Abstract
The recognition that human tumors stimulate the production of autoantibodies against autologous cellular proteins called tumor-associated antigens (TAAs) has opened the door to the possibility that autoantibodies could be exploited as serological tools for the early diagnosis and management of cancer. Cancer-associated autoantibodies are often driven by intracellular proteins that are mutated, modified, or aberrantly expressed in tumor cells and hence are regarded as immunological reporters that could help uncover molecular events underlying tumorigenesis. Emerging evidence suggests that each type of cancer might trigger unique autoantibody signatures that reflect the nature of the malignant process in the affected organ. The advent of novel genomic, proteomic, and high throughput approaches has accelerated interest in the serum autoantibody repertoire in human cancers for the discovery of candidate TAAs. The use of individual anti-TAA autoantibodies as diagnostic or prognostic tools has been tempered by their low frequency and heterogeneity in most human cancers. However, TAA arrays comprising several antigens significantly increase this frequency and hold great promise for the early detection of cancer, monitoring cancer progression, guiding individualized therapeutic interventions, and identification of novel therapeutic targets. Our recent studies suggest that the implementation of TAA arrays in screening programs for the diagnosis of prostate cancer and other cancers should be preceded by the optimization of their sensitivity and specificity through the careful selection of the most favorable combinations of TAAs.
Cancer remains a major public health problem in the United States and developing countries and is responsible for one in four deaths with cancers of the lung, breast, prostate, and colon topping the list of estimated new cancer cases and deaths in 2006 (1). Despite the availability of a plethora of genes and their protein products that could serve as cancer biomarkers, it is widely recognized that their combined use with the available clinical information is still insufficient for early cancer diagnosis and for guiding individualized therapeutic interventions and predicting outcomes. There remains a need for the development of innovative diagnostic and prognostic tools that effectively exploit biomarkers for the management of human cancers. Autoantibodies to TAAs1 have generated increasing interest as one such biomarker (2, 3).
Presently there is a growing enthusiasm for applying proteomic approaches to the identification of serum biomarkers for the early, non-invasive diagnosis of cancer and for monitoring tumor progression. These approaches include direct profiling of human sera, using two-dimensional (2D) gel electrophoresis and MS to identify distinctive protein signatures characteristic of different tumor types (4–7), and the exploitation of the serum autoantibody repertoire from cancer patients for the identification of TAAs and the design of TAA panels or arrays (2, 3, 7–12). This review emphasizes the utility of such arrays for serological diagnosis in cancer. The first part will focus on the biological and clinical significance of serum autoantibodies to TAAs and novel approaches for the identification and validation of these antigens. This will be followed by a discussion of recent advances and current issues in the design of TAA arrays for cancer diagnosis.
CANCER-ASSOCIATED AUTOANTIBODIES AS REPORTERS OF TUMORIGENESIS
Our knowledge of the biology of serum autoantibodies and their target autoantigens is derived primarily from the intense investigations conducted during the past 4 decades on the nature of autoantibody responses in patients with systemic autoimmune diseases such as systemic lupus erythematosus (SLE), scleroderma, rheumatoid arthritis (RA), and Sjögren syndrome. Several principles have been established from these investigations. First, autoantibodies in systemic autoimmune diseases are directed predominantly against intracellular antigens that participate in important cellular functions, including DNA replication and transcription, RNA processing and splicing, translation, and mitosis (13, 14). Second, these autoantibodies appear to be antigen-driven as evidenced by their recognition of both linear and conformational epitopes in protein and nucleic acid components of nuclear and cytoplasmic organelles or particles (e.g. ribosomes, nucleosomes, spliceosomes, coiled bodies, and nucleoli) (13, 14). Third, autoantibodies in systemic autoimmune diseases have diagnostic and prognostic value as they often precede the onset of clinical symptoms and are associated with disease severity (15–17). Furthermore their utility in diagnosis is underscored by the fact that each systemic autoimmune disease is associated with a particular set of autoantibodies (13, 14). Fourth, with a few exceptions, these autoantibodies are not considered pathogenic but rather immunological “fingerprints” of pathological processes associated with the development of systemic autoimmunity (18). In recent years, it has become clear that circulating serum autoantibodies that are reactive with intracellular proteins (generically termed antinuclear autoantibodies) are not a unique feature of systemic autoimmune or rheumatic diseases but are also present in a variety of human disease conditions where there is immune system involvement, including but not limited to atopic dermatitis (19, 20), chronic fatigue syndrome (21), interstitial cystitis (20), inflammatory diseases of the eye (22, 23), and cancer (24–38).
It is not entirely clear how intracellular proteins become targets of autoantibodies, but it has been suggested that posttranslational modifications (e.g. proteolytic cleavage, phosphorylation, and oxidation) associated with aberrant cell death may enhance their immunogenicity under a proinflammatory environment (39–43). Other possibilities are that specific autoantigens are fetal proteins aberrantly expressed in tumor cells (44) or expressed in abnormally high amounts in tissues affected by autoimmune disease, inflammatory disease, or cancer, contributing to loss of immune tolerance to these antigens (45).
The elicitation of serum autoantibodies to autologous cellular antigens expressed in tumors is a well recognized form of cancer-related autoimmunity. These autoantibodies have been detected in several human cancers, and significant advances have been made in the identification of their target antigens, particularly in lung cancer (28, 30), colorectal cancer (36), breast cancer (29), prostate cancer (27, 37), leukemia (26), non-Hodgkin lymphoma (24), hepatocellular carcinoma (25, 32, 34), ovarian cancer (31), pancreatic cancer (33, 38), and paraneoplastic neurological syndromes (35). Although the mechanisms leading to autoantibody production in cancer patients are not clearly understood, emerging evidence indicates that most TAAs are cellular proteins whose aberrant regulation or function could be linked to malignancy (3). For instance, TAAs include known oncoproteins such as HER-2/Neu and c-MYC (46–49); tumor suppressor proteins such as p53 (50); survival proteins such as survivin and lens epithelium-derived growth factor (LEDGF/p75) (27, 51); cell cycle regulatory proteins such as cyclin B1 (52); mitosis-associated proteins such as centromere protein F (CENP-F) (25, 53, 54); chromatin-associated proteins such as topoisomerases (29, 55); mRNA-binding proteins such as p62, IMP1, and Koc (56, 57); and differentiation and cancer testis antigens such as NY-ESO-1 (58).
The oncoprotein nature of most TAAs has led to the hypothesis that cancer-associated autoantibodies are immunological “reporters” or “sentinels” identifying aberrant cellular mechanisms associated with tumorigenesis (2, 3). This was first hinted by the observation that these autoantibodies often arise during the transition from conditions that pose a high risk for cancer to the development of malignancy (59, 60). For instance, the transition from chronic liver disease to hepatocellular carcinoma is associated with the appearance of autoantibodies to proteins linked to cell survival and proliferation that are also highly expressed in tumors, such as CENP-F and p62; the latter is a developmentally regulated protein that binds the mRNA of insulin-like growth factor II (IGF-II) (60). This transition is also associated with increasing titers and changes in the specificities of antibodies to specific nuclear proteins (59).
Additional evidence that cancer-associated autoantibodies function as “red flags” indicating the presence of a malignant transformation process can be found in studies on the autoantibody response to p53, one of the best characterized in cancer. p53 is a multifunctional transcription factor that promotes tumor cell death by regulating the expression of genes involved in cell cycle control, apoptosis, DNA repair, and angiogenesis (61). Mutation or down-regulation of p53 prevents it from exerting its tumor suppressor function, contributing to cancer development and progression (61). p53 mutations in cancer patients are often associated with poor prognosis (61). The presence of serum autoantibodies to p53 has been reported in patients with various types of cancers at frequencies ranging from 4 to 30% depending on the cancer type (50), and in some cancers, autoantibodies to p53 are associated with poor prognosis (62). A strong correlation between the presence of these autoantibodies and p53 mutation and/or accumulation has been reported (50). Only missense mutations have been associated with this autoantibody response (63, 64). Although these mutations could alter the native conformation of p53, which may result in aberrant processing and presentation by antigen-presenting cells, there is some controversy with regard to whether the anti-p53 autoantibodies are driven by the p53 mutations themselves or by accumulation of non-functional p53 protein in the nucleus.
An emerging concept is that autoantibodies associated with a specific type of cancer are directed against aberrantly regulated or activated protein components of molecular pathways involved in the malignant transformation process in that particular type of cancer. For example, the rapamycin-sensitive mTOR (mammalian target of rapamycin) phosphorylation pathway has been implicated in the pathogenesis of breast cancer, and members of this pathway are targeted by autoantibodies in patients with breast cancer (29). These include ribosomal protein S6, eukaryotic elongation factor 2, eukaryotic elongation factor 2 kinase, and heat shock protein 90 (HSP90). Autoantibodies in breast cancer also recognize components of the DNA repair pathway such as the Ku protein, topoisomerase I, and the 32-kDa subunit of replication protein A (29). It should be noted that replication protein A interacts with the DNA repair and tumor suppressor proteins BRCA1 and BRCA2 (65, 66). Loss of BRCA function has been linked to development of breast cancer (67).
The autoantibody response in prostate cancer may also reflect the activation of genes and their protein products that play an important role in prostate tumorigenesis, including several proteins that participate in the cellular response to oxidative stress (Table I). There is growing evidence that an augmented state of cellular oxidative stress (ASCOS), associated with a proinflammatory environment, plays a major role in prostate carcinogenesis (68, 69). This evidence is derived from the following observations: (a) Clinical trials with antioxidants such as lycopene, vitamin E, and selenium suggest that antioxidants may be beneficial in limiting the progression of prostate cancer (70–73), (b) androgens stimulate prostate cell growth by increasing oxidative stress (74, 75), (c) prostate cancer tissues manifest increased oxidative stress (76, 77), and (d) prostate cancer development is associated with early loss of the antioxidant enzyme glutathione S-transferase protein 1 (GSTP1), leaving prostate cells vulnerable or tolerant to oxidative DNA damage (68, 69). The cellular response to increased oxidative stress can lead to either activation of the apoptotic machinery if oxidative stress levels are too high or activation of a protective stress/survival response if the levels are not excessive enough to induce massive damage and rapid cell death but sufficient to induce sublethal oxidative damage (78). Cancer cells appear to develop a high threshold of resistance to oxidative stress-induced cell death as evidenced by their ability to survive under conditions of increased oxidative stress and other stressors such as hypoxia, heat, and lack of growth and survival factors (78–81). This resistance is associated with overexpression of survival and antioxidant proteins in tumors, which could lead to loss of immune tolerance and provoke autoantibody responses.
Table I.
Candidate TAAs recognized by autoantibodies from prostate cancer patients
| TAA | Function | Frequency of autoantibodies | Ref. |
|---|---|---|---|
| 5-α-Reductase | Reduction of testosterone to DHT | ND | 112 |
| AKRIA1 | Alcohol dehydrogenase | ND | 98 |
| AMACR | Branched chain fatty acid β oxidation | ND | 113 |
| Brd2 | Mitogen-activated kinase | ND | 37 |
| C17 orf 25 | Chromosome 17 open reading frame 25 | ND | 98 |
| CAPZA1 | Actin-capping protein | ND | 98 |
| Clusterin | Antioxidant defense | ND | 98, 103 |
| c-MYC | Oncogene | 3.9 | 12, 111, 144, 150 |
| Cyclin A | Cell cycle regulation | 7.5 | 150 |
| Cyclin B1 | Cell cycle regulation | 14.1 | 12, 111, 144, 150 |
| Cyclin D1 | Cell cycle regulation | 11.3 | 150 |
| DJ-1 | Oncoprotein and ROS scavenger | ND | 98 |
| Drebrin | Actin-binding protein | ND | 122 |
| eIF4G1 | Translation initiation | ND | 37 |
| GRP78 | Stress response protein | ND | 108 |
| HIP1 | Clathrin-binding protein involved in growth factor receptor trafficking | 46.4 | 152 |
| HSP70 protein 1 | Stress response protein | ND | 98 |
| HSPA8 | Stress response protein | ND | 98 |
| HSPB1 | Same as HSP27, stress response protein | ND | 98 |
| IMP1 | IGF-II mRNA-binding protein | 8.7 | 12, 111, 144, 150 |
| Koc | IGF-II mRNA-binding protein | 9.2 | 12, 111, 144, 150 |
| Lactoylglutathione lyase | Known as glyoxalase 1 (GLO1); catalyzes detoxification of α-oxoaldehydes | ND | 98 |
| LEDGF/p75 | Stress response transcription co-activator | 20 | 27 |
| MAD-CT-1 | Condensation and packaging of sperm chromatin | ND | 153 |
| MAD-CT-2 | Unknown | ND | 153 |
| No55 | Nucleolar protein | ND | 122 |
| P53 | Tumor suppressor | 4.9 | 12, 111, 144, 150 |
| P62 | IGF-II mRNA-binding protein | 25.2 | 12, 111, 144, 150 |
| P90 | Unknown | 30.8 | 150 |
| PP4R | Regulatory subunit of serine/threonine phosphatase 4 | ND | 122 |
| PRDX6 | Antioxidant defense | ND | 98 |
| PIP | Prolactin inducible protein | ND | 98 |
| PSA | Prostate-specific antigen | ND | 98 |
| RPL13a | Translation control | ND | 37 |
| RPL22 | Ribosomal protein | ND | 37 |
| SOD1 | Antioxidant defense | ND | 98 |
| Survivin | Inhibitor of apoptosis | 2.9 | 12, 111, 144, 150 |
| Syntenin 1 | Interactor with the syndecans | ND | 98 |
| TDP-43 | Splicing factor | ND | 122 |
| VCP | Transitional endoplasmic reticulum ATPase | ND | 98 |
| vWF | Mediates platelet aggregation | ND | 154 |
ROS, reactive oxygen species; DHT, dihydrotestosterone; ND, not determined; AMACR, α -methylacyl-coenzyme A racemase; vWF, von Willebrand factor; VCP, valosin-containing protein.
