Abstract
Replication-competent retrovirus vectors based on murine leukemia virus (MLV) have been shown to effectively transfer therapeutic genes over multiple serial infections in cell culture and through solid tumors in vivo with a high degree of genomic stability. While simple retroviruses possess a natural tumor selectivity in that they can transduce only actively dividing cells, additional tumor-targeting strategies would nevertheless be advantageous, since tumor cells are not the only actively dividing cells. In this study, we used the promiscuous murine cytomegalovirus promoter, a chimeric regulatory sequence consisting of the hepatitis B virus enhancer II and the human α1-antitrypsin (EII-Pa1AT) promoter, and a synthetic regulatory sequence consisting of a series of T-cell factor binding sites named the CTP4 promoter to generate replicating MLV vectors, whereby the last two are transcriptionally restricted to liver- and β-catenin/T-cell factor-deregulated cells, respectively. When the heterologous promoters were used to replace almost the entire MLV U3 region, including the MLV TATA box, vector replication was inefficient since nascent virus particle production from infected cells was greatly decreased. Fusion of the heterologous promoters lacking the TATA box to the MLV TATA box, however, generated vectors which replicated with almost-wild-type kinetics throughout permissive cells while exhibiting low or negligible spread in nonpermissive cells. The genomic stability of the vectors was shown to be comparable to that of a similar vector containing wild-type MLV long terminal repeats, and tropism analysis over repeated infection cycles showed that the targeted vectors retained their original specificity.
Because simple retroviruses can transduce only actively dividing cells (3, 26, 36, 44), their use in cancer gene therapy has been extensively investigated, and over the last decade, numerous preclinical in vivo studies and clinical trials have been carried out using replication-defective retroviral (RDR) vectors (13). Although promising results have been obtained with animal models, therapeutic benefit in clinical settings has remained elusive, especially for cancer gene therapy, since the infection efficiency of solid tumors is too low (34). Of late, therefore, the use of replication-competent retroviral (RCR) vectors has been advocated, and it has been demonstrated by various groups that these are much more efficacious than their RDR counterparts (15, 23, 26-29, 40-42, 45). Mitotic cells, of course, are not unique to tumors, and although it may be expected that RCR vectors would not replicate efficiently outside of the immune-privileged environment of a solid tumor in a healthy individual, the possibility of spread occurring in dividing cells outside of the tumor mass must nevertheless be considered (7, 33, 35). Moreover, not least due to recent events demonstrating that retroviral vectors are capable, albeit in rare circumstances, of inducing oncogenesis in humans (19, 30), safety is a primary concern in retroviral vector design (46).
To date, the most effective targeting strategies for RDR vectors are transcriptional targeting (11, 14, 18, 31), whereby promiscuous viral promoter elements are exchanged for more-tightly regulated cellular, viral, or synthetic promoter elements, and, to a lesser extent, modifications of the envelope protein such that infection is restricted to certain cell types (infection targeting) (21). It has recently been demonstrated that, by using a highly active synthetic variant of the probasin promoter, the expression targeting strategy can also be applied to RCR vectors such that both transgene expression and vector replication are strictly confined to prostate cells (27). Although transcriptional targeting of RCR vectors, in contrast to infection targeting, is not designed to prevent infection of nontarget cells, it should prevent subsequent spread following an initial infection event. Moreover, transcriptional targeting should minimize the risk of oncogenesis via insertional mutagenesis since the promoter/enhancer elements may not be able to activate cellular genes (37).
In this study, we have investigated whether a transcriptional targeting strategy could be applied to RCR vectors to target specific cell types. In contrast to the transcriptional targeting of replication-deficient retroviral vectors, in which only expression of the transgene is required, in RCR vectors, sufficient amounts of Gag, Pol, and Env proteins must also be made in order to facilitate efficient virus spread, necessitating a promoter which drives high levels of transcription (27). Moreover, since the promoter is inserted into such vectors in addition to the full complement of viral genes and it is known that lengthening of the viral genome can lead to large decreases in replication efficiency (38), suitable candidate promoters should be of similar size to or smaller than the murine leukemia virus (MLV) elements which they replace and should mediate strong transcription in permissive cells. Following an extensive literature search, we therefore selected the liver-specific chimeric promoter EII-Pa1AT (24), the synthetic, beta-catenin/T-cell factor (TCF)-dependent promoter CTP4 (25), and the promiscuous murine cytomegalovirus (mCMV) promoter (1), all of which are relatively short and have been shown to drive high levels of transcription in permissive cells (1, 24, 25). The 380-bp CTP4 promoter consists of a minimal TATA box preceded by 10 TCF binding sites and has been shown to allow strict expression targeting of adenoviral vectors to cells deregulated for β-catenin, including those derived from prostate, ovarian, liver, and colorectal cancers, and to be highly active in biopsy specimens from primary human colon and colorectal cancers (25). The 460-bp EII-Pa1AT promoter, which consists of enhancer II (EII) of the human hepatitis B virus (HBV) fused to the human α1-antitrypsin promoter (Pa1AT), has been described as having strong transcriptional activity in a liver-selective manner, both in vitro and in vivo (24).
The above-described regulatory sequences were used to create two types of RCR vectors. TATA fusion (TF) vectors were constructed by fusing the heterologous promoters lacking the TATA box precisely to the TATA box in the MLV U3 region, such that transcription initiation should occur at the 5′ end of the MLV R region, as is the case for wild-type MLV. TATA replacement (TR) vectors, on the other hand, were designed such that the entire heterologous promoter region, including the TATA box and transcriptional start site (TSS), was used to replace almost the entire MLV U3 region, including the TATA box.
Our results show that although both TF and TR vectors restricted transgene expression to permissive cell lines, only the TF design allowed efficient replication of targeted vectors within these cells. TF vectors replicate in a wide range of permissive cells, with kinetics similar to that of the parental nonspecific RCR vectors, while retaining their genomic stability and transcriptional specificity over multiple infection cycles.
MATERIALS AND METHODS
Construction of plasmids.
To generate nonviral expression constructs harboring the heterologous regulatory sequences to be tested, the mCMV, EII-Pa1AT, and CTP4 promoters were cloned into the promoterless expression vector pEGFP1 (Clontech), upstream of the enhanced green fluorescent protein (eGFP) gene. To generate targeted RCR vectors in which the MLV TATA box was retained (TF), heterologous promoters were PCR amplified using forward primers binding to their 5′ ends and reverse primers binding to the 5′ borders of their TATA boxes. For the generation of vectors in which the MLV TATA box was replaced by the heterologous TATA box (TR), the sequence encompassing most of the 3′ long terminal repeat (LTR) and part of the downstream plasmid backbone was excised from pACE-GFP, using NheI and SspI, and replaced with the corresponding NheI-SspI fragment from vector pPCEWmCMVΔSNori (22), which contains a unique MluI site 21 bp downstream of the MLV TATA box in the 3′ LTR, generating plasmid pACE-GFP-ProCon, into which the heterologous promoters were cloned.
