from this change in thermocycler platform, but in an effort to indirectly evaluate the robustness of the QRT-PCR analysis, the classification algorithms from the secondary screen were applied to the validation set data without any correction factors.

FIGURE 2. A, Data obtained from the validation set of SLN on individual gene expression observed in negative and positive lymph nodes (green o = histologically negative lymph node; red + = histologically positive lymph node). The horizontal line indicates the decision rule calculated from data obtained from the secondary screening set. The line equalizes the predicted probability that a lymph node is positive or negative. B, Validation set data on gene expression for 2-marker combinations using linear discriminator decision rule for all potential marker pairs. Classification characteristics of the marker combinations are reported in Table 3. C, Validation set data analyzed using the equal probability contours generated from the secondary screening set data. The relative levels of expression observed for the positive lymph nodes were beyond the 0.999 probability contours for 3 2-marker combinations. Some of the positive nodes are positive for only one marker (red crosses located in the left upper or right lower quadrants), demonstrating how a 2-marker assay improves sensitivity while maintaining high specificity.
Pathologic review identified 73 negative SLN and 17 SLN positive for metastasis with a median tumor percentage in the positive lymph nodes of 60% (range, 5%–95%). The relative expression data for each of the 4 selected markers and marker combinations is shown in Figure 2, and prospective classification accuracy for individual markers and all potential marker pairs is reported in Table 3. When cutoff values (individual markers) or linear prediction rules (marker combinations) from the secondary screen were applied to the validation set data, overall classification accuracy ranged from 89% (MGB1 alone) to 98% (TACSTD1 alone, TACSTD1/PIP, PIP/CK19 and MGB1/CK19). When probability contours from the secondary screen were applied, several marker combinations identified 16 of 17 (94%) positive nodes as positive with respect to the 0.999 probability contour of the benign nodes. All negative nodes fell within the 0.99 probability contour. One histologically positive sample was characterized as negative with >0.95 probability by analysis with all 4 markers. In a postanalysis review, this specimen was found to have been discordantly interpreted by the 2 pathologists. A consensus opinion had been reached, agreeing that a minute focus of tumor was present in the first 2 serial sections, but was not present in the remaining 8 slides. Thus, our finding that this specimen was consistently classified as negative by QRT-PCR may represent sampling error.
TABLE 3. Validation Set Results: Prospective Classification Characteristics of QRT-PCR Assays Using Single or Paired Markers

From our data, we conclude there are a number of mRNA markers and marker combinations capable of accurately detecting metastatic breast cancer in lymph nodes. However, there are at least 3 pseudogenes for CK19 within the human genome that lack intronic sequence. Thus, an mRNA-specific primer set cannot be designed for CK19, and failure of DNAse treatment to completely digest contaminating genomic DNA within the sample could produce a false-positive result. Thus, the combination that produced the highest accuracy without other potentially negative attributes was the marker pair of TACSTD1 and PIP.
Automated Lymph Node Analysis With the GeneXpert
Eighteen lymph node specimens from additional, individual patients were evaluated with fully automated, QRT-PCR assays for the markers TACSTD1 and PIP (Fig. 3). Histologic review confirmed that this set consisted of 9 positive (60%–95% tumor) and 9 negative lymph nodes. When prospectively analyzed by either a linear decision rule or equal probability contour analysis using decision rules based on data from the secondary screen set, the multiplex GeneXpert assay accurately (100%) characterized all 18 specimens within 35 minutes per assay (this does not include time for specimen delivery to the surgical pathology suite, imbedding in OCT, or cryostat cutting). We conclude that a fully automated, rapid QRT-PCR assay accurately characterizes lymph nodes for the presence of metastatic breast cancer.

FIGURE 3. Results of a fully automated, 2-marker QRT-PCR analysis of lymph nodes. By either linear decision rule analysis (left panel) or equal probability contour analysis (right panel), the assay accurately characterized all 18 lymph nodes (9 negative, 9 positive) evaluated (green o = histologically negative lymph node; red + = histologically positive lymph node).