We reported recently that LEDGF/p75 (also known as the DFS70 autoantigen), a transcription co-activator that is up-regulated by increased oxidative stress, is overexpressed in prostate tumors and targeted by autoantibodies in ~20% of patients with prostate cancer compared with a frequency of 5% in age- and gender-matched individuals with no history of prostate cancer (27). Although originally identified as a transcription co-activator and a growth and survival factor in lens epithelial cells (82, 83), LEDGF/p75 is now known to be ubiquitously expressed in tumor cells and is emerging as a key regulator of the cellular response to stress (84, 85). LEDGF/p75 promotes protection against oxidative stress-induced cell death by transcriptionally activating stress proteins, such as HSP27, αB-crystallin, and anti-oxidant protein 2 (AOP2; also known as peroxiredoxin 6 (PRDX6)) (85–88). This transactivation is facilitated by the binding of LEDGF/p75 to heat shock elements and stress-related elements in promoter regions of target genes (88). LEDGF/p75 has been also identified as a target autoantigen in leukemia by serological analysis of recombinant expression libraries (SEREX) (89). Consistent with this finding, the LEDGF gene is involved in chromosomal translocations in patients with various types of leukemia, resulting in a fusion with nucleoprotein-98 that retains the C terminus of LEDGF/p75 (90–92). This domain has been implicated in DNA binding and transcription function (93), survival function (93, 94), binding to human immunodeficiency virus integrase (95), and autoantibody recognition (96). It should be emphasized that anti-LEDGF/p75 antibodies are not specific for prostate cancer because they have been also detected, albeit at relatively low frequencies and titers, in “healthy individuals” and patients with various human inflammatory conditions (97). We suggested that autoantibodies to LEDGF/p75 could be considered as reporters of the up-regulation of this protein by inflammation and ASCOS in cancer and other inflammatory conditions (27, 97).
Using a combination of proteomic and immunological approaches, Ronquist et al. (98) identified various proteins present in prostasomes (submicron prostate-derived particles) that were capable of eliciting autoantibody responses in prostate cancer patients. Intriguingly, several of these candidate TAAs are proteins that, like LEDGF/p75, regulate the cellular redox environment and protect cancer cells against the increased oxidative stress and other stresses present within the tumor microenvironment. Among these candidate TAAs were PRDX6/AOP2, clusterin, DJ-1, superoxide dismutase, alcohol dehydrogenase, heat shock proteins, and lactoylglutathione lyase. As mentioned above, PRDX6/AOP2 is transcriptionally regulated by LEDGF/p75 and plays an important role in anti-oxidant defense (86, 99). Interestingly, loss of the homeobox gene Nkx3.1 in mice is associated with loss of PRDX6/AOP2 and promotes increased oxidative damage in prostate carcinogenesis (100). Clusterin (also known as apolipoprotein J) is a ubiquitously expressed secreted glycoprotein whose over-expression in prostate cancer cells protects against death induced by high levels of oxidative stress (101, 102). DJ-1 is a novel oncoprotein that is the causative gene for the familial form of Parkinson disease and antagonizes oxidative stress by eliminating reactive oxygen species (103). Superoxide dismutase is an antioxidant defense enzyme that plays an important role in protecting the prostate against oxidative damage (104). Heat shock protein 70 is known to inhibit stress-induced apoptosis in prostate cancer cells (105). HSP27 (also called HSPB1) is also a stress response antiapoptotic protein that is regulated by LEDGF/p75 (88, 105–107). αB-crystallin, another heat shock protein implicated in resistance to cell death in various cancer types, is also regulated by LEDGF/p75 (88, 106), but to our knowledge the presence of autoantibodies to this protein in prostate cancer sera has not been reported. Another stress response protein, glucose-regulated protein-78 kDa (GRP78), was shown to be a target of autoantibodies in advanced prostate cancer but not in lung, breast, and ovarian cancers (108). Overexpression of this protein was detected in metastatic prostate cancer but not in normal prostate (108). We have also demonstrated the presence of serum autoantibodies in prostate cancer patients against p62, Koc, and IMP1, proteins that bind the mRNA of IGF-II, an important global regulator of cell survival in breast and prostate cancers (109–111). It has been suggested that overexpression of these proteins in prostate and other tumors may stabilize IGF-II mRNA, leading to its up-regulation and, consequently, to tumor cell survival (57). α-Methylacyl-coenzyme A racemase and 5-α-reductase are two enzymes involved in redox reactions that are also targeted by autoantibodies in prostate cancer (112, 113); however, their role in prostate tumor cell survival is not clear.
Based on the limited information available, it seems that the autoantibody response to TAAs in prostate cancer appears to be directed preferentially against proteins that participate in redox and stress/survival pathways and are highly expressed in prostate tumors. However, further studies are needed to confirm this association. It would be also important to determine whether different cancers have unique signature antibody repertoires that target TAAs preferentially associated with specific molecular or metabolic pathways activated during malignant transformation.
IDENTIFICATION AND VALIDATION OF TAA
Although several approaches have been used during the past 20 years for the identification of TAAs, the most successful have been the serological screening of cDNA expression libraries and phage display libraries and more recently proteomics-based approaches. In the early 1990s, our group used sera from patients with hepatocellular carcinoma to immunoscreen commercially available HepG2 cDNA expression libraries for the identification of putative TAAs (114). This approach, which had been previously used to identify autoantigens in systemic autoimmune diseases (115), yielded several putative TAAs targeted by autoantibodies in liver cancer, including HCC1, a protein that plays a role in alternative splicing (114); SG2NA, a cell cycle-regulated protein with WD-40 motifs (116); CENP-F, a mitosis-associated protein previously identified as an autoantigen in various malignancies (53, 54, 60); and p62, an IGF-II mRNA-binding protein (60). A later variant of this approach, termed SEREX, used cDNA expression libraries that were constructed from a patient’s tumors and screened with autologous sera (117, 118). SEREX has been exploited by several groups for the identification of hundreds of candidate TAAs in various human cancer types (119), including but not limited to lung cancer (28), liver cancer (120), breast cancer (121), prostate cancer (122), ovarian cancer (123), renal cancer (124), head and neck cancer (125), esophageal cancer (126), lymphoma (127), and leukemia (89). Although a powerful approach for the identification of candidate TAAs, SEREX has several limitations. It has an inherent bias toward selecting antigens that are highly expressed in the particular tumor used to construct the cDNA library as these are overrepresented in the library (128). This may preclude the selection of underrepresented antigens that might be involved in tumorigenesis (128). It is also possible that antigens that are highly expressed in a tumor from a particular patient may not be overexpressed in a tumor from another patient, which presents the challenge of having to develop several cDNA libraries from different patients with the same tumor type to maximize the number of identifiable candidate TAAs. Another complication is that cDNA libraries derived from patient cancer tissue contain genes of different cell types. Given these limitations, later studies using SEREX have reverted to the use of cDNA libraries from established tumor cell lines (129). SEREX is also labor-intensive and difficult to be adapted for automated high throughput analysis of large numbers of patient sera and tumor libraries. The limitations of the SEREX approach and the development of improved methods to maximize its utility in the identification of TAAs have been discussed recently in detail by Fernandez Madrid et al. (128).
A second approach for the serological identification of putative TAAs with the possibility of high throughput analysis of patient sera and tumor libraries is the cDNA or peptide phage surface display technology (130). This approach involves the construction of a cDNA phage display library from a specific tumor or cancer cell line. The candidate TAAs in these libraries are expressed and displayed on the surface of a phage, such as the T7 phage, as peptides fused to the capsid protein of the phage. This surface complex is then used as a bait to capture autoantibodies to TAAs using pools of cancer and control sera. After several rounds of biopanning to enrich the library for peptides that bind specifically to autoantibodies in the cancer sera, phage clones are selected, and their cDNA inserts are sequenced to identify the encoded candidate TAAs. This approach has been used for the identification of candidate TAAs in various cancers, including colorectal (131), breast (132), prostate (133), and ovarian cancers (9). Recently Chatterjee et al. (9) designed protein microarrays containing 480 antigen clones derived from biopanning a T7 phage display cDNA library from an ovarian cancer cell line and used them for immunoscreening against sera from ovarian cancer patients, healthy women, and women with other gynecologic diseases. This approach yielded 65 clones that interacted with autoantibodies in sera from the ovarian cancer patients but not in the control sera. Sequence analysis of the 65 clones revealed 62 different antigens, including several known TAAs.
The combination of standard proteomic tools of 2D gels and MS with serological screening of 2D gel-derived Western blots is an emerging approach that promises to yield a high number of candidate TAAs (8, 134, 135). This approach has been termed “serological proteome analysis” (SERPA) (134). In this approach, proteins in lysates prepared from tumors or cancer cell lines are separated by 2D gel electrophoresis and transferred to membranes that are then incubated with individual or pooled sera from cancer patients or healthy controls. Immunoreactive proteins that exhibit specific reactivity with cancer sera are then identified by MS. Various groups have used SERPA to identify candidate TAAs associated with breast cancer, including the RNA-binding protein regulatory subunit (RS), DJ-1 oncogene, glucose-6-phosphate dehydrogenase, heat shock 70-kDa protein 1 (HS71), and dihydrolipoamide dehydrogenase (8, 29, 134, 135). Interestingly, although glucose-6-phosphate dehydrogenase is widely expressed in normal cells, the breast cancer sera reacted preferentially with some isoforms of the protein detected by MS, suggesting that autoantibodies in these sera might be driven by posttranslational modifications (8), which is consistent with previous observations that intracellular proteins targeted by autoantibodies in systemic autoimmune diseases often undergo these modifications (41). The SERPA approach has also been used to identify calreticulin and DEAD-box protein 48 (DDX48) as target autoantigens in pancreatic cancer (33, 38) and the Rho GDP dissociation inhibitor 2 as a major candidate TAA in leukemia (26). Although a powerful approach, SERPA also presents some drawbacks, including the difficulty in generating reproducible gels, which leads to variations in the migration of spots of interest from gel to gel; the difficulty in resolving certain classes of proteins; the requirement of highly expensive and sophisticated MS and imaging equipment and appropriately trained personnel; and the dependence on Western blotting for the serological screening, which has an inherent bias toward the detection of denatured epitopes (8, 135, 136).