Cell lines, transfections, and virus production.
293 (ATCC CRL-1573), 293T (ATCC CRL-11268), NIH 3T3 (ATCC CRL-1658), HeLa (ATCC CCL-2), HepG2 (ATCC HB-8065), HuH-7 (ATCC CCL-185), and SW480 (ATCC CCL-228) cells, as well as cell lines AKH12 and AKH13 which were obtained from human primary liver tumors, were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). DLD-1 cells (ATCC CCL-221) were maintained in RPMI medium supplemented with 10% FCS.
A total of 8 × 105 293 or 293T cells, 2 × 105 HeLa cells, or 3 × 105 HepG2 cells were seeded per well of six-well plates and transfected 24 h later with 3 μg plasmid DNA per well, using the calcium phosphate coprecipitation technique (17). Stably transfected cell populations were generated by selection in DMEM with 10% FCS supplemented with 400 μg/ml G418 for a minimum of 3 weeks. Subsequently, fluorescence-activated cell sorting (FACS) analysis to measure eGFP expression was performed three times within an interval of 2 to 3 days. Viral stocks were generated by transient transfection of 293 or 293T cells, from which the supernatant was harvested at 48 h posttransfection, filtered through a 0.45-μm filter, and stored at −80°C.
β-Catenin/TCF activity assay for AKH12 and AKH13 cells.
A total of 2 × 106 cells of cell lines AKH12 and AKH13 were transfected with 3 μg of plasmids pEGFP1, pEGFP1-CTP4, pEGFP1-E2, and pEGFP1-mCMV, followed by selection with 400 μg/ml G418 and expansion to T75 flasks. Stably transfected cells were analyzed by FACS at 3 weeks posttransfection.
Infections and determination of viral titers.
Infections were carried out in a total volume of 1 ml medium per well in the presence of 8 μg/ml polybrene, followed by the addition of DMEM with 10% FCS or RPMI medium with 10% FCS after 6 h. The infected cells were passaged at regular intervals, and FACS analysis was performed at each passage. For titer determination, cells were infected with serially diluted virus stocks and 50 μM azidothymidine was added 24 h after infection to avoid secondary spread of the vector. Two days after infection, the number of infected cells was measured by FACS analysis.
Flow cytometry (FACS) analysis.
Cells transfected with expression constructs were washed with phosphate-buffered saline (PBS) and resuspended in a solution of 10% FCS in PBS. Cells transfected or infected with RCR vectors were fixed by being incubated in a solution of 4% formaldehyde in PBS for 30 min, and then the cells were washed and resuspended in PBS. Resuspended cells were subjected to FACS analysis, which was carried out with a FACSCalibur apparatus (Becton Dickinson).
Quantification of viral RNA and proviral DNA.
Viral RNA from the supernatant of infected cells and genomic DNA from infected cells were extracted using the QIAGEN viral RNA kit and the QIAGEN DNeasy kit (QIAGEN), respectively, according to the manufacturer's instructions. Real-time PCR and real-time reverse transcription (RT)-PCR were performed using primers 5′-GCAGTGCTTCAGCCGCTAC-3′ and 5′-AAGAAGATGGTGCGCTCCTG-3′ and probe 5′-6-carboxyfluorescein-ACCACATGAAGCAGCACGACTT-6-carboxytetramethylrhodamine-3′, all binding within the eGFP gene, or primers 5′-GTAGCGGTCGTGGGCACTTATA-3′ and 5′-CTTATGTTGGGAAGTGGCCGTA-3′ and probe 5′-6-carboxyfluorescein-CATTCCACCGCTCCGGCCAACT-6-carboxytetramethylrhodamine-3′, which bind specifically in env RNA.
PCR analysis and sequencing.
For molecular analysis of the genomic stability of the vectors, DNA was extracted from infected cells of each infection cycle by using the QIAGEN DNeasy kit. A fragment encompassing the transgenic cassette of the integrated vector genome was PCR amplified using forward primer 5′-GGCCAAGGATGGTTCGAAGG-3′, binding to the MLV env gene, and reverse primer 5′-GCGAGACCACAAGTCGGATG-3′, binding to the MLV 3′ LTR. PCR was carried out using Advantage genomic PCR polymerase mix (BD Biosciences) according to the manufacturer's recommendations. The PCR products were subjected to agarose gel electrophoresis to determine length variations. To analyze the promoters of integrated proviruses, the promoter region in the 3′ LTR was PCR amplified using forward primer 5′-GGCCGCGCCATAGATAAAAT-3′, binding just downstream of the eGFP gene, and reverse primer 5′-CGGGTAGTCAATCACTCAGA-3′, binding in the 3′ LTR U5 region, or forward primer 5′-CTCGGCATGGACGAGCTGTA-3′, binding in the eGFP gene, and reverse primer 5′-AAGGAACAGCGAGACCACAA-3′, binding in the 3′ LTR U5 region. The PCR products were subjected to agarose gel electrophoresis and sequencing, using the same primers as those used for the PCR, to detect length variations and mutations, respectively.
RESULTS
Transcriptional activity of mCMV, EII-Pa1AT, and CTP4 promoters in human cell lines.
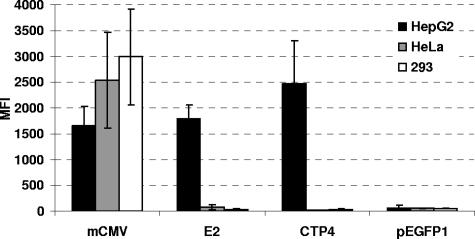
Based on an extensive literature search for short, highly active, and preferably tissue- or tumor-specific promoters which would be promising candidates for the generation of transcriptionally targeted RCR vectors, the 380-bp CTP4 promoter fragment, the 460-bp HBV enhancer II/human α1-antitrypsin promoter (E2), and the 300-bp murine CMV immediate-early enhancer/promoter region (mCMV) were inserted into the promoterless eGFP expression vector pEGFP1 and the resulting vectors pEGFP1-CTP4, pEGFP1-E2, and pEGFP1-mCMV were used to generate populations of stably transfected HepG2, HeLa, and 293 cells. The hepatic carcinoma cell line HepG2 is highly deregulated for β-catenin (4, 10) and has been shown to facilitate high-level HBV enhancer II activity (2); hence, both CTP4 and EII-Pa1AT promoters, as well as the promiscuous mCMV promoter, were expected to be active in these cells. FACS analysis of the stably transfected cell populations revealed that while the mCMV promoter was highly active in all cell lines, EII-Pa1AT or CTP4 promoter activity was restricted to HepG2 cells and was similar to or even higher than that of the mCMV promoter, respectively (Fig. 1).