DISCUSSION
We found excellent accuracy detecting metastatic disease within the SLN of breast cancer patients using a 2-marker QRT-PCR assay compared with the current methods of complete SLN analysis, including histologic and IHC review. We also demonstrated that accurate classification of the lymph node specimens was obtained when the assay was fully automated using the GeneXpert instrument. Thus, this assay surpassed the accuracy of current frozen section analysis of SLNB specimens and is potentially superior to complete histologic and IHC analysis for reasons including: 1) the assay is fully automated, reducing the potential for human error; 2) objective criteria are used, removing subjective analysis and improving standardization; and 3) the time from tissue acquisition to result is less than 35 minutes, facilitating intraoperative use and reducing anxious apprehension for the patient and the possibility of a second surgical procedure.
Previous studies have aimed to determine if RT-PCR analysis of lymph nodes is more sensitive than IHC and thus capable of further improving the clinical staging of breast cancer patients.31–34 Our study differs from these studies in that our present aim was not to determine if QRT-PCR identified metastatic disease in definitively analyzed, histologically negative SLN, but to identify metastatic disease with high accuracy while surpassing current methods of analysis with regards to timeliness, reproducible objectivity, and automation. However, based on the published literature regarding sensitivity of QRT-PCR analyses, the interim analysis from the Minimally Invasive Molecular Staging of Breast Cancer Trial trial, and the ability of this automated assay to improve sampling by evaluating a larger percentage of the LN (current SLNB analysis examines <1.5% of the specimen), we think that this assay may prove to be capable of surpassing current techniques in this regard.35 Multicenter clinical trials will be required to determine if this new assay can produce superior prognostic information.
This assay may ultimately prove to be superior to conventional histologic analysis because of the objective nature of the test. The accurate histologic analysis of lymph nodes for micrometastatic disease is challenging under ideal conditions, by nature subjective, and the interpretation of microscopic foci of tumor cells has eclipsed clinical outcome data. The AJCC Cancer Staging Manual, 6th edition, has established definitions to facilitate consistency in interpretation of these materials, yet these definitions make further demands on the pathologist's subjective interpretation of the lymph node. In the only published study examining this problem, Roberts et al found that, when 10 pathologists evaluated 25 cases of breast cancer SLN specimens, only 12% of the cases were correctly classified by all the pathologists, and 80% of the IHC-positive cases had at least one pathologist incorrectly characterize the case.21 In contrast, we demonstrate here, and in a separate report that describes the GeneXpert in extensive detail including interassay variability data, that the fully automated QRT-PCR assay is robust and objective. Thus, a reproducible, fully automated, objective analysis of SLN has the potential to be superior to current methods of analysis, and a multicenter, prospective trial designed to make this comparison is currently in development.
While this assay may have the potential to eventually replace current methods of lymph node analysis, further investigation is necessary and it is currently applicable only as a research tool. The implementation of this assay into routine clinical practice represents a paradigm shift in cancer diagnostics and will require a period where the assay is performed concurrently with standard methods of analysis. Concomitant use of these techniques requires a dedicated surgical pathologist and a thoughtful approach to specimen preparation. We propose a method of sample preparation that combines formalin fixation of the majority of the SLNB specimen for histologic analysis while providing fresh-frozen tissue for rapid histologic analysis and QRT-PCR analysis (Fig. 4). This approach will allow pathologists to perform the 2 methods of analysis simultaneously in an intraoperative time frame and still have paraffin-embedded material for full histologic analysis.

FIGURE 4. Proposed method of lymph node preparation that facilitates thorough histologic analysis and isolation of high-quality RNA. The lymph node is cut into 5 sections. Three sections are processed into paraffin for subsequent histologic analysis. Two sections are embedded in OCT and cryostat sections obtained for histologic analysis and RNA isolation. The remaining uncut tissue may be stored frozen or paraffin-embedded for future analysis.