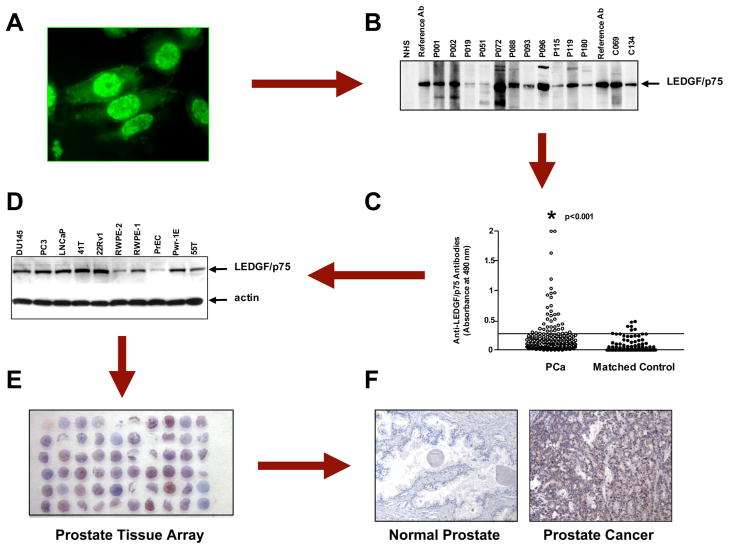
As these approaches continue to be optimized for enhancing the identification of candidate TAAs, it is also necessary to develop methods to optimize the validation of these TAAs. For a candidate TAA to be considered useful in diagnosis, it has to be preferentially recognized by sera from patients with the particular tumor type in which the antigen was identified when compared with sera from control individuals, other tumors, or autoimmune diseases. A caveat, however, is that some TAAs such as p53 and survivin lack specificity with regard to their tumor association because they are almost universally involved in the process of malignant transformation and are targeted by the immune system not only in various cancer types but also in autoimmune diseases (50, 51). In our experience, once a TAA is identified, the combined use of various immunoassays (e.g. ELISA, immunoblotting, and immunoprecipitation) is required to enhance the detection of specific autoantibodies to that particular TAA in the cancer of interest and determine more accurately the frequency of these autoantibodies in patient sera (27). The reason for this is that there is often an incomplete correlation in the detection of autoantibodies to TAAs between the different immunoassays (27, 51). This divergence could be caused by differences in the sensitivity of the various assays and in changes in antigen conformation from one assay platform to another. Cancer-associated autoantibodies might also display heterogeneity in epitope recognition within a given antigen. Consequently some patients would produce autoantibodies against non-denatured epitopes, whereas others may produce antibodies against denatured epitopes. It is also important to establish that candidate TAAs are indeed expressed in the tumor type associated with the anti-TAA autoantibody response. This could be facilitated by the availability of tissue microarrays specific for various kinds of tumors (Fig. 1).
Fig. 1. The path to the identification of LEDGF/p75 as a candidate TAA in prostate cancer.
A, autoantibodies with a nuclear fluorescence staining pattern resembling that of the known autoantigen LEDGF/p75 (also called DFS70) were initially detected in prostate cancer sera by screening the sera in Hep-2 slides and LnCaP prostate cancer cells growing on coverslips via immunofluorescence microscopy. B, the presence of these autoantibodies in prostate cancer sera (p) and some positive matched control sera (c) was detected by immunoblotting of LnCaP cell lysates using reference anti-LEDGF/p75 sera as positive controls. C, the frequency of these autoantibodies in prostate cancer was determined in an ELISA system using purified recombinant LEDGF/p75 by testing against sera from prostate cancer patients and age- and gender-matched controls. Significantly high frequencies were obtained in the prostate cancer group. D, validation of LEDGF/p75 as a protein associated with prostate cancer was first achieved by analyzing its expression in a panel of prostate cancer cell lines by immunoblotting. High expression was observed in most tumor and transformed cell lines. The lowest expression was in the primary normal cell line PrEC. E, the availability of the human prostate cancer Histo-Array™ facilitated immunohistochemical analysis of LEDGF/p75 in tumor tissue. F, LEDGF/p75 was found to be highly expressed in the majority of prostate tumor tissue specimens but not in normal prostate tissue. For details see Ref. 27.
AUTOANTIGEN ARRAYS: LESSONS FROM AUTOIMMUNE DISEASES
The utility of autoantigen arrays to profile serum autoantibody responses in human disease is generating much interest because of their potential for improving early diagnosis, monitoring disease progression, and guiding interventions for disease prevention or early treatment (136). As mentioned earlier, serum autoantibodies in systemic autoimmune diseases such as SLE and RA have diagnostic value because these diseases are generally associated with distinctive sets of autoantibodies. Unfortunately when considered alone as diagnostic tools, most autoantibodies show poor sensitivity and/or specificity for their associated diseases. For example, autoantibodies to topoisomerase I, although associated with diffuse cutaneous involvement and pulmonary fibrosis in patients with scleroderma, are present only at frequencies of ~30%, indicating that a relatively large subset of patients would be missed if the presence of these autoantibodies were to be used as a diagnostic test (137). Other autoantibodies primarily associated with a particular autoimmune disease may also appear in other diseases. For instance, autoantibodies to the SSA/Ro and SSB/La ribonucleoprotein particles, which are present in patients with Sjögren syndrome, are also present in SLE and other autoimmune diseases (138). It has been recognized that to achieve the highest degree of sensitivity and specificity in the diagnosis of systemic autoimmune diseases, multiple autoantibody reactivities, not a single autoantibody reactivity, should be used in a diagnostic test consisting of a well defined antigen array or panel (136, 139).
The protein microarray technology was first adapted for the profiling of serum autoantibodies by Joos et al. (140) and Robinson et al. (141). Joos et al. (140) used an ELISA-based microarray containing serial dilutions of 18 known autoantigens targeted in systemic autoimmune diseases for accurate determination of autoantibody titer using minimal amounts of serum. These microarrays were very sensitive and showed little cross-reactivity with nonspecific proteins. The array constructed by Robinson et al. (141) used the conventional DNA microarray technology and consisted of 196 known autoantigens that were spotted onto derivatized microscope glass slides. Using over 100 different serum samples from patients with eight different autoimmune diseases, these investigators demonstrated that comprehensive autoantigen microarrays could be used to profile autoantibodies in these diseases with high sensitivity and antigen specificity. These autoantigen microarrays also provided a tool for the identification of multiple cellular targets of the autoimmune response in individual patients. The results were validated using traditional immunoassays such as ELISA and immunoblotting. In a recent study, Robinson and co-workers (142) used this technology to develop a synovial proteome microarray containing 225 peptides and proteins that represent candidate and control antigens. These microarrays were used to profile autoantibodies in over 60 patients with RA and 38 controls. The results from this study provided useful diagnostic and prognostic information, which allowed the stratification of patients with early RA into clinically relevant disease subsets. For example, autoantibodies targeting citrunilated epitopes were present in a subset of patients with early stage RA who had features predictive of the development of severe RA. In contrast, autoantibodies targeting autoantigens native to the synovium, such as gp39 and type II collagen, delineated a subpopulation of RA patients who had laboratory and clinical features predictive of less severe RA.
The development of autoantigen microarrays for autoantibody profiling in systemic autoimmune diseases has paved the way for the use of this technology in other diseases, including cancer, multiple sclerosis, infectious diseases, and diabetes (136, 139). The potential applications of autoantigen microarrays have been discussed recently by Balboni et al. (136) and include (a) improved diagnosis, (b) monitoring disease progression and response to therapy, (c) development of individualized therapies, (d) identification of autoantibody signatures that might be associated with a particular subgroup within the disease or have prognostic value, (e) development of antigen-specific therapies, and (f) identification of new autoantigens that could serve as disease biomarkers. Current challenges in the use of this technology include (a) establishing optimal conditions to minimize variations observed when using different slide surfaces for autoantigen spotting and printing conditions, (b) minimizing epitope alterations following attachment of antigens to a planar surface, which may result in inefficient or no detection of autoantigens in the array, (c) development of efficient internal controls, and (d) validation of the arrays with well characterized serum samples before this technology is ready for use in clinical settings (136, 139). An issue that needs further attention is whether the use of microarrays comprised of hundreds of autoantigens provides an advantage for disease diagnosis and prognosis over smaller arrays consisting of carefully selected antigen biomarkers.
TAA ARRAYS FOR AUTOANTIBODY PROFILING IN CANCER
As mentioned earlier, cancer patients often produce autoantibodies to TAAs that are either mutated, overexpressed, posttranslationally modified, misfolded, aberrantly cleaved, or aberrantly localized in tumor cells. Given that cancer-associated autoantibodies are considered as reporters or sentinels of the immune response to a developing tumor, they are excellent candidates for cancer biomarkers. Their persistence and stability in the serum of cancer patients is an advantage over other potential markers, including the TAAs themselves, that are released by tumors but rapidly degraded or cleared after circulating in the serum for a limited time (2). The widespread availability of methods and reagents to detect serum autoantibodies facilitates their characterization in cancer patients and assay development. In contrast to systemic autoimmune diseases where the presence of a particular autoantibody may have diagnostic value (e.g. anti-Sm antibodies in SLE or anti-centromere antibodies in the CREST (calcinosis, Raynaud, esophageal dysmotility, sclerodactyly, and telangiectasia) syndrome), cancer-associated autoantibodies when evaluated individually have little diagnostic value for several reasons. First, the frequencies of autoantibodies specific for a particular TAA (e.g. p53) in a given cancer population are often relatively low, typically about 10% and up to 20–30% in the best case scenarios (27, 50, 51, 111). Second, because certain TAAs play a role in tumorigenesis in multiple cancer types, a particular antibody may appear in different cancer types (50, 111). Although the presence of such antibodies could be indicative of a developing tumor, their use in diagnosis is impractical because of their inadequacy in discriminating between different cancer types. Third, certain autoantibodies also lack specificity because they may arise as a consequence of molecular events that are commonly associated with both cancer and other diseases. This is illustrated by our own studies on autoimmunity to the stress-regulated survival protein LEDGF/p75 in which the autoantibodies were detected not only in a subset of patients with prostate cancer but also in chronic inflammatory conditions associated with aberrant cell death and ASCOS and even in healthy individuals (27, 84, 97). This led to the hypothesis that accumulation of LEDGF/p75 and its truncated variants in these diseases, due perhaps to a combination of up-regulation by oxidative stress and cleavage during cell death under a proinflammatory environment, could trigger loss of immune tolerance against the protein with ensuing production of autoantibodies (27, 84, 97). We and others also reported that autoantibodies to the mitosis and cell proliferation autoantigen CENP-F are present not only in certain patients with cancer but also in patients with non-malignant disorders associated with increased cell proliferation (53, 54). Fourth, patients with cancer are usually aging patients with other underlying illnesses, including inflammatory and rheumatic diseases that are also associated with autoantibody production, which would be a significant confounding factor in the analysis of specific cancer-associated antibodies. Therefore, implementation of the antigen microarray technology for cancer diagnosis would require well defined and validated TAA arrays that ensure the highest sensitivity and specificity.
Several TAA arrays have been designed and evaluated for autoantibody profiling in cancer patients using a variety of formats, including glass slide-based microarrays, ELISA multiplex, and membrane-based immunoassays. Using an ELISA system, Stockert et al. (58) evaluated autoantibody responses in patients with various cancers to seven protein cancer testis and differentiation antigens (NY-ESO-1, Melan-A, SSX2, MAGE-1, MAGE-3, tyrosinase, and carbonic anhydrase). A survey of 234 cancer sera showed frequencies of antibodies specific to NY-ESO-1 of ~10% in melanoma and ovarian cancer; however, the majority of the other antigens were recognized by the cancer sera at very low frequencies (<2%). Normal serum reactivity was not observed for any of the antigens. Although the main finding in this study was that NY-ESO-1 is an important autoantigen in melanoma, the study also provided the first survey of the human immune response to a panel of candidate TAAs. Subsequently Scanlan et al. (143) screened colon cancer sera against a panel of 77 SEREX-defined TAAs using nitrocellulose-based spot immunoassays and identified a panel of 13 TAAs that reacted exclusively with sera from colon cancer patients but not with sera from normal blood donors. In this study, it was found that 46% of sera from colon cancer patients detected one or more of these 13 antigens, whereas sera from normal blood donors were not reactive with this subset of antigens. This study confirmed the need for using TAA panels for increasing specific autoantibody detection in cancer patients.