FIG. 1.
Transcriptional activities of mCMV, EII-Pa1AT, and CTP4 promoters in human cell lines. HepG2, HeLa, and 293 cells were stably transfected with plasmids driving eGFP expression under the control of the mCMV, EII-Pa1AT (E2), and CTP4 promoters, and expression levels were quantified by FACS analysis. The MFI values depicted are the means from three independent FACS measurements. Plasmid pEGFP1 is the promoterless plasmid control.
Replication kinetics of targeted retroviral vectors.
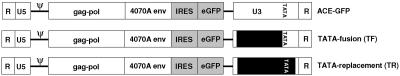
The mCMV, EII-Pa1AT, and CTP4 promoters were then inserted into the 3′ LTR U3 region of the MLV-based replication-competent retroviral vector ACE-GFP (27). Upon infection and reverse transcription, these promoters are duplicated to the 5′ LTR U3 region, thereby driving expression of the viral genes as well as of the heterologous transgene inserted following a heterologous internal ribosome entry site element downstream of the env gene (Fig. 2). Two different designs of promoter insertions were applied. On the one hand, in so-called TF vectors, most of the 3′ LTR U3 region up to the 5′ border of the MLV TATA box is removed and replaced with the mCMV, EII-Pa1AT, and CTP4 promoter sequences from the 5′ end to the 5′ border of their respective TATA boxes, such that transcription initiation in the respective vectors mCMV-TF, E2-TF, and CTP4-TF should occur at the 5′ end of the MLV R region, as is the case in wild-type MLV (Fig. 2). On the other hand, in TR vectors, almost the entire MLV U3 region, including the TATA box, is deleted and replaced by the full-length heterologous promoter, including its TATA box and TSS, generating vectors mCMV-TR, E2-TR, and CTP4-TR (Fig. 2).
FIG. 2.
Vector construction. All constructs are based on the parental vector ACE-GFP, which is comprised of Moloney MLV containing the amphotropic 4070A env gene and from which eGFP expression is mediated by the encephalomyocarditis virus internal ribosome entry site (IRES) fused to the 3′ end of the 4070A env gene. The position of the inserted heterologous promoters is depicted by a black box, and its insertion is designed such that either the heterologous promoter lacking its TATA box is fused to the MLV TATA box (TF) or almost the entire MLV U3 region is deleted and replaced by the heterologous promoter, including its TATA box and transcriptional start site (TR). As a result of these modifications, the original 448-bp MLV U3 region in the parental vector ACE-GFP is either shortened to 299 bp and 402 bp in the case of vectors CTP4-TF and CTP4-TR, respectively, or lengthened to 492 bp and 489 bp, in the case of vectors E2-TF and E2-TR, respectively, and to 538 bp and 615 bp, in the case of vectors mCMV-TF and mCMV-TR, respectively.
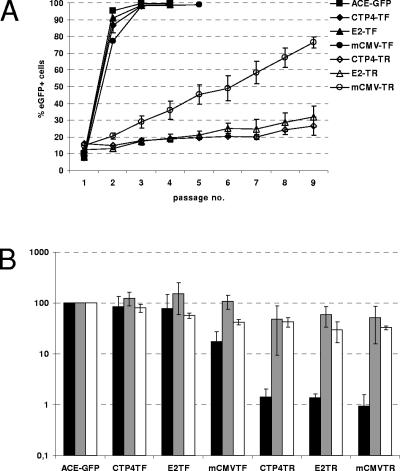
HepG2 cells were infected with these vectors generated from transiently transfected 293 cells, and replication kinetics were monitored by passaging the cells every 3 to 4 days and performing FACS analysis at each passage. The parental vector ACE-GFP as well as the TATA fusion vectors mCMV-TF, E2-TF, and CTP4-TF spread rapidly and infected 100% of the target cells after two to three passages (Fig. 3A). The respective TATA replacement vectors, mCMV-TR, E2-TR, and CTP4-TR, on the other hand, spread much more slowly and in a linear, rather than exponential, manner (Fig. 3A). These results cannot simply be explained by differences in promoter activity between TF and TR vectors, since the differences in replication kinetics did not correspond to the differences in intensity of eGFP expression of HepG2 cells infected with TF and TR vectors (Fig. 3B).
FIG. 3.
Replication kinetics, virus production, infectivity, and transgene expression of targeted vectors in HepG2 cells. (A) HepG2 cells were infected with RCR vectors, and spread of virus was monitored by serial passaging and FACS analysis at each passage. The mean values from three independent experiments of the percentage of eGFP-positive cells at each passage are depicted. (B) Virus-containing supernatant was harvested from infected HepG2 cells and used to infect new HepG2 cells, and 50 μM azidothymidine was applied at 24 h postinfection to prevent secondary infection events. The quantity of integrated proviral copies per infected producer cell and the quantity of nascent virus released into the supernatant from each infected producer cell were determined by real-time PCR and real-time RT-PCR, respectively, using primers and probes binding in the rRNA genes and eGFP genes. Values shown indicate the number of nascent virions released per proviral copy in infected producer cells relative to the value obtained for ACE-GFP (black bars). The infectivity of nascent virus particles was quantified by FACS analysis at 2 days postinfection of HepG2 cells. The quantity of eGFP-positive cells generated per virus particle was calculated for each vector relative to the value obtained for ACE-GFP (gray bars). White bars represent the MFI of cells at 48 h postinfection relative to the values obtained for ACE-GFP.
We thus compared the levels of virus production from infected cells and the infectivities of these viruses. To this end, infected HepG2 cell populations and the supernatants thereof were harvested, and the number of proviral copies in the infected cells and the number of virions released into the supernatant were quantified by real-time PCR and real-time RT-PCR, respectively. Compared to vector ACE-GFP, TF vectors produced between 2-fold and 10-fold fewer genome-containing virus particles per proviral copy in the producer cells (Fig. 3B). TR vectors, on the other hand, produced about 100-fold fewer genome-containing virus particles per provirus copy (Fig. 3B). Subsequent infection of HepG2 cells with the same amount of vector particles revealed roughly equal infectivities of vector ACE-GFP and of the TF and TR vectors. Thus, transcription of vector RNA from a heterologous promoter in the TR design does not lead to a decrease in the infectivity of such vectors per se. However, the amount of nascent virus particles produced from cells infected with TR vectors is severely retarded in comparison to the amount produced from ACE-GFP- or TF vector-infected cells, indicating a possible reason for the differences in replication kinetics between the TR and TF vectors.