A number of controversies currently exist regarding the surgical management of patients with breast cancer. This QRT-PCR assay was initially conceived, in part, to resolve the clinical dilemma that arose from a lack of an accurate intraoperative analysis of SLNB specimens that led to the need for a separate, second surgical procedure to complete the ALND in a substantial proportion of patients. Some data suggest that completion of lymph node dissection may not be necessary for either therapeutic or staging purposes, thus potentially eliminating this particular need for a rapid assay.20 Large, randomized trials (ACOSOG Z-0011 and NSABP 32) are designed to settle this important clinical controversy. Another issue is that some leaders in breast cancer surgery are currently determining the need for ALND based on the size of the focus of metastatic disease or other histologic parameters, and this practice has basis in published data.36 This QRT-PCR assay cannot produce such information presently, but the assay is quantitative, and such capabilities are certainly a future possibility.
A molecular method of analyzing SLNB specimens is only one of a number of potential advances in the staging of cancer patients. Recent studies have been able to identify patterns of gene expression in primary breast carcinomas that are associated with a poor prognosis.37 It is important to note that these results were obtained from a selected subset of patients that were node-negative. While future studies may show that gene expression profiling is independently sufficient for determining prognosis and subsequent therapies, SLN status continues to be an essential component of clinical staging.
Finally, this technology holds promise for improving patient care for a number of other malignancies. The same clinical dilemma presented for breast cancer exists for the intraoperative analysis of SLN specimens in melanoma and potentially squamous cancers of the oropharynx.13 The accurate assessment of lymph node involvement in an intraoperative time frame may also impact the surgical management of lung cancer.13 Similar intraoperative applications exist in the surgical management of metastatic colorectal cancer. Assays for all of these clinical applications are currently in development. The automated, objective nature of this assay may also facilitate use in other less time-dependent analyses such as the routine analysis of lymph nodes in gastrointestinal malignancies. Current analysis of these lymph nodes does not typically include IHC analysis or sufficient attention to sampling, and multiple studies have suggested this likely results in the understaging of a significant percentage of patients.12
CONCLUSION
We have shown that a 2-marker, QRT-PCR assay that is fully automated and completed in under 35 minutes can accurately characterize lymph nodes for the presence of metastatic breast cancer. This assay is clearly superior to current methods of intraoperative analysis and is as accurate as current methods of complete histologic analysis with immunohistochemistry. Theoretical advantages to such an assay include improved standardization across varying healthcare environments, more thorough sampling of the lymph node, and reduced human error.
ACKNOWLEDGMENTS
The authors thank Dr. Lee Wilke for her thoughtful review of the manuscript.
Footnotes
Supported in part by NIH CA-01958 (S.J.H.), NIH CA-099123 (J.C., T.E.G., S.J.H.), Department of Defense N00014-99-1-0784 (D.J.C., W.E.G., K.M.), and a cooperative research and development grant from Cepheid (T.E.G.).
Reprints: Steven J. Hughes, MD, Department of Surgery, University of Pittsburgh, 497 Scaife Hall, 3550 Terrace Street, Pittsburgh, PA 15261. E-mail: Hughess2@upmc.edu.