Our group further refined a miniarray comprised of multiple TAAs to enhance antibody detection and explore their utility in cancer detection and diagnosis (111). This miniarray of TAAs included full-length recombinant proteins from cDNA encoding c-MYC, p53, cyclin B1, p62, Koc, IMP1, and survivin. These TAAs were selected based on previous observations that they were targeted by autoantibodies in cancer patients and not in normal controls or in other disease conditions. They were evaluated in an ELISA multiplex system to detect antibodies in 527 sera from six different types of cancer: breast, lung, colorectal, gastric, hepatocellular, and prostate. Consistent with previous observations, we observed that although antibody frequencies to any individual TAA were variable and rarely exceeded 15–20%, the successive addition of TAAs to the array to a total of seven antigens was associated with a stepwise increase of positive antibody reactions up to a range of 44–68% (Table II). However, there was no increase in positive antibody reactions in sera from normal blood donors or from autoimmune diseases, indicating that the miniarray had high specificity for the detection of cancer-associated antibodies. These results provided evidence that detection of autoantibodies in cancer sera could be enhanced if TAA miniarrays, rather than individual TAAs, were used. We also observed that there was preferential reactivity against certain TAAs in some cancers, with some TAAs targeted at higher frequencies or increasing antibody reactivity when added to the array. Interestingly, for some cancers addition of a particular TAA to the array did not further increase the antibody reactivity. This was consistent with the notion that not all cellular proteins recognized as antigens by autoantibodies in cancer sera are specific for cancer or for a particular type of cancer. This study underscored a general principle that should be given serious consideration when applying the TAA microarray technology for autoantibody profiling in cancer. That is, to optimize sensitivity and specificity, it is extremely important to evaluate first different combinations of TAAs against sera from both normal matched controls and other disease conditions. This would determine whether the inclusion of a particular TAA in the array would enhance or diminish the effectiveness of the array.
Table II.
Frequency of autoantibodies in relationship to number of antigen-antibody systems tested
| Type of cancer |
||||||
|---|---|---|---|---|---|---|
| TAAs | Breast 64 | Lung 56 | Colon 45 | Prostate 206 | Gastric 91 | HCCa 65 |
| IMP1 | 5 (7.8)b | 4 (7.1) | 6 (13.3) | 18 (8.7) | 15 (16.5) | 10 (15.4) |
| IMP1 or p62 | 8 (12.5) | 15 (26.8) | 8 (17.8) | 64 (31.1) | 18 (19.8) | 15 (23.1) |
| IMP1 or p62 or Koc | 14 (21.9) | 19 (33.9) | 10 (22.2) | 72 (35.0) | 28 (30.8) | 20 (30.8) |
| IMP1 or p62 or Koc or p53 | 17 (26.6) | 25 (44.6) | 16 (35.6) | 78 (37.9) | 37 (40.7) | 24 (36.9) |
| IMP1 or p62 or Koc or p53 or c-MYC | 25 (39.1) | 27 (48.2) | 17 (37.8) | 81 (39.3) | 40 (44.0) | 32 (49.2) |
| IMP1 or p62 or Koc or p53 or c-MYC or cyclin B1 | 26 (40.6) | 36 (64.3) | 23 (51.1) | 95 (46.1) | 47 (51.6) | 35 (53.8) |
| IMP1 or p62 or Koc or p53 or c-MYC or cyclin B1 or survivin | 28 (43.8) | 38 (67.9) | 23 (51.1) | 95 (46.1) | 48 (52.7) | 37 (56.9) |
Hepatocellular carcinoma.
Numbers in parentheses are percentage of positive reactors in that cancer category. Positive reactors are defined as above the mean of 82 normal human sera + 3 standard deviations.
In a follow-up study, we used a multivariate statistical approach, recursive partitioning, to assess whether we could accurately classify individuals as either cancer patients or normal individuals on the basis of the profile of autoantibody reactivity to the seven TAAs used in the ELISA-based miniarray (144). The recursive partitioning program determines for each variable (in this case each of the seven TAAs) a cut point that optimally splits all the individuals into cases (cancer) and controls (normal) and selects the variable that performs best. It then takes the resulting subpopulations and repeats the process on each until no additional partitioning is warranted: meaning that either the subpopulation contains one class of individuals or is too small to subdivide further. This approach yielded classification trees for the selection of subsets of the seven-TAA panel that differentiated between tumors and controls, and these subsets were unique to each cancer cohort. For instance, cyclin B1 was the first determinant in the classification trees (constructed using +3 and +2 standard deviations as standard cutoffs for immunoassay positivity) for lung and gastric cancers, c-MYC was the initial determinant in the classification trees for breast cancer, and the IGF-II mRNA-binding protein p62 was the initial determinant in the classification trees for prostate cancer. Interestingly, survivin did not appear in any tree, which could have been influenced by the relative low frequencies (<10%) of autoantibodies found for this protein in the six cancer types. The classification trees had sensitivities ranging from 0.77 to 0.92 and specificities ranging from 0.85 to 0.91 in the cancer cohorts using +2 standard deviations as cutoffs. No more than three of the seven TAAs were needed for any cancer cohort to arrive at these levels of sensitivity and specificity. It should be emphasized that these relatively high sensitivities and specificities have to be considered in the context of the seven TAAs that were used in the analysis. These TAAs are known targets of autoantibody responses in multiple types of cancers and therefore might not have been the best possible candidate TAAs for the cancers that were analyzed. It also follows that the inclusion of a specific TAA may increase the sensitivity and specificity of an array in a particular cancer but decrease them in another type of cancer. As mentioned above, the sensitivity and specificity of TAA arrays could be dramatically improved upon a careful design of larger arrays in which additional TAAs are added or deleted. To achieve this goal, it is important to continue mining the serum autoantibody repertoire of cancer patients to identify new and more specific TAAs.
TAA ARRAYS IN PROSTATE CANCER DIAGNOSIS
Although antibody responses to candidate TAAs have been characterized in a wide variety of cancers, information about these responses in prostate cancer was scarce until very recently. Early studies by Ablin (145, 146) demonstrated the presence of serum autoantibodies in patients with prostate cancer, but because the significance of these autoantibodies in cancer was not fully appreciated at the time they were considered representative of a nonspecific immune response. During the past 5 years, however, there has been a renewed interest in the characterization of autoantibody responses in prostate cancer. This has led to the identification and characterization of a number of candidate TAAs, using SEREX, serological screening of phage display libraries, and proteomic approaches. As pointed out earlier, several of these candidate TAAs are proteins that participate in stress and survival pathways and are highly expressed in prostate tumors (Table I). Their availability provides a unique opportunity for designing highly sensitive and specific TAA arrays that could be used to supplement prostate-specific antigen (PSA) tests in the early diagnosis of prostate cancer. However, as shown in Table I, the frequency of many autoantibodies in prostate cancer and even more importantly analysis of the frequency in normal individuals and other conditions were not determined.
Prostate cancer is the most frequently diagnosed cancer in men and the second leading cause of male cancer deaths in the United States with an estimated 234,460 cases and 27,350 deaths in 2006 (1). It is also becoming evident that prostate cancer presents the greatest racial disparity of any cancer in the United States with an incidence and mortality in African-American men that is 3 times higher than in White and Latino men (147). An alarming fact is that African-American men are more likely to present with advanced stage, hormone-refractory prostate cancer than men from other ethnic/racial groups (147). Reducing these disparities would require a multifaceted approach that includes developing more effective screening interventions for the early detection of prostate cancer. Early detection of prostate cancer using the PSA blood test has increased the proportion of patients with lower tumor stage at the time of diagnosis (148). However, although the sensitivity of PSA testing is exceptional, its specificity, particularly at lower PSA levels, remains controversial (148, 149). The gradual lowering over the past few years of the cutoff PSA level, which has lead to recommendations for prostate biopsy, has resulted in confusion as to how to interpret low PSA values, leading to an increasing number of unnecessary biopsies (149). Furthermore serum PSA is not specific for prostate cancer but rather a marker of prostate disease given that it can be also be detected in serum from patients with benign prostatic hyperplasia (BPH) and prostatitis (149). It was suggested that because of the heterogeneity of prostate cancer and other diseases of the prostate, multiple biomarkers would be needed to discriminate between the various stages of prostate cancer and differentiate prostate cancer from BPH and prostatitis (149). TAA arrays could be useful tools for supplementing PSA screening in the diagnosis of prostate cancer.
We tested our miniarray of seven TAAs (Table II) in 206 patients diagnosed with prostate cancer at Loma Linda University Medical Center and observed that the frequency of serum autoantibodies reacting with the TAAs increased in relationship to the number of TAAs in the array, going from 8.7% using IMP1 alone to 46% using the seven TAAs (111). We noticed that addition of just p62 to IMP1 in a two-antigen array increased the total frequency to 31.1% due to the fact that the frequency of autoantibodies to p62 was the highest among the seven TAAs. However, the addition of the remaining five antigens to the array gradually increased the frequency from 31.1 to 46.1% (Table II). In a subsequent study, we were able to dramatically increase the frequency of positively reacting sera to 92.5% by deleting from the miniarray three TAAs that were targeted by autoantibodies in prostate cancer patients at frequencies below 5% (p53, MYC, and survivin) and adding a new candidate TAA, p90, which was targeted at a frequency of 30.8% (Table III) (150). It should be noted that this miniarray was relatively specific for cancer because the total autoantibody frequencies against the combined TAAs were less than 15% for sera from normal controls, patients with autoimmune diseases, and patients with BPH (111).
Table III.
Stepwise increase in rate of antibody positivity with successive addition of TAAs
| TAAs | Number/percentage of autoantibodies in prostate cancer | Number/percentage of autoantibodies in NHSa |
|---|---|---|
| p90 | 41 (30.8) | 3 (3.1) |
| p90 or p62 | 70 (53.4) | 6 (6.2) |
| p90 or p62 or Koc | 81 (61.7) | 8 (8.4) |
| p90 or p62 or Koc or IMP1 | 92 (70.0) | 10 (10.6) |
| p90 or p62 or Koc or IMP1 or cyclin B1 | 111 (84.3) | 12 (12.6) |
| p90 or p62 or Koc or IMP1 or cyclin B1 or cyclin D1 | 122 (92.5) | 14 (14.8) |
Normal human serum.
In the study using recursive partitioning (which did not include p90) we determined that p62, cyclin B1, and IMP1 were the most relevant antigens to achieve the highest sensitivity and specificity, 0.79 and 0.86, respectively, in the prostate cancer cohort (144). These results reinforced the notion that optimizing the composition of a TAA array is essential for achieving high sensitivity and specificity. For instance, when our TAA array was used for autoantibody profiling in prostate cancer, p53, c-MYC, and survivin acted as detracting TAAs that reduced sensitivity. On the other hand, IMP1, cyclin B1, p62, and p90 contributed to a higher sensitivity and specificity. However, whether an array containing these TAAs will provide high specificity with regard to discriminating between prostate cancer and other cancers remains to be determined. It should also be noted that cyclin B1 and p62 are targeted at frequencies above 20% in patients with lung cancer (111), suggesting the possibility that limited arrays containing these two TAAs, although useful for discriminating between patients with cancer and individuals with no diagnosis of cancer, may not be adequate for discriminating between different cancers. This underscores the need for identifying additional TAAs that are targeted in prostate cancer at higher frequencies than in other cancer types. These TAAs should then be carefully arranged in various combinations to optimize arrays that are highly sensitive and specific for a diagnosis of prostate cancer and could be used to discriminate between prostate cancer and other diseases of the prostate and, potentially, aid in monitoring prostate cancer progression.
In a recent study, Wang et al. (120) used a phage display library derived from prostate cancer tissue to develop a phage protein microarray for the analysis of serum samples from 119 patients with prostate cancer and 138 individuals with no history of prostate cancer. These investigators identified a panel of 22 phage-peptide clones that could distinguish serum samples from patients with prostate cancer from those of controls with high sensitivity and specificity, 81.6 and 88.2%, respectively. An interesting finding was that the peptide array not only performed better than PSA in discriminating between the two groups but also provided additional discriminatory power (120). It should be noted, however, that of the 22 phage-peptide clones, only five peptides were derived from in-frame coding sequences corresponding to known proteins: bromo domain-containing protein 2 (BRD2), eukaryotic translation initiation factor eIF4G1, ribosomal proteins L22 and L13a, and a hypothetical protein designated XP_373908 (120). The remaining 17 peptides could be mimotopes, i.e. epitopes that are structurally similar to other peptides expressed in proteins but are unrelated or weakly related at the protein sequence level. Although this was a promising study that confirmed the power of TAA arrays for discriminating between cancer and non-cancer populations, it had some limitations that prevent its adoption in a screening program for prostate cancer in the near future. For instance, it was not established whether this peptide array is specific for prostate cancer when evaluated against sera from patients with other cancers, other diseases of the prostate, and autoimmune diseases. Some of the antigens identified in this peptide array, such as eIF4G1, might be overexpressed in other tumor tissues (151) and elicit autoantibody responses in various cancer cell types. Another limitation, which is prevalent in most studies using TAA arrays for cancer diagnosis, including our own studies, was the lack of biopsies in the control group to rule out the absence of prostate cancer (120). As pointed out by Wang et al. (120), prospective and multi-institutional studies need to be conducted to determine the utility of this and other TAA arrays in evaluating autoantibody signatures associated with prostate cancer, a required step to establish these arrays as screening tools for prostate cancer.