Cell-specific replication of targeted TF vectors.
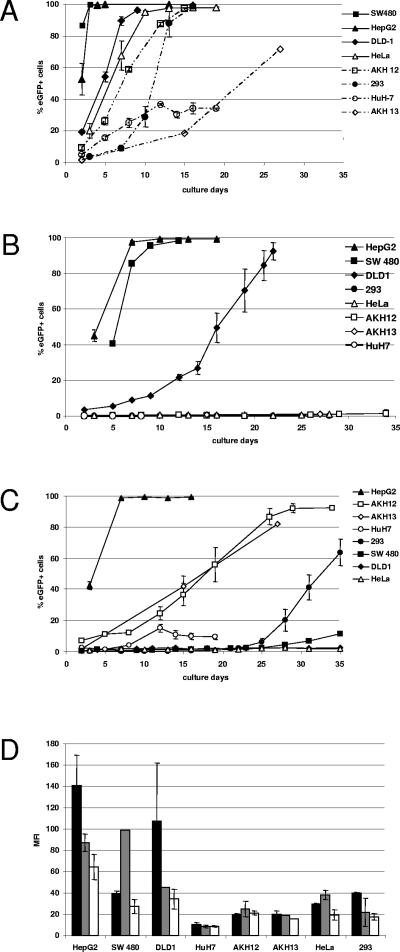
TF vectors were subsequently used to infect a wider range of human cell lines. HeLa and 293 cells, the liver carcinoma-derived cell lines HepG2 and HuH-7, the primary liver tumor-derived cells AKH12 and AKH13, as well as the colon carcinoma cell lines SW480 and DLD-1, were infected with vectors CTP4-TF, E2-TF, and ACE-GFP, and vector spread was followed by serial passaging and FACS analysis of the infected cells every few days. As expected, vector ACE-GFP spread in all cells, although replication kinetics differed considerably among cell lines and apparent initial spread in HuH-7 cells over the first few passages did not continue (Fig. 4A).
FIG. 4.
Replication kinetics and transgene expression of targeted vectors. HepG2, SW480, DLD-1, AKH12, AKH13, HuH-7, HeLa, and 293 cells were infected with vectors (A) ACE-GFP, (B) CTP4-TF, and (C) E2-TF. Virus-containing supernatant (500 μl) was used to infect each cell line. Infected cells were passaged at regular intervals, and FACS analysis was performed at each passage. The values shown are the percentage of eGFP-positive cells at each passage. (D) The MFIs of eGFP-positive cells infected with ACE-GFP (black bars), CTP4-TF (gray bars), and E2-TF (white bars) are shown as the means from three independent experiments. Error bars represent the standard deviations between experiments.
Vector CTP4-TF replicated with kinetics similar to those of ACE-GFP in HepG2 and SW480 cells, which are known to be deregulated for β-catenin, infecting nearly all HepG2 cells within two passages and all SW40 cells within four passages. Considerable spread of CTP4-TF could also be observed in DLD-1 cells, also deregulated in the β-catenin pathway, although at a much lower rate than for ACE-GFP, taking eight passages to reach over 80% of the cells. The CTP4-TF vector did not spread at all in cell lines which are not known to be deregulated for β-catenin, such as HuH-7, 293, and HeLa cells (Fig. 4B). The β-catenin status of the AKH12 and AKH13 cell lines, in which vector CTP4-TF also did not spread, was determined according to a commonly employed assay (6). To this end, cells were stably transfected with plasmids pEGFP1, pEGFP1-CTP4, pEGFP1-E2, and pEGFP1-mCMV, and FACS analysis of stably transfected cell populations at 3 weeks posttransfection revealed that they were not deregulated in the β-catenin/TCF pathway (data not shown). Vector E2-TF replicated very efficiently in HepG2 cells, exhibiting kinetics similar to that of ACE-GFP and rapidly infecting almost 100% of the cells within two passages (Fig. 4C). Considerable, albeit slower, spread was also observed in AKH12 and AKH13 cells (Fig. 4C) and was also comparable to the rate of spread of ACE-GFP in these cells (Fig. 4A). In the liver-derived HuH-7 cells, however, only very little spread could be observed and only for the first few passages. Little or no spread could be detected in the nonliver-derived cell lines SW480, DLD-1, 293, and HeLa, at least during the first 20 days of passaging (Fig. 4C). To a certain extent, there is a relationship between the mean fluorescence intensity (MFI) of the infected cells (Fig. 4D) and the ability of the respective vector to spread in these cells; in cell lines where very rapid spread is observed, the MFI of the infected cells is usually much higher than in cell lines where no or only slow spread is observed. In several cases, however, there is no discernible difference in MFI between cells infected with a vector which replicates moderately well and those infected with the same vector which does not spread at all. Moreover, vector CTP4-TF does not replicate at all in AKH12 or AKH13 cells, even though the MFI of the infected cells is the same as that when the cells are infected with vector ACE-GFP or E2-TF, which replicate in both cell lines.
In 293 and SW480 cells, spread of vector E2-TF could not be observed for the first 20 days of passaging but was thereafter quite robust in spite of the fact that these cell lines are not liver derived (Fig. 4B). Analysis of the 3′ LTR of integrated proviruses in genomic DNA isolated from a late passage of the 293 cells in which the E2-TF vector had begun to spread showed that mutant vectors had emerged which contained sequences derived from the human CMV promoter which was originally present in the 5′ LTR of the vector on the plasmid DNA, indicating that a rare recombination event had taken place at the plasmid DNA level during production of vector stocks by transient transfection (data not shown). The recombination junction occurred between sequences 85 bp downstream of the 5′ end of the human CMV promoter and 248 bp downstream of the 5′ end of the EII-Pa1AT promoter.
Genomic stability of targeted RCR vectors.
The parental vector, ACE-GFP, has been shown to allow stable transgene propagation over multiple serial infection cycles in several different cell lines (28; M. Paar, personal communication). To investigate the potential effect on genomic stability of the presence of heterologous promoters in the transcriptionally targeted vectors, we followed the replication kinetics of the vectors E2-TF, CTP4-TF, and ACE-GFP through multiple serial-diluting infection cycles and analyzed vector stability at different time points.