REFERENCES
- 1.Fisher B, Bauer M, Wickerham DL, et al. Relation of number of positive axillary nodes to the prognosis of patients with primary breast cancer: an NSABP update. Cancer. 1983;52:1551–1557. [DOI] [PubMed] [Google Scholar]
- 2.Fisher ER, Sass R, Fisher B. Pathologic findings from the National Surgical Adjuvant Project for Breast Cancers (protocol no. 4): X. Discriminants for tenth year treatment failure. Cancer. 1984;53:712–723. [DOI] [PubMed] [Google Scholar]
- 3.Fisher ER, Costantino J, Fisher B, et al. Pathologic findings from the National Surgical Adjuvant Breast Project (Protocol 4): Discriminants for 15-year survival. National Surgical Adjuvant Breast and Bowel Project Investigators. Cancer. 1993;71:2141–2150. [DOI] [PubMed] [Google Scholar]
- 4.Siziopikou KP, Schnitt SJ, Connolly JL, et al. Detection and significance of occult axillary metastatic disease in breast cancer patients. Breast J. 1999;5:221–229. [DOI] [PubMed] [Google Scholar]
- 5.Turner RR, Ollila DW, Krasne DL, et al. Histopathologic validation of the sentinel lymph node hypothesis for breast carcinoma. Ann Surg. 1997;226:271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cote RJ, Peterson HF, Chaiwun B, et al. Role of immunohistochemical detection of lymph-node metastases in management of breast cancer. Lancet. 1999;354:896–900. [DOI] [PubMed] [Google Scholar]
- 7.de Mascarel I, Bonichon F, Coindre JM, et al. Prognostic significance of breast cancer axillary lymph node micrometastases assessed by two special techniques: reevaluation with longer follow-up. Br J Cancer. 1992;66:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giuliano AE, Dale PS, Turner RR, et al. Improved axillary staging of breast cancer with sentinel lymphadenectomy. Ann Surg. 1995;222:394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leong AS. The prognostic dilemma of nodal micrometastases in breast carcinoma. Gan To Kagaku Ryoho. 2000;27(suppl 2):315–320. [PubMed] [Google Scholar]
- 10.Noguchi M. Therapeutic relevance of breast cancer micrometastases in sentinel lymph nodes. Br J Surg. 2002;89:1505–1515. [DOI] [PubMed] [Google Scholar]
- 11.Weaver DL. Sentinel lymph nodes and breast carcinoma: which micrometastases are clinically significant? Am J Surg Pathol. 2003;27:842–845. [DOI] [PubMed] [Google Scholar]
- 12.Saha S, Dan AG, Bilchik AJ, et al. Historical review of lymphatic mapping in gastrointestinal malignancies. Ann Surg Oncol. 2004;11(suppl):245–249. [DOI] [PubMed] [Google Scholar]
- 13.Goyal A, Mansel RE. Current status of sentinel lymph node biopsy in solid malignancies. World J Surg Oncol. 2004;2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross GL, Soutar DS, MacDonald DG, et al. Improved staging of cervical metastases in clinically node-negative patients with head and neck squamous cell carcinoma. Ann Surg Oncol. 2004;11:213–218. [DOI] [PubMed] [Google Scholar]
- 15.Wawroschek F, Vogt H, Weckermann D, et al. The sentinel lymph node concept in prostate cancer: first results of gamma probe-guided sentinel lymph node identification. Eur Urol. 1999;36:595–600. [DOI] [PubMed] [Google Scholar]
- 16.Giuliano AE, Haigh PI, Brennan MB, et al. Prospective observational study of sentinel lymphadenectomy without further axillary dissection in patients with sentinel node-negative breast cancer. J Clin Oncol. 2000;18:2553–2559. [DOI] [PubMed] [Google Scholar]
- 17.Chao C, Wong SL, Ackermann D, et al. Utility of intraoperative frozen section analysis of sentinel lymph nodes in breast cancer. Am J Surg. 2001;182:609–615. [DOI] [PubMed] [Google Scholar]
- 18.Gulec SA, Su J, O'Leary JP, et al. Clinical utility of frozen section in sentinel node biopsy in breast cancer. Am Surg. 2001;67:529–532. [PubMed] [Google Scholar]
- 19.Gemignani ML, Cody HS III, Fey JV, et al. Impact of sentinel lymph node mapping on relative charges in patients with early-stage breast cancer. Ann Surg Oncol. 2000;7:575–580. [DOI] [PubMed] [Google Scholar]