CONCLUSIONS AND FUTURE PERSPECTIVES
Exploiting the immune response to tumors provides a unique opportunity for developing new tools for the serological detection of cancer. The analysis of uniquely designed TAA arrays for autoantibody profiling in cancer should lead in the future to the development of diagnostic chips that could be used for early detection of cancer with high sensitivity and specificity. Given that the presence of serum autoantibodies to TAAs might signal molecular events associated with tumorigenesis, the use of highly sensitive and specific TAA chips for screening populations at high risk of developing cancer may lead to early preventive or therapeutic interventions aimed at suppressing or slowing the appearance of a tumor. These interventions should potentially contribute to a decrease in the relatively high incidence and mortality that some ethnic and racial groups experience with specific cancer types, thereby translating into a reduction of cancer health disparities. The use of TAA arrays to profile the antitumor autoantibody response in a particular cancer patient could also provide key information on disease progression, which, combined with other available clinical information, could guide physicians and patients in the making of important decisions regarding treatment options. A TAA array specific for a particular cancer type could be also exploited for the development of cancer vaccines designed to boost the patient’s immune system to fight the tumor. Furthermore, specific TAA arrays could provide fingerprints or “signatures” indicating that a particular cellular signaling pathway is being preferentially targeted by the immune system in a given cancer type, suggesting that it might play an important role in the development of malignancy. This knowledge would be useful for identifying novel targets for therapeutic interventions and for evaluating the effects of molecular targeting of components of that particular pathway in the sensitization of tumor cells to death induced by already established therapeutic agents such as antitumor drugs and radiation.
In conclusion, TAA arrays provide promising and powerful tools for enhancing cancer detection and treatment, but their utility in a clinical setting is currently in its infancy. Before TAA arrays could be widely implemented in screening programs for cancer diagnosis or as tools for monitoring cancer progression and guiding therapeutic interventions, it would be important to maximize their sensitivity and specificity by defining systematically the optimal combination of TAAs. Different array platforms (e.g. glass slide microarray, spot-based membranes, and multiplex ELISA) would also have to be evaluated to determine which one yields the highest sensitivity with minimal experimental variation. Prospective studies in multiple centers would then be required to ensure the reproducibility of these arrays.
Footnotes
The abbreviations used are: TAA, tumor-associated antigen; AOP2, antioxidant protein 2; ASCOS, augmented state of cellular oxidative stress; BPH, benign prostatic hyperplasia; BRCA, breast cancer; CENP-F, centromere protein F; DFS70, dense fine speckled protein of 70 kDa; eIF4G1, eukaryotic translation initiation factor 4 γ1; HSP, heat shock protein; IGF-II, insulin-like growth factor 2; IMP1, IGF-II mRNA-binding protein 1; LEDGF/p75, lens epithelium-derived growth factor p75; PRDX6, peroxiredoxin 6; PSA, prostate-specific antigen; RA, rheumatoid arthritis; SEREX, serological analysis of recombinant expression libraries; SERPA, serological proteome analysis; SLE, systemic lupus erythematosus; 2D, two-dimensional.
This work was supported by grants from the National Institutes of Health (NCMHD 5P20MD001632, NIGMS 2R25GM60507, and NCI CA56956), and the National Medical Technology Test Bed/United States Army Medical Research and Materiel Command (Subagreement DAMD 17-97-2-7016).
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Anderson KS, LaBaer J. The sentinel within: exploiting the immune system for cancer biomarkers. J Proteome Res. 2005;4:1123–1133. doi: 10.1021/pr0500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan EM. Autoantibodies as reporters identifying aberrant cellular mechanisms in tumorigenesis. J Clin Investig. 2001;108:1411–1415. doi: 10.1172/JCI14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chignard N, Beretta L. Proteomics for hepatocellular carcinoma marker discovery. Gastroenterology. 2004;127:S120–S125. doi: 10.1053/j.gastro.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 5.Mian S, Ugurel S, Parkinson E, Schlenzka I, Dryden I, Lancashire L, Ball G, Creaser C, Rees R, Schadendorf D. Serum proteomic fingerprinting discriminates between clinical stages and predicts disease progression in melanoma patients. J Clin Oncol. 2005;23:5088–5093. doi: 10.1200/JCO.2005.03.164. [DOI] [PubMed] [Google Scholar]
- 6.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC, Liotta LA. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 7.Shin BK, Wang H, Hanash S. Proteomics approaches to uncover the repertoire of circulating biomarkers for breast cancer. J Mammary Gland Biol Neoplasia. 2002;7:407–413. doi: 10.1023/a:1024038132381. [DOI] [PubMed] [Google Scholar]
- 8.Canelle L, Bousquet J, Pionneau C, Deneux L, Imam-Sghiouar N, Caron M, Joubert-Caron R. An efficient proteomics-based approach for the screening of autoantibodies. J Immunol Methods. 2005;299:77–89. doi: 10.1016/j.jim.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee M, Mohapatra S, Ionan A, Bawa G, Ali-Fehmi R, Wang X, Nowak J, Ye B, Nahhas FA, Lu K, Witkin SS, Fishman D, Munkarah A, Morris R, Levin NK, Shirley NN, Tromp G, Abrams J, Draghici S, Tainsky MA. Diagnostic markers of ovarian cancer by high-throughput antigen cloning and detection on arrays. Cancer Res. 2006;66:1181–1190. doi: 10.1158/0008-5472.CAN-04-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imafuku Y, Omenn GS, Hanash S. Proteomics approaches to identify tumor antigen directed autoantibodies as cancer biomarkers. Dis Markers. 2004;20:149–153. doi: 10.1155/2004/829450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu J, Madoz-Gurpide J, Misek DE, Kuick R, Brenner DE, Michailidis G, Haab BB, Omenn GS, Hanash S. Development of natural protein microarrays for diagnosing cancer based on an antibody response to tumor antigens. J Proteome Res. 2004;3:261–267. doi: 10.1021/pr049971u. [DOI] [PubMed] [Google Scholar]
- 12.Zhang JY. Tumor-associated antigen arrays to enhance antibody detection for cancer diagnosis. Cancer Detect Prev. 2004;28:114–118. doi: 10.1016/j.cdp.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Casiano CA, Tan EM. Recent developments in the understanding of antinuclear autoantibodies. Int Arch Allergy Immunol. 1996;111:308–313. doi: 10.1159/000237385. [DOI] [PubMed] [Google Scholar]
- 14.Tan EM. Autoantibodies and autoimmunity: a three-decade perspective. A tribute to Henry G Kunkel. Ann N Y Acad Sci. 1997;815:1–14. doi: 10.1111/j.1749-6632.1997.tb52040.x. [DOI] [PubMed] [Google Scholar]
- 15.Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, James JA, Harley JB. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349:1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 16.Hu PQ, Fertig N, Medsger TA, Jr, Wright TM. Correlation of serum anti-DNA topoisomerase I antibody levels with disease severity and activity in systemic sclerosis. Arthritis Rheum. 2003;48:1363–1373. doi: 10.1002/art.10977. [DOI] [PubMed] [Google Scholar]
- 17.Rantapaa-Dahlqvist S, de Jong BA, Berglin E, Hallmans G, Wadell G, Stenlund H, Sundin U, van Venrooij WJ. Antibodies against cyclic citrullinated peptide and IgA rheumatoid factor predict the development of rheumatoid arthritis. Arthritis Rheum. 2003;48:2741–2749. doi: 10.1002/art.11223. [DOI] [PubMed] [Google Scholar]
- 18.Tan EM. Antinuclear antibodies: diagnostic markers and clues to the basis of systemic autoimmunity. Pediatr Infect Dis J. 1988;7:S3–S9. [PubMed] [Google Scholar]
- 19.Mittermann I, Aichberger KJ, Bunder R, Mothes N, Renz H, Valenta R. Autoimmunity and atopic dermatitis. Curr Opin Allergy Clin Immunol. 2004;4:367–371. doi: 10.1097/00130832-200410000-00007. [DOI] [PubMed] [Google Scholar]
- 20.Ochs RL, Muro Y, Si Y, Ge H, Chan EK, Tan EM. Autoantibodies to DFS 70 kd/transcription coactivator p75 in atopic dermatitis and other conditions. J Allergy Clin Immunol. 2000;105:1211–1220. doi: 10.1067/mai.2000.107039. [DOI] [PubMed] [Google Scholar]
- 21.Vernon SD, Reeves WC. Evaluation of autoantibodies to common and neuronal cell antigens in Chronic Fatigue Syndrome. J Autoimmune Dis. 2005;2:1–5. doi: 10.1186/1740-2557-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanc F, Fleury M, Talmant V, Deroide N, Szwarcberg J, Tranchant C. Vogt-Koyanagi-Harada syndrome. Rev Neurol (Paris) 2005;161:1079–1090. doi: 10.1016/s0035-3787(05)85175-6. [DOI] [PubMed] [Google Scholar]
- 23.Shinohara T, Singh DP, Chylack LT., Jr Review: Age-related cataract: immunity and lens epithelium-derived growth factor (LEDGF) J Ocul Pharmacol Ther. 2000;16:181–191. doi: 10.1089/jop.2000.16.181. [DOI] [PubMed] [Google Scholar]
- 24.Bencimon C, Salles G, Moreira A, Guyomard S, Coiffier B, Bienvenu J, Fabien N. Prevalence of anticentromere F protein autoantibodies in 347 patients with non-Hodgkin’s lymphoma. Ann N Y Acad Sci. 2005;1050:319–326. doi: 10.1196/annals.1313.034. [DOI] [PubMed] [Google Scholar]
- 25.Covini G, von Muhlen CA, Pacchetti S, Colombo M, Chan EK, Tan EM. Diversity of antinuclear antibody responses in hepatocellular carcinoma. J Hepatol. 1997;26:255–265. doi: 10.1016/s0168-8278(97)80460-6. [DOI] [PubMed] [Google Scholar]
- 26.Cui JW, Li WH, Wang J, Li AL, Li HY, Wang HX, He K, Li W, Kang LH, Yu M, Shen BF, Wang GJ, Zhang XM. Proteomics-based identification of human acute leukemia antigens that induce humoral immune response. Mol Cell Proteomics. 2005;4:1718–1724. doi: 10.1074/mcp.M400165-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Daniels T, Zhang J, Gutierrez I, Elliot ML, Yamada B, Heeb MJ, Sheets SM, Wu X, Casiano CA. Antinuclear autoantibodies in prostate cancer: immunity to LEDGF/p75, a survival protein highly expressed in prostate tumors and cleaved during apoptosis. Prostate. 2005;62:14–26. doi: 10.1002/pros.20112. [DOI] [PubMed] [Google Scholar]
- 28.Diesinger I, Bauer C, Brass N, Schaefers HJ, Comtesse N, Sybrecht G, Meese E. Toward a more complete recognition of immunoreactive antigens in squamous cell lung carcinoma. Int J Cancer. 2002;102:372–378. doi: 10.1002/ijc.10714. [DOI] [PubMed] [Google Scholar]