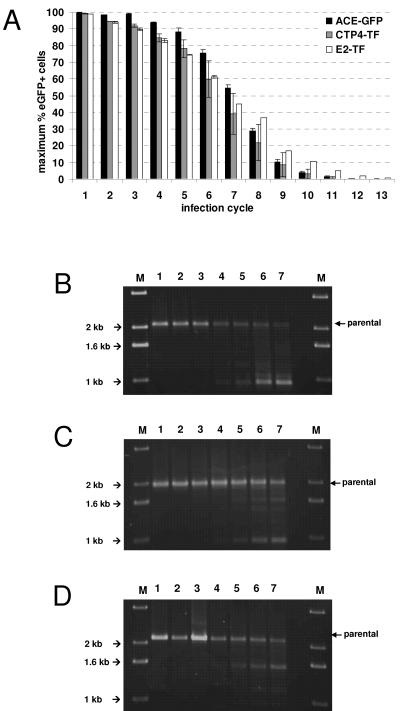
HepG2 cells were initially infected at a multiplicity of infection of 0.01 and subsequently passaged and subjected to FACS analysis every 3 days, until a maximum percentage of cells in the population was expressing eGFP. For every second passage, the supernatant from infected cells was harvested, diluted 100-fold, and used to infect fresh HepG2 cells. This was repeated for 13 consecutive infection cycles (Fig. 5A).
FIG. 5.
Genomic stability of targeted vectors. HepG2 cells were infected with vectors ACE-GFP, CTP4-TF, and E2-TF at a multiplicity of infection of 0.01 and passaged until a maximum percentage of eGFP-positive cells was reached. The supernatant was harvested from infected cells at the second passage, diluted 100-fold, and used to initiate a new infection cycle. This was repeated for 13 serial infection cycles. (A) The values shown indicate the maximum percentage of eGFP-expressing cells reached in each infection cycle. (B to D) Genomic DNA was extracted from HepG2 cells infected with ACE-GFP (B), CTP4-TF (C), and E2-TF (D) at cycles 1 to 7, and PCR was performed using primers binding in the 3′ end of the env gene and 3′ U3 regions flanking the transgene cassette. Lane M, marker.
Virions which have lost the transgene cassette do not express eGFP but can still infect cells and prevent superinfection of these cells by other virions via superinfection resistance (32). Therefore, when the vector becomes unstable, it is expected that the maximum number of eGFP-positive cells in the population should gradually decrease with each infection cycle. This process had already become evident by the second cycle, and by the 13th cycle, almost all virions had lost their insert. However, the stability of the vectors containing heterologous promoters was not less than that of the parental vector ACE-GFP.
In addition, the genomic stability of the vectors was monitored on a molecular level by performing PCR on genomic DNA extracted from infected HepG2 cells at cycles 1 to 7, using primers designed to bind to sequences flanking the transgene cassette in the integrated proviruses. Intact proviral genomes of ACE-GFP (Fig. 5B), CTP4-TF (Fig. 5C), or E2-TF (Fig. 5D) give rise to PCR products of 2,093, 1,944, or 2,137 bp in length, respectively. For the first three infection cycles, only PCR products corresponding to intact genomes were detectable, but from cycle 4 onward, several shorter products, representing deletion mutants, could be observed (Fig. 5B to D), which is in accordance with the FACS data showing that the maximum percentage of eGFP-positive cells began to decrease from the fourth cycle. The main mutant vector arising in cells infected with vector ACE-GFP carries a deletion of about 1,100 bp, while the major mutant vectors arising in CTP-TF- and E2-TF-infected cells carry deletions of about 900 bp and 600 bp, respectively (Fig. 5B to D). It is most likely that these deletions confine themselves to the transgene cassette and confer a replicative advantage on the vector mutants such that they become increasingly dominant in the viral population. Supporting this hypothesis, real-time RT-PCR performed on viral RNA extracted from filtered supernatants from infected cells from infection cycles 1, 5, 9, and 13, using primers and probes which bind in envelope-specific RNA, demonstrated that the production of infectious retrovirus particles did not decrease over successive infection cycles, even though they no longer led to eGFP expression in infected cells (data not shown).
Analysis of promoter sequence and specificity over multiple infection cycles.
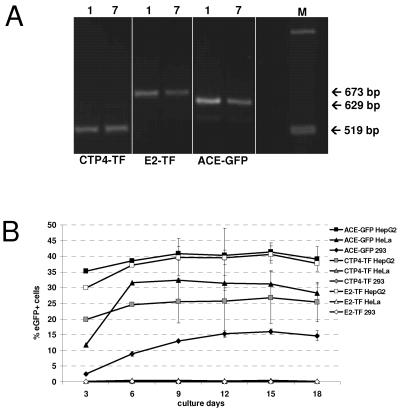
To monitor the genetic integrity of the promoter following several serial diluting infection cycles, the 3′ promoter regions of integrated proviruses from the first and seventh infection cycles were PCR amplified using primers binding to sequences flanking the U3 region. No change in length of promoters between the first and seventh cycles could be observed by gel electrophoretic analysis of the PCR products (Fig. 6A), and subsequent sequence analyses of the obtained PCR fragments revealed that no mutations occurred in the CTP4 promoter, while in a fraction of the virus populations, two point mutations occurred in the EII-Pa1AT promoter of vector E2-TF and the U3 region of vector ACE-GFP. Neither of the point mutations in the EII-Pa1AT promoter is located in any known transcription factor binding site.
FIG. 6.
Promoter sequence and specificity over multiple infection cycles. (A) PCR was performed on genomic DNA extracted from HepG2 cells infected with ACE-GFP, CTP4-TF, and E2-TF at cycles 1 and 7, using primers flanking the heterologous promoter in the MLV LTR. The expected fragment sizes are indicated. Lane M, marker. (B) Vector tropism remains unchanged following multiple serial infection cycles. HepG2, 293, and HeLa cells were infected with the supernatant from the seventh infection cycle of HepG2 cells infected with ACE-GFP, CTP4-TF, and E2-TF. The cells were passaged every 3 days, and spread of virus was monitored by FACS analysis. The percentage of eGFP-positive cells at each passage is shown.
To analyze whether the replicative ability of the vectors in different cell lines was maintained following several serial diluting infection cycles in HepG2 cells, vectors ACE-GFP, E2-TF, and CTP4-TF were harvested from cells of the seventh infection cycle, diluted 10-fold, and used to inoculate fresh HepG2, HeLa, and 293 cells. Newly infected cells were passaged every 3 days, and FACS analysis was performed at each passage (Fig. 6B). Vector ACE-GFP spread in all cell lines, whereas replication of vectors CTP4-TF and E2-TF remained restricted to the permissive HepG2 cells. Despite virus spread, the maximum percentage of eGFP-expressing cells did not reach more than 50% for any of the vectors, which correlates with data from the serial infection cycles (Fig. 5A) and is explained by the fact that by infection cycle 7, mutant vectors with deletions in the transgene cassette are becoming dominant in the virus population. Moreover, analysis of the MFI of infected cells over the course of the experiment demonstrated that transgene expression in nonpermissive cells remained low (data not shown).