- 20.Wilke LG, Giuliano A. Sentinel lymph node biopsy in patients with early-stage breast cancer: status of the National Clinical Trials. Surg Clin North Am. 2003;83:901–910. [DOI] [PubMed] [Google Scholar]
- 21.Roberts CA, Beitsch PD, Litz CE, et al. Interpretive disparity among pathologists in breast sentinel lymph node evaluation. Am J Surg. 2003;186:324–329. [DOI] [PubMed] [Google Scholar]
- 22.Hunt JL, Baloch ZW, LiVolsi VA. Sentinel lymph node evaluation for tumor metastasis. Semin Diagn Pathol. 2002;19:263–277. [PubMed] [Google Scholar]
- 23.Godfrey TE, Raja S, Finkelstein SD, et al. Prognostic value of quantitative reverse transcription-polymerase chain reaction in lymph node-negative esophageal cancer patients. Clin Cancer Res. 2001;7:4041–4048. [PubMed] [Google Scholar]
- 24.Liefers GJ, Cleton-Jansen AM, van de Velde CJ, et al. Micrometastases and survival in stage II colorectal cancer. N Engl J Med. 1998;339:223–228. [DOI] [PubMed] [Google Scholar]
- 25.Raja S, Luketich JD, Kelly LA, et al. Rapid, quantitative reverse transcriptase-polymerase chain reaction: application to intraoperative molecular detection of occult metastases in esophageal cancer. J Thorac Cardiovasc Surg. 2002;123:475–482. [DOI] [PubMed] [Google Scholar]
- 26.Xi L, Raja S, Gooding W, et al. Molecular staging of lymph nodes from patient's with esophageal adenocarcinoma. Clin Cancer Res. in press. [PubMed]
- 27.Tassone F, Hagerman RJ, Taylor AK, et al. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet. 2000;66:6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raja S, El Hefnawy T, Kelly LA, et al. Temperature-controlled primer limit for multiplexing of rapid, quantitative reverse transcription-PCR assays: application to intraoperative cancer diagnostics. Clin Chem. 2002;48:1329–1337. [PubMed] [Google Scholar]
- 29.Efron B. An Introduction to the Bootstrap. Boca Raton, FL: Chapman and Hall, 1993:247–252. [Google Scholar]
- 30.Mitas M, Mikhitarian K, Walters C, et al. Quantitative real-time RT-PCR detection of breast cancer micrometastasis using a multigene marker panel. Int J Cancer. 2001;93:162–171. [DOI] [PubMed] [Google Scholar]
- 31.Bostick PJ, Huynh KT, Sarantou T, et al. Detection of metastases in sentinel lymph nodes of breast cancer patients by multiple-marker RT-PCR. Int J Cancer. 1998;79:645–651. [DOI] [PubMed] [Google Scholar]
- 32.Bostick PJ, Chatterjee S, Chi DD, et al. Limitations of specific reverse-transcriptase polymerase chain reaction markers in the detection of metastases in the lymph nodes and blood of breast cancer patients. J Clin Oncol. 1998;16:2632–2640. [DOI] [PubMed] [Google Scholar]
- 33.Mori M, Mimori K, Inoue H, et al. Detection of cancer micrometastases in lymph nodes by reverse transcriptase-polymerase chain reaction. Cancer Res. 1995;55:3417–3420. [PubMed] [Google Scholar]
- 34.Schoenfeld A, Luqmani Y, Smith D, et al. Detection of breast cancer micrometastases in axillary lymph nodes by using polymerase chain reaction. Cancer Res. 1994;54:2986–2990. [PubMed] [Google Scholar]
- 35.Gillanders WE, Mikhitarian K, Hebert R, et al. Molecular detection of micrometastatic breast cancer in histopathology-negative axillary lymph nodes correlates with traditional predictors of prognosis: an interim analysis of a prospective multi-institutional cohort study. Ann Surg. 2004;239:828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Zee KJ, Manasseh DM, Bevilacqua JL, et al. A nomogram for predicting the likelihood of additional nodal metastases in breast cancer patients with a positive sentinel node biopsy. Ann Surg Oncol. 2003;10:1140–1151. [DOI] [PubMed] [Google Scholar]
- 37.van de Vijver M, Van't Veer LJ, Dai H, et al. Gene expression profiling of breast cancer accurately predicts clinical outcome of disease. Mod Pathol. 2002;15:55A. [Google Scholar]