- 29.Fernandez Madrid F. Autoantibodies in breast cancer sera: candidate biomarkers and reporters of tumorigenesis. Cancer Lett. 2005;230:187–198. doi: 10.1016/j.canlet.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 30.Fernandez-Madrid F, VandeVord PJ, Yang X, Karvonen RL, Simpson PM, Kraut MJ, Granda JL, Tomkiel JE. Antinuclear antibodies as potential markers of lung cancer. Clin Cancer Res. 1999;5:1393–1400. [PMC free article] [PubMed] [Google Scholar]
- 31.Forges T, Monnier-Barbarino P, Faure GC, Bene MC. Autoimmunity and antigenic targets in ovarian pathology. Hum Reprod Update. 2004;10:163–175. doi: 10.1093/humupd/dmh014. [DOI] [PubMed] [Google Scholar]
- 32.Himoto T, Kuriyama S, Zhang JY, Chan EK, Kimura Y, Masaki T, Uchida N, Nishioka M, Tan EM. Analyses of autoantibodies against tumor-associated antigens in patients with hepatocellular carcinoma. Int J Oncol. 2005;27:1079–1085. [PubMed] [Google Scholar]
- 33.Hong SH, Misek DE, Wang H, Puravs E, Giordano TJ, Greenson JK, Brenner DE, Simeone DM, Logsdon CD, Hanash SM. An autoantibody-mediated immune response to calreticulin isoforms in pancreatic cancer. Cancer Res. 2004;64:5504–5510. doi: 10.1158/0008-5472.CAN-04-0077. [DOI] [PubMed] [Google Scholar]
- 34.Imai H, Kiyosawa K, Chan EK, Tan EM. Autoantibodies in viral hepatitis-related hepatocellular carcinoma. Intervirology. 1993;35:73–85. doi: 10.1159/000150297. [DOI] [PubMed] [Google Scholar]
- 35.Inuzuka T. Autoantibodies in paraneoplastic neurological syndrome. Am J Med Sci. 2000;319:217–226. doi: 10.1097/00000441-200004000-00004. [DOI] [PubMed] [Google Scholar]
- 36.Scanlan MJ, Chen YT, Williamson B, Gure AO, Stockert E, Gordan JD, Tureci O, Sahin U, Pfreundschuh M, Old LJ. Characterization of human colon cancer antigens recognized by autologous antibodies. Int J Cancer. 1998;76:652–658. doi: 10.1002/(sici)1097-0215(19980529)76:5<652::aid-ijc7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Yu J, Sreekumar A, Varambally S, Shen R, Giacherio D, Mehra R, Montie JE, Pienta KJ, Sanda MG, Kantoff PW, Rubin MA, Wei JT, Ghosh D, Chinnaiyan AM. Autoantibody signatures in prostate cancer. N Engl J Med. 2005;353:1224–1235. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- 38.Xia Q, Kong XT, Zhang GA, Hou XJ, Qiang H, Zhong RQ. Proteomics-based identification of DEAD-box protein 48 as a novel autoantigen, a prospective serum marker for pancreatic cancer. Biochem Biophys Res Commun. 2005;330:526–532. doi: 10.1016/j.bbrc.2005.02.181. [DOI] [PubMed] [Google Scholar]
- 39.Hall JC, Casciola-Rosen L, Rosen A. Altered structure of autoantigens during apoptosis. Rheum Dis Clin North Am. 2004;30:455–471. doi: 10.1016/j.rdc.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 40.Pollard KM, Lee DK, Casiano CA, Bluthner M, Johnston MM, Tan EM. The autoimmunity-inducing xenobiotic mercury interacts with the autoantigen fibrillarin and modifies its molecular and antigenic properties. J Immunol. 1997;158:3521–3528. [PubMed] [Google Scholar]
- 41.Utz PJ, Anderson P. Posttranslational protein modifications, apoptosis, and the bypass of tolerance to autoantigens. Arthritis Rheum. 1998;41:1152–1160. doi: 10.1002/1529-0131(199807)41:7<1152::AID-ART3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 42.Rosen A, Casciola-Rosen C, Wigley F. Role of metal-catalyzed oxidation reactions in the early pathogenesis of scleroderma. Curr Opin Rheumatol. 1997;9:538–543. doi: 10.1097/00002281-199711000-00010. [DOI] [PubMed] [Google Scholar]
- 43.Wu X, Molinaro C, Johnson N, Casiano CA. Secondary necrosis is a source of proteolytically modified forms of specific intra-cellular autoantigens: implications for systemic autoimmunity. Arthritis Rheum. 2001;44:2642–2652. doi: 10.1002/1529-0131(200111)44:11<2642::aid-art444>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 44.Lu M, Nakamura RM, Dent ED, Zhang JY, Nielsen FC, Christiansen J, Chan EK, Tan EM. Aberrant expression of fetal RNA-binding protein p62 in liver cancer and liver cirrhosis. Am J Pathol. 2001;159:945–953. doi: 10.1016/S0002-9440(10)61770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Casciola-Rosen L, Nagaraju K, Plotz P, Wang K, Levine S, Gabrielson E, Corse A, Rosen A. Enhanced autoantigen expression in regenerating muscle cells in idiopathic inflammatory myopathy. J Exp Med. 2005;201:591–601. doi: 10.1084/jem.20041367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ben-Mahrez K, Thierry D, Sorokine I, Danna-Muller A, Kohiyama M. Detection of circulating antibodies against c-myc protein in cancer patient sera. Br J Cancer. 1988;57:529–534. doi: 10.1038/bjc.1988.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Disis ML, Calenoff E, McLaughlin G, Murphy AE, Chen W, Groner B, Jeschke M, Lydon N, McGlynn E, Livingston RB, Moe R, Cheever MA. Existent T-cell and antibody immunity to HER-2/neu protein in patients with breast cancer. Cancer Res. 1994;54:16–20. [PubMed] [Google Scholar]
- 48.Disis ML, Cheever MA. Oncogenic proteins as tumor antigens. Curr Opin Immunol. 1996;8:637–642. doi: 10.1016/s0952-7915(96)80079-3. [DOI] [PubMed] [Google Scholar]
- 49.Yamamoto A, Shimizu E, Ogura T, Sone S. Detection of auto-antibodies against L-myc oncogene products in sera from lung cancer patients. Int J Cancer. 1996;69:283–289. doi: 10.1002/(SICI)1097-0215(19960822)69:4<283::AID-IJC8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 50.Soussi T. p53 Antibodies in the sera of patients with various types of cancer: a review. Cancer Res. 2000;60:1777–1788. [PubMed] [Google Scholar]
- 51.Rohayem J, Diestelkoetter P, Weigle B, Oehmichen A, Schmitz M, Mehlhorn J, Conrad K, Rieber EP. Antibody response to the tumor-associated inhibitor of apoptosis protein survivin in cancer patients. Cancer Res. 2000;60:815–817. [PubMed] [Google Scholar]
- 52.Covini G, Chan EK, Nishioka M, Morshed SA, Reed SI, Tan EM. Immune response to cyclin B1 in hepatocellular carcinoma. Hepatology. 1997;25:75–80. doi: 10.1002/hep.510250114. [DOI] [PubMed] [Google Scholar]
- 53.Casiano CA, Humbel RL, Peebles C, Covini G, Tan EM. Autoimmunity to the cell cycle-dependent centromere protein p330d/CENP-F in disorders associated with cell proliferation. J Autoimmun. 1995;8:575–586. doi: 10.1016/0896-8411(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 54.Rattner JB, Rees J, Whitehead CM, Casiano CA, Tan EM, Humbel RL, Conrad K, Fritzler MJ. High frequency of neoplasia in patients with autoantibodies to centromere protein CENP-F. Clin Investig Med. 1997;20:308–319. [PubMed] [Google Scholar]
- 55.Imai H, Furuta K, Landberg G, Kiyosawa K, Liu LF, Tan EM. Autoantibody to DNA topoisomerase II in primary liver cancer. Clin Cancer Res. 1995;1:417–424. [PubMed] [Google Scholar]
- 56.Himoto T, Kuriyama S, Zhang JY, Chan EK, Nishioka M, Tan EM. Significance of autoantibodies against insulin-like growth factor II mRNA-binding proteins in patients with hepatocellular carcinoma. Int J Oncol. 2005;26:311–317. [PubMed] [Google Scholar]
- 57.Zhang JY, Chan EK, Peng XX, Lu M, Wang X, Mueller F, Tan EM. Autoimmune responses to mRNA binding proteins p62 and Koc in diverse malignancies. Clin Immunol. 2001;100:149–156. doi: 10.1006/clim.2001.5048. [DOI] [PubMed] [Google Scholar]
- 58.Stockert E, Jager E, Chen YT, Scanlan MJ, Gout I, Karbach J, Arand M, Knuth A, Old LJ. A survey of the humoral immune response of cancer patients to a panel of human tumor antigens. J Exp Med. 1998;187:1349–1354. doi: 10.1084/jem.187.8.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Imai H, Nakano Y, Kiyosawa K, Tan EM. Increasing titers and changing specificities of antinuclear antibodies in patients with chronic liver disease who develop hepatocellular carcinoma. Cancer. 1993;71:26–35. doi: 10.1002/1097-0142(19930101)71:1<26::aid-cncr2820710106>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 60.Zhang JY, Zhu W, Imai H, Kiyosawa K, Chan EK, Tan EM. De-novo humoral immune responses to cancer-associated autoantigens during transition from chronic liver disease to hepatocellular carcinoma. Clin Exp Immunol. 2001;125:3–9. doi: 10.1046/j.1365-2249.2001.01585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oliveira AM, Ross JS, Fletcher JA. Tumor suppressor genes in breast cancer: the gatekeepers and the caretakers. Am J Clin Pathol. 2005;124(suppl):S16–S28. doi: 10.1309/5XW3L8LU445QWGQR. [DOI] [PubMed] [Google Scholar]
- 62.Houbiers JG, van der Burg SH, van de Watering LM, Tollenaar RA, Brand A, van de Velde CJ, Melief CJ. Antibodies against p53 are associated with poor prognosis of colorectal cancer. Br J Cancer. 1995;72:637–641. doi: 10.1038/bjc.1995.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.von Brevern MC, Hollstein MC, Cawley HM, De Benedetti VM, Bennett WP, Liang L, He AG, Zhu SM, Tursz T, Janin N, Trivers GE. Circulating anti-p53 antibodies in esophageal cancer patients are found predominantly in individuals with p53 core domain mutations in their tumors. Cancer Res. 1996;56:4917–4921. [PubMed] [Google Scholar]
- 64.Winter SF, Minna JD, Johnson BE, Takahashi T, Gazdar AF, Carbone DP. Development of antibodies against p53 in lung cancer patients appears to be dependent on the type of p53 mutation. Cancer Res. 1992;52:4168–4174. [PubMed] [Google Scholar]
- 65.Choudhary SK, Li R. BRCA1 modulates ionizing radiation-induced nuclear focus formation by the replication protein A p34 subunit. J Cell Biochem. 2002;84:666–674. doi: 10.1002/jcb.10081. [DOI] [PubMed] [Google Scholar]
- 66.Wong JM, Ionescu D, Ingles CJ. Interaction between BRCA2 and replication protein A is compromised by a cancer-predisposing mutation in BRCA2. Oncogene. 2003;22:28–33. doi: 10.1038/sj.onc.1206071. [DOI] [PubMed] [Google Scholar]
- 67.Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004;95:866–871. doi: 10.1111/j.1349-7006.2004.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De Marzo AM, DeWeese TL, Platz EA, Meeker AK, Nakayama M, Epstein JI, Isaacs WB, Nelson WG. Pathological and molecular mechanisms of prostate carcinogenesis: implications for diagnosis, detection, prevention, and treatment. J Cell Biochem. 2004;91:459–477. doi: 10.1002/jcb.10747. [DOI] [PubMed] [Google Scholar]
- 69.Nelson WG, De Marzo AM, DeWeese TL, Isaacs WB. The role of inflammation in the pathogenesis of prostate cancer. J Urol. 2004;172:S6–11. doi: 10.1097/01.ju.0000142058.99614.ff. [DOI] [PubMed] [Google Scholar]
- 70.Fleshner NE, Kucuk O. Antioxidant dietary supplements: Rationale and current status as chemopreventive agents for prostate cancer. Urology. 2001;57:90–94. doi: 10.1016/s0090-4295(00)00949-3. [DOI] [PubMed] [Google Scholar]