Thus, in summary, we have generated replication-competent MLV-based vectors which can be used to specifically target either liver-derived cells or cells deregulated in the β-catenin pathway. The targeted vectors display wild-type replication kinetics in permissive cells, are genomically stable, and maintain their transcriptional targeting profile over serial infection cycles.
DISCUSSION
Through the generation of a series of MLV-based RCR vectors whose replication and transgene expression are driven by a heterologous promoter which replaces viral promoter/enhancer elements in the U3 region of the viral LTR, we demonstrated that the expression of such vectors can be targeted in a liver-specific manner or to cells which are deregulated for β-catenin by using the chimeric EII-Pa1AT promoter or the synthetic CTP4 promoter, respectively. TF vectors, in which the EII-Pa1AT or CTP4 promoter is inserted into the U3 region such that transcription is controlled by the MLV TATA box and transcription initiation at the 5′ end of the R region is maintained, replicated with wild-type kinetics in a largely liver-specific (Fig. 3A and 4C) or strictly β-catenin deregulation-dependent (Fig. 3A and 4B) manner, respectively. Insertion of the mCMV promoter in this fashion gave rise to a vector which also demonstrated near-wild-type replication kinetics (Fig. 3A). TR vectors, on the other hand, in which promoters were inserted into the MLV U3 such that transcription was controlled by the heterologous TATA box and in which transcription should be initiated at the TSS of the heterologous promoter, demonstrated greatly attenuated replication kinetics in spite of having transgene expression levels similar to those of the TF vectors (Fig. 3A and B).
Detailed analyses revealed that particle production but not the infectivity of TR vectors was greatly reduced compared to that of ACE-GFP and TF vectors (Fig. 3B). The similar MFIs of cells infected with TR and TF vectors indicate that transcription levels are equal in the two vector designs (Fig. 3B). Since virus particles which are produced from cells infected with TR vectors are just as infectious as those produced from cells infected with ACE-GFP or TF vectors (Fig. 3B), the TR design does not seem to impinge on reverse transcription or integration, despite the fact that transcription initiation from the heterologous transcriptional start sites in TR vectors would lead to an elongation in the R region as the TSS defines the borders of the R region in retroviruses (9). Our data were not unanticipated, however, since there appears to be no particular upper size constraint on the retroviral R region, the length of which varies widely among different retroviruses, up to 247 nucleotides in the case of human T-cell leukemia virus type 2 (39). Moreover, previous work has shown that the precise sequence of the R region is of no importance during reverse transcription, provided that there are repeated sequences available for minus-strand DNA transfer (5). The MLV R region has also been proposed to act as a constitutive transport element (43). This function is based on the formation of a stem-loop structure, and disruption of this structure can greatly decrease viral gene expression (8). It is possible therefore that 5′ elongation of the R region could influence the structure of the natural R region. This would also account for the fact that only virus particle production but not eGFP expression is reduced, since eGFP expression is not dependent on full-length viral RNA as the transgene cassette is also present on the spliced env message which is exported efficiently from the nucleus in the absence of the proposed constitutive transport element.
Logg and colleagues (27) also made a series of transcriptionally targeted vectors in which the TATA box of the heterologous promoter is used but in which the TSS of the heterologous promoter is fused to the MLV TSS, such that the sequence between the heterologous TATA box and its TSS is maintained and transcription is initiated from the 5′ border of the MLV R region and, thus, the sequence and size of the R region do not change during virus replication (27). However, these vectors also replicate as poorly as our TR vectors.
Subsequent infection of a wider range of cells with the TF vectors revealed that vectors CTP4-TF and E2-TF are tightly restricted to β-catenin-deregulated cells and preferentially active in liver cells, respectively. Although there are some correlations between the MFI of cells infected with a particular vector and the ability of the vector to spread in these cells, in many instances, such a correlation does not exist. Clearly then, it is not always possible to make predictions about the ability of transcriptionally targeted retroviral vectors to replicate in a particular cell line based solely upon the expression of the transgene in these cells. One explanation could be that since eGFP expression can occur from both full-length and subgenomic viral RNAs in our vectors, the MFI of infected cells does not always mirror the expression level of Gag proteins, which could be a limiting factor for vector production.
In the case of HuH-7 cells, in which spread of both ACE-GFP and E2-TF was detectable for the first few passages but then ceased, it is possible that the vector becomes rapidly unstable in this cell line and loses the ability to express eGFP in infected cells. Alternatively, it is possible that due to the extremely low MFI of the infected cells, vectors did actually spread in these cells but could not be detected in all of the cells because the level of eGFP expression was below the FACS detection threshold.
In the case of the E2-TF vector, which showed spreading in 293 cells after about 25 days, it seems that a rare recombination event which took place at the plasmid DNA level during production of vector stocks by transient transfection created a vector whose transcription was driven by the human CMV promoter and that this vector emerged only following extensive passaging. Although we did not analyze the genomic DNA of SW480 cells infected with E2-TF which also showed spreading after about 25 days, it is possible that a similar event occurred here, too. This problem could be easily avoided by generating virus stocks from a characterized, stably transfected producer clone instead of by transient transfection.
The efficacies of transgene propagation of vectors E2-TF and CTP4-TF were not reduced compared to that of vector ACE-GFP (Fig. 5A), and molecular analysis by PCR amplification of the transgenic cassette revealed that deletions of the transgene region of vectors E2-TF and CTP4-TF are not detectable any earlier than in ACE-GFP (Fig. 5B to D), confirming that replacing the MLV promoter with these heterologous sequences was not detrimental to genomic stability. Real-time RT-PCR performed on viral RNA extracted from filtered supernatants from infected cells demonstrated that the production of infectious retrovirus particles did not decrease over successive infection cycles, formally ruling out the possibility that the decrease in eGFP-expressing cells was due to the vectors somehow becoming nonfunctional. Molecular analysis of the promoter regions of the targeted vectors detected mutations only in the EII-Pa1AT promoter of E2-TF, and these were not in any known transcription factor binding sites (data not shown). Moreover, the targeting specificities of vectors CTP4-TF and E2-TF were maintained over time (Fig. 6B). In accordance with FACS results from the stability analysis (Fig. 5A), no more than 50% of permissive cells infected with vectors from the seventh infection cycle expressed eGFP following passaging. However, this is expected since by this stage, deletion mutants have already arisen (Fig. 5B to D). Interestingly, the stability of the vectors in HepG2 cells seems to be much reduced compared to that in NIH 3T3 and 293 cells (28; M. Paar, personal communication). One possible explanation is that the difference in genomic stability of the RCR vectors in different cell lines stems from rearrangements and recombination of integrated proviruses due to intrinsic differences in the genetic stability of the cell lines (12, 16, 20). The stability profiles of the targeted vectors imply, however, that even in HepG2 cells, they should be able to transduce upwards of 1011 cells following an initial transduction of only 5,000 cells, which should be sufficient to allow complete transduction of any solid tumor before becoming unstable.
These vectors therefore combine high replicative potential with the ability of targeted replication and expression of the inserted transgene, providing a valuable tool for specific, highly efficient cancer gene therapy.
Acknowledgments
We are grateful to Maria Eisenbauer (Institute of Cancer Research of the Medical University of Vienna, Vienna, Austria) for providing the primary liver tumor cell lines AKH12 and AKH13 and to Kai Lipinski (ML Research, Keele, United Kingdom) and Gabriela Kramer (University of Navarra, Pamplona, Spain) for kindly providing the CTP4 and EII-Pa1AT promoters, respectively. We also thank Noriyuki Kasahara (UCLA) for providing the vector ACE-GFP. Special thanks to Elzbieta Knapp, Reinhard Ertl, and Magdalena Pusch for help with FACS analysis and real-time RT-PCR and to Daria Deitermann for excellent technical support. We also specially thank Chris Logg (UCLA) and Kai Lipinski for their helpful advice and expert opinions during this work.
This study was supported by the Christian-Doppler Forschungsgesellschaft, Austria, and Daniela Mischek was funded by the Gen-AU (Genomforschung in Austria) research program.
REFERENCES
- 1.Addison, C. L., M. Hitt, D. Kunsken, and F. L. Graham. 1997. Comparison of the human versus murine cytomegalovirus immediate early gene promoters for transgene expression by adenoviral vectors. J. Gen. Virol. 78:1653-1661. [DOI] [PubMed] [Google Scholar]
- 2.Antonucci, T. K., and W. J. Rutter. 1989. Hepatitis B virus (HBV) promoters are regulated by the HBV enhancer in a tissue-specific manner. J. Virol. 63:579-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boettiger, D., and H. M. Temin. 1970. Light inactivation of focus formation by chicken embryo fibroblasts infected with avian sarcoma virus in the presence of 5-bromodeoxyuridine. Nature 228:622-624. [DOI] [PubMed] [Google Scholar]
- 4.Carruba, G., M. Cervello, M. D. Miceli, R. Farruggio, M. Notarbartolo, L. Virruso, L. Giannitrapani, R. Gambino, G. Montalto, and L. Castagnetta. 1999. Truncated form of beta-catenin and reduced expression of wild-type catenins feature HepG2 human liver cancer cells. Ann. N. Y. Acad. Sci. 886:212-216. [DOI] [PubMed] [Google Scholar]
- 5.Cheslock, S. R., J. A. Anderson, C. K. Hwang, V. K. Pathak, and W. S. Hu. 2000. Utilization of nonviral sequences for minus-strand DNA transfer and gene reconstitution during retroviral replication. J. Virol. 74:9571-9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coghlan, M. P., A. A. Culbert, D. A. Cross, S. L. Corcoran, J. W. Yates, N. J. Pearce, O. L. Rausch, G. J. Murphy, P. S. Carter, L. Roxbee Cox, D. Mills, M. J. Brown, D. Haigh, R. W. Ward, D. G. Smith, K. J. Murray, A. D. Reith, and J. C. Holder. 2000. Selective small molecule inhibitors of glycogen synthase kinase-3 modulate glycogen metabolism and gene transcription. Chem. Biol. 7:793-803. [DOI] [PubMed] [Google Scholar]
- 7.Cornetta, K., R. A. Morgan, A. Gillio, S. Sturm, L. Baltrucki, R. O'Reilly, and W. F. Anderson. 1991. No retroviremia or pathology in long-term follow-up of monkeys exposed to a murine amphotropic retrovirus. Hum. Gene Ther. 2:215-219. [DOI] [PubMed] [Google Scholar]
- 8.Cupelli, L., S. A. Okenquist, A. Trubetskoy, and J. Lenz. 1998. The secondary structure of the R region of a murine leukemia virus is important for stimulation of long terminal repeat-driven gene expression. J. Virol. 72:7807-7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cupelli, L. A., and J. Lenz. 1991. Transcriptional initiation and postinitiation effects of murine leukemia virus long terminal repeat R-region sequences. J. Virol. 65:6961-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de La Coste, A., B. Romagnolo, P. Billuart, C. A. Renard, M. A. Buendia, O. Soubrane, M. Fabre, J. Chelly, C. Beldjord, A. Kahn, and C. Perret. 1998. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc. Natl. Acad. Sci. USA 95:8847-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz, R. M., T. Eisen, I. R. Hart, and R. G. Vile. 1998. Exchange of viral promoter/enhancer elements with heterologous regulatory sequences generates targeted hybrid long terminal repeat vectors for gene therapy of melanoma. J. Virol. 72:789-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duval, A., and R. Hamelin. 2002. Genetic instability in human mismatch repair deficient cancers. Ann. Genet. 45:71-75. [DOI] [PubMed] [Google Scholar]
- 13.Edelstein, M. L., M. R. Abedi, J. Wixon, and R. M. Edelstein. 2004. Gene therapy clinical trials worldwide 1989-2004—an overview. J. Gene Med. 6:597-602. [DOI] [PubMed] [Google Scholar]
- 14.Ferrari, G., G. Salvatori, C. Rossi, G. Cossu, and F. Mavilio. 1995. A retroviral vector containing a muscle-specific enhancer drives gene expression only in differentiated muscle fibers. Hum. Gene Ther. 6:733-742. [DOI] [PubMed] [Google Scholar]
- 15.Finger, C., Y. Sun, L. Sanz, L. Alvarez-Vallina, C. J. Buchholz, and K. Cichutek. 2005. Replicating retroviral vectors mediating continuous production and secretion of therapeutic gene products from cancer cells. Cancer Gene Ther. 12:464-474. [DOI] [PubMed] [Google Scholar]
- 16.Fodde, R., R. Smits, and H. Clevers. 2001. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer 1:55-67. [DOI] [PubMed] [Google Scholar]
- 17.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 18.Gunzburg, W. H., A. Fleuchaus, R. Saller, and B. Salmons. 1996. Retroviral vector targeting for gene therapy. Cytokines Mol. Ther. 2:177-184. [PubMed] [Google Scholar]
- 19.Hacein-Bey-Abina, S., C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302:415-419. [DOI] [PubMed] [Google Scholar]
- 20.Harfe, B. D., and S. Jinks-Robertson. 2000. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34:359-399. [DOI] [PubMed] [Google Scholar]
- 21.Haynes, C., O. Erlwein, and B. S. Schnierle. 2003. Modified envelope glycoproteins to retarget retroviral vectors. Curr. Gene Ther. 3:405-410. [DOI] [PubMed] [Google Scholar]