- 71.Nelson WG, DeWeese TL, DeMarzo AM. The diet, prostate inflammation, and the development of prostate cancer. Cancer Metastasis Rev. 2002;21:3–16. doi: 10.1023/a:1020110718701. [DOI] [PubMed] [Google Scholar]
- 72.Sikka SC. Role of oxidative stress response elements and anti-oxidants in prostate cancer pathobiology and chemoprevention—a mechanistic approach. Curr Med Chem. 2003;10:2679–2692. doi: 10.2174/0929867033456341. [DOI] [PubMed] [Google Scholar]
- 73.Wertz K, Siler U, Goralczyk R. Lycopene: modes of action to promote prostate health. Arch Biochem Biophys. 2004;430:127–134. doi: 10.1016/j.abb.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 74.Ripple MO, Henry WF, Rago RP, Wilding G. Prooxidant-antioxidant shift induced by androgen treatment of human prostate carcinoma cells. J Natl Cancer Inst. 1997;89:40–48. doi: 10.1093/jnci/89.1.40. [DOI] [PubMed] [Google Scholar]
- 75.Ripple MO, Henry WF, Schwarze SR, Wilding G, Weindruch R. Effect of antioxidants on androgen-induced AP-1 and NF-κB DNA-binding activity in prostate carcinoma cells. J Natl Cancer Inst. 1999;91:1227–1232. doi: 10.1093/jnci/91.14.1227. [DOI] [PubMed] [Google Scholar]
- 76.Bostwick DG, Alexander EE, Singh R, Shan A, Qian J, Santella RM, Oberley LW, Yan T, Zhong W, Jiang X, Oberley TD. Antioxidant enzyme expression and reactive oxygen species damage in prostatic intraepithelial neoplasia and cancer. Cancer. 2000;89:123–134. [PubMed] [Google Scholar]
- 77.Giovannini C, Chieco P, Bertaccini A, Gramantieri L, Lacchini M, Martorana G. Checkpoint effectors CDKN1A and Gadd45 correlate with oxidative DNA damage in human prostate carcinoma. Anticancer Res. 2004;24:3955–3960. [PubMed] [Google Scholar]
- 78.Ueda S, Masutani H, Nakamura H, Tanaka T, Ueno M, Yodoi J. Redox control of cell death. Antioxid Redox Signal. 2002;4:405–414. doi: 10.1089/15230860260196209. [DOI] [PubMed] [Google Scholar]
- 79.Behrend L, Henderson G, Zwacka RM. Reactive oxygen species in oncogenic transformation. Biochem Soc Trans. 2003;31:1441–1444. doi: 10.1042/bst0311441. [DOI] [PubMed] [Google Scholar]
- 80.Benhar M, Engelberg D, Levitzki A. ROS, stress-activated kinases and stress signaling in cancer. EMBO Rep. 2002;3:120–125. doi: 10.1093/embo-reports/kvf094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Toyokuni S, Okamoto K, Yodoi J, Hiai H. Persistent oxidative stress in cancer. FEBS Lett. 1995;358:1–3. doi: 10.1016/0014-5793(94)01368-b. [DOI] [PubMed] [Google Scholar]
- 82.Ge H, Si Y, Roeder RG. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 1998;17:6723–6729. doi: 10.1093/emboj/17.22.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Singh DP, Ohguro N, Chylack LT, Jr, Shinohara T. Lens epithelium-derived growth factor: increased resistance to thermal and oxidative stresses. Investig Ophthalmol Vis Sci. 1999;40:1444–1451. [PubMed] [Google Scholar]
- 84.Ganapathy V, Daniels T, Casiano CA. LEDGF/p75: a novel nuclear autoantigen at the crossroads of cell survival and apoptosis. Autoimmun Rev. 2003;2:290–297. doi: 10.1016/s1568-9972(03)00063-6. [DOI] [PubMed] [Google Scholar]
- 85.Shinohara T, Singh DP, Fatma N. LEDGF, a survival factor, activates stress-related genes. Prog Retin Eye Res. 2002;21:341–358. doi: 10.1016/s1350-9462(02)00007-1. [DOI] [PubMed] [Google Scholar]
- 86.Fatma N, Singh DP, Shinohara T, Chylack LT., Jr Transcriptional regulation of the antioxidant protein 2 gene, a thiol-specific antioxidant, by lens epithelium-derived growth factor to protect cells from oxidative stress. J Biol Chem. 2001;276:48899–48907. doi: 10.1074/jbc.M100733200. [DOI] [PubMed] [Google Scholar]
- 87.Sharma P, Singh DP, Fatma N, Chylack LT, Jr, Shinohara T. Activation of LEDGF gene by thermal-and oxidative-stresses. Biochem Biophys Res Commun. 2000;276:1320–1324. doi: 10.1006/bbrc.2000.3606. [DOI] [PubMed] [Google Scholar]
- 88.Singh DP, Fatma N, Kimura A, Chylack LT, Jr, Shinohara T. LEDGF binds to heat shock and stress-related element to activate the expression of stress-related genes. Biochem Biophys Res Commun. 2001;283:943–955. doi: 10.1006/bbrc.2001.4887. [DOI] [PubMed] [Google Scholar]
- 89.Krackhardt AM, Witzens M, Harig S, Hodi FS, Zauls AJ, Chessia M, Barrett P, Gribben JG. Identification of tumor-associated antigens in chronic lymphocytic leukemia by SEREX. Blood. 2002;100:2123–2131. doi: 10.1182/blood-2002-02-0513. [DOI] [PubMed] [Google Scholar]
- 90.Ahuja HG, Hong J, Aplan PD, Tcheurekdjian L, Forman SJ, Slovak ML. t(9;11)(p22;p15) in acute myeloid leukemia results in a fusion between NUP98 and the gene encoding transcriptional coactivators p52 and p75-lens epithelium-derived growth factor (LEDGF) Cancer Res. 2000;60:6227–6229. [PubMed] [Google Scholar]
- 91.Grand FH, Koduru P, Cross NC, Allen SL. NUP98-LEDGF fusion and t(9;11) in transformed chronic myeloid leukemia. Leuk Res. 2005;29:1469–1472. doi: 10.1016/j.leukres.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 92.Morerio C, Acquila M, Rosanda C, Rapella A, Tassano E, Micalizzi C, Panarello C. t(9;11)(p22;p15) with NUP98-LEDGF fusion gene in pediatric acute myeloid leukemia. Leuk Res. 2005;29:167–170. doi: 10.1016/j.leukres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 93.Singh DP, Kubo E, Takamura Y, Shinohara T, Kumar A, Chylack LT, Jr, Fatma N. DNA binding domains and nuclear localization signal of LEDGF: contribution of two helix-turn-helix (HTH)-like domains and a stretch of 58 amino acids of the N-terminal to the trans-activation potential of LEDGF. J Mol Biol. 2006;355:379–394. doi: 10.1016/j.jmb.2005.10.054. [DOI] [PubMed] [Google Scholar]
- 94.Wu X, Daniels T, Molinaro C, Lilly MB, Casiano CA. Caspase cleavage of the nuclear autoantigen LEDGF/p75 abrogates its pro-survival function: implications for autoimmunity in atopic disorders. Cell Death Differ. 2002;9:915–925. doi: 10.1038/sj.cdd.4401063. [DOI] [PubMed] [Google Scholar]
- 95.Van Maele B, Busschots K, Vandekerckhove L, Christ F, Debyser Z. Cellular co-factors of HIV-1 integration. Trends Biochem Sci. 2006;31:98–105. doi: 10.1016/j.tibs.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 96.Ogawa Y, Sugiura K, Watanabe A, Kunimatsu M, Mishima M, Tomita Y, Muro Y. Autoantigenicity of DFS70 is restricted to the conformational epitope of C-terminal α-helical domain. J Autoimmun. 2004;23:221–231. doi: 10.1016/j.jaut.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 97.Ganapathy V, Casiano CA. Autoimmunity to the nuclear autoantigen DFS70 (LEDGF): what exactly are the autoantibodies trying to tell us? Arthritis Rheum. 2004;50:684–688. doi: 10.1002/art.20095. [DOI] [PubMed] [Google Scholar]
- 98.Ronquist KG, Carlsson L, Ronquist G, Nilsson S, Larsson A. Prostasome-derived proteins capable of eliciting an immune response in prostate cancer patients. Int J Cancer. 2006;119:847–853. doi: 10.1002/ijc.21895. [DOI] [PubMed] [Google Scholar]
- 99.Manevich Y, Fisher AB. Peroxiredoxin 6, a 1-Cys peroxiredoxin, functions in antioxidant defense and lung phospholipid metabolism. Free Radic Biol Med. 2005;38:1422–1432. doi: 10.1016/j.freeradbiomed.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 100.Ouyang X, DeWeese TL, Nelson WG, Abate-Shen C. Loss-of-function of Nkx3.1 promotes increased oxidative damage in prostate carcinogenesis. Cancer Res. 2005;65:6773–6779. doi: 10.1158/0008-5472.CAN-05-1948. [DOI] [PubMed] [Google Scholar]
- 101.Miyake H, Hara I, Gleave ME, Eto H. Protection of androgen-dependent human prostate cancer cells from oxidative stress-induced DNA damage by overexpression of clusterin and its modulation by androgen. Prostate. 2004;61:318–323. doi: 10.1002/pros.20087. [DOI] [PubMed] [Google Scholar]
- 102.Trougakos IP, So A, Jansen B, Gleave ME, Gonos ES. Silencing expression of the clusterin/apolipoprotein j gene in human cancer cells using small interfering RNA induces spontaneous apoptosis, reduced growth ability, and cell sensitization to genotoxic and oxidative stress. Cancer Res. 2004;64:1834–1842. doi: 10.1158/0008-5472.can-03-2664. [DOI] [PubMed] [Google Scholar]
- 103.Sekito A, Koide-Yoshida S, Niki T, Taira T, Iguchi-Ariga SM, Ariga H. DJ-1 interacts with HIPK1 and affects H2O2-induced cell death. Free Radic Res. 2006;40:155–165. doi: 10.1080/10715760500456847. [DOI] [PubMed] [Google Scholar]
- 104.Suzuki Y, Kondo Y, Himeno S, Nemoto K, Akimoto M, Imura N. Role of antioxidant systems in human androgen-independent prostate cancer cells. Prostate. 2000;43:144–149. doi: 10.1002/(sici)1097-0045(20000501)43:2<144::aid-pros9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 105.Jones EL, Zhao MJ, Stevenson MA, Calderwood SK. The 70 kilodalton heat shock protein is an inhibitor of apoptosis in prostate cancer. Int J Hyperth. 2004;20:835–849. doi: 10.1080/02656730410001721807. [DOI] [PubMed] [Google Scholar]
- 106.Parcellier A, Schmitt E, Brunet M, Hammann A, Solary E, Garrido C. Small heat shock proteins HSP27 and αB-crystallin: cytoprotective and oncogenic functions. Antioxid Redox Signal. 2005;7:404–413. doi: 10.1089/ars.2005.7.404. [DOI] [PubMed] [Google Scholar]
- 107.Rocchi P, So A, Kojima S, Signaevsky M, Beraldi E, Fazli L, Hurtado-Coll A, Yamanaka K, Gleave M. Heat shock protein 27 increases after androgen ablation and plays a cytoprotective role in hormone-refractory prostate cancer. Cancer Res. 2004;64:6595–6602. doi: 10.1158/0008-5472.CAN-03-3998. [DOI] [PubMed] [Google Scholar]
- 108.Mintz PJ, Kim J, Do KA, Wang X, Zinner RG, Cristofanilli M, Arap MA, Hong WK, Troncoso P, Logothetis CJ, Pasqualini R, Arap W. Fingerprinting the circulating repertoire of antibodies from cancer patients. Nat Biotechnol. 2003;21:57–63. doi: 10.1038/nbt774. [DOI] [PubMed] [Google Scholar]
- 109.Cardillo MR, Monti S, Di Silverio F, Gentile V, Sciarra F, Toscano V. Insulin-like growth factor (IGF)-I, IGF-II and IGF type I receptor (IGFR-I) expression in prostatic cancer. Anticancer Res. 2003;23:3825–3835. [PubMed] [Google Scholar]
- 110.Vyas S, Asmerom Y, De Leon DD. Resveratrol regulates insulin-like growth factor-II in breast cancer cells. Endocrinology. 2005;146:4224–4233. doi: 10.1210/en.2004-1344. [DOI] [PubMed] [Google Scholar]
- 111.Zhang JY, Casiano CA, Peng XX, Koziol JA, Chan EK, Tan EM. Enhancement of antibody detection in cancer using panel of recombinant tumor-associated antigens. Cancer Epidemiol Biomark Prev. 2003;12:136–143. [PubMed] [Google Scholar]
- 112.Liang T, Hiipakka RA, Stebbins J, Liao S. Anti-5α-reductase autoantibodies in the serum of patients with prostatic cancer. J Clin Endocrinol Metab. 1990;71:1666–1668. doi: 10.1210/jcem-71-6-1666. [DOI] [PubMed] [Google Scholar]
- 113.Sreekumar A, Laxman B, Rhodes DR, Bhagavathula S, Harwood J, Giacherio D, Ghosh D, Sanda MG, Rubin MA, Chinnaiyan AM. Humoral immune response to α-methylacyl-CoA racemase and prostate cancer. J Natl Cancer Inst. 2004;96:834–843. doi: 10.1093/jnci/djh145. [DOI] [PubMed] [Google Scholar]