- 22.Hlavaty, J., D. Portsmouth, A. Stracke, B. Salmons, W. H. Gunzburg, and M. Renner. 2004. Effects of sequences of prokaryotic origin on titer and transgene expression in retroviral vectors. Virology 330:351-360. [DOI] [PubMed] [Google Scholar]
- 23.Jespersen, T., M. Duch, M. L. Carrasco, S. Warming, and F. S. Pedersen. 1999. Expression of heterologous genes from an IRES translational cassette in replication competent murine leukemia virus vectors. Gene 239:227-235. [DOI] [PubMed] [Google Scholar]
- 24.Kramer, M. G., M. Barajas, N. Razquin, P. Berraondo, M. Rodrigo, C. Wu, C. Qian, P. Fortes, and J. Prieto. 2003. In vitro and in vivo comparative study of chimeric liver-specific promoters. Mol. Ther. 7:375-385. [DOI] [PubMed] [Google Scholar]
- 25.Lipinski, K. S., H. A. Djeha, J. Gawn, S. Cliffe, N. J. Maitland, D. H. Palmer, A. Mountain, A. S. Irvine, and C. J. Wrighton. 2004. Optimization of a synthetic beta-catenin-dependent promoter for tumor-specific cancer gene therapy. Mol. Ther. 10:150-161. [DOI] [PubMed] [Google Scholar]
- 26.Logg, C. R., and N. Kasahara. 2004. Retrovirus-mediated gene transfer to tumors: utilizing the replicative power of viruses to achieve highly efficient tumor transduction in vivo. Methods Mol. Biol. 246:499-525. [DOI] [PubMed] [Google Scholar]
- 27.Logg, C. R., A. Logg, R. J. Matusik, B. H. Bochner, and N. Kasahara. 2002. Tissue-specific transcriptional targeting of a replication-competent retroviral vector. J. Virol. 76:12783-12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Logg, C. R., A. Logg, C.-K. Tai, P. M. Cannon, and N. Kasahara. 2001. Genomic stability of murine leukemia viruses containing insertions at the Env-3′ untranslated region boundary. J. Virol. 75:6989-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Logg, C. R., C. K. Tai, A. Logg, W. F. Anderson, and N. Kasahara. 2001. A uniquely stable replication-competent retrovirus vector achieves efficient gene delivery in vitro and in solid tumors. Hum. Gene Ther. 12:921-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCormack, M. P., A. Forster, L. Drynan, R. Pannell, and T. H. Rabbitts. 2003. The LMO2 T-cell oncogene is activated via chromosomal translocations or retroviral insertion during gene therapy but has no mandatory role in normal T-cell development. Mol. Cell. Biol. 23:9003-9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore, K. A., M. Scarpa, S. Kooyer, A. Utter, C. T. Caskey, and J. W. Belmont. 1991. Evaluation of lymphoid-specific enhancer addition or substitution in a basic retrovirus vector. Hum. Gene Ther. 2:307-315. [DOI] [PubMed] [Google Scholar]
- 32.Nethe, M., B. Berkhout, and A. C. van der Kuyl. 2005. Retroviral superinfection resistance. Retrovirology 2:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pardoll, D. 2003. Does the immune system see tumors as foreign or self? Annu. Rev. Immunol. 21:807-839. [DOI] [PubMed] [Google Scholar]
- 34.Rainov, N. G., and H. Ren. 2003. Clinical trials with retrovirus mediated gene therapy—what have we learned? J. Neurooncol. 65:227-236. [DOI] [PubMed] [Google Scholar]
- 35.Restifo, N. P., P. A. Antony, S. E. Finkelstein, W. W. Leitner, D. P. Surman, M. R. Theoret, and C. E. Touloukian. 2002. Assumptions of the tumor ‘escape’ hypothesis. Semin. Cancer Biol. 12:81-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubin, H., and H. M. Temin. 1959. A radiological study of cell-virus interaction in the Rous sarcoma. Virology 7:75-91. [DOI] [PubMed] [Google Scholar]
- 37.Sadelain, M. 2004. Insertional oncogenesis in gene therapy: how much of a risk? Gene Ther. 11:569-573. [DOI] [PubMed] [Google Scholar]
- 38.Shin, N. H., D. Hartigan-O'Connor, J. K. Pfeiffer, and A. Telesnitsky. 2000. Replication of lengthened Moloney murine leukemia virus genomes is impaired at multiple stages. J. Virol. 74:2694-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sodroski, J., M. Trus, D. Perkins, R. Patarca, F. Wong-Staal, E. Gelmann, R. Gallo, and W. A. Haseltine. 1984. Repetitive structure in the long-terminal-repeat element of a type II human T-cell leukemia virus. Proc. Natl. Acad. Sci. USA 81:4617-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solly, S. K., S. Trajcevski, C. Frisen, G. W. Holzer, E. Nelson, B. Clerc, E. Abordo-Adesida, M. Castro, P. Lowenstein, and D. Klatzmann. 2003. Replicative retroviral vectors for cancer gene therapy. Cancer Gene Ther. 10:30-39. [DOI] [PubMed] [Google Scholar]
- 41.Sun, Y., C. Finger, L. Alvarez-Vallina, K. Cichutek, and C. J. Buchholz. 2005. Chronic gene delivery of interferon-inducible protein 10 through replication-competent retrovirus vectors suppresses tumor growth. Cancer Gene Ther. 12:900-912. [DOI] [PubMed] [Google Scholar]
- 42.Tai, C. K., W. J. Wang, T. C. Chen, and N. Kasahara. 2005. Single-shot, multicycle suicide gene therapy by replication-competent retrovirus vectors achieves long-term survival benefit in experimental glioma. Mol. Ther. 12:842-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trubetskoy, A. M., S. A. Okenquist, and J. Lenz. 1999. R region sequences in the long terminal repeat of a murine retrovirus specifically increase expression of unspliced RNAs. J. Virol. 73:3477-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varmus, H. E., T. Padgett, S. Heasley, G. Simon, and J. M. Bishop. 1977. Cellular functions are required for the synthesis and integration of avian sarcoma virus-specific DNA. Cell 11:307-319. [DOI] [PubMed] [Google Scholar]
- 45.Wang, W. J., C. K. Tai, N. Kasahara, and T. C. Chen. 2003. Highly efficient and tumor-restricted gene transfer to malignant gliomas by replication-competent retroviral vectors. Hum. Gene Ther. 14:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yi, Y., S. H. Hahm, and K. H. Lee. 2005. Retroviral gene therapy: safety issues and possible solutions. Curr. Gene Ther. 5:25-35. [DOI] [PubMed] [Google Scholar]