- 114.Imai H, Chan EK, Kiyosawa K, Fu XD, Tan EM. Novel nuclear autoantigen with splicing factor motifs identified with antibody from hepatocellular carcinoma. J Clin Investig. 1993;92:2419–2426. doi: 10.1172/JCI116848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chan EK, Imai H, Hamel JC, Tan EM. Human autoantibody to RNA polymerase I transcription factor hUBF. Molecular identity of nucleolus organizer region autoantigen NOR-90 and ribosomal RNA transcription upstream binding factor. J Exp Med. 1991;174:1239–1244. doi: 10.1084/jem.174.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Muro Y, Chan EK, Landberg G, Tan EM. A cell-cycle nuclear autoantigen containing WD-40 motifs expressed mainly in S and G2 phase cells. Biochem Biophys Res Commun. 1995;207:1029–1037. doi: 10.1006/bbrc.1995.1288. [DOI] [PubMed] [Google Scholar]
- 117.Tureci O, Sahin U, Pfreundschuh M. Serological analysis of human tumor antigens: molecular definition and implications. Mol Med Today. 1997;3:342–349. doi: 10.1016/s1357-4310(97)01081-2. [DOI] [PubMed] [Google Scholar]
- 118.Tureci O, Sahin U, Zwick C, Neumann F, Pfreundschuh M. Exploitation of the antibody repertoire of cancer patients for the identification of human tumor antigens. Hybridoma. 1999;18:23–28. doi: 10.1089/hyb.1999.18.23. [DOI] [PubMed] [Google Scholar]
- 119.Tureci O, Usener D, Schneider S, Sahin U. Identification of tumor-associated autoantigens with SEREX. Methods Mol Med. 2005;109:137–154. doi: 10.1385/1-59259-862-5:137. [DOI] [PubMed] [Google Scholar]
- 120.Wang Y, Han KJ, Pang XW, Vaughan HA, Qu W, Dong XY, Peng JR, Zhao HT, Rui JA, Leng XS, Cebon J, Burgess AW, Chen WF. Large scale identification of human hepatocellular carcinoma-associated antigens by autoantibodies. J Immunol. 2002;169:1102–1109. doi: 10.4049/jimmunol.169.2.1102. [DOI] [PubMed] [Google Scholar]
- 121.Qian F, Odunsi K, Blatt LM, Scanlan MJ, Mannan M, Shah N, Montgomery J, Haddad F, Taylor M. Tumor associated antigen recognition by autologous serum in patients with breast cancer. Int J Mol Med. 2005;15:137–144. [PubMed] [Google Scholar]
- 122.Fossa A, Siebert R, Aasheim HC, Maelandsmo GM, Berner A, Fossa SD, Paus E, Smeland EB, Gaudernack G. Identification of nucleolar protein No55 as a tumour-associated autoantigen in patients with prostate cancer. Br J Cancer. 2000;83:743–749. doi: 10.1054/bjoc.2000.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stone B, Schummer M, Paley PJ, Thompson L, Stewart J, Ford M, Crawford M, Urban N, O’Briant K, Nelson BH. Serologic analysis of ovarian tumor antigens reveals a bias toward antigens encoded on 17q. Int J Cancer. 2003;104:73–84. doi: 10.1002/ijc.10900. [DOI] [PubMed] [Google Scholar]
- 124.Devitt G, Meyer C, Wiedemann N, Eichmuller S, Kopp-Schneider A, Haferkamp A, Hautmann R, Zoller M. Serological analysis of human renal cell carcinoma. Int J Cancer. 2006;118:2210–2219. doi: 10.1002/ijc.21626. [DOI] [PubMed] [Google Scholar]
- 125.Vaughan HA, St Clair F, Scanlan MJ, Chen YT, Maraskovsky E, Sizeland A, Old LJ, Cebon J. The humoral immune response to head and neck cancer antigens as defined by the serological analysis of tumor antigens by recombinant cDNA expression cloning. Cancer Immun. 2004;4:1–16. [PubMed] [Google Scholar]
- 126.Shimada H, Nakashima K, Ochiai T, Nabeya Y, Takiguchi M, Nomura F, Hiwasa T. Serological identification of tumor antigens of esophageal squamous cell carcinoma. Int J Oncol. 2005;26:77–86. [PubMed] [Google Scholar]
- 127.Liggins AP, Guinn BA, Banham AH. Identification of lymphoma-associated antigens using SEREX. Methods Mol Med. 2005;115:109–128. doi: 10.1385/1-59259-936-2:109. [DOI] [PubMed] [Google Scholar]
- 128.Fernandez Madrid F, Tang N, Alansari H, Karvonen RL, Tomkiel JE. Improved approach to identify cancer-associated autoantigens. Autoimmun Rev. 2005;4:230–235. doi: 10.1016/j.autrev.2004.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Miles AK, Matharoo-Ball B, Li G, Ahmad M, Rees RC. The identification of human tumour antigens: current status and future developments. Cancer Immunol Immunother. 2006;55:996–1003. doi: 10.1007/s00262-005-0115-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sioud M, Hansen M, Dybwad A. Profiling the immune responses in patient sera with peptide and cDNA display libraries. Int J Mol Med. 2000;6:123–128. doi: 10.3892/ijmm.6.2.123. [DOI] [PubMed] [Google Scholar]
- 131.Somers VA, Brandwijk RJ, Joosten B, Moerkerk PT, Arends JW, Menheere P, Pieterse WO, Claessen A, Scheper RJ, Hoogenboom HR, Hufton SE. A panel of candidate tumor antigens in colorectal cancer revealed by the serological selection of a phage displayed cDNA expression library. J Immunol. 2002;169:2772–2780. doi: 10.4049/jimmunol.169.5.2772. [DOI] [PubMed] [Google Scholar]
- 132.Sioud M, Hansen MH. Profiling the immune response in patients with breast cancer by phage-displayed cDNA libraries. Eur J Immunol. 2001;31:716–725. doi: 10.1002/1521-4141(200103)31:3<716::aid-immu716>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 133.Fossa A, Alsoe L, Crameri R, Funderud S, Gaudernack G, Smeland EB. Serological cloning of cancer/testis antigens expressed in prostate cancer using cDNA phage surface display. Cancer Immunol Immunother. 2004;53:431–438. doi: 10.1007/s00262-003-0458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Klade CS, Voss T, Krystek E, Ahorn H, Zatloukal K, Pummer K, Adolf GR. Identification of tumor antigens in renal cell carcinoma by serological proteome analysis. Proteomics. 2001;1:890–898. doi: 10.1002/1615-9861(200107)1:7<890::AID-PROT890>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 135.Naour FL, Brichory F, Beretta L, Hanash SM. Identification of tumor-associated antigens using proteomics. Technol Cancer Res Treat. 2002;1:257–262. doi: 10.1177/153303460200100406. [DOI] [PubMed] [Google Scholar]
- 136.Balboni I, Chan SM, Kattah M, Tenenbaum JD, Butte AJ, Utz PJ. Multiplexed protein array platforms for analysis of autoimmune diseases. Annu Rev Immunol. 2006;24:391–418. doi: 10.1146/annurev.immunol.24.021605.090709. [DOI] [PubMed] [Google Scholar]
- 137.Steen VD. Autoantibodies in systemic sclerosis. Semin Arthritis Rheum. 2005;35:35–42. doi: 10.1016/j.semarthrit.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 138.Franceschini F, Cavazzana I. Anti-Ro/SSA and La/SSB antibodies. Autoimmunity. 2005;38:55–63. doi: 10.1080/08916930400022954. [DOI] [PubMed] [Google Scholar]
- 139.Chan SM, Utz PJ. The challenge of analyzing the proteome in humans with autoimmune diseases. Ann N Y Acad Sci. 2005;1062:61–68. doi: 10.1196/annals.1358.009. [DOI] [PubMed] [Google Scholar]
- 140.Joos TO, Schrenk M, Hopfl P, Kroger K, Chowdhury U, Stoll D, Schorner D, Durr M, Herick K, Rupp S, Sohn K, Hammerle H. A microarray enzyme-linked immunosorbent assay for autoimmune diagnostics. Electrophoresis. 2000;21:2641–2650. doi: 10.1002/1522-2683(20000701)21:13<2641::AID-ELPS2641>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 141.Robinson WH, DiGennaro C, Hueber W, Haab BB, Kamachi M, Dean EJ, Fournel S, Fong D, Genovese MC, de Vegvar HE, Skriner K, Hirschberg DL, Morris RI, Muller S, Pruijn GJ, van Venrooij WJ, Smolen JS, Brown PO, Steinman L, Utz PJ. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat Med. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- 142.Hueber W, Kidd BA, Tomooka BH, Lee BJ, Bruce B, Fries JF, Sonderstrup G, Monach P, Drijfhout JW, van Venrooij WJ, Utz PJ, Genovese MC, Robinson WH. Antigen microarray profiling of autoantibodies in rheumatoid arthritis. Arthritis Rheum. 2005;52:2645–2655. doi: 10.1002/art.21269. [DOI] [PubMed] [Google Scholar]
- 143.Scanlan MJ, Welt S, Gordon CM, Chen YT, Gure AO, Stockert E, Jungbluth AA, Ritter G, Jager D, Jager E, Knuth A, Old LJ. Cancer-related serological recognition of human colon cancer: identification of potential diagnostic and immunotherapeutic targets. Cancer Res. 2002;62:4041–4047. [PubMed] [Google Scholar]
- 144.Koziol JA, Zhang JY, Casiano CA, Peng XX, Shi FD, Feng AC, Chan EK, Tan EM. Recursive partitioning as an approach to selection of immune markers for tumor diagnosis. Clin Cancer Res. 2003;9:5120–5126. [PubMed] [Google Scholar]
- 145.Ablin RJ. Malignancy associated with antinuclear antibodies. Lancet. 1972;2:1253–1254. doi: 10.1016/s0140-6736(72)92308-2. [DOI] [PubMed] [Google Scholar]
- 146.Ablin RJ. Serum antibody in patients with prostatic cancer. Br J Urol. 1976;48:355–361. doi: 10.1111/j.1464-410x.1976.tb06652.x. [DOI] [PubMed] [Google Scholar]
- 147.Hoffman RM, Gilliland FD, Eley JW, Harlan LC, Stephenson RA, Stanford JL, Albertson PC, Hamilton AS, Hunt WC, Potosky AL. Racial and ethnic differences in advanced-stage prostate cancer: the Prostate Cancer Outcomes Study. J Natl Cancer Inst. 2001;93:388–395. doi: 10.1093/jnci/93.5.388. [DOI] [PubMed] [Google Scholar]
- 148.Barry MJ. Clinical practice. Prostate-specific-antigen testing for early diagnosis of prostate cancer. N Engl J Med. 2001;344:1373–1377. doi: 10.1056/NEJM200105033441806. [DOI] [PubMed] [Google Scholar]
- 149.Mikolajczyk SD, Song Y, Wong JR, Matson RS, Rittenhouse HG. Are multiple markers the future of prostate cancer diagnostics? Clin Biochem. 2004;37:519–528. doi: 10.1016/j.clinbiochem.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 150.Shi FD, Zhang JY, Liu D, Rearden A, Elliot M, Nachtsheim D, Daniels T, Casiano CA, Heeb MJ, Chan EK, Tan EM. Preferential humoral immune response in prostate cancer to cellular proteins p90 and p62 in a panel of tumor-associated antigens. Prostate. 2005;63:252–258. doi: 10.1002/pros.20181. [DOI] [PubMed] [Google Scholar]
- 151.Kida Y. Autoantibodies in prostate cancer. N Engl J Med. 2005;353:2815–2817. [PubMed] [Google Scholar]
- 152.Bradley SV, Oravecz-Wilson KI, Bougeard G, Mizukami I, Li L, Munaco AJ, Sreekumar A, Corradetti MN, Chinnaiyan AM, Sanda MG, Ross TS. Serum antibodies to huntingtin interacting protein-1: a new blood test for prostate cancer. Cancer Res. 2005;65:4126–4133. doi: 10.1158/0008-5472.CAN-04-4658. [DOI] [PubMed] [Google Scholar]
- 153.Hoeppner LH, Dubovsky JA, Dunphy EJ, McNeel DG. Humoral immune responses to testis antigens in sera from patients with prostate cancer. Cancer Immun. 2006;6:1–7. [PubMed] [Google Scholar]
- 154.Miller JC, Zhou H, Kwekel J, Cavallo R, Burke J, Butler EB, The BS, Haab BB. Antibody microarray profiling of human prostate cancer sera: antibody screening and identification of potential biomarkers. Proteomics. 2003;3:56–63. doi: 10.1002/pmic.200390009. [DOI] [PubMed] [Google Scholar